Introduction
Colorectal cancer (CRC) is a common human malignancy
worldwide and its incidence has significantly increased in recent
years, accounting for 8% of all new cancer diagnoses in 2016; it
has become the third most common cancer in the United States
(1), and the fifth most common cause
of cancer-associated mortality in China, with 191,000 CRC deaths in
2015 (2). Early detection through
screening has improved the 5-year survival of patients with CRC;
however, the overall prognosis of advanced CRC remains poor due to
lack of effective therapeutic strategies (3). Further investigation of the complex
molecular networks in the pathogenesis of CRC is required to
facilitate the development of novel therapeutic approaches and
improve the clinical management of CRC, particularly for patients
with advanced stages of the disease.
Previous studies have demonstrated the essential
roles of genetic alterations and aberrant expressions of several
oncogenes or tumor suppressor genes in the development of CRC
(4,5). Whole-exome and genome-scale
transcriptome sequencing of seven liver metastases, along with
their matched primary tumors and normal tissues showed that,
mutations in KRAS, APC, POLE, PTPRT, PLXND1, CELSR3, BAHD1 and
PNPLA6 are associated with CRC metastasis (6). Several tumor suppressor genes are
associated to CRC carcinogenic process, with the APC, PTEN, FBXW7
and p53 being the common mutated ones in CRC; while oncogenes,
including KRAS, CDK8, BRAF, PIK3CA, EGFR, have been shown to have
mutations in patients with CRC (7).
In addition to the protein-coding genes, microRNAs (miRNAs/miRs), a
class of non-protein coding small RNAs, have also been reported to
play a critical role in CRC tumorigenesis (8).
Mature miRNAs are a class of small, non-coding RNAs,
with a length of ~22 nucleotides. miRNAs mediate the binding of the
RNA-induction silencing complex with the 3′-untranslated region
(UTR) of target mRNAs and regulate their degradation or
translational inhibition (9). miRNAs
are involved in several physiological processes, such as cell
proliferation, differentiation, apoptosis, individual growth and
development, neoplasia and metabolism (10). Thus, altered expression levels of
miRNAs have been demonstrated to influence several oncogenic
signaling pathways. For example, miR-210-3p promotes prostate
cancer cell epithelial-mesenchymal transition and bone metastasis
via the NF-κB signaling pathway (11). Furthermore, miR-365a-3p regulates
JAK-STAT signaling to suppress the proliferation and metastasis of
colorectal cancer cells (12). Thus,
miRNAs are considered to play critical roles in the pathogenesis of
different types of human cancer, including CRC (13). Deregulated expression levels of
several miRNAs, including miR-134, miR-320b, miR-766 and miR-19a,
have been reported in CRC. A previous study demonstrated that
downregulation of miR-134 in CRC leads to overexpression of
metastasis-associated genes in colon cancer-1, an oncogenic protein
promoting tumor metastasis via deregulation of the hepatocyte
growth factor/Met signaling pathway (13). The tumor suppressor role of miR-320b
in CRC by inhibition of c-Myc expression has also been reported
(14). Furthermore, oncogenic roles
of miR-766 and miR-19b have been demonstrated in the pathogenesis
of CRC. Overexpression of miR-766 and miR-19b levels have been
reported to promote tumor epithelial-to-mesenchymal transitions and
tumor metastasis in CRC (15,16). A
previous study indicated that miR-126 expression is deregulated in
several CRC cancer cell lines, including SW480, SW620, HT-29 and
HCT-116 (17). Subsequent studies
have reported similar findings, that miR-126 is downregulated in
several types of cancer, including CRC (18–23).
Further functional analysis revealed that exogenous overexpression
of miR-126 suppresses AKT (also known as protein kinase B or PKB)
and extracellular signal-regulated kinase-1 and −2 (ERK1/2)
activation, and inhibits proliferation, migration and invasion of
CRC cancer cells (24). Furthermore,
in vitro knockdown experiments demonstrated that decreased
miR-126 expression in HCT-116 cells leads to AKT and ERK1/2
activation by upregulating insulin receptor substrate-1 (IRS-1)
protein expression (24), suggesting
a tumor suppressor role of miR-126. However, evidence of altered
miR-126 expression and its interplay with IRS-1 in primary CRC
tumors remains limited.
IRS-1, a member of the IRS family, is the principle
substrate of the insulin-like growth factor I receptor and the
insulin receptor, and plays an essential role in cytokine signaling
(25). IRS-1 mediates a range of
biological responses, including apoptosis and cell differentiation,
by binding to and activating enzymes or adapter molecules (26,27).
Previous studies have reported that IRS-1 plays an important role
in the tumorigenesis of different types of human cancer, including
CRC (24). Overexpression of IRS-1
has been detected in CRC and appears to inversely correlate with
CRC differentiation, progression and liver metastasis (28). Increasing evidence suggests that the
interplay between miR-126 and IRS-1 may notably contribute to
colorectal carcinogenesis (29,30).
The present study aimed to investigate the
expression statuses of miR-126 and IRS-1 in primary CRC tumors, and
to determine the clinical significance of altered miR-126
expression in patients with CRC. The findings would help to
determine whether the downregulation of miR-126 expression may play
an important role in the pathogenesis and progression of human
CRC.
Materials and methods
Patient specimens
A total of 126 colorectal specimens were collected
from patients who underwent surgical resection or colonoscopy
biopsy at the Affiliated Hospital of Guangdong Medical University
(Zhanjiang, China) between July 2011 and December 2012. The
specimens included samples from primary CRC and adjacent normal
tissues from 40 patients with CRC, colorectal adenoma tissues from
26 patients who underwent resection of adenomas, and colonoscopy
biopsy samples from 20 healthy volunteers. Among the 40 patients
with CRC, 20 were men and 20 were women, with a mean age of 56.2
years (age range, 32–73 years). There were 16 men and 10 women,
with a mean age of 46 years (age range, 29–65 years) in the
colorectal adenoma group. A total of 20 healthy people, including
11 men and 9 women were selected as the control group, with a mean
age of 47.5 years (age range, 28–60 years). Adjacent normal
colorectal epithelial tissues were 5 cm away from the edge of the
primary tumor. Diagnoses were confirmed via pathological analysis.
The specimens were immediately frozen in liquid nitrogen and
subsequently stored at −80°C until further experimentation. All
specimens were obtained prior to chemotherapy or radiotherapy, and
the clinicopathological data of all patients, including sex, age,
tumor size, tumor location, pathological data, distant metastasis,
Tumor-Node-Metastasis (TNM) staging (31), were recorded and complete. A 5-year
follow-up was carried out for all patients, with an average
follow-up time of 30.83 months (15–52 months). Follow-up data were
conducted using hospital medical records and telephone interviews.
The present study was approved by the Medical Ethics Committee of
the Affiliated Hospital of Guangdong Medical University, and
written informed consent was provided by all participants prior to
the study.
Bioinformatics prediction of miR-126
target
Targetscan (http://www.targetscan.org), and PicTar (http://pictar.mdc-berlin.de) databases were used to
predict miR-126 targets. The 3′-UTR of IRS-1 mRNA has a putative
miR-126 binding site (24).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from tissue specimens,
including primary CRC and adjacent normal tissues, colorectal
adenoma tissues and normal samples, using RNAiso Plus (Takara Bio,
Inc.), according to the manufacturer's protocol, and the purified
RNA was stored at −80°C until further use. Total miRNA and mRNA
were reverse transcribed into cDNA using the Mir-X miRNA
First-Strand Synthesis kit (cat. no. 638315; Clontech Laboratories,
Inc.) and PrimeScript® RT Master Mix Perfect Real Time
(Takara Bio, Inc.), respectively. The following temperature
protocol was used for RT: The tube was incubated for 1 h (miR-126)
or 15 min (IRS-1 mRNA) at 37°C, then terminated at 85°C for 5 min
(miR-126) or 5 sec (IRS-1 mRNA) to inactivate the enzymes. qPCR was
subsequently performed using TB Green Premix Ex Taq II (Tli RNaseH
Plus; Takara Bio, Inc.), on a LightCycler® (Roche
Diagnostics). The universal reverse primer for miR-126 and U6 PCR
amplification, mRQ 3′ Primer, was included in the Mir-X miRNA
First-Strand Synthesis kit, while the remaining primer sequences
for miRNA and mRNA were synthesized by Sangon Biotech Co., Ltd. The
primer sequences used for qPCR are listed in Table I. The following thermocycling
conditions were used for qPCR: Pre-denaturation (95°C for 30 sec),
and 40 cycles of denaturation (95°C for 5 sec), annealing (60°C for
20 sec) and extension (65°C for 15 sec). Relative miR-126 and IRS-1
mRNA expression levels were calculated using the 2−∆∆Cq
method (32) and normalized to the
internal reference genes U6 (miR-126) and β-actin (IRS-1),
respectively. All experiments were performed in triplicate.
 | Table I.Primer sequences used for
quantitative PCR. |
Table I.
Primer sequences used for
quantitative PCR.
| Name | Direction | Primer (5′→3′) |
|---|
| miRNA qPCR |
|
miR-126 | Forward |
TCGTACCGTGAGTAATAATGCG |
| U6
snRNA | Forward |
CTCGCTTCGGCAGCACA |
| Universal qPCR | Reverse | mRQ 3′ Primer
included in the Mir-X miRNA First-Strand Synthesis kit |
| mRNA qPCR |
|
IRS-1 | Forward |
AGTCCTAACCGCAACCAGAGT |
|
| Reverse |
CCTCAGCCACACATTCTCAA |
|
β-actin | Forward |
GGCGGCAACACCATGTACCCT |
|
| Reverse |
AGGGGCCGGACTCGTCATACT |
Western blotting
Western blotting was performed according to the
standard protocol (33). Briefly,
total protein was extracted from tissues and cells, using RIPA
lysis buffer containing 1% phenylmethylsulphonyl fluoride (Beyotime
Institute of Biotechnology), and concentration was measured using a
BCA protein assay kit (Beyotime Institute of Biotechnology). A
total of 20 µg protein/sample was separated via 10% SDS-PAGE and
subsequently transferred onto polyvinylidene difluoride membranes.
The membranes were blocked with 5% fat-free milk for 1 h at room
temperature and then incubated with rabbit anti-human IRS-1
(1:1,000; cat. no. 2390S; Cell Signaling Technology, Inc.) and
mouse anti-human GAPDH (1:1,000; cat. no. AG019; Beyotime Institute
of Biotechnology) primary antibodies overnight at 4°C, prior to
incubation with horseradish peroxidase-labeled goat anti-rabbit IgG
(1:1,000; cat. no. A0208; Beyotime Institute of Biotechnology)
secondary antibody for 1 h at room temperature. Protein bands were
visualized using the enhanced chemiluminescence kit (cat. no.
P0018S; Beyotime Institute of Biotechnology) and analyzed using
Quantity One image analysis software (version 4.6.6; Bio-Rad
Laboratories, Inc.).
Immunohistochemistry (IHC)
IHC staining was performed according to the standard
protocol (Santa Cruz Biotechnology, Inc.). All tissues were fixed
in 4% paraformaldehyde for at least 24 h at 4°C and embedded in
paraffin. Paraffin-embedded tissues were cut into 4-µm thick
sections and placed onto positively charged slides pretreated with
polylysine (Wuhan Boster Biological Technology, Ltd.). The tissue
sections were subsequently deparaffinized in xylene, rehydrated in
a descending ethanol series (90, 80 and 70% for 5 min,
respectively) and distilled water, and subjected to heat-induced
antigen retrieval in 10 mM citrate buffer (pH=6.0) (Beijing
Solarbio Science and Technology Co., Ltd.) for 5 min at room
temperature. The slides were blocked with 10% goat serum (Wuhan
Boster Biological Technology, Ltd.) for 10 min at room temperature,
prior to incubation with 3% H2O2 for 10 min
at room temperature to inhibit endogenous peroxidase activity.
Tissue sections were incubated with rabbit anti-human IRS-1 primary
antibody (1:50; cat. no. SC-559; Santa Cruz Biotechnology, Inc.)
overnight at 4°C. The slides were washed three times with PBS
(Wuhan Boster Biological Technology, Ltd.), prior to incubation
with the MaxVision™ HRP-polymer goat anti-rabbit
secondary antibody in the IHC kit (Fuzhou Maixin Biotech Co., Ltd.)
for 30 min at room temperature. The slides were subsequently
stained with 3,3′-diaminobenzidine for 10 min at room temperature
to visualize IRS-1 expression and counterstained with hematoxylin
for 2 min at room temperature, prior to mounting in permanent
mounting medium. The stained tissue sections were observed under a
confocal microscope (magnification, ×400; Leica Microsystems,
Ltd.). Tissues incubated with PBS instead of the primary antibody
were used as negative controls. All experiments were performed in
triplicate.
Statistical analysis
Statistical analysis was performed using SPSS
software version 13.0 (SPSS, Inc.). All experiments were performed
in triplicate and data are presented as the mean ± standard
deviation. One-way analysis of variance, followed by Bonferroni's
post-hoc test, was used for multiple intergroup comparisons between
miR-126 and IRS-1 mRNA or protein expression levels. Associations
between miR-126 or IRS-1 expression and clinicopathological
characteristics of patients with CRC were analyzed using the
χ2 test. Kaplan-Meier curves and the log-rank test were
generated to assess the survival data, while Pearson's correlation
analysis was performed to determine the correlation between miR-126
and IRS-1 protein expression. P<0.05 was considered to indicate
a statistically significant difference.
Results
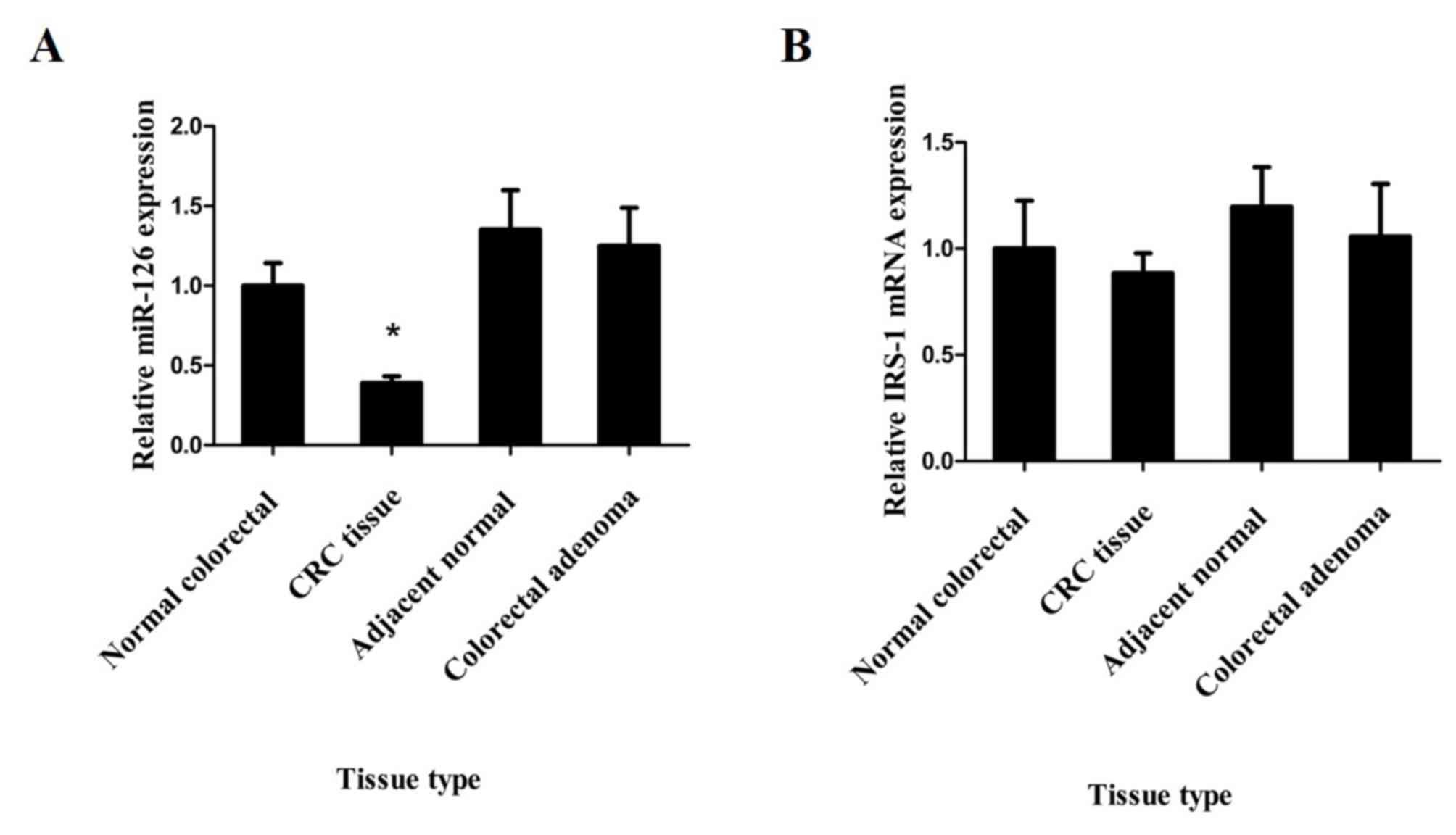
miR-126 is downregulated in CRC
In order to determine the status of miR-126
expression in CRC, RT-qPCR analysis was performed to assess miR-126
expression in 40 primary CRC tissues and their adjacent normal
tissues, in 26 colorectal adenomas and in 20 normal colorectal
tissues without concomitant CRC. The results demonstrated that
miR-126 expression was significantly downregulated in CRC tissues
compared with that in matched adjacent normal tissues, colorectal
adenoma tissues and normal colorectal tissues, respectively
(P<0.05; Fig. 1A). However, no
significant differences in miR-126 expression were observed between
adjacent normal tissues, colorectal adenoma tissues and normal
colorectal tissues, respectively (P>0.05; Fig. 1A).
Correlation between IRS-1 and miR-126
expression levels in colorectal tissues
The in silico miRNA target prediction
analysis demonstrated that IRS-1 is one of the potential downstream
targets for miR-126. In order to determine the status of IRS-1 in
CRC, and its correlation with miR-126 expression, RT-qPCR and
western blot analyses were performed to assess the mRNA and protein
expression levels of IRS-1, respectively. No significant
differences in IRS-1 mRNA expression were observed between CRC,
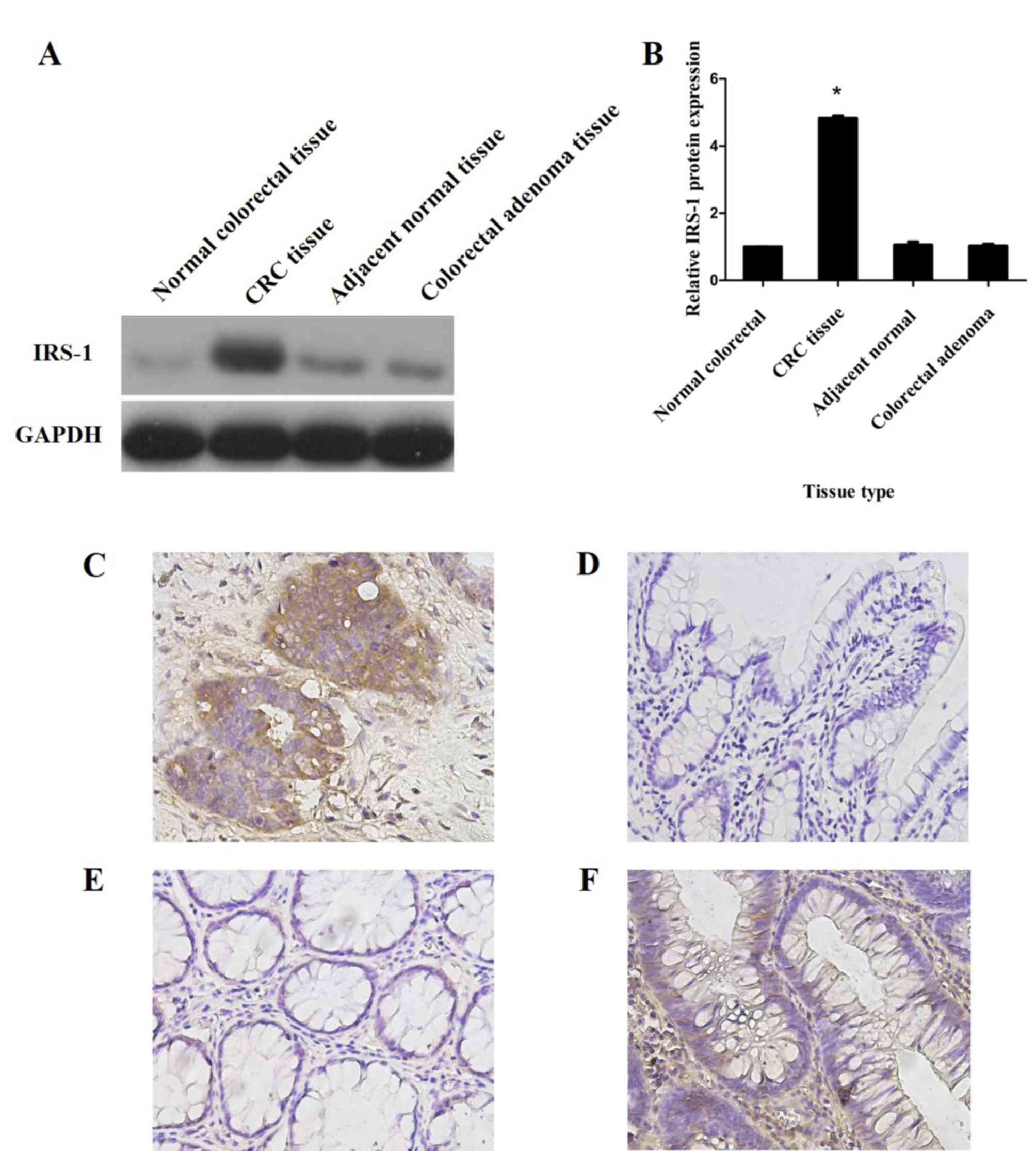
adjacent tissues, adenoma and normal colorectal tissues (Fig. 1B). However, western blot analysis
demonstrated significantly increased IRS-1 protein expression in
CRC tissues compared with that in adjacent normal tissues,
colorectal adenoma tissues and normal colorectal tissues
(P<0.05; Fig. 2A and B).
Furthermore, IHC analysis indicated that IRS-1 protein was strongly
expressed in the cytoplasm of CRC tissues (Fig. 2C), but only weakly expressed or
absent in the adjacent normal, colorectal adenoma and normal
colorectal tissue specimens (Fig.
2D-F).
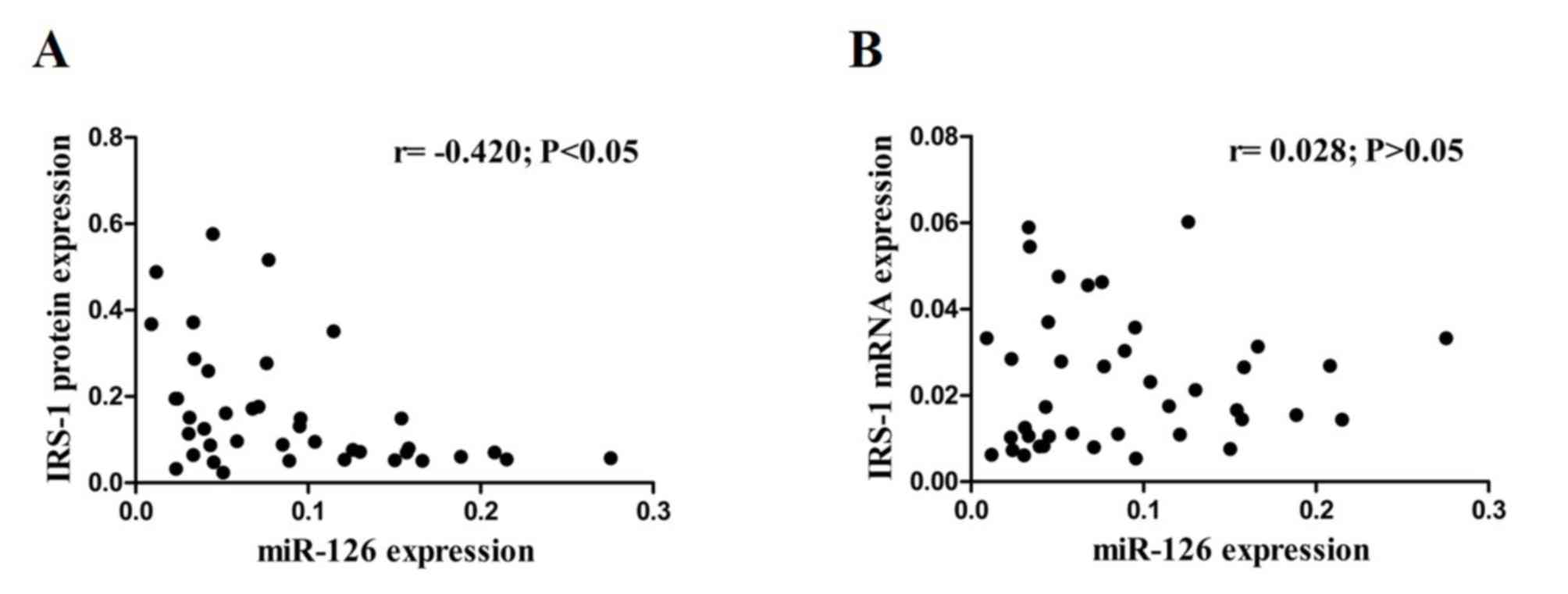
Pearson's correlation analysis was performed to
demonstrate the potential regulatory role of miR-126 in IRS-1
expression. The results demonstrated a significantly inverse
correlation between miR-126 and IRS-1 protein expression levels
(r=−0.420; P<0.05; Fig. 3A),
while no significant correlation was observed between miR-126 and
IRS-1 mRNA expression levels (r=0.028; P>0.05; Fig. 3B) in CRC tissues.
Association between miR-126 and IRS-1
expression levels and clinicopathological characteristics in
patients with CRC
In order to understand the clinical significance of
altered miR-126 expression in patients with CRC, the association
between miR-126 and IRS-1 protein expression levels and
clinicopathological characteristics of patients with CRC was
assessed. Of the 40 patients with CRC, decreased miR-126 expression
and increased IRS-1 expression levels were observed in 29 and 27
cases compared with the levels in the matched adjacent normal
tissues, respectively. As presented in Table II, downregulation of miR-126
expression was significantly associated with lymph node (P=0.012)
and distant metastasis (P=0.020). Similarly, upregulation of IRS-1
protein expression was significantly associated with lymph node
metastasis (P=0.002) and distant metastasis (P=0.025). Furthermore,
low levels of miR-126 expression and high levels of IRS-1
expression were significantly associated with advanced TNM stage
(both P<0.05; Table II).
However, no significant associations were observed between miR-126
and IRS-1 protein expression levels and other clinicopathological
characteristics in patients with CRC.
 | Table II.Association between miR-126 and IRS-1
protein expression levels and clinicopathological characteristics
in patients with CRC (n=40). |
Table II.
Association between miR-126 and IRS-1
protein expression levels and clinicopathological characteristics
in patients with CRC (n=40).
|
| miR-126 expression,
n |
| IRS-1 expression,
n |
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Upregulated | Downregulated | P-value | Upregulated | Downregulated | P-value |
|---|
| Total number of
patients | 11 | 29 |
| 27 | 13 |
|
| Age, years |
|
|
|
|
|
|
| 60 | 7 | 16 | 0.900 | 14 | 9 | 0.484 |
|
<60 | 4 | 13 |
| 13 | 4 |
|
| Sex |
|
|
|
|
|
|
|
Male | 5 | 15 | 0.723 | 15 | 5 | 0.311 |
|
Female | 6 | 14 |
| 12 | 8 |
|
| TNM stage |
|
|
|
|
|
|
| Tis,
T1, T2 | 8 | 8 | 0.025 | 5 | 11 | <0.001 |
| T3,
T4 | 3 | 21 |
| 22 | 2 |
|
| Diameter, cm |
|
|
|
|
|
|
| ≥5 | 4 | 14 | >0.999 | 12 | 6 | 0.919 |
|
<5 | 7 | 15 |
| 15 | 7 |
|
| Tumor location |
|
|
|
|
|
|
| C, A,
T | 5 | 16 | 0.583 | 16 | 5 | 0.217 |
| D, S,
R | 6 | 13 |
| 11 | 8 |
|
| Histological
type |
|
|
|
|
|
|
| Well
and moderate | 3 | 12 | 0.648 | 13 | 2 | 0.098 |
|
Poor | 8 | 17 |
| 14 | 11 |
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
Positive | 2 | 20 | 0.012 | 20 | 2 | 0.002 |
|
Negative | 9 | 9 |
| 7 | 11 |
|
| Distant
metastasis |
|
|
|
|
|
|
|
Positive | 2 | 19 | 0.020 | 18 | 3 | 0.025 |
|
Negative | 9 | 10 |
| 9 | 10 |
|
Association between miR-126 expression
and prognosis of patients with CRC
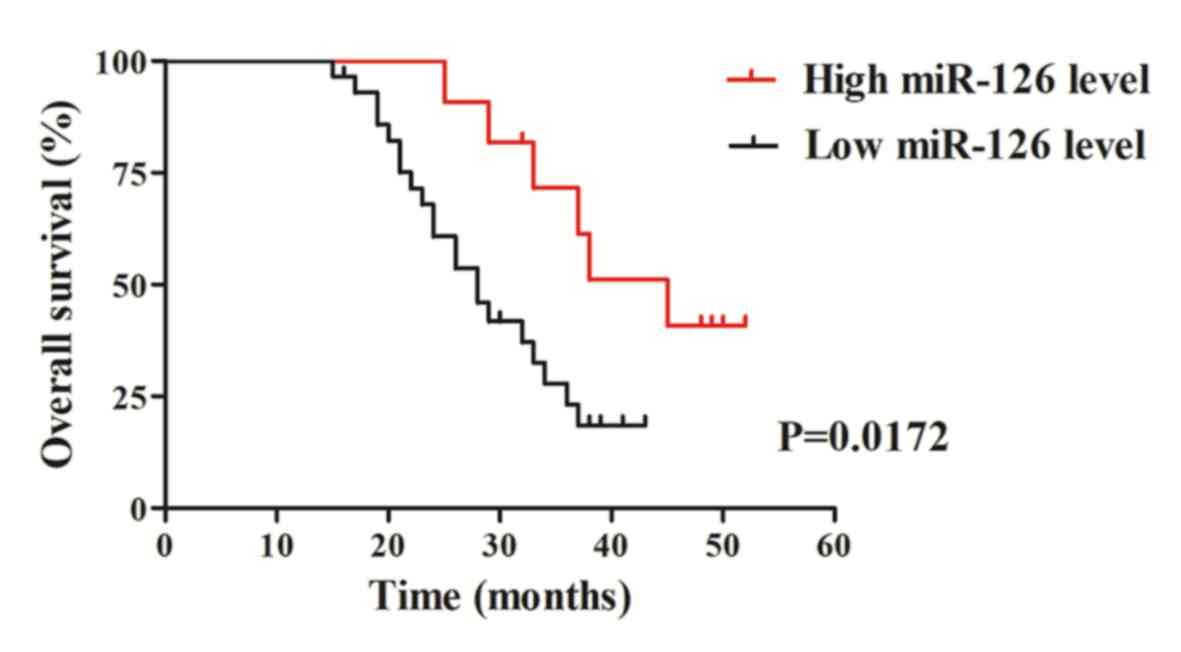
The association between miR-126 expression and
overall survival time was analyzed using the Kaplan-Meier method to
determine the prognostic value of miR-126 in patients with CRC.
Follow-up survival data was recorded up to 60 months for all 40
patients with CRC. The results demonstrated that patients with CRC,
with downregulated miR-126 expression exhibited a significantly
shorter overall survival time than those with high miR-126
expression (P=0.0172; Fig. 4). Taken
together, these results suggest that downregulated miR-126
expression may be associated with CRC progression, thus have the
potential to act as a candidate molecular marker predictive of a
poor prognosis for patients with CRC.
Discussion
Altered expression of miRNAs has been reported in
several types of human cancer, including CRC (34). miRNAs represent a set of biological
modulators that can function as either oncogenes or tumor
suppressor genes, and play an essential role in complex molecular
networks leading to the initiation, progression and metastasis of
different types of human cancer (35). For example, miR-92a, miR-140-5p,
miR-331-3p, miR-223 and miR-374a were identified as circulating
miRNA biomarkers for early diagnosis and monitoring of lung cancer
(36). Furthermore, miR-193a-5p and
miR-146a-5p have been demonstrated to induce G1 arrest
in CRC (37). Understanding the
roles of miRNA in different types of cancer will provide an
important basis for elucidation of molecular mechanisms for human
tumorigenesis and the development of novel strategies for clinical
management of human cancer.
Diagnostic and prognostic molecular biomarkers have
revolutionized clinical strategies for the management of patients
with cancer. While the majority of previous studies have primarily
focused on genetic and expression markers of protein-coding genes
(38–40), only a few studies have investigated
the potential roles of miRNA as biomarkers for clinical assessments
of different types of human cancer (41–43). A
number of studies have performed differential expression analyses
of miRNAs in several types of cancer to investigate the role of
miRNAs as clinical biomarkers and identified some miRNAs whose
aberrant expression levels are associated with pathological and
clinical phenotypes of patients with cancer (44–46).
Increased miR-210 expression has been reported in several types of
cancer, including non-small cell lung cancer (47), glioma (48), CRC (49) and higher expression of miR-210
correlated with worse recurrence free survival or disease free
survival (RFS/DFS), overall survival (OS), metastasis free survival
or distant relapse free survival (MFS/DRFS) in breast cancer. For
RFS/DFS, OS and MFS/DRFS, the combined hazard ratio (HR) and 95%
confidence interval (95% CI) of higher miR-210 expression were 3.36
(2.30, 4.93), 3.29 (1.65, 6.58) and 2.85 (1.76, 4.62) in patients
with breast cancer, respectively (50). Furthermore, increased miR-29c
expression, a miRNA molecule regulating T-cell co-regulatory
molecule B7-H3, is associated with a shorter overall survival time
of patients with breast cancer (51). In non-small cell lung cancer, altered
expression levels of miR-429, miR-19b and miR-146a have been
reported to be associated with higher TNM stage, lymph node
metastasis, unfavorable survival outcomes and poor patient
prognosis (52,53). Similarly, altered expression levels
of miRNAs have also been demonstrated in CRC. Several miRNAs,
including miR-200a (54), miR-16
(55) and miR-30b (56), have been demonstrated to be
associated with a number of clinical features in patients with CRC,
including poor differentiation of tumor, high TNM stages, lymph
node status and distant metastasis. Furthermore, altered expression
levels of these miRNAs were also demonstrated to independently
predict clinical outcomes (recurrence and survival) of patients
with CRC (55,57,58).
miR-126 is regarded as an important tumor suppressor, whereby its
downregulation has been detected in several types of cancer,
including CRC, lung cancer and breast cancer (59–62).
However, the associations between miR-126 expression and
pathological features and clinical outcomes of patients with CRC
have not been extensively investigated.
The present study assessed miR-126 expression in a
small cohort of patients with CRC, with recorded pathological
characteristics and clinical outcomes. The results demonstrated
that miR-126 was downregulated in CRC tumors, which was
significantly associated with advanced TNM and Dukes stages of
tumor, positive lymph node status and distant metastasis.
Furthermore, Kaplan-Meier survival analysis indicated that
downregulation of miR-126 expression was significantly associated
with a shorter overall survival time of patients with CRC. Taken
together, the results of the present study suggest that miR-126
expression may be a potential candidate molecular marker predictive
of the progression and prognosis of patients with CRC.
Previous studies have investigated the functional
and mechanical roles of miRNAs in different types of human cancer
(18,48,52). In
CRC, several miRNAs have been identified to be critical molecular
regulators interplaying with important molecular pathways, and thus
play key roles in the initiation and progression of CRC (24,54,56). A
previous study demonstrated that miR-30b targets oncogenic KRAS,
PIK3CD and BCL2, and regulates functional activities of the MAPK
and PI3K signaling pathways (58).
Thus, downregulation of miR-30b expression may serve as an
alternative mechanism for the activation of its downstream
oncogenic signaling pathways (58).
Previous studies have indicated the underlying molecular mechanisms
of miR-126 as a tumor suppressor in different types of human
cancer. For example, in vitro analysis of thyroid cancer
cells demonstrated that overexpression of miR-126 inhibits cell
proliferation, migration and invasion by targeting the C-X-C motif
chemokine receptor 4 (61). Another
in vitro experiment indicated that overexpression of miR-126
suppresses cell cycle progression from G1/G0
phase to S phase in the breast cancer MSF-7 cell line (60).
The mechanistic roles of miR-126 in CRC
tumorigenesis and progression have not yet been fully investigated.
A previous study used in vitro CRC cell line models and
demonstrated that miR-126 functionally targets the IRS-1 gene and
regulates IRS-1 expression in CRC cells (24). IRS-1 has been demonstrated to possess
an oncogenic property, which can lead to activation of the AKT and
MAPK signaling pathways (63).
Increased IRS-1 expression is functionally associated with enhanced
cell proliferation and migration, loss of differentiation and liver
metastasis in CRC (28,63–66).
However, to the best of our knowledge, there is currently no
evidence of the interplay between miR-126 and IRS-1 in primary CRC.
Thus, the present study simultaneously analyzed the expression
levels of miR-126 and IRS-1 mRNA, as well as IRS-1 protein levels
in primary CRC tumors, and successfully provided the first in
vivo evidence demonstrating the potential interplay between
miR-126 and IRS-1 in CRC tumorigenesis. The results of the present
study demonstrated that downregulation of miR-126 expression was
significantly associated with increased IRS-1 protein levels in
primary CRC tumors, whereas no significant association was observed
with IRS-1 mRNA expression levels. These results suggest that
miR-126 mechanistically regulates IRS-1 expression by blocking
IRS-1 protein translation, rather than targeting IRS-1 mRNA
degradation in primary CRC tumors. Taken together, the results of
the present study suggest that miR-126 is a candidate tumor
suppressor for CRC and may serve as a potential biomarker for
clinical assessment of patients with CRC. However, due to the small
sample size used in the present study, multivariate analysis could
not be performed, thus the effects of potential confounding factors
was not assessed. Therefore, prospective studies will include
larger sample sizes in order to provide sufficient evidence to
determine the role of miR-126 in CRC.
In conclusion, the results of the present study
demonstrated that miR-126 expression was significantly
downregulated in CRC tissues and inversely correlated with IRS-1
protein expression. Furthermore, downregulation of miR-126
expression was associated with advanced stages of the disease and a
poor prognosis in patients with CRC. Taken together, these results
suggest that miR-126 may function as a tumor suppressor and play an
important role in CRC carcinogenesis and progression.
Acknowledgements
The authors of the present study would like to thank
Professor Zhe Wang from the Department of Gastrointestinal Surgery
at the Affiliated Hospital of Guangdong Medical University
(Zhanjiang, China) for providing the surgical specimens and
collecting the clinical information.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YZ designed the present study. SY, CY, GZ, FS, YC,
JY and WW performed the experiments. SY, CY, WW and YZ performed
the statistical analysis and drafted the initial manuscript, while
WW and YZ critically revised the manuscript for important
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of the Affiliated Hospital of Guangdong Medical
University (Zhanjiang, China; approval no. NPJ2011005), and written
informed consent was provided by all participants prior to the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ganesh K, Stadler ZK, Cercek A, Mendelsohn
RB, Shia J, Segal NH and Diaz LA Jr: Immunotherapy in colorectal
cancer: Rationale, challenges and potential. Nat Rev Gastroenterol
Hepatol. 16:361–375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harris TJ and McCormick F: The molecular
pathology of cancer. Nat Rev Clin Oncol. 7:251–265. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mármol I, Sánchez-de-Diego C, Pradilla
Dieste A, Cerrada E and Rodriguez Yoldi MJ: Colorectal carcinoma: A
general overview and future perspectives in colorectal cancer. Int
J Mol Sci. 18:1972017. View Article : Google Scholar
|
|
6
|
Goryca K, Kulecka M, Paziewska A,
Dabrowska M, Grzelak M, Skrzypczak M, Ginalski K, Mroz A, Rutkowski
A, Paczkowska K, et al: Exome scale map of genetic alterations
promoting metastasis in colorectal cancer. BMC Genet. 19:852018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Valeri N, Gasparini P, Fabbri M, Braconi
C, Veronese A, Lovat F, Adair B, Vannini I, Fanini F, Bottoni A, et
al: Modulation of mismatch repair and genomic stability by miR-155.
Proc Natl Acad Sci USA. 107:6982–6987. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van den Berg A, Mols J and Han J:
RISC-target interaction: Cleavage and translational suppression.
Biochim Biophys Acta. 1779:668–677. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ren D, Yang Q, Dai Y, Guo W, Du H, Song L
and Peng X: Oncogenic miR-210-3p promotes prostate cancer cell EMT
and bone metastasis via NF-κB signaling pathway. Mol Cancer.
16:1172017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hong YG, Xin C, Zheng H, Huang ZP, Yang Y,
Zhou JD, Gao XH, Hao L, Liu QZ, Zhang W and Hao LQ: miR-365a-3p
regulates ADAM10-JAK-STAT signaling to suppress the growth and
metastasis of colorectal cancer cells. J Cancer. 11:3634–3644.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Wang Z, Chen M, Peng L, Wang X,
Ma Q, Ma F and Jiang B: MicroRNA-143 targets MACC1 to inhibit cell
invasion and migration in colorectal cancer. Mol Cancer. 11:232012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Cao F, Li X, Miao H, E J, Xing J
and Fu CG: miR-320b suppresses cell proliferation by targeting
c-Myc in human colorectal cancer cells. BMC Cancer. 15:7482015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li YC, Li CF, Chen LB, Li DD, Yang L, Jin
JP and Zhang B: MicroRNA-766 targeting regulation of SOX6
expression promoted cell proliferation of human colorectal cancer.
Onco Targets Ther. 8:2981–2988. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang L, Wang X, Wen C, Yang X, Song M,
Chen J, Wang C, Zhang B, Wang L, Iwamoto A, et al: Hsa-miR-19a is
associated with lymph metastasis and mediates the TNF-α induced
epithelial-to-mesenchymal transition in colorectal cancer. Sci Rep.
5:133502015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Zhou Y, Feng X, An P, Quan X, Wang
H, Ye S, Yu C, He Y and Luo H: MicroRNA-126 functions as a tumor
suppressor in colorectal cancer cells by targeting CXCR4 via the
AKT and ERK1/2 signaling pathways. Int J Oncol. 44:203–210. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng R, Chen X, Yu Y, Su L, Yu B, Li J,
Cai Q, Yan M, Liu B and Zhu Z: miR-126 functions as a tumour
suppressor in human gastric cancer. Cancer Lett. 298:50–63. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Shen Y, Ichikawa H, Antes T and
Goldberg GS: Regulation of miRNA expression by Src and contact
normalization: Effects on nonanchored cell growth and migration.
Oncogene. 28:4272–4283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Crawford M, Brawner E, Batte K, Yu L,
Hunter MG, Otterson GA, Nuovo G, Marsh CB and Nana-Sinkam SP:
MicroRNA-126 inhibits invasion in non-small cell lung carcinoma
cell lines. Biochem Biophys Res Commun. 373:607–612. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saito Y, Friedman JM, Chihara Y, Egger G,
Chuang JC and Liang G: Epigenetic therapy upregulates the tumor
suppressor microRNA-126 and its host gene EGFL7 in human cancer
cells. Biochem Biophys Res Commun. 379:726–731. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li XM, Wang AM, Zhang J and Yi H:
Down-regulation of miR-126 expression in colorectal cancer and its
clinical significance. Med Oncol. 28:1054–1057. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Z, Li N, Wu M, Li X, Luo Z and Wang X:
Expression of miR-126 suppresses migration and invasion of colon
cancer cells by targeting CXCR4. Mol Cell Biochem. 381:233–242.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou Y, Feng X, Liu YL, Ye SC, Wang H, Tan
WK, Tian T, Qiu YM and Luo HS: Down-regulation of miR-126 is
associated with colorectal cancer cells proliferation, migration
and invasion by targeting IRS-1 via the AKT and ERK1/2 signaling
pathways. PLoS One. 8:e812032013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dumpati R, Ramatenki V, Vadija R, Vellanki
S and Vuruputuri U: Structural insights into suppressor of cytokine
signaling 1 protein-identification of new leads for type 2 diabetes
mellitus. J Mol Recognit. 31:e27062018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
White MF: The IRS-signalling system: A
network of docking proteins that mediate insulin action. Mol Cell
Biochem. 182:3–11. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang Q, Li Y, White MF, Fletcher JA and
Xiao S: Constitutive activation of insulin receptor substrate 1 is
a frequent event in human tumors: Therapeutic implications. Cancer
Res. 62:6035–6038. 2002.PubMed/NCBI
|
|
28
|
Esposito DL, Aru F, Lattanzio R, Morgano
A, Abbondanza M, Malekzadeh R, Bishehsari F, Valanzano R, Russo A,
Piantelli M, et al: The insulin receptor substrate 1 (IRS1) in
intestinal epithelial differentiation and in colorectal cancer.
PLoS One. 7:e361902012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Esposito DL, Verginelli F, Toracchio S,
Mammarella S, De Lellis L, Vanni C, Russo A, Mariani-Costantini R
and Cama A: Novel insulin receptor substrate 1 and 2 variants in
breast and colorectal cancer. Oncol Rep. 30:1553–1560. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li N, Li X, Huang S, Shen S and Wang X:
miR-126 inhibits colon cancer proliferation and invasion through
targeting IRS1, SLC7A5 and TOM1 gene. Zhong Nan Da Xue Xue Bao Yi
Xue Ban. 38:809–817. 2013.(In Chinese). PubMed/NCBI
|
|
31
|
Sorbin LH, Gospodarowicz MK and Wittekind
C: International Union against Cancer (UICC). TNM classification of
malignant tumors. 7. West Sussex; UK, Wiley-Blackwell: 2009
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kurien BT and Scofield RH: Western
blotting: An introduction. Methods Mol Biol. 1312:17–30. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen WS, Chen TW, Yang TH, Hu LY, Pan HW,
Leung CM, Li SC, Ho MR, Shu CW, Liu PF, et al: Co-modulated
behavior and effects of differentially expressed miRNA in
colorectal cancer. BMC Genomics. 14 (Suppl 5):S122013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang YH, Jin M, Li J and Kong X:
Identifying circulating miRNA biomarkers for early diagnosis and
monitoring of lung cancer. Biochim Biophys Acta Mol Basis Dis.
1658472020.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Noorolyai S, Baghbani E, Aghebati Maleki
L, Baghbanzadeh Kojabad A, Shanehbansdi D, Khaze Shahgoli V,
Mokhtarzadeh A and Baradaran B: Restoration of miR-193a-5p and
miR-146 a-5p expression induces G1 arrest in colorectal cancer
through targeting of MDM2/p53. Adv Pharm Bull. 10:130–134. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li XQ, Li L, Xiao CH and Feng YM: NEFL
mRNA expression level is a prognostic factor for early-stage breast
cancer patients. PLoS One. 7:e311462012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Salvatore L, Calegari MA, Loupakis F,
Fassan M, Di Stefano B, Bensi M, Bria E and Tortora G: PTEN in
colorectal cancer: Shedding light on its role as predictor and
target. Cancers (Basel). 11:17652019. View Article : Google Scholar
|
|
40
|
Li Z, Xue TQ, Yang C, Wang YL, Zhu XL and
Ni CF: EGFL7 promotes hepatocellular carcinoma cell proliferation
and inhibits cell apoptosis through increasing CKS2 expression by
activating Wnt/β-catenin signaling. J Cell Biochem.
119:10327–10337. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tachibana H, Sho R, Takeda Y, Zhang X,
Yoshida Y, Narimatsu H, Otani K, Ishikawa S, Fukao A, Asao H and
Iino M: Circulating miR-223 in oral cancer: Its potential as a
novel diagnostic biomarker and therapeutic target. PLoS One.
11:e01596932016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang J, Zhou Y, Fei X, Chen X and Zhu Z:
Regulator of G-protein signaling 3 targeted by miR-126 correlates
with poor prognosis in gastric cancer patients. Anticancer Drugs.
28:161–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang H, Mao F, Shen T, Luo Q, Ding Z,
Qian L and Huang J: Plasma miR-145, miR-20a, miR-21 and miR-223 as
novel biomarkers for screening early-stage non-small cell lung
cancer. Oncol Lett. 13:669–676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ruan L and Qian X: MiR-16-5p inhibits
breast cancer by reducing AKT3 to restrain NF-κB pathway. Biosci
Rep. 39:BSR201916112019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi XY, Wang H, Wang W and Gu YH:
MiR-98-5p regulates proliferation and metastasis of MCF-7 breast
cancer cells by targeting Gab2. Eur Rev Med Pharmacol Sci.
23:2847–2855. 2019.PubMed/NCBI
|
|
47
|
Yang F, Yan Y, Yang Z, Hong X, Wang M,
Yang Y, Ye L and Liu B: miR-210 in exosomes derived from CAFs
promotes non-small cell lung cancer migration and invasion through
PTEN/PI3K/AKT pathway. Cell Signal. 73:1096752020.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lan F, Yue X and Xia T: Exosomal
microRNA-210 is a potentially non-invasive biomarker for the
diagnosis and prognosis of glioma. Oncol Lett. 19:1967–1974.
2020.PubMed/NCBI
|
|
49
|
Wang W, Qu A, Liu W, Liu Y, Zheng G, Du L,
Zhang X, Yang Y, Wang C and Chen X: Circulating miR-210 as a
diagnostic and prognostic biomarker for colorectal cancer. Eur J
Cancer Care. 22:Feb;2016.(Epub ahead of print).
|
|
50
|
Li M, Ma X, Li M, Zhang B, Huang J, Liu L
and Wei Y: Prognostic role of microRNA-210 in various carcinomas: A
systematic review and meta-analysis. Dis Markers. 2014:1061972014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nygren MK, Tekle C, Ingebrigtsen VA,
Mäkelä R, Krohn M, Aure MR, Nunes-Xavier CE, Perälä M, Tramm T,
Alsner J, et al: Identifying microRNAs regulating B7-H3 in breast
cancer: The clinical impact of microRNA-29c. Br J Cancer.
110:2072–2080. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhu W, He J, Chen D, Zhang B, Xu L, Ma H,
Liu X, Zhang Y and Le H: Expression of miR-29c, miR-93, and miR-429
as potential biomarkers for detection of early stage non-small lung
cancer. PLoS One. 9:e877802014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wu C, Cao Y, He Z, He J, Hu C, Duan H and
Jiang J: Serum levels of miR-19b and miR-146a as prognostic
biomarkers for non-small cell lung cancer. Tohoku J Exp Med.
232:85–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Y, Sun J, Cai Y, Jiang Y, Wang X, Huang
X, Yin Y and Li H: MiR-200a acts as an oncogene in colorectal
carcinoma by targeting PTEN. Exp Mol Pathol. 101:308–313. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Qian J, Jiang B, Li M, Chen J and Fang M:
Prognostic significance of microRNA-16 expression in human
colorectal cancer. World J Surg. 37:2944–2949. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhao H, Xu Z, Qin H, Gao Z and Gao L:
miR-30b regulates migration and invasion of human colorectal cancer
via SIX1. Biochem J. 460:117–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pichler M, Ress AL, Winter E, Stiegelbauer
V, Karbiener M, Schwarzenbacher D, Scheideler M, Ivan C, Jahn SW,
Kiesslich T, et al: MiR-200a regulates epithelial to mesenchymal
transition-related gene expression and determines prognosis in
colorectal cancer patients. Br J Cancer. 110:1614–1621. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY,
Cui YM, Qi L, Wu P, Jiao HL, Xie YJ, et al: MicroRNA-30b functions
as a tumour suppressor in human colorectal cancer by targeting
KRAS, PIK3CD and BCL2. J Pathol. 232:415–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu Y, Zhou Y, Feng X, Yang P, Yang J, An
P, Wang H, Ye S, Yu C, He Y and Luo H: Low expression of
microRNA-126 is associated with poor prognosis in colorectal
cancer. Genes Chromosomes Cancer. 53:358–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dong Y, Fu C, Guan H, Zhang Z, Zhou T and
Li B: Prognostic significance of miR-126 in various cancers: A
meta-analysis. Onco Targets Ther. 9:2547–2555. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Qian Y and Wang X, Lv Z, Guo C, Yang Y,
Zhang J and Wang X: MicroRNA-126 is downregulated in thyroid cancer
cells, and regulates proliferation, migration and invasion by
targeting CXCR4. Mol Med Rep. 14:453–459. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ebrahimi F, Gopalan V, Wahab R, Lu CT,
Smith RA and Lam AK: Deregulation of miR-126 expression in
colorectal cancer pathogenesis and its clinical significance. Exp
Cell Res. 339:333–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wu L, Shi B, Huang K and Fan G:
MicroRNA-128 suppresses cell growth and metastasis in colorectal
carcinoma by targeting IRS1. Oncol Rep. 34:2797–2805. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yin Y, Yan ZP, Lu NN, Xu Q, He J, Qian X,
Yu J, Guan X, Jiang BH and Liu LZ: Downregulation of miR-145
associated with cancer progression and VEGF transcriptional
activation by targeting N-RAS and IRS1. Biochim Biophys Acta.
1829:239–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Li H, Meng F, Ma J, Yu Y, Hua X, Qin J and
Li Y: Insulin receptor substrate-1 and Golgi phosphoprotein 3 are
downstream targets of miR-126 in esophageal squamous cell
carcinoma. Oncol Rep. 32:1225–1233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Meyer K, Albaugh B, Schoenike B and Roopra
A: Type 1 insulin-like growth factor receptor/insulin receptor
substrate 1 signaling confers pathogenic activity on breast tumor
cells lacking REST. Mol Cell Biol. 35:2991–3004. 2015. View Article : Google Scholar : PubMed/NCBI
|