Gastric cancer (GC) is the fourth most commonly
diagnosed cancer and the third leading cause of cancer-related
mortality worldwide (1). The
pathogenic mechanisms and progression of GC are complex. Currently,
well-known pathogenic factors include poor eating habits, chronic
Helicobacter pylori infection and the misregulation of
oncogenes or tumor suppressors (2).
The 5-year survival rate of patients with GC is only 27.4% in China
(3). The poor prognosis of GC is due
to tumor invasion and metastasis (4,5), which
are complex processes involving a series of cell biological
regulations and requiring multi-step genetic mutations (6). The genes and their products involved in
each step of tumor progression are potential prognostic markers and
therapeutic targets. Among these biomarkers, E3 ligases play a
crucial role in the proliferation, invasion and metastasis of GC
(7).
The ubiquitin-proteasome system (UPS) is a common
post-translational modification pathway involved in the regulation
of cell survival and differentiation (8). The ubiquitination pathway is catalyzed
by three types of key enzymes: Ubiquitin-activating enzyme (E1),
ubiquitin-conjugating enzyme (E2) and ubiquitin ligases (E3)
(9). E3 ligase is the most important
component in the UPS owing to its specific ability to recognize
target proteins and transfer ubiquitin to substrates for
degradation (9,10). To date, >650 E3 ubiquitin ligases
have been described in humans, and they can be subdivided into
three different families: Homologous to E6-associated protein
C-terminus (HECT), really interesting new gene (RING) and
RING-in-between-RING (RBR) E3 ligases (11,12).
However, RBR E3 ligases, an emerging group of E3 ligases that
feature with characteristics of RING and HECT, have not been
discovered in GC (13). Hence, RING
and HECT E3 ligases are dysregulated in GC cells. Owing to the
notable role of E3 ligases on the ubiquitination of
tumor-associated signaling molecules, such as the AKT pathway,
targeting E3 ligases could be an efficient approach in cancer
treatment (14). Recently, many new
types of E3 ligases have been increasingly detected in GC, such as
RNF6 and RNF38 (15,16). Herein, a comprehensive review of
studies is presented to summarize the latest progress and treatment
prospects.
HECTs, the second largest E3 ligase family in
humans, comprises 28 members and can be categorized into three
subfamilies: Neural precursor cell expressed developmentally
downregulated protein (NEDD)4 family, HECT domain-containing
protein (HERC) family and ‘other’ HECT ligases (22). Studies to date have suggested that
only the NEDD4 family and a member of the Other HECT ligase
families are related to GC. The NEDD4 subfamily is the most
characteristic family, including nine members in humans: NEDD4-1
(also known as NEDD4), NEDD4-2, Itchy E3 ubiquitin-protein ligase,
SMAD ubiquitin regulatory factor (Smurf)1, Smurf2, WW
domain-containing E3 ubiquitin protein ligase (WWP)1, WWP2,
NEDD4-like 1 (NEDL1) and NEDL2 (23). The Other HECT ligase family includes
13 members, and they all contain different domains in addition to
the HECT domain.
The structure and functional significance of RINGs
in GC will be described in the following paragraphs in the order of
monomer, dimer and multiple subunit E3 ligases, and the HECTs will
be described in the order of the NEDD4 family, HERC family and
other HECT ligases.
The RNF protein family contains a large number of
members that are associated with several types of digestive system
tumors, such as colorectal cancer and hepatocellular carcinoma
(24,25). RNFs also play a vital role in the
occurrence of GC. RNF6 encodes a 685-amino-acid protein with a
coiled-coil domain at the N-terminus and a RING-H2 finger at the
C-terminus (26). RNF38 shares a
similar structure with RNF6 (27),
and previous studies have shown that RNF6 and RNF38 are
overexpressed in GC and regulate GC cell growth. Both RNF6 and
RNF38 induce polyubiquitination of tyrosine-protein phosphatase
non-receptor type 6 and subsequently enhance STAT3 signaling, which
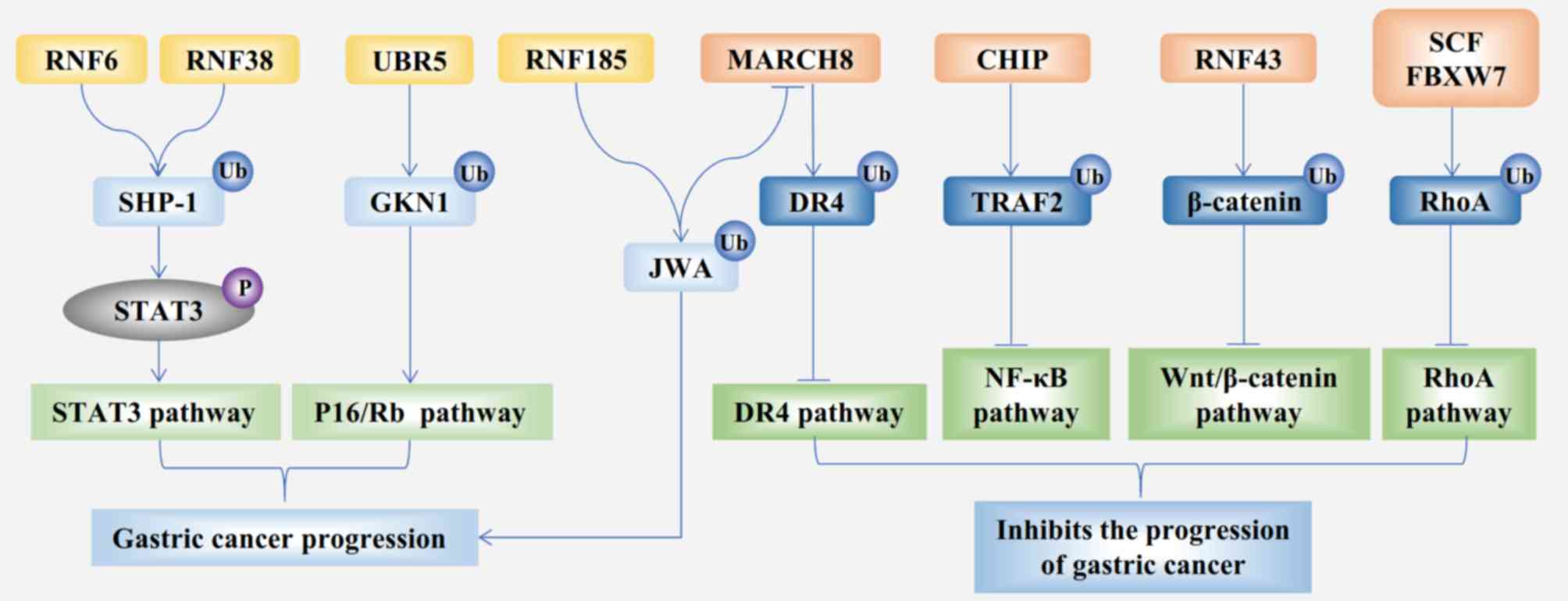
promotes the proliferation of GC cells (Fig. 1) (15,16).
RNF26, which is located in the endoplasmic
reticulum, is a polypeptide of 433 amino acids with an N-terminal
leucine zipper domain and a C-terminal RING finger domain (28,29). The
expression level of RNF26 is upregulated in several types of human
cancer cell lines, including HL-60, HeLa S3, SW480 and MKN7 cells
(28). As the substrate protein of
RNF26, the mediator of interferon regulatory factor 3 can be
ubiquitinated and regulate the innate antiviral response (30). However, the functional mechanism of
RNF26 in GC has not been fully elucidated.
Similar to RNF6, RNF26 and RNF38, RNF185 also acts
as an oncogene in GC, but with distinct subcellular localization
and a distinct mechanism of action. RNF185 localizes to the
mitochondria, contains a C3HC4-type RING domain and two
transmembrane domains (31). PRA1
family protein 3 (JWA) is a multifunctional cytoskeleton-binding
protein induced by all-trans retinoic acid. The function of JWA
involves enhancing intracellular defenses against
H2O2-induced oxidative stress and reducing
cell apoptosis (32). RN F185 can
downregulate the expression of JWA and promote GC cell migration
(Fig. 1) (33). Generally, higher expression of RNF185
is associated with a worse prognosis of GC.
RNF43 and zinc/RING finger 3 (ZNRF3) are homologous
proteins that have antagonistic effects in combination with
leucine-rich repeat-containing G-protein-coupled receptor 5 (Lgr5)
(34). The Lgr5 protein, a member of
the G-protein coupled receptor family of proteins, was identified
as a novel gastrointestinal stem cell marker (35). Moreover, Lgr5-positive gastric stem
cells are cancer-initiating cells able to drive GC cell
self-renewal, which contributes to malignant progression (35–37).
RNF43 negatively regulates the Wnt/β-catenin pathway by recognizing
Lgr5 and markedly downregulating the expression of Lgr5 protein
(34). A previous study demonstrated
that ZNRF3 acts as a tumor suppressor by downregulating the
expression levels of β-catenin and transcription factor 4 protein
(38). RNF43/ZNRF3 mediates the
ubiquitylation of seven transmembrane domains of frizzled receptors
and subsequently inhibits the proliferation of GC cells (39). A few large-scale genomic analyses
have reported RNF43 mutations in different cancer types, including
GC (40). Whole-genome sequencing
revealed that RNF43 is mutated in 4.8% of microsatellite-stable and
54.6% of microsatellite-unstable tumors (41). The mutational landscape of RNF43 may
provide a new approach to facilitate genome-guided personalized
therapy in GC. Overall, RNF43/ZNRF3 is a tumor suppressor and a
potential therapeutic target for GC. Therefore, five members of the
RNF subfamily participate in regulating the development of GC,
where RNF6, RNF26, RNF38 and RNF185 function as carcinogenic
factors, while RNF43/ZNRF3 acts as a tumor suppressor.
MARCH8 is a member of the MARCH subfamily. A recent
study identified 11 E3 ligases that contained the RING-CH domain
and were named the MARCH1-11 subfamily (42). The structure of MARCH8 contains the
RING-CH domain and transmembrane domains. JWA is a downstream
protein of RNF185 ligase (33); it
promotes the ubiquitination of death receptor 4 by increasing the
expression of MARCH8 in GC cells, thereby reducing tumor necrosis
factor-related apoptosis-inducing ligand-mediated apoptosis
(Fig. 1) (43). Moreover, MARCH8 induces the apoptosis
of GC cell lines by inactivating the PI3K and β-catenin/STAT3
signaling pathways (44). In
summary, MARCH8 is a tumor suppressor in GC.
TRIM proteins form a subfamily that regulates
multiple cellular processes. TRIMs share a common N-terminal
tripartite domain, a RING domain, one or two B-boxes and
coiled-coil domains (45). To date,
three E3 ligases have been shown to be involved in the development
and metastasis of GC. They are TRIM28 (also known as KAP1 or
TIF1-β), TRIM29 and TRIM59 (46–49).
TRIM28 is a universal transcriptional corepressor (50). The overexpression of TRIM28 causes
peritoneal carcinomatosis and poor prognosis in GC (47). Similar to TRIM28, the expression of
TRIM29 is also upregulated in GC; the high expression of TRIM29
mRNA may be an independent predictor of lymph node metastasis and
depth of invasion (48). TRIM59 has
been reported in several human tumors and acts on diverse signaling
pathways, such as the focal adhesion kinase/AKT/matrix
metalloproteinases pathway in epithelial ovarian cancer (51), the PI3K/AKT/mTOR pathway in human
cholangiocarcinoma (52), the
Wnt/β-catenin signaling pathway in neuroblastoma (53), the NF-κB pathway in non-small cell
lung cancer (54) and the p53
signaling pathway in GC (49). The
p53 signaling pathway can be negatively regulated by the E3 ligase
TRIM59, which enhances the ubiquitination and subsequent
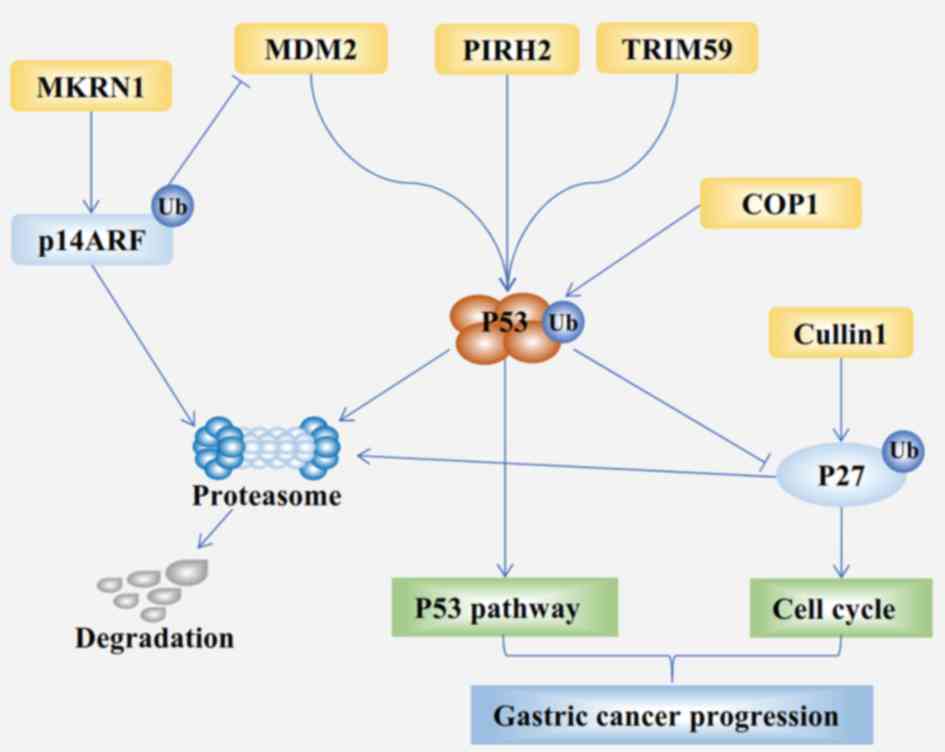
degradation of p53 (Fig. 2). TRIM59
might promote gastric carcinogenesis through this mechanism
(53). In summary, TRIM28, TRIM29
and TRIM59 play oncogenic roles in gastric tumorigenesis, but only
the regulatory mechanism of TRIM59 has been elucidated.
CBL proteins contain two highly conserved domains:
The N-terminal tyrosine kinase-binding domain and the C3HC4 RING
finger domain (60). The mammalian
CBL family consists of Cbl, Cbl-b and c-Cbl ligases (61). The three members have similar
functions, perhaps due to the specific structure. The c-Cbl protein
was the first CBL family protein discovered, followed by Cbl-b and
Cbl (62). Previous studies have
confirmed that all three CBL proteins are closely associated with
GC. In 2000, a study reported that the c-Cbl protein was frequently
tyrosine phosphorylated in a tumor-specific manner in human GC
tissues (63). Subsequently, c-Cbl
was found to be involved in stomach carcinogenesis by connecting
with the EGFR system (64).
Cbl-b has a significant impact on the prognosis and
drug sensitivity of GC, and a previous study showed that Cbl-b is
an oncogene (64). However,
subsequent studies provide different perspectives, in which Cbl-b
is found to enhance the sensitivity to 5-fluorouracil and cetuximab
in GC cells through the ubiquitination pathway (65,66).
Other studies have also indicated that Cbl-b inhibits tumor
metastasis and growth in multiple drug-resistant (MDR) gastric and
breast cancer cells, as well as increasing the sensitivity of MDR
cells to anticancer drugs (67–69).
These findings provide a new research direction for the
chemotherapy and targeted therapy of GC.
Cbl, in conjunction with the EGFR system, might be
related to gastric carcinogenesis and metastasis (70). In summary, Cbl and c-Cbl are
oncogenes in GC, whereas Cbl-b can act as an oncogene or tumor
suppressor, and only its regulatory mechanism has been well
clarified among the three members.
Previous studies have reported that the aberrant
methylation of CHFR promotes the development of GC (71,72). As
an E3 ligase, CHFR contains an N-terminal forkhead-associated
domain, a central RING domain and a C-terminal cysteine-rich region
(73). PARP-1 may be a substrate by
CHFR for ubiquitination and degradation in GC; this process leads
to cell cycle arrest before entering mitosis and inhibits the
proliferation of GC cells (74).
Thus, CHFR functions as a tumor suppressor in GC.
These two E3 ligases are rarely mentioned in GC.
MKRN1 was first identified owing to its interaction with human
telomerase reverse transcriptase (TERT) and modulation of telomere
length homeostasis (75). The
MKRN1-mediated ubiquitination of tumor suppressor ARF (p14ARF) was
described in 2011 (76). MKRN1
induces the ubiquitination and degradation of p14ARF and
downregulated p14ARF expression (Fig.
2) (76). Furthermore, MKRN1
overexpression was associated with well-differentiated gastric
carcinoma, whereas p14ARF overexpression was associated with poorly
differentiated gastric carcinoma. FIGC, a novel FGF-induced
ubiquitin-protein ligase, consists of an N-terminal RING finger
module and proline-rich region at the C-terminus. Only one study
has shown that FIGC probably functions as an E3 ligase and is
implicated in carcinogenesis through the dysregulation of
fibroblast growth modulator (77).
In brief, further research is needed to confirm the mechanism of
these two E3 ligases in GC.
Dimeric RING domain E3 ligases can be classified
into homodimers and heterodimers. MDM2, a heterodimeric RING
ligase, was originally identified as a ubiquitin ligase E3 that
promotes the degradation of tumor suppressor p53 (Fig. 2) (78). Subsequent studies suggest that MDM2
can ubiquitinate and degrade multiple signaling molecules in GC,
including p53, forkhead box protein O3A (FOXO3A) and runt-related
transcription factor 3 (RUNX3) (78–81).
Human hTERT can interact with MDM2 and dramatically increase the
ubiquitination of FOXO3A, resulting in the invasion of GC cells
(79). RUNX3 is known as a tumor
suppressor (82). MDM2 ligases can
recognize Lys94 and Lys148 of RUNX3 and decrease the expression
levels of RUNX3 (80). Overall, MDM2
acts as a carcinogenic factor in GC by affecting different
signaling proteins.
CHIP includes a C-terminal U-box domain and an
N-terminal tetratricopeptide repeat domain, which have E3 ubiquitin
ligase activity and interact with the molecular chaperones
Hsc70-Hsp70 and Hsp90, respectively (83). CHIP is characterized as a homologous
dimeric RING ligase and antioncogene in human cancer (15,84). The
U-box domain of CHIP facilitates TNF receptor-associated factor 2
ubiquitination for degradation and then inactivates NF-κB signaling
(Fig. 1). A previous study also
showed that CHIP expression prevents the angiogenesis and
metastasis of GC (85). Above all,
CHIP overexpression is correlated with good prognosis in GC
patients, and targeting CHIP may be a new approach in GC
therapy.
The CRL1 complex comprises SKP1, CUL1, RING box1
(RBX1) and a member of the F-box protein family (86). The F-box protein family can be
further categorized into three subclasses: i) FBXW; ii) F-box and
leucine-rich repeat (FBXL); and iii) F-boxes containing other
domain motifs (FBXO) proteins. Each subunit of SCF has unique
features as follows: i) CUL1 serves as a rigid molecular scaffold
protein; ii) RBX1 contains a RING finger domain for the recruitment
of E2 enzyme; iii) SKP1 functions as an adaptor; iv) F-box proteins
act as a substrate-determining component (86,87),
many of them function as E3 ligase and will be discussed in detail
below.
The WD repeat domain comprises a 44–60 residue
sequence that typically contains the GH dipeptide 11–24 residues
from its N-terminus and the WD dipeptide at the C-terminus
(88). This class of E3 ligases
mainly recognizes proteins involved in cell cycle regulation and
tumorigenesis, thereby regulating cancerous growth. FBXW7 and
β-transducin repeat-containing protein (β-TRCP) are directly
correlated with the progression of GC in the form of E3 ligases
(89,90).
FBXW7 is a well-characterized SCF in GC and
facilitates the destruction of oncogenic proteins, such as c-Myc,
transforming protein RhoA (RhoA), transcription activator BRG1
(Brg1) and zinc-finger protein GFI-1 (GFI1) (90–93).
These targeted proteins all govern gastric tumorigenesis; for
example, Brg1 promotes the metastasis of GC (92), RhoA has been implicated in gastric
tumorigenesis (Fig. 1) (94), and GFI1 promotes GC cell
proliferation as an oncoprotein (93). FBXW7 is also regulated by several
upstream signaling molecules. Previous studies indicated that
microRNAs and long noncoding RNAs are involved in the occurrence
and development of GC by altering the expression of FBXW7 (95,96).
Therefore, FBXW7 is a complex tumor suppressor in GC because of its
involvement in numerous upstream and downstream signals.
β-TRCP has two distinct isoforms, β-TRCP1 and
β-TRCP2, which share similar biochemical properties (97). It was reported that β-TRCP1 and
β-TRCP2 were predominantly expressed in the stomach and the small
intestine, respectively. A previous study showed that β-TRCP2 might
promote gastric carcinogenesis through the activation of the Wnt
signaling pathway (98). Moreover,
β-TRCPs participate in the regulation of the AKT signaling pathway
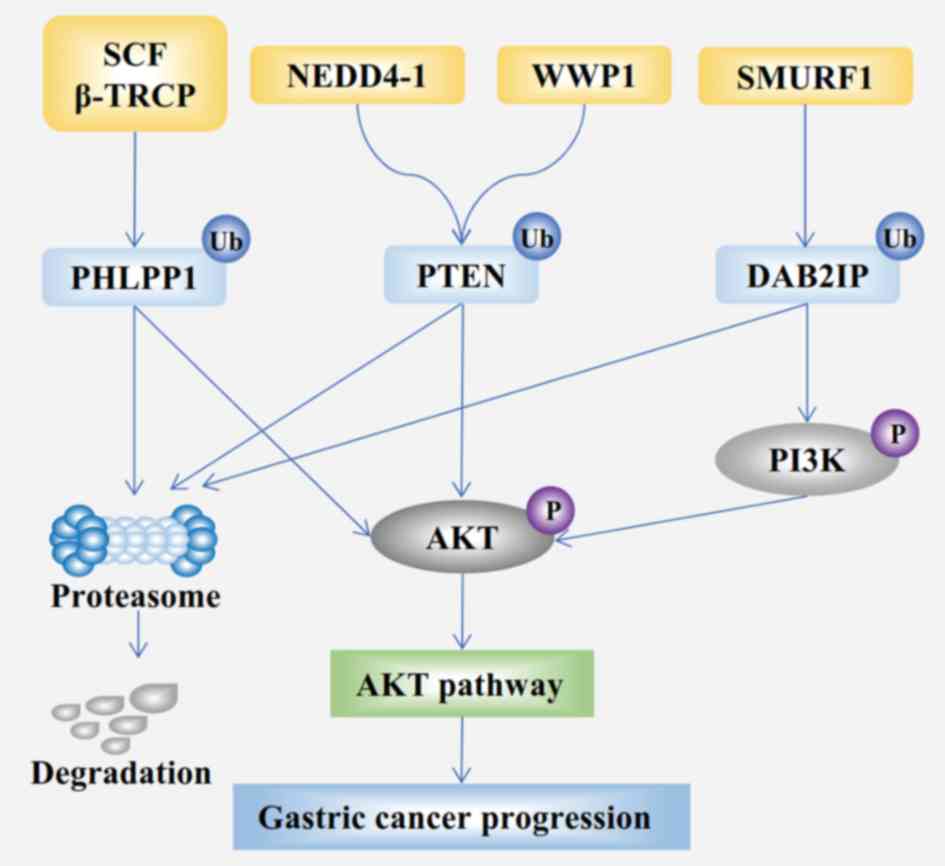
(Fig. 3) and epidermal growth factor
receptor/glycogen synthase kinase-3β/FOXP3 axis through the
ubiquitination of PH domain leucine-rich repeat-containing protein
phosphatase 1 and FOX3, respectively (99,100).
One previous study has reported that β-TRCP can serve as a tumor
suppressor or oncoprotein in the etiology of a variety of cancers
depending on the type of tumor tissue (99). Nevertheless, β-TRCP might serve as an
oncoprotein in GC.
FBXL proteins contain leucine-rich repeat sequences.
FBXL2 and FBXL5 exhibit similar characteristics in GC (101,102).
FBXL2 promotes the ubiquitination and degradation of FOXM1, which
then inhibits GC proliferation (101). Similarly, FBXL5 can also suppress
GC cell migration by the ubiquitination-mediated destruction of
Cortactin and Snail1 (102,103). FBXO31, a member of the third class
of the F-box protein family, can also target Snail1 for its
ubiquitylation and degradation (104). Hence, FBXL2, FBXL5 and FBXO31 exert
tumor inhibitory roles in GC.
CUL1 is a member of the CUL family and acts as a
scaffold protein of the SCF ubiquitin E3 ligase (105). CUL1 is modified by the
ubiquitin-like protein NEDD8 and enhances the activity of SCF
ligases to p27 (106). Several
studies have shown that diverse types of human malignant tumors are
related to CUL1. CUL1 can facilitate cell proliferation in
osteosarcoma, breast cancer, prostate cancer and lung cancer in
vitro and in vivo (107–110).
In addition, the co-expression of CUL1 and CUL2 induces the
initiation of carcinogenesis in colorectal cancer by arresting
p53-positive colon cells in the G1 phase of the cell cycle
(111). Before these findings,
immunohistochemistry results suggested that high expression levels
of CUL1 were detected in 60% of all GC tissues, in a study of 792
patients (112). Further in
vitro studies showed that increased CUL1 expression was
correlated with poor patient survival by decreasing p27 expression
(112) (Fig. 2). Therefore, CUL1, an oncoprotein,
can be regarded as a prognostic biomarker of GC.
Von Hippel-Lindau disease was first considered to be
a heritable cancer syndrome characterized by retinal and neuronal
hemangioblastoma owing to a mutation in the VHL gene (113). Although the pVHL protein itself
displays no enzymatic activity, pVHL functions as a substrate
recognition subunit in CRL2 E3 ligases after binding with the
elongin and CUL proteins (114).
pVHL is a tumor suppressor in renal cell carcinoma (RCC) (115,116).
Serine/threonine-protein kinase NEK8 is a serine/threonine-specific
protein kinase family member that serves a role in the progression
of mitosis and carcinogenesis (117). NEK8 is reported to be a novel
target of pVHL in the regulation of RCC and GC (118,119);
pVHL promotes NEK8 protein degradation via the proteasome-ubiquitin
pathway in GC cells (119). In
summary, the E3 ligase pVHL targets NEK8 and inhibits the
proliferation, colony formation and migration of GC (119).
COP1 protein structure comprises an N-terminal RING
finger region, a coiled-coil domain and seven WD40 repeats at its
C-terminus (120). COP1 is well
studied in plants, but it has been little studied in humans. COP1
is known as a central repressor in the light signaling pathway and
forms a complex with suppressor of PHYA-1 (COP1-SPA1) (121). This complex interacts with the
CUL4-DDB1 ligase, which belongs to the CRL4 family, to form
CUL4-DDB1COP1-SPA1 ligases in plants (122). However, the catalytic mechanism of
COP1 has not been fully elucidated in humans. Previous studies
revealed that the ubiquitin ligase COP1 promoted the progression of
multiple cancer types in vitro, including GC, by
downregulating the expression of p53 (Fig. 2) (123–125).
The role of COP1 in GC is controversial (123–126).
One previous study indicates that low COP1 expression resulted in
poorer prognoses in patients with GCs (126); however, more studies have suggested
that COP1 functions as an oncogene (123–125).
In summary, there are eight multi-subunit RING
domain E3 ligases associated with GC, including six SCFs, one CRL2
ligase and one CRL4 ligase. Among these eight CRLs, β-TRCP, CUL1
and COP1 are oncoproteins, whereas FBXW7, FBXL2, FBXL5, FBXO31 and
pVHL act as tumor suppressors.
The E3 ligases of the NEDD4 subfamily are
characterized by an N-terminal C2 domain responsible for
subcellular localization, between two and four WW domains that
recruit substrates and a HECT domain at the C-terminus (127). Three NEDD4 subfamily members might
be related to GC, including NEDD4-1, WWP1 and Smurf1, and they will
be elaborated below.
NEDD4-1 contains four WW domains and is an ancestral
member of the NEDD4 family (23).
Previous studies have indicated that NEDD4-1 is frequently
overexpressed in several types of human cancers, including
hepatocellular carcinoma, lung cancer and gastrointestinal cancer
(128–130). A range of tumor suppressors can be
ubiquitinated by NEDD4-1, including PTEN, c-Myc and large tumor
suppressor kinase 1 (128,131). Immunohistochemical analysis
conducted by Kim et al (130) showed that NEDD4-1 is overexpressed
in colorectal and gastric carcinomas. NEDD4-1 was also found to
promote GC cell migration and invasion (129). As a carcinogenic factor, the
targets of NEDD4-1 remain unclear in human GC, and further studies
are needed to explore this research topic.
WWP1 is another GC-related member of the NEDD4
subfamily that also has four WW domains. Similar to NEDD4-1, WWP1
has been revealed as a versatile E3 ligase with a large repertoire
of substrates (23). In GC cell
lines, WWP1 is overexpressed, and is closely associated with worse
survival by regulating the PTEN-AKT signaling pathway in patients
with GC (Fig. 3) (132). The overall survival rate of
patients who were positive for WWP1 protein was 25.9%, whereas it
was 66.0% in patients who without WWP1 protein in China in 2015
(132). Subsequent studies further
confirmed that WWP1 might play a role as an oncogene in GC.
miR-584-5p, miR-129-5p and miR-129-3p were found to suppress WWP1
protein expression and inhibit the proliferation of GC in
vivo and in vitro (133,134).
These findings suggested that WWP1 might be a valuable prognostic
marker and potential target in the treatment of GC.
Smurf1 was first recognized in selective
interactions with receptor-regulated SMADs, which led to its
initial naming (135). Smurf1
contains two WW domains and negatively regulates the transforming
growth factor-β/bone morphogenetic protein signaling (136). DAB2-interacting protein (DAB2IP), a
tumor suppressor, is known to be downregulated in GC (137); Smurf1 significantly promotes the
ubiquitination-dependent degradation of DAB2IP (Fig. 3) (138). In addition, a subsequent study
concluded that the Smurf1/DAB2IP signaling axis has an important
impact in GC (139). Overall,
Smurf1 might act an oncoprotein in GC.
UBR5 is the only member of Other HECT ligase family
that serves a role in regulating GC cell growth (140). Structurally, UBR5 is composed of an
N-terminal ubiquitin-associated domain, two nuclear localization
signals, a ubiquitin recognin box domain, a C-terminal
poly(A)-binding domain and a HECT domain at its far C-terminus
(141). A previous study showed
that UBR5 gene mutations occur in 27.8% of GC and 23.3% of
colorectal cancer (142).
Gastrokine 1, a gastric tumor suppressor, can be downregulated by
UBR5 E3 ligase (Fig. 1), and the
overexpression of UBR5 is associated with poor overall and
disease-free survival (140). Thus,
UBR5 may serve as a carcinogenic agent and a prognostic factor in
GC.
E3 ligases have potent effects on the origin,
progression and prognosis of GC through a series of signaling
pathways. E3 ligase-targeting molecules and drugs may provide a new
approach to GC treatment. Bortezomib was the first proteasome
inhibitor approved by the US Food and Drug Administration in
multiple myeloma (143). In
vitro, bortezomib has a significant negative effect on the
growth of GC cells (144). It is
possible that bortezomib may become a common adjuvant therapeutic
target in GC because it has a significant negative effect on the
proliferation of GC cells (144).
However, only a few E3 ligase-targeting molecules have the ability
to suppress the progression of GC.
APG115 has been identified as a novel inhibitor of
MDM2 ligase, and its potential for treating GC has been shown in
vitro and in vivo (145). In vitro, APG115 inhibited
the proliferation of GC cell lines that harbored MDM2 expression by
downregulating the mRNA expression of MDM2. In a xenograft mouse
model, APG115 contributed to a smaller GC tumor size and enhanced
the effect of radiotherapy. As previously mentioned, MDM2
downregulates several tumor suppressors in GC. Consequently, the
MDM2 inhibitor APG115 may be applied for GC treatment in the
future.
Nutlin proteins were identified in 2004 as the first
selective small molecules of MDM2, which could antagonize p53-MDM2
binding (146). Among the nutlins,
only nutlin-3 represents a promising therapeutic candidate for drug
development in human cancer (147).
Over the past decade, it has been confirmed that nutlin-3 can
induce cell growth arrest and apoptosis in a number of cancer cell
types (148,149). In p53-defective cancer cells, there
is a synergistic effect between nutlin-3 and bortezomib;
cotreatment with bortezomib and nutlin-3 significantly induce
paraptosis and cell death (150).
Nutlin-3 has not been used in the treatment of GC; however, the
antitumor activity of nutlin-3 against GC cells has been
demonstrated in vivo and in vitro (151). It has been reported that nutlin-3
induces G1 arrest in MKN-45 and SNU-1 gastric adenocarcinoma cell
lines in vitro, and the activation of p53 by nutlin-3
effectively increased the incidence of apoptosis in wild-type p53
GC cells. In vivo, the combined treatment of nutlin-3 and
fluorouracil led to a more potent inhibitory effect on the tumor
growth of experimental animals compared with treatment with each
agent alone. Overall, nutlin-3 is a broad-spectrum antitumor agent
and has the potential to be used in the treatment of GC by
targeting the E3 ligase MDM2.
Triptolide is a compound purified from tripterygium
wilfordii that exhibits antitumor effects in GC (152–155).
In 1991, Chinese scholars found that triptolide had antitumor
activity in a variety of cancer cell lines, including GC (152). A subsequent study demonstrated that
triptolide treatment of GC cells containing wild-type p53 gene
resulted in a significant inhibitory effect on cell growth, whereas
GC cells with mutant p53 did not exhibit this effect (153). Another study indicated that this
p53-dependent antitumor activity was achieved by inhibiting the
overexpression of MDM2 (154).
Moutan cortex is another Traditional Chinese Medicine that can also
induce apoptosis through the MDM2-p53-dependent pathway in GC cells
(155). Therefore, Chinese herbs,
such as triptolide and moutan cortex, might be potential anticancer
agents for GC.
MLN4924, a neddylation inhibitor, is an indirect
inhibitor of CRL E3 ligases. MLN4924 acts as a promoter of
apoptosis and is a potential anticancer drug in diverse types of
human cancers, including GC (156).
It has been reported that MLN4924 downregulates the expression of
CRLs and then suppresses the growth of GC cells. There are some
other small molecules that can also target E3 ligase. miR-223 can
target FBXW7 and downregulate its expression, so drugs that promote
degradation of miR-223 may be useful in patients with GC (157). As aforementioned, WWP1 is an
oncoprotein in GC, and miR-584-5p, miR-129-5p and miR-129-3p
suppress WWP1 protein expression and inhibit the proliferation of
GC (133,134). Thus, miRNAs may also be a research
direction for E3-targeted therapy for GC. In summary, there are
still no drugs targeting E3 used in the clinical treatment of GC,
and the effects of the compounds mentioned above are still in the
research stage.
Over the past few years, an increasing number of E3
ligases have been described as tumor regulators in GC. The present
review summarizes approximately thirty types of E3 ligases that
play essential roles in regulating the development of GC, including
RING and HECT ligases. The function and significance of E3 ligases
in GC has been well examined, but several E3 ligases, such as COP1,
need further studies to elucidate their mechanisms. It has been
shown that many synthetic and natural compounds targeting E3
ligases could regulate the level of various signaling proteins
through UPS-mediated degradation in human cancers (158). Compounds and small molecules
targeting E3 ligases may become underlying templates for the
synthesis of targeted therapeutic drugs in GC. However, there are
still many obstacles to overcome before the application of
compounds targeting E3 ligases in GC, such as the detection and
analysis of their complex functional mechanisms and molecular
structures. Therefore, further studies should aim to reveal the
molecular mechanism of individual E3 ligases in different subtypes
of GC, and determine the structure of these targeting compounds to
facilitate further synthesis of such targeted therapy drugs. In
conclusion, E3 ligases are crucial tumor regulatory factors and
potential therapeutic targets in GC.
Not applicable.
This research was funded by The National Natural
Science Foundation of China (grant no. 30871207), The National
Natural Science Foundation of China (grant nos. 81874063 and
81672389) and Anhui Province Science and Technology Key Project
(grant no. 1704a0802176).
Not applicable.
MW and YL developed the concept of the manuscript.
MW, WD and ZK were responsible for writing, reviewing and editing
the manuscript. WD participated in revising the manuscript. ZK
supervised the project. YL was involved with the project
administration. All authors read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu Y, Fan Y, Jiang Y, Wang Y, Liu H and
Wei M: Analysis of risk factors associated with precancerous lesion
of gastric cancer in patients from eastern China: A comparative
study. J Cancer Res Ther. 9:205–209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Woo Y, Goldner B, Son T, Song K, Noh SH,
Fong Y and Hyung WJ: Western validation of a novel gastric cancer
prognosis prediction model in US gastric cancer patients. J Am Coll
Surg. 226:252–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wen T, Wang Z, Li Y, Li Z, Che X, Fan Y,
Wang S, Qu J, Yang X, Hou K, et al: A four-factor immunoscore
system that predicts clinical outcome for stage II/III gastric
cancer. Cancer Immunol Res. 5:524–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abramov IS, Emelyanova MA, Ryabaya OO,
Krasnov GS, Zasedatelev AS and Nasedkina TV: Somatic mutations
associated with metastasis in acral melanoma. Mol Biol (Mosk).
53:648–653. 2019.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hou YC and Deng JY: Role of E3 ubiquitin
ligases in gastric cancer. World J Gastroenterol. 21:786–793. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hickey CM, Xie Y and Hochstrasser M: DNA
binding by the MATα2 transcription factor controls its access to
alternative ubiquitin-modification pathways. Mol Biol Cell.
29:542–556. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bulatov E, Valiullina A, Sayarova R and
Rizvanov A: Promising new therapeutic targets for regulation of
inflammation and immunity: RING-type E3 ubiquitin ligases. Immunol
Lett. 202:44–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu W, Yang C, He H and Liu H: The
CARM1-p300-c-Myc-Max (CPCM) transcriptional complex regulates the
expression of CUL4A/4B and affects the stability of CRL4 E3 ligases
in colorectal cancer. Int J Biol Sci. 16:1071–1085. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu L, Wong CC, Gong B and Yu J:
Functional significance and therapeutic implication of ring-type E3
ligases in colorectal cancer. Oncogene. 37:148–159. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uchida C and Kitagawa M: RING-, HECT-, and
RBR-type E3 ubiquitin ligases: Involvement in human cancer. Curr
Cancer Drug Targets. 16:157–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johansson H, Isabella Tsai YC, Fantom K,
Chung CW, Kümper S, Martino L, Thomas DA, Eberl HC, Muelbaier M,
House D and Rittinger K: Fragment-based covalent ligand screening
enables rapid discovery of inhibitors for the RBR E3 ubiquitin
ligase HOIP. J Am Chem Soc. 141:2703–2712. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu JM, Sun W, Wang ZH, Liang X, Hua F, Li
K, Lv XX, Zhang XW, Liu YY, Yu JJ, et al: TRIB3 supports breast
cancer stemness by suppressing FOXO1 degradation and enhancing SOX2
transcription. Nat Commun. 10:57202019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Wu H, Yi B, Zhou J, Wei L, Chen Y
and Zhang L: RING finger protein 38 induces gastric cancer cell
growth by decreasing the stability of the protein tyrosine
phosphatase SHP-1. FEBS Lett. 592:3092–3100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang Z, Cai Y, Yang C, Chen Z, Sun H, Xu
Y, Chen W, Xu D, Tian W and Wang H: Knockdown of RNF6 inhibits
gastric cancer cell growth by suppressing STAT3 signaling. Onco
Targets Ther. 11:6579–6587. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Berndsen CE and Wolberger C: New insights
into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol.
21:301–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ohi MD, Vander Kooi CW, Rosenberg JA,
Chazin WJ and Gould KL: Structural insights into the U-box, a
domain associated with multi-ubiquitination. Nat Struct Biol.
10:250–255. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Leslie PL, Ke H and Zhang Y: The MDM2 RING
domain and central acidic domain play distinct roles in MDM2
protein homodimerization and MDM2-MDMX protein heterodimerization.
J Biol Chem. 290:12941–12950. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Genschik P, Sumara I and Lechner E: The
emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): Cellular
functions and disease implications. EMBO J. 32:2307–2320. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kelsall IR, Kristariyanto YA, Knebel A,
Wood NT, Kulathu Y and Alpi AF: Coupled monoubiquitylation of the
co-E3 ligase DCNL1 by Ariadne-RBR E3 ubiquitin ligases promotes
cullin-RING ligase complex remodeling. J Biol Chem. 294:2651–2664.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Weber J, Polo S and Maspero E: HECT E3
ligases: A tale with multiple facets. Front Physiol. 10:370–377.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lorenz S: Structural mechanisms of
HECT-type ubiquitin ligases. Biol Chem. 399:127–145. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Geng R, Tan X, Wu J, Pan Z, Yi M, Shi W,
Liu R, Yao C, Wang G, Lin J, et al: RNF183 promotes proliferation
and metastasis of colorectal cancer cells via activation of
NF-κB-IL-8 axis. Cell Death Dis. 8:e29942017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng R, Zhang PF, Yang X, Wei CY, Huang
XY, Cai JB, Lu JC, Gao C, Sun HX, Gao Q, et al: Overexpression of
RNF38 facilitates TGF-β signaling by Ubiquitinating and degrading
AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res.
38:1132019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Macdonald DH, Lahiri D, Sampath A, Chase
A, Sohal J and Cross NC: Cloning and characterization of RNF6, a
novel RING finger gene mapping to 13q12. Genomics. 58:94–97. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eisenberg I, Hochner H, Levi T, Yelin R,
Kahan T and Mitrani-Rosenbaum S: Cloning and characterization of a
novel human gene RNF38 encoding a conserved putative protein with a
RING finger domain. Biochem Biophys Res Commun. 294:1169–1176.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Katoh M: Molecular cloning and
characterization of RNF26 on human chromosome 11q23 region,
encoding a novel RING finger protein with leucine zipper. Biochem
Biophys Res Commun. 282:1038–1044. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jongsma ML, Berlin I, Wijdeven RH, Janssen
L, Janssen GM, Garstka MA, Janssen H, Mensink M, van Veelen PA,
Spaapen RM and Neefjes J: An ER-associated pathway defines
endosomal architecture for controlled cargo transport. Cell.
166:152–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo
L, Zhong B and Shu HB: RNF26 temporally regulates virus-triggered
type I interferon induction by two distinct mechanisms. PLoS
Pathog. 10:e10043582014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang R, Zhao X, Xu J, Wen Y, Li A, Lu M
and Zhou J: Astrocytic JWA deletion exacerbates dopaminergic
neurodegeneration by decreasing glutamate transporters in mice.
Cell Death Dis. 9:3522018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu T, Chen R, Li A, Liu J, Gu D, Liu Q, C
Chang H and Zhou J: JWA as a novel molecule involved in oxidative
stress-associated signal pathway in myelogenous leukemia cells. J
Toxicol Environ Health A. 69:1399–1411. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiu D, Wang Q, Wang Z, Chen J, Yan D, Zhou
Y, Li A, Zhang R, Wang S and Zhou J: RNF185 modulates JWA
ubiquitination and promotes gastric cancer metastasis. Biochim
Biophys Acta Mol Basis Dis. 1864:1552–1561. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao Y, Cai A, Xi H, Li J, Xu W, Zhang Y,
Zhang K, Cui J, Wu X, Wei B and Chen L: Ring finger protein 43
associates with gastric cancer progression and attenuates the
stemness of gastric cancer stem-like cells via the Wnt-β/catenin
signaling pathway. Stem Cell Res Ther. 8:982017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xi HQ, Cai AZ, Wu XS, Cui JX, Shen WS,
Bian SB, Wang N, Li JY, Lu CR, Song Z, et al: Leucine-rich
repeat-containing G-protein-coupled receptor 5 is associated with
invasion, metastasis, and could be a potential therapeutic target
in human gastric cancer. Br J Cancer. 110:2011–2020. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li XB, Yang G, Zhu L, Tang YL, Zhang C, Ju
Z, Yang X and Teng Y: Gastric Lgr5(+) stem cells are the cellular
origin of invasive intestinal-type gastric cancer in mice. Cell
Res. 26:838–849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barker N, Huch M, Kujala P, van de
Wetering M, Snippert HJ, van Es JH, Sato T, Stange DE, Begthel H,
van den Born M, et al: Lgr5(+ve) stem cells drive self-renewal in
the stomach and build long-lived gastric units in vitro. Cell Stem
Cell. 6:25–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Y, Lan J, Wang W, Shi Q, Lan Y, Cheng
Z and Guan H: ZNRF3 acts as a tumour suppressor by the Wnt
signalling pathway in human gastric adenocarcinoma. J Mol Histol.
44:555–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nanki K, Toshimitsu K, Takano A, Fujii M,
Shimokawa M, Ohta Y, Matano M, Seino T, Nishikori S, Ishikawa K, et
al: Divergent routes toward Wnt and R-spondin niche independency
during human gastric carcinogenesis. Cell. 174:856–869.e17. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hao HX, Jiang X and Cong F: Control of Wnt
receptor turnover by R-spondin-ZNRF3/RNF43 signaling module and its
dysregulation in cancer. Cancers (Basel). 8(pii): E542016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi
ST, Siu HC, Deng S, Chu KM, Law S, et al: Whole-genome sequencing
and comprehensive molecular profiling identify new driver mutations
in gastric cancer. Nat Genet. 46:573–582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu H, Mintern JD and Villadangos JA:
MARCH ligases in immunity. Curr Opin Immunol. 58:38–43. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Q, Chen Q, Zhu L, Chen M, Xu W,
Panday S, Wang Z, Li A, Røe OD, Chen R, et al: JWA regulates
TRAIL-induced apoptosis via MARCH8-mediated DR4 ubiquitination in
cisplatin-resistant gastric cancer cells. Oncogenesis. 6:e3532017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yin J, Ji Z, Hong Y, Song Z, Hu N, Zhuang
M, Bian B, Liu Y and Wu F: Sh-MARCH8 inhibits tumorigenesis via
PI3K pathway in gastric cancer. Cell Physiol Biochem. 49:306–321.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun M, Li S, Yu K, Xiang J and Li F: An E3
ubiquitin ligase TRIM9 is involved in WSSV infection via
interaction with β-TrCP. Dev Comp Immunol. 97:57–63. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun Y, Keown JR, Black MM, Raclot C,
Demarais N, Trono D, Turelli P and Goldstone DC: A dissection of
oligomerization by the TRIM28 tripartite motif and the interaction
with members of the Krab-ZFP family. J Mol Biol. 431:2511–2527.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yokoe T, Toiyama Y, Okugawa Y, Tanaka K,
Ohi M, Inoue Y, Mohri Y, Miki C and Kusunoki M: KAP1 is associated
with peritoneal carcinomatosis in gastric cancer. Ann Surg Oncol.
17:821–828. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kosaka Y, Inoue H, Ohmachi T, Yokoe T,
Matsumoto T, Mimori K, Tanaka F, Watanabe M and Mori M: Tripartite
motif-containing 29 (TRIM29) is a novel marker for lymph node
metastasis in gastric cancer. Ann Surg Oncol. 14:2543–2549. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH
and Gao WQ: TRIM59 is up-regulated in gastric tumors, promoting
ubiquitination and degradation of p53. Gastroenterology.
147:1043–1054. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ma X, Zhang S, Zhang M, Zhu Y, Ma P, Yang
S, Su L, Li Z, Lv W and Luan W: TRIM28 down-regulation on
methylation imprints in bovine preimplantation embryos. Zygote.
26:449–456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang P, Zhang H, Wang Y, Zhang P and Qi
Y: Tripartite motif-containing protein 59 (TRIM59) promotes
epithelial ovarian cancer progression via the focal adhesion
kinase(FAK)/AKT/matrix metalloproteinase (MMP) pathway. Med Sci
Monit. 25:3366–3373. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shen H, Zhang J, Zhang Y, Feng Q, Wang H,
Li G, Jiang W and Li X: Knockdown of tripartite motif 59 (TRIM59)
inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR
signaling pathway. Gene. 698:50–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen G, Chen W, Ye M, Tan W and Jia B:
TRIM59 knockdown inhibits cell proliferation by down-regulating the
Wnt/β-catenin signaling pathway in neuroblastoma. Biosci Rep.
39(pii): BSR201812772019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Cui Z, Liu Z, Zeng J, Chen L, Wu Q, Mo J,
Zhang G, Song L, Xu W, Zhang S and Guo X: Eugenol inhibits
non-small cell lung cancer by repressing expression of
NF-κB-regulated TRIM59. Phytother Res. 33:1562–1569. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Halaby MJ, Hakem R and Hakem A: Pirh2: An
E3 ligase with central roles in the regulation of cell cycle, DNA
damage response, and differentiation. Cell Cycle. 12:2733–2737.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bao Y, Wu X, Yuan D, Shi W and Shi J: High
expression of Pirh2 is associated with poor prognosis in glioma.
Cell Mol Neurobiol. 37:1501–1509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Daks A, Petukhov A, Fedorova O, Shuvalov
O, Merkulov V, Vasileva E, Antonov A and Barlev NA: E3 ubiquitin
ligase Pirh2 enhances tumorigenic properties of human non-small
cell lung carcinoma cells. Genes Cancer. 7:383–393. 2016.PubMed/NCBI
|
|
58
|
Yang S, Chen Y, Sun F, Ni Q, Wang H, Huang
Y, Zhang C, Liu K, Wang S, Qiu J, et al: Downregulated pirh2 can
decrease the proliferation of breast cancer cells. Arch Med Res.
47:186–195. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang G, Gong Y, Wang Q, Wang L and Zhang
X: miR-100 antagonism triggers apoptosis by inhibiting
ubiquitination-mediated p53 degradation. Oncogene. 36:1023–1037.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Eichinger L, Pachebat JA, Glöckner G,
Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski
K, Xu Q, et al: The genome of the social amoeba Dictyostelium
discoideum. Nature. 435:43–57. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Langdon WY, Hyland CD, Grumont RJ and
Morse HC III: The c-cbl proto-oncogene is preferentially expressed
in thymus and testis tissue and encodes a nuclear protein. J Virol.
63:5420–5424. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lee H and Tsygankov AY: Cbl-family
proteins as regulators of cytoskeleton-dependent phenomena. J Cell
Physiol. 228:2285–2293. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kamei T, Machida K, Nimura Y, Senga T,
Yamada I, Yoshii S, Matsuda S and Hamaguchi M: C-Cbl protein in
human cancer tissues is frequently tyrosine phosphorylated in a
tumor-specific manner. Int J Oncol. 17:335–339. 2000.PubMed/NCBI
|
|
64
|
Dong Q, Liu YP, Qu XJ, Hou KZ and Li LL:
Expression of c-Cbl, Cbl-b, and epidermal growth factor receptor in
gastric carcinoma and their clinical significance. Chin J Cancer.
29:59–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Feng D, Ma Y, Liu J, Xu L, Zhang Y, Qu J,
Liu Y and Qu X: Cbl-b enhances sensitivity to 5-fluorouracil via
EGFR- and mitochondria-mediated pathways in gastric cancer cells.
Int J Mol Sci. 14:24399–24411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yu P, Fan Y, Qu X, Zhang J, Song N, Liu J
and Liu Y: Cbl-b regulates the sensitivity of cetuximab through
ubiquitin-proteasome system in human gastric cancer cells. J BUON.
21:867–873. 2016.PubMed/NCBI
|
|
67
|
CHe X, Zhang Y, Qu X, Guo T, Ma Y, Li C,
Fan Y, Hou K, Cai Y, Yu R, et al: The E3 ubiquitin ligase Cbl-b
inhibits tumor growth in multidrug-resistant gastric and breast
cancer cells. Neoplasma. 64:887–892. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xu L, Zhang Y, Qu X, Che X, Guo T, Cai Y,
Li A, Li D, Li C, Wen T, et al: E3 ubiquitin ligase Cbl-b prevents
tumor metastasis by maintaining the epithelial phenotype in
multiple drug-resistant gastric and breast cancer cells. Neoplasia.
19:374–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang Y, Qu X, Hu X, Yang X, Hou K, Teng
Y, Zhang J, Sada K and Liu Y: Reversal of P-glycoprotein-mediated
multi-drug resistance by the E3 ubiquitin ligase Cbl-b in human
gastric adenocarcinoma cells. J Pathol. 218:248–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lai AZ, Durrant M, Zuo D, Ratcliffe CD and
Park M: Met kinase-dependent loss of the E3 ligase Cbl in gastric
cancer. J Biol Chem. 287:8048–8059. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gao YJ, Xin Y, Zhang JJ and Zhou J:
Mechanism and pathobiologic implications of CHFR promoter
methylation in gastric carcinoma. World J Gastroenterol.
14:5000–5007. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Li Y, Yang Y, Lu Y, Herman JG, Brock MV,
Zhao P and Guo M: Predictive value of CHFR and MLH1 methylation in
human gastric cancer. Gastric Cancer. 18:280–287. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Satoh A, Toyota M, Itoh F, Sasaki Y,
Suzuki H, Ogi K, Kikuchi T, Mita H, Yamashita T, Kojima T, et al:
Epigenetic inactivation of CHFR and sensitivity to microtubule
inhibitors in gastric cancer. Cancer Res. 63:8606–8613.
2003.PubMed/NCBI
|
|
74
|
Kashima L, Idogawa M, Mita H, Shitashige
M, Yamada T, Ogi K, Suzuki H, Toyota M, Ariga H, Sasaki Y and
Tokino T: CHFR protein regulates mitotic checkpoint by targeting
PARP-1 protein for ubiquitination and degradation. J Biol Chem.
287:12975–12984. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kim JH, Park SM, Kang MR, Oh SY, Lee TH,
Muller MT and Chung IK: Ubiquitin ligase MKRN1 modulates telomere
length homeostasis through a proteolysis of hTERT. Genes Dev.
19:776–781. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Ko A, Shin JY, Seo J, Lee KD, Lee EW, Lee
MS, Lee HW, Choi IJ, Jeong JS, Chun KH and Song J: Acceleration of
gastric tumorigenesis through MKRN1-mediated posttranslational
regulation of p14ARF. J Natl Cancer Inst. 104:1660–1672. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jang JH: FIGC, a novel FGF-induced
ubiquitin-protein ligase in gastric cancers FEBS. Lett. 578:21–25.
2004.
|
|
78
|
Wu CE, Esfandiari A, Ho YH, Wang N, Mahdi
AK, Aptullahoglu E, Lovat P and Lunec J: Targeting negative
regulation of p53 by MDM2 and WIP1 as a therapeutic strategy in
cutaneous melanoma. Br J Cancer. 118:495–508. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hu C, Ni Z, Li BS, Yong X, Yang X, Zhang
JW, Zhang D, Qin Y, Jie MM, Dong H, et al: hTERT promotes the
invasion of gastric cancer cells by enhancing FOXO3a ubiquitination
and subsequent ITGB1 upregulation. Gut. 66:31–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chi XZ, Kim J, Lee YH, Lee JW, Lee KS, Wee
H, Kim WJ, Park WY, Oh BC, Stein GS, et al: Runt-related
transcription factor RUNX3 is a target of MDM2-mediated
ubiquitination. Cancer Res. 69:8111–8119. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jung CR, Lim JH, Choi Y, Kim DG, Kang KJ,
Noh SM and Im DS: Enigma negatively regulates p53 through MDM2 and
promotes tumor cell survival in mice. J Clin Invest. 120:4493–4506.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Feng Y, Gao S, Gao Y, Song D, Wang X and
Chen Z: Runx3 expression in rectal cancer cells and its effect on
cell invasion and proliferation. Oncol Lett. 18:3290–3294.
2019.PubMed/NCBI
|
|
83
|
Connell P, Ballinger CA, Jiang J, Wu Y,
Thompson LJ, Höhfeld J and Patterson C: The co-chaperone CHIP
regulates protein triage decisions mediated by heat-shock proteins.
Nat Cell Biol. 3:93–96. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Xiao M, Yan M, Zhang J, Xu Q and Chen W:
Carboxy-terminus Hsc70 interacting protein exerts a tumor
inhibition function in head and neck cancer. Oncol Rep.
38:1629–1636. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Liu F, Zhou J, Zhou P, Chen W and Guo F:
The ubiquitin ligase CHIP inactivates NF-κB signaling and impairs
the ability of migration and invasion in gastric cancer cells. Int
J Oncol. 46:2096–2106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zheng N, Schulman BA, Song L, Miller JJ,
Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al:
Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase
complex. Nature. 416:703–709. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lisztwan J, Marti A, Sutterlüty H,
Gstaiger M, Wirbelauer C and Krek W: Association of human CUL-1 and
ubiquitin-conjugating enzyme CDC34 with the F-box protein
p45(SKP2): Evidence for evolutionary conservation in the subunit
composition of the CDC34-SCF pathway. EMBO J. 17:368–383. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Uddin S, Bhat AA, Krishnankutty R, Mir F,
Kulinski M and Mohammad RM: Involvement of F-BOX proteins in
progression and development of human malignancies. Semin Cancer
Biol. 36:18–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Jiang ZH, Peng T, Qian HL, Lu CD, Qiu F
and Zhang SZ: DNA damage-induced activation of ATM promotes
β-TRCP-mediated ARID1A ubiquitination and destruction in gastric
cancer cells. Cancer Cell Int. 19:1622019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Milne AN, Leguit R, Corver WE, Morsink FH,
Polak M, de Leng WW, Carvalho R and Offerhaus GJ: Loss of
CDC4/FBXW7 in gastric carcinoma. Cell Oncol. 32:347–359.
2010.PubMed/NCBI
|
|
91
|
Li H, Wang Z, Zhang W, Qian K, Xu W and
Zhang S: Fbxw7 regulates tumor apoptosis, growth arrest and the
epithelial-to-mesenchymal transition in part through the RhoA
signaling pathway in gastric cancer. Cancer Lett. 370:39–55. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Huang LY, Zhao J, Chen H, Wan L, Inuzuka
H, Guo J, Fu X, Zhai Y, Lu Z, Wang X, et al:
SCFFBW7-mediated degradation of Brg1 suppresses gastric
cancer metastasis. Nat Commun. 9:35692018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kuai X, Li L, Chen R, Wang K, Chen M, Cui
B, Zhang Y, Li J, Zhu H, Zhou H, et al:
SCFFBXW7/GSK3β-Mediated GFI1 Degradation Suppresses
Proliferation of Gastric Cancer Cells. Cancer Res. 79:4387–4398.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhou J, Hayakawa Y, Wang TC and Bass AJ:
RhoA mutations identified in diffuse gastric cancer. Cancer Cell.
26:9–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y
and Sun H: MicroRNA-25 promotes gastric cancer proliferation,
invasion, and migration by directly targeting F-box and WD-40
domain protein 7, FBXW7. Tumour Biol. 36:7831–7840. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lv Z, Zhang Y, Yu X, Lin Y and Ge Y:
RETRACTED: The function of long non-coding RNA MT1JP in the
development and progression of gastric cancer. Pathol Res Pract.
214:1218–1223. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Frescas D and Pagano M: Deregulated
proteolysis by the F-box proteins SKP2 and beta-TrCP: Tipping the
scales of cancer. Nat Rev Cancer. 8:438–449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Saitoh T and Katoh M: Expression profiles
of betaTRCP1 and betaTRCP2, and mutation analysis of betaTRCP2 in
gastric cancer. Int J Oncol. 18:959–964. 2001.PubMed/NCBI
|
|
99
|
Gao G, Kun T, Sheng Y, Qian M, Kong F, Liu
X, Yu Z, Zhang H, Zhang Q, Gu J and Zhang X: SGT1 regulates Akt
signaling by promoting beta-TrCP-dependent PHLPP1 degradation in
gastric cancer cells. Mol Biol Rep. 40:2947–2953. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang S, Zhang Y, Wang Y, Ye P, Li J, Li H,
Ding Q and Xia J: Amphiregulin confers regulatory T cell
suppressive function and tumor invasion via the EGFR/GSK-3β/Foxp3
axis. J Biol Chem. 291:21085–21095. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Li LQ, Pan D, Chen H, Zhang L and Xie WJ:
F-box protein FBXL2 inhibits gastric cancer proliferation by
ubiquitin-mediated degradation of forkhead box M1. FEBS Lett.
590:445–452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cen G, Ding HH, Liu B and Wu WD: FBXL5
targets cortactin for ubiquitination-mediated destruction to
regulate gastric cancer cell migration. Tumour Biol. 35:8633–8638.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wu W, Ding H, Cao J and Zhang W: FBXL5
inhibits metastasis of gastric cancer through suppressing Snail1.
Cell Physiol Biochem. 35:1764–1772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zou S, Ma C, Yang F, Xu X, Jia J and Liu
Z: FBXO31 Suppresses gastric cancer EMT by targeting Snail1 for
proteasomal degradation. Mol Cancer Res. 16:286–295. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Petroski MD and Deshaies RJ: Function and
regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol.
6:9–20. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Morimoto M, Nishida T, Honda R and Yasuda
H: Modification of cullin-1 by ubiquitin-like protein Nedd8
enhances the activity of SCF(skp2) toward p27(kip1). Biochem
Biophys Res Commun. 270:1093–1096. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Cheng Q and Yin G: Cullin-1 regulates MG63
cell proliferation and metastasis and is a novel prognostic marker
of osteosarcoma. Int J Biol Markers. 32:e202–e209. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zhou YH, Xia J, Xu WH, Zhu X, Wu XH, Hua D
and Xing C: Cullin-1 promotes cell proliferation in human breast
cancer and is related to diabetes. Int J Biol Markers.
31:e375–e381. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Jiang H, He D, Xu H, Liu J, Qu L and Tong
S: Cullin-1 promotes cell proliferation via cell cycle regulation
and is a novel in prostate cancer. Int J Clin Exp Pathol.
8:1575–1583. 2015.PubMed/NCBI
|
|
110
|
Chen TJ, Gao F, Yang T, Thakur A, Ren H,
Li Y, Zhang S, Wang T and Chen MW: CDK-associated Cullin 1 promotes
cell proliferation with activation of ERK1/2 in human lung cancer
A549 cells. Biochem Biophys Res Commun. 437:108–113. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Michail O, Moris D, Theocharis S and
Griniatsos J: Cullin-1 and −2 protein expression in colorectal
cancer: Correlation with clinicopathological variables. In Vivo.
32:391–396. 2018.PubMed/NCBI
|
|
112
|
Bai J, Zhou Y, Chen G, Zeng J, Ding J, Tan
Y, Zhou J and Li G: Overexpression of Cullin1 is associated with
poor prognosis of patients with gastric cancer. Hum Pathol.
42:375–383. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Clifford SC, Cockman ME, Smallwood AC,
Mole DR, Woodward ER, Maxwell PH, Ratcliffe PJ and Maher ER:
Contrasting effects on HIF-1alpha regulation by disease-causing
pVHL mutations correlate with patterns of tumourigenesis in von
Hippel-Lindau disease. Hum Mol Genet. 10:1029–1038. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yokoe S, Nakagawa T, Kojima Y, Higuchi K
and Asahi M: Indomethacin-induced intestinal epithelial cell damage
is mediated by pVHL activation through the degradation of collagen
I and HIF-1α. Biochem Biophys Res Commun. 468:671–676. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Clifford SC, Astuti D, Hooper L, Maxwell
PH, Ratcliffe PJ and Maher ER: The pVHL-associated SCF ubiquitin
ligase complex: molecular genetic analysis of elongin B and C, Rbx1
and HIF-1alpha in renal cell carcinoma. Oncogene. 20:5067–5074.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Gao L, Wu GJ, Liu B, Shen MZ, Pan TJ, Yu
CG, Wang QH, Ru Y, Liu XP, Niu TS, et al: Up-regulation of pVHL
along with down-regulation of HIF-1α by NDRG2 expression attenuates
proliferation and invasion in renal cancer cells. PLoS One.
8:e841272013. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Bowers AJ and Boylan JF: Nek8, a NIMA
family kinase member, is overexpressed in primary human breast
tumors. Gene. 328:135–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Ding XF, Zhou J, Hu QY, Liu SC and Chen G:
The tumor suppressor pVHL down-regulates never-in-mitosis A-related
kinase 8 via hypoxia-inducible factors to maintain cilia in human
renal cancer cells. J Biol Chem. 290:1389–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Ding XF, Chen J, Zhou J, Chen G and Wu YL:
Never-in-mitosis A-related kinase 8, a novel target of
von-Hippel-Lindau tumor suppressor protein, promotes gastric cancer
cell proliferation. Oncol Lett. 16:5900–5906. 2018.PubMed/NCBI
|
|
120
|
Bianchi E, Denti S, Catena R, Rossetti G,
Polo S, Gasparian S, Putignano S, Rogge L and Pardi R:
Characterization of human constitutive photomorphogenesis protein
1, a RING finger ubiquitin ligase that interacts with Jun
transcription factors and modulates their transcriptional activity.
J Biol Chem. 278:19682–19690. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhu D, Maier A, Lee JH, Laubinger S, Saijo
Y, Wang H, Qu LJ, Hoecker U and Deng XW: Biochemical
characterization of Arabidopsis complexes containing CONSTITUTIVELY
PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control
of plant development. Plant Cell. 20:2307–2323. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Saijo Y, Zhu D, Li J, Rubio V, Zhou Z,
Shen Y, Hoecker U, Wang H and Deng XW: Arabidopsis COP1/SPA1
complex and FHY1/FHY3 associate with distinct phosphorylated forms
of phytochrome A in balancing light signaling. Mol Cell.
31:607–613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ka WH, Cho SK, Chun BN, Byun SY and Ahn
JC: The ubiquitin ligase COP1 regulates cell cycle and apoptosis by
affecting p53 function in human breast cancer cell lines. Breast
Cancer. 25:529–538. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zou S, Zhu Y, Wang B, Qian F, Zhang X,
Wang L, Fu C, Bao H, Xie M, Gao S, et al: The ubiquitin ligase COP1
promotes glioma cell proliferation by preferentially downregulating
tumor suppressor p53. Mol Neurobiol. 54:5008–5016. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Li YF, Wang DD, Zhao BW, Wang W, Huang CY,
Chen YM, Zheng Y, Keshari RP, Xia JC and Zhou ZW: High level of
COP1 expression is associated with poor prognosis in primary
gastric cancer. Int J Biol Sci. 8:1168–1177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sawada G, Ueo H, Matsumura T, Uchi R,
Ishibashi M, Mima K, Kurashige J, Takahashi Y, Akiyoshi S, Sudo T,
et al: Loss of COP1 expression determines poor prognosis in
patients with gastric cancer. Oncol Rep. 30:1971–1975. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Rotin D and Kumar S: Physiological
functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell
Biol. 10:398–409. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zheng H, Ke X, Li D and Wang Q, Wang J,
Liu X, Deng M, Deng X, Xue Y, Zhu Y and Wang Q: NEDD4 promotes cell
growth and motility in hepatocellular carcinoma. Cell Cycle.
17:728–738. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Shao G, Wang R, Sun A, Wei J, Peng K, Dai
Q, Yang W and Lin Q: The E3 ubiquitin ligase NEDD4 mediates cell
migration signaling of EGFR in lung cancer cells. Mol Cancer.
17:242018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Kim SS, Yoo NJ, Jeong EG, Kim MS and Lee
SH: Expression of NEDD4-1, a PTEN regulator, in gastric and
colorectal carcinomas. APMIS. 116:779–784. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zou X, Levy-Cohen G and Blank M: Molecular
functions of NEDD4 E3 ubiquitin ligases in cancer. Biochim Biophys
Acta. 1856:91–106. 2015.PubMed/NCBI
|
|
132
|
Zhang L, Wu Z, Ma Z, Liu H, Wu Y and Zhang
Q: WWP1 as a potential tumor oncogene regulates PTEN-Akt signaling
pathway in human gastric carcinoma. Tumour Biol. 36:787–798. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Ma L, Chen X, Li C, Cheng R, Gao Z, Meng
X, Sun C, Liang C and Liu Y: miR-129-5p and −3p co-target WWP1 to
suppress gastric cancer proliferation and migration. J Cell
Biochem. Nov 11–2018.(Epub ahead of print).
|
|
134
|
Li Q, Li Z, Wei S, Wang W, Chen Z, Zhang
L, Chen L, Li B, Sun G, Xu J, et al: Overexpression of miR-584-5p
inhibits proliferation and induces apoptosis by targeting WW
domain-containing E3 ubiquitin protein ligase 1 in gastric cancer.
J Exp Clin Cancer Res. 36:592017. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhu H, Kavsak P, Abdollah S, Wrana JL and
Thomsen GH: A SMAD ubiquitin ligase targets the BMP pathway and
affects embryonic pattern formation. Nature. 400:687–693. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Koganti P, Levy-Cohen G and Blank M:
Smurfs in protein homeostasis, signaling, and cancer. Front Oncol.
8:2952018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Dote H, Toyooka S, Tsukuda K, Yano M, Ota
T, Murakami M, Naito M, Toyota M, Gazdar AF and Shimizu N: Aberrant
promoter methylation in human DAB2 interactive protein (hDAB2IP)
gene in gastrointestinal tumour. Br J Cancer. 92:1117–1125. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Li X, Dai X, Wan L, Inuzuka H, Sun L and
North BJ: Smurf1 regulation of DAB2IP controls cell proliferation
and migration. Oncotarget. 7:26057–26069. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Tao Y, Sun C, Zhang T and Song Y: SMURF1
promotes the proliferation, migration and invasion of gastric
cancer cells. Oncol Rep. 38:1806–1814. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Yang M, Jiang N, Cao QW, Ma MQ and Sun Q:
The E3 ligase UBR5 regulates gastric cancer cell growth by
destabilizing the tumor suppressor GKN1. Biochem Biophys Res
Commun. 478:1624–1629. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Kozlov G, Nguyen L, Lin T, De Crescenzo G,
Park M and Gehring K: Structural basis of ubiquitin recognition by
the ubiquitin-associated (UBA) domain of the ubiquitin ligase EDD.
J Biol Chem. 282:35787–35795. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Kim MS, Oh JE, Eom HS, Yoo NJ and Lee SH:
Mutational analysis of UBR5 gene encoding an E3 ubiquitin ligase in
common human cancers. Pathology. 42:93–94. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Richardson PG, Hideshima T and Anderson
KC: Bortezomib (PS-341): A novel, first-in-class proteasome
inhibitor for the treatment of multiple myeloma and other cancers.
Cancer Control. 10:361–369. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Zhang B and Gu Y: Bortezomib inhibits
gastric carcinoma HGC-27 cells through the phospho-Jun N-terminal
kinase (p-JNK) pathway in vitro. Gene. 559:164–171. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Yi H, Yan X, Luo Q, Yuan L, Li B, Pan W,
Zhang L, Chen H, Wang J, Zhang Y, et al: A novel small molecule
inhibitor of MDM2-p53 (APG-115) enhances radiosensitivity of
gastric adenocarcinoma. J Exp Clin Cancer Res. 37:972018.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Vassilev LT, Vu BT, Graves B, Carvajal D,
Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et
al: In vivo activation of the p53 pathway by small-molecule
antagonists of MDM2. Science. 303:844–848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Impicciatore G, Sancilio S, Miscia S and
Di Pietro R: Nutlins and ionizing radiation in cancer therapy. Curr
Pharm Des. 16:1427–1442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Yee-Lin V, Pooi-Fong W and Soo-Beng AK:
Nutlin-3, A p53-Mdm2 antagonist for nasopharyngeal carcinoma
treatment. Mini Rev Med Chem. 18:173–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Meijer A, Kruyt FA, van der Zee AG,
Hollema H, Le P, ten Hoor KA, Groothuis GM, Quax WJ, de Vries EG
and de Jong S: Nutlin-3 preferentially sensitises wild-type
p53-expressing cancer cells to DR5-selective TRAIL over rhTRAIL. Br
J Cancer. 109:2685–2695. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Lee DM, Kim IY, Seo MJ, Kwon MR and Choi
KS: Nutlin-3 enhances the bortezomib sensitivity of p53-defective
cancer cells by inducing paraptosis. Exp Mol Med. 49:e3652017.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Endo S, Yamato K, Hirai S, Moriwaki T,
Fukuda K, Suzuki H, Abei M, Nakagawa I and Hyodo I: Potent in vitro
and in vivo antitumor effects of MDM2 inhibitor nutlin-3 in gastric
cancer cells. Cancer Sci. 102:605–613. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Wei YS and Adachi I: Inhibitory effect of
triptolide on colony formation of breast and stomach cancer cell
lines. Zhongguo Yao Li Xue Bao. 12:406–410. 1991.PubMed/NCBI
|
|
153
|
Jiang XH, Wong BC, Lin MC, Zhu GH, Kung
HF, Jiang SH, Yang D and Lam SK: Functional p53 is required for
triptolide-induced apoptosis and AP-1 and nuclear factor-kappaB
activation in gastric cancer cells. Oncogene. 20:8009–8018. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Wang BY, Cao J, Chen JW and Liu QY:
Triptolide induces apoptosis of gastric cancer cells via inhibiting
the overexpression of MDM2. Med Oncol. 31:2702014. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Choi HS, Seo HS, Kim JH, Um JY, Shin YC
and Ko SG: Ethanol extract of paeonia suffruticosa Andrews (PSE)
induced AGS human gastric cancer cell apoptosis via fas-dependent
apoptosis and MDM2-p53 pathways. J Biomed Sci. 19:822012.
View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Lan H, Tang Z, Jin H and Sun Y:
Neddylation inhibitor MLN4924 suppresses growth and migration of
human gastric cancer cells. Sci Rep. 6:242182016. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Eto K, Iwatsuki M, Watanabe M, Ishimoto T,
Ida S, Imamura Y, Iwagami S, Baba Y, Sakamoto Y, Miyamoto Y, et al:
The sensitivity of gastric cancer to trastuzumab is regulated by
the miR-223/FBXW7 pathway. Int J Cancer. 136:1537–1545. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Schneekloth JS Jr and Crews CM: Natural
product inhibitors of the ubiquitin-proteasome pathway. Curr Drug
Targets. 12:1581–1594. 2011. View Article : Google Scholar : PubMed/NCBI
|