In recent years, there has been an upsurge in cancer
burden worldwide, and cancer has become the leading cause of death,
following cardiovascular diseases, in both men and women globally
(1). There are nearly 18 million
cases of cancer registered worldwide; among them, 268,600 are
breast cancer patients (2). Among
the different cancer types, breast cancer is one of the major
causes of death in the female population (3). Compelling evidence suggests that
specific genetic, epigenetic and environmental factors play a
critical role in the development of breast cancer. The prevalence
of breast cancer is caused by many factors, including unhealthy
lifestyle, excessive consumption of red meat, alcohol, smoking and
genetics (4). Nowadays,
high-throughput technologies, such as next-generation sequencing
have begun to elucidate tumor heterogeneity and has brought us
closer towards devising new diagnostic and therapeutic strategies
(5). Advanced experimental
methodologies have started to categorize proteome into sub-classes
of pro-apoptotic and anti-apoptotic proteins (5). This has led to characterization of
tumor necrosis factor (TNF)-related apoptosis-inducing ligand
(TRAIL) sub-proteomes (5).
Alterations in the TRAIL-mediated signaling pathway are associated
with the proliferation of breast cancer (6). Translation and functional studies have
clarified the underlying mechanisms and biomolecular signatures
responsible for impeding cancer treatment (7,8) Thus,
the search for better diagnostic and management of breast cancer is
needed.
Positive and negative regulation of apoptosis has
been a subject of extensive study over the past decades (9). There has been an increase in new
regulators of apoptosis that have deepened our understanding of the
process (10). A number of studies
have investigated the mechanisms that impede the TRAIL signaling
cascade (11–13). Knowledge of the association between
different pro-survival and cell death pathways in cancer is vital
for devising therapeutic strategies for cancer. TRAIL belongs to a
small subset of pro-apoptotic protein ligands in the TNF
superfamily, which also includes TNF and cluster of differentiation
(CD)95L (FasL/APO-1L) (14). TRAIL
has been investigated since 1997, when it was observed that
TRAIL-mediated apoptosis was responsible for death in cancer cells,
leaving normal cells intact (15).
This was followed by a number of studies documenting the molecular
characteristics of TRAIL-mediated apoptosis in various cancer
types, such as breast (16), thyroid
(17), colorectal (18), renal (19), bladder, prostate (20) and ovarian cancer (21). Parallel studies revealed that in
cancer cells, TRAIL was underexpressed, leading to loss of
TRAIL-induced apoptosis (22–24).
TRAIL-induced apoptosis is triggered through the activation of
death receptors (DRs), specifically DR4 and DR5 (25). This interaction in turn facilitates
the attachment of the apoptosis antigen 1 (Fas)-associated death
domain containing protein (FADD) (26). FADD attachment results in the
recruitment of adapter proteins to the cytoplasmic domain of DR
(26). Recruitment of adapter
proteins facilitates the activation of pro-caspases 8 and 10, which
then trigger the activation of caspase 3 (27). Activation of caspase 3 in turn leads
to activation of either the extrinsic pathway (caspase 8-mediated)
or the intrinsic pathway, which involves the release of cytochrome
c (28). Cytochrome
c-mediated activation of procaspase 9 to caspase 9 promotes
activation of the intrinsic pathway, which involves the
translocation of the BH3 interacting-domain death agonist to the
mitochondria (29). This facilitates
recruitment of Bax/Bak, which aid in the transportation of
cytochrome c and second mitochondria-derived activator of
caspases/Diablo homolog through the formation of the mitochondrial
pore (30,31).
Bioinformatics studies and RNA sequencing have been
used to delineate the role of lncRNA in breast cancer (43). Genetic heterogeneity of the
individual tumor is a crucial factor that triggers activation of
certain lncRNAs (44).
X inactive specific transcript (XIST) is an
oncogenic lncRNA that plays a significant role in the progression
of breast cancer. XIST RNA directs transcriptional changes by
binding to poly comb repressive complex 2 (PRC2). Deregulated XIST
promotes tumor progression (45).
XIST activation has been reported to accelerate tumor growth of
breast cancer gene 1-deficient ovarian cell lines (46). Accumulation of XIST promotes the
expression of X-linked oncogenes, including the V-RAF murine
sarcoma 3611 oncogene homolog 1 and member of ETS oncogene family,
which triggers the growth of tumor cells (47). Several factors are prerequisite for
triggering XIST. A recent study has reported that scaffold
attachment factor A, also known as heterogenous ribonucleoprotein
U, aids lncRNA attachment to the X chromosome. This promotes
activation of SMART/histone deacetylase (HDAC)1-associated
repressor protein, which recruits HDAC C3 and PRC2 components to
formulate histone repressive complex (48). In addition, high-throughput
sequencing has revealed that several XIST interactors serve a role
in the activation of XIST. In a recent study, lower expression of
XIST was reported in triple-negative breast cancer (TNBC). The
restored expression of XIST reduces the epithelial-mesenchymal
transition (EMT) property of cancer cells and cell proliferation,
and induces apoptosis (49). XIST in
TNBC functions by inhibiting microRNA (miR)-454 (49). XIST expression is also reported to be
downregulated in estrogen receptor-negative (ER−) and
progesterone receptor-negative (PR−) breast cancer
(50). However, XIST is highly
expressed in human epidermal growth factor receptor 2
(HER2)-positive breast cancer (51).
Few studies have been performed to indicate breast
cancer subtype-specific expression of lncRNAs (78); however, the underlying mechanism for
the tumorigenicity in breast cancer remains to be elucidated.
The contribution of lncRNAs to the growth,
proliferation and survival of different types of cancer has been
studied (36,46,79–83)
Several studies have emphasized the potential of lncRNAs in
promoting metastasis in breast cancer cell lines and tissues
(84,85). Dysregulation of lncRNA inhibiting
proliferation and metastasis (NLIPMT) has been reported to
enhance growth and metastasis in breast cancer tissue. Restoration
of NLIPMT expression in the breast cancer MDA-MB-231 cell
line inhibits cellular proliferation by suppressing glycogen
synthase kinase 3β phosphorylation (86). Some lncRNAs are overexpressed in
breast cancer cells, which facilitates tumor growth, spread and
survival by targeting the transcription of proteins. Such lncRNAs
are associated with cell growth suppression and apoptosis (87). High expression of lncRNA
FOXD3-AS1 in cancer tissues has a direct correlation with
tumor size increase and distant metastasis (88). A high level of lncRNA AWPPH in
patients' plasma is associated with enhanced cell growth in early
stage TNBC (89). Overexpression of
lncRNA AWPPH causes resistance to carboplatin treatment
(89).
Dysregulation of some lncRNAs is also associated
with the potential of breast tumor cells to metastasize to
different organ sites (90). The
majority of studies have reported an lncRNA role in the metastasis
of breast cancer to the lungs (91,92).
LINC00478-associated cytoplasmic RNA (lacRNA) is a cleaved
version of lncRNA LINC00478. LINC00478 is
significantly downregulated in metastatic breast tumors and
promotes active transcription of MYC proto-oncogene
(MYC)-activated genes (93).
lncRNA overexpression suppresses the metastatic and invasive
potential of breast cancer cells by stabilizing prohibitin-2 (PHB2)
protein. PHB2 then brings about transcriptional inhibition of
MYC target genes (93).
Furthermore, its overexpression inhibits lung metastasis in mouse
models (93). lncRNA
HOXA11-AS is also reported to be associated with breast
cancer metastasis to the lungs; it modulates EMT by downregulating
E-cadherin and vimentin expression. In mouse models treated with
shHOXA11-AS, the expression of HOXA11-AS is decreased
in both primary and secondary tumors (94). lncRNA ANCR is downregulated in
breast tumor cells and induces metastasis via active signal
transduction through the TGF-β pathway (95). Upon introduction of
ANCR-deficient MDA-MB-231-ANCR cells into BABL/c nude
mice, these cells metastasize to the lungs (95).
The role of lncRNAs in promoting metastasis in
breast cancer subtypes with different molecular signatures, such as
luminal A, luminal B, HER2-type, normal-like and triple-negative,
has yet to be properly studied. This indicates the need for further
studies in the area to better understand the role of lncRNAs in
breast cancer according to the different subtypes. This may be
helpful in designing more effective therapeutics for this
disease.
lncRNAs have a dual role in cellular homeostasis.
Depending on their interactive molecular landscape they can either
favor survival of the cancer cells or apoptosis (84). TRAIL-mediated apoptosis is one such
pathway and the alteration in the expression of its members shifts
the balance of the cell in favor of survival (96,97).
Recent advances in biomolecular studies have hinted towards the
association of the interplay of TRAIL and lncRNA with breast cancer
development (77). The activity of
caspases is a chief factor that is modulated by most lncRNAs in
breast cancer to ensure the rapid multiplication and growth of
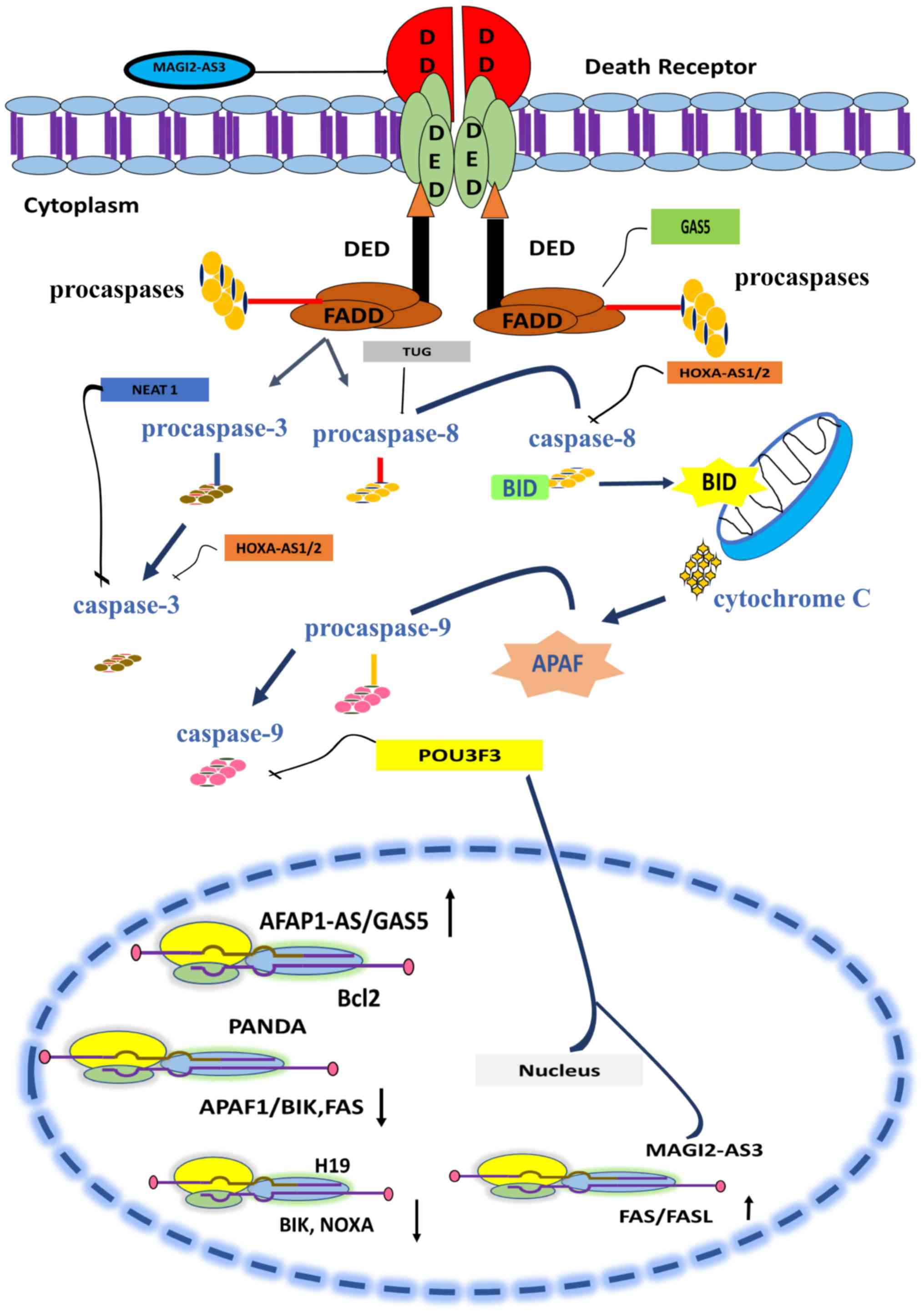
cancerous cells (98). Table I contains a list of lncRNAs whose
dysregulation in breast cancer disrupts the TRAIL-induced apoptosis
pathway by modulating the activity of caspases. The modulatory role
of lncRNA in the extrinsic pathway is illustrated in Fig. 2.
The extrinsic and intrinsic apoptotic pathways are
both regulated by the various lncRNAs (101). Death receptor triggers the
activation of caspases. Several lncRNAs serve pivotal roles in the
regulation of caspase activity (101). HOXAS1/2 is involved in
inhibition of caspase 8 and 3 (102). NEAT1 inhibits the activity
of caspase 3 (65) and TUG
promotes the activity of caspase 8. Signals from caspases are
transferred to mitochondria and lead to apoptosis. lncRNAs such as
GAS5/AFAP1-AS and MAG12-AS3 promote the upregulation
of BCL-2 and FAS genes and facilitate apoptosis
(103–105). lncRNA PANDA (p21-associated
ncRNA DNA damage activated) downregulates the expression of
proapoptotic proteins such as the Fas cell surface death receptor
(FAS)/BCL-2 interacting killer (BIK) and apoptotic protease
activating factor (APAF)1, thus inhibiting apoptosis and promoting
cell growth in breast cancer cells (106).
Suppression subtractive hybridization in combination
with reverse dot-blotting suggests the correlation between high
expression of lncRNA Z38 and tumorigenesis in breast cancer
cells. Suppression of Z38 expression via shRNA causes
inhibition of in vivo tumorigenesis and a reduction in cell
viability. In addition, the TUNEL assay performed after
administration of Z38 siRNA reveals induction of apoptosis
in cancerous cells (109). This
study indicated that the administration of Z38 siRNA
mechanistically activates the intrinsic apoptotic pathway.
Knockdown of Z38 negatively influences cell proliferative
rate together with the induction of apoptosis in gastric cancer in
a similar way in breast cancer (109). Z38 acts through the
activation of caspase 3 and 9 to initiate the apoptotic pathway
(110). High expression of
Z38 and its oncogenic influence makes it prognostically
significant in cases of breast cancer (111).
Cancer cells manage to grow and survive after
hijacking TRAIL-mediated apoptosis (116). Tumor cells use Fas receptor as a
reserve route to initiate activation of caspase 8 via proteolytic
cleavage and hence induce apoptosis (117,118).
In breast cancer, the expression levels of Fas and FasL are also
downregulated, eliminating all apoptotic threats for cancerous
cells and making their proliferation possible (119). In breast cancer tissue, the
expression of Fas and FasL is reported to be positively correlated
with the expression of lncRNA MAGI2-AS3 (105). Using transcript transfection and
lentiviral approaches, Yang et al (120) reported that MAGI2-AS3
expression facilitates the upregulation of Fas and FasL expression
in MDA-MB-231 and MCF-7 cell lines. CCK-8 assay and flow cytometry
further demonstrated that the lentivirus-induced expression of
MAGI2-AS3 reduces cell viability and promotes cell death via
activation of the Fas/FasL-induced apoptotic pathway (120).
A few identified lncRNAs negatively modulate the
TRAIL-induced apoptotic pathway by affecting the transcription of
pro-apoptotic proteins whose expression is triggered by TRAIL
signaling (Table II). Among them;
overexpression of H19 is reported in ERα+ breast
cancer cells, where it halts apoptotic signaling of the cell by
suppressing transcription of BIK and NOXA genes
(121). Due to its aberrant levels
in ERα+ breast cancer tissues and patients' plasma, it
has the potential to be used as a diagnostic marker for this breast
cancer type (122–124). lncRNA H19, with the help of
epigenetic modification, brings about the silencing of the
BIK gene; it blocks the promoter region of BIK by
facilitating the recruitment of EZH2, which then induces
trimethylation of histone H3 at lysine 27 (121). A recent development has revealed
that the expression of lncRNA H19 is modulated by lncRNA
PTCSC3 in TNBC (77). The
high H19 level in TNBC tumor tissues is inversely correlated with
PTCSC3 expression. Wang et al (121,122)
transfected the BT-549 and HCC70 cell lines with PTCSC3
vectors and reported that overexpression of PTCSC3
attenuates the expression of lncRNA H19 and consequently
suppresses cancer cell proliferation. Considering the role of
lncRNA H19 in the rapid proliferation and chemo-resistance
of breast cancer (125), treatment
with PTCSC3 could be a potential strategy to counter the
oncogenic effects of H19 in breast cancer.
A few more involvements of lncRNA in breast cancer
have been demonstrated to modulate the TRAIL-mediated apoptotic
pathway by regulating downstream factors of the TGF-β signaling
pathway. It has been well established by various studies that TGF-β
induces TRAIL expression, which is necessary for preventing
cancerous cell growth (28,134) By contrast, the tumor-suppressive
role of TGF-β reverses in advanced types of cancer, including in
breast cancer, where it promotes cancer advancement and metastasis
by downregulating the expression of TRAIL (135). Long intergenic non-protein coding
RNA regulator of reprograming (linc-ROR) lncRNA plays a crucial
role in the upregulation of TGF-β expression in advanced stages of
cancer (136); it is highly
expressed in tumor tissue and also in the highly invasive breast
cancer MCF-7 and MDA-MB-231 cell lines. Knockdown of linc-ROR
through siRNA in MCF-7 and MDA-MB-231 cells showed that linc-ROR
silencing negatively regulates TGF-β and the expression of its
downstream factors, which consequently attenuates aggressive tumor
growth (136). Unlike lncRNA
linc-ROR, the expression of lncRNA CASC2 is downregulated,
which facilitates TGF-β pathway activation in advanced breast
cancer (137). Induced expression
of CASC2 in MCF-7 and LCC-9 cell lines via transfection with
pcDNA-CASC2 results in CASC2 overexpression in these cell
lines. Furthermore, CASC2 inhibits cell metastasis and
promotes cell death by targeting smad-2 (a downstream factor of the
TGF-β pathway) and triggering TRAIL apoptosis (137).
TGF-β needs to halt the apoptotic pathway in order
to ensure tumor proliferation and metastasis (138). Through the application of northern
blotting and qPCR, it has been determined in several mouse breast
cancer cell lines, that to prompt suppression of the apoptotic
pathway, TGF-β induces the expression of a ~3-kb long transcript of
lncRNA Smad7 (139). The
results from TUNEL staining and RT-qPCR have established that
lncRNA Smad7 functions as a downstream anti-apoptotic factor
of TGF-β signaling, the overexpression of which halts apoptosis by
inhibiting Bim expression and upregulating anti-apoptotic protein
differentiated embryonic chondrocyte-expressed gene 1 expression in
invasive breast cancer cell lines (139,140).
Breast cancer is a highly complex disease involving
a number of types and genetics. Thus, an efficient and precise
therapeutic regimen for breast cancer patients can only be achieved
by rapid and comprehensive prognosis and diagnosis. lncRNAs have
crucial implementations in different cancer types; they have
established themselves as important regulators of transcription, as
well as activators of various signaling cascades. These non-coding
RNA molecules are tissue-specific and have the potential to serve
as biomarkers for breast cancer. However, few studies have
elucidated the involvement of these micromanagers in regulating
apoptosis and even fewer have addressed their interplay with
TRAIL-mediated apoptosis. Technological advances in bioinformatics,
sequencing and mass spectrometry have, to some extent, delineated
the role of lncRNA in tumor biology. Identifying lncRNA as
non-invasive biomarkers that can be robustly detected in liquid
biopsies could revolutionize the way breast cancer is detected.
Unearthing the many functions of ncRNAs in cancer development
delves into the genomic complexity of cancer and further highlights
the extensive interplay between various genetic elements in the
cells.
Not applicable.
No funding was received.
Data sharing is not applicable to this article, as
no datasets were generated or analyzed during the current
study.
ZJ, KK and BS wrote the manuscript. MI, QR, TA, BS
and SR revised the review. HS, JR and WC conceptualized the study
and revised it critically. All authors have read and approved the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Fedewa SA, Goding Sauer A,
Kramer JL, Smith RA and Jemal A: Breast cancer statistics, 2015:
Convergence of incidence rates between black and white women. CA
Cancer J Clin. 66:31–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zur Hausen H, Bund T and de Villiers EM:
Specific nutritional infections early in life as risk factors for
human colon and breast cancers several decades later. Int J Cancer.
144:1574–1583. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Naoum GE, Buchsbaum DJ, Tawadros F,
Farooqi A and Arafat WO: Journey of TRAIL from bench to bedside and
its potential role in immuno-oncology. Oncol Rev.
11:3322017.PubMed/NCBI
|
|
6
|
Johnstone RW, Frew AJ and Smyth MJ: The
TRAIL apoptotic pathway in cancer onset, progression and therapy.
Nat Rev Cancer. 8:782–798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi X, Li Y, Sun Y, Zhao X, Sun X, Gong T,
Liang Z, Ma Y and Zhang X: Genome-wide analysis of lncRNAs, miRNAs,
and mRNAs forming a prognostic scoring system in esophageal
squamous cell carcinoma. PeerJ. 8:e83682020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tirosh I and Suvà ML: Deciphering human
tumor biology by single-cell expression profiling. Ann Rev Cancer
Biol. 3:151–166. 2019. View Article : Google Scholar
|
|
9
|
Farooqi AA, Mukhtar S, Riaz AM, Waseem S,
Minhaj S, Dilawar BA, Malik BA, Nawaz A and Bhatti S: Wnt and SHH
in prostate cancer: Trouble mongers occupy the TRAIL towards
apoptosis. Cell Prolif. 44:508–515. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Walensky LD: Cheating death: New molecules
block BAX. Trends Mol Med. 25:259–261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mazurek N, Byrd JC, Sun Y, Hafley M,
Ramirez K, Burks J and Bresalier RS: Cell-surface galectin-3
confers resistance to TRAIL by impeding trafficking of death
receptors in metastatic colon adenocarcinoma cells. Cell Death
Differ. 19:523–533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seyrek K, Richter M and Lavrik IN:
Decoding the sweet regulation of apoptosis: The role of
glycosylation and galectins in apoptotic signaling pathways. Cell
Death Differ. 26:981–993. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ivanova S, Polajnar M, Narbona-Perez AJ,
Hernandez-Alvarez MI, Frager P, Slobodnyuk K, Plana N, Nebreda AR,
Palacin M, Gomis RR, et al: Regulation of death receptor signaling
by the autophagy protein TP53INP2. EMBO J. 38:e993002019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaufmann T, Strasser A and Jost PJ: Fas
death receptor signalling: Roles of Bid and XIAP. Cell Death
Differ. 19:42–50. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Walczak H, Degli-Esposti MA, Johnson RS,
Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA,
Smith CA, et al: TRAIL-R2: A novel apoptosis-mediating receptor for
TRAIL. EMBO J. 16:5386–5397. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rahman M, Davis SR, Pumphrey JG, Bao J,
Nau MM, Meltzer PS and Lipkowitz S: TRAIL induces apoptosis in
triple-negative breast cancer cells with a mesenchymal phenotype.
Br Cancer Res Treat. 113:217–230. 2009. View Article : Google Scholar
|
|
17
|
Ahmad M and Shi Y: TRAIL-induced apoptosis
of thyroid cancer cells: Potential for therapeutic intervention.
Oncogene. 19:3363–3371. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao L, Dong A, Gu J, Liu Z, Zhang Y,
Zhang W, Wang Y, He L, Qian C, Qian Q and Liu X: The antitumor
activity of TRAIL and IL-24 with replicating oncolytic adenovirus
in colorectal cancer. Cancer Gene Ther. 13:1011–1022. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brooks AD and Sayers TJ: Reduction of the
antiapoptotic protein cFLIP enhances the susceptibility of human
renal cancer cells to TRAIL apoptosis. Cancer Immunol Immunother.
54:499–505. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Voelkel-Johnson C: TRAIL-mediated
signaling in prostate, bladder and renal cancer. Nat Rev Urol.
8:417–427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cuello M, Ettenberg SA, Nau MM and
Lipkowitz S: Synergistic induction of apoptosis by the combination
of trail and chemotherapy in chemoresistant ovarian cancer cells.
Gynecol Oncol. 81:380–390. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Finnberg NK and El-Deiry WS: TRAIL death
receptors as tumor suppressors and drug targets. Cell Cycle.
7:1525–1528. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y and Zhang B: TRAIL resistance of
breast cancer cells is associated with constitutive endocytosis of
death receptors 4 and 5. Mol Cancer Res. 6:1861–1871. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tollefson AE, Toth K, Doronin K,
Kuppuswamy M, Doronina OA, Lichtenstein DL, Hermiston TW, Smith CA
and Wold WS: Inhibition of TRAIL-induced apoptosis and forced
internalization of TRAIL receptor 1 by adenovirus proteins. J
Virol. 75:8875–8887. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suliman A, Lam A, Datta R and Srivastava
RK: Intracellular mechanisms of TRAIL: Apoptosis through
mitochondrial-dependent and-independent pathways. Oncogene.
20:2122–2133. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Screaton RA, Kiessling S, Sansom OJ,
Millar CB, Maddison K, Bird A, Clarke AR and Frisch SM:
Fas-associated death domain protein interacts with methyl-CpG
binding domain protein 4: A potential link between genome
surveillance and apoptosis. Proc Natl Acad Sci USA. 100:5211–5216.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Aggarwal BB, Bhardwaj U and Takada Y:
Regulation of TRAIL-induced apoptosis by ectopic expression of
antiapoptotic factors. Vitamins & Hormones Elsevier. 453–483.
2004. View Article : Google Scholar
|
|
28
|
Jang CW, Chen CH, Chen CC, Chen JY, Su YH
and Chen RH: TGF-Beta induces apoptosis through Smad-mediated
expression of DAP-kinase. Nat Cell Biol. 4:51–58. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kruidering M and Evan GI: Caspase-8 in
apoptosis: The beginning of ‘the end’? IUBMB Life. 50:85–90. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Farooqi AA and De Rosa G: TRAIL and
microRNAs in the treatment of prostate cancer: Therapeutic
potential and role of nanotechnology. Appl Microbiol Biotechnol.
97:8849–8857. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Falschlehner C, Emmerich CH, Gerlach B and
Walczak H: TRAIL signalling: Decisions between life and death. Int
J Biochem Cell Biol. 39:1462–1475. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han Li C and Chen Y: Small and long
non-coding RNAs: Novel targets in perspective cancer therapy. Curr
Genomics. 16:319–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Müller V, Oliveira-Ferrer L, Steinbach B,
Pantel K and Schwarzenbach H: Interplay of lncRNA H19/miR-675 and
lncRNA NEAT1/miR-204 in breast cancer. Mol Oncol. 13:1137–1149.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Javed Z, Ahmed Shah F, Rajabi S, Raza Q,
Iqbal Z, Ullah M, Ahmad T, Salehi B, Sharifi-Rad M, Pezzani R, et
al: LncRNAs as potential therapeutic targets in thyroid cancer.
Asian Pac J Cancer Prev. 21:281–287. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi
J, Liu B, Sun S, Yang F, Wang L and Qu L: Long noncoding RNA
EGFR-AS1 promotes cell growth and metastasis via affecting HuR
mediated mRNA stability of EGFR in renal cancer. Cell Death Dis.
10:1542019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Luo H, Xu C, Le W, Ge B and Wang T: lncRNA
CASC11 promotes cancer cell proliferation in bladder cancer through
miRNA-150. J Cell Biochem. 120:13487–13493. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao
Y, Xu W, Niu Y, Cheng L, Maity SN, et al: LncRNA-p21 alters the
antiandrogen enzalutamide-induced prostate cancer neuroendocrine
differentiation via modulating the EZH2/STAT3 signaling. Nat
Commun. 10:25712019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y and Ruan F: LncRNA LEF1-AS1
promotes ovarian cancer development through interacting with
miR-1285-3p. Cancer Manag Res. 12:687–694. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He RZ, Luo DX and Mo YY: Emerging roles of
lncRNAs in the post-transcriptional regulation in cancer. Genes
Dis. 6:62019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lau E: Non-coding RNA: Zooming in on
lncRNA functions. Nat Rev Genet. 15:574–575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
van Leeuwen S and Mikkers H: Long
non-coding RNAs: Guardians of development. Differentiation.
80:175–183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ye N, Wang B, Quan ZF, Cao SJ, Wen XT,
Huang Y, Huang XB, Wu R, Ma XP, Yan QG, et al: Functional roles of
long non-coding RNA in human breast cancer. Asian Pac J Cancer
Prev. 15:5993–5997. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cheetham S, Gruhl F, Mattick J and Dinger
M: Long noncoding RNAs and the genetics of cancer. Br J Cancer.
108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee JT and Bartolomei MS: X-inactivation,
imprinting, and long noncoding RNAs in health and disease. Cell.
152:1308–1323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang TH, Liang LZ, Liu XL, Wu JN, Su K,
Chen JY and Zheng QY: LncRNA UCA1/miR-124 axis modulates
TGFβ1-induced epithelial-mesenchymal transition and invasion of
tongue cancer cells through JAG1/Notch signaling. J Cell Biochem.
120:10495–10504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kawakami T, Zhang C, Taniguchi T, Kim CJ,
Okada Y, Sugihara H, Hattori T, Reeve AE, Ogawa O and Okamoto K:
Characterization of loss-of-inactive X in Klinefelter syndrome and
female-derived cancer cells. Oncogene. 23:6163–6169. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Postlmayr A and Wutz A: Insights into the
establishment of chromatin states in pluripotent cells from studies
of X inactivation. J Mol Biol. 429:1521–1531. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li X, Hou L, Yin L and Zhao S: LncRNA XIST
interacts with miR-454 to inhibit cells proliferation, epithelial
mesenchymal transition and induces apoptosis in triple-negative
breast cancer. J Biosci. 45:452020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng R, Lin S, Guan L, Yuan H, Liu K, Liu
C, Ye W, Liao Y, Jia J and Zhang R: Long non-coding RNA XIST
inhibited breast cancer cell growth, migration, and invasion via
miR-155/CDX1 axis. Biochem Biophys Res Commun. 498:1002–1008. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao L, Zhao Y, He Y, Li Q and Mao Y: The
functional pathway analysis and clinical significance of miR-20a
and its related lncRNAs in breast cancer. Cell Signal. 51:152–165.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen Y and Li CH: Novel therapeutic
targets for hepatocellular carcinoma treatment. Hepatocellular
Carcinoma Basic Res. 352012.doi: 10.5772/28894.
|
|
54
|
Battistelli C, Sabarese G, Santangelo L,
Montaldo C, Gonzalez FJ, Tripodi M and Cicchini C: The lncRNA
HOTAIR transcription is controlled by HNF4α-induced chromatin
topology modulation. Cell Death Differ. 26:890–901. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Qu X, Alsager S, Zhuo Y and Shan B: HOX
transcript antisense RNA (HOTAIR) in cancer. Cancer Lett.
454:90–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bhan A and Mandal SS: LncRNA HOTAIR: A
master regulator of chromatin dynamics and cancer. Biochim Biophys
Acta. 1856:151–164. 2015.PubMed/NCBI
|
|
57
|
Cai B, Song X, Cai J and Zhang S: HOTAIR:
A cancer-related long non-coding RNA. Neoplasma. 61:379–391. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hajjari M and Salavaty A: HOTAIR: An
oncogenic long non-coding RNA in different cancers. Cancer Bio Med.
12:1–9. 2015.
|
|
60
|
Bhan A, Hussain I, Ansari KI, Kasiri S,
Bashyal A and Mandal SS: Antisense transcript long noncoding RNA
(lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol
Biol. 425:3707–3722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Amândio AR, Necsulea A, Joye E, Mascrez B
and Duboule D: Hotair is dispensible for mouse development. PLoS
Genet. 12:e10062322016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sørensen KP, Thomassen M, Tan Q, Bak M,
Cold S, Burton M, Larsen MJ and Kruse TA: Long non-coding RNA
HOTAIR is an independent prognostic marker of metastasis in
estrogen receptor-positive primary breast cancer. Breast cancer Res
Treat. 142:529–536. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Tao S, He H and Chen Q: Estradiol induces
HOTAIR levels via GPER-mediated miR-148a inhibition in breast
cancer. J Transl Med. 13:1312015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lv R, Zhang J, Zhang W, Huang Y, Wang N,
Zhang Q and Qu S: Circulating HOTAIR expression predicts the
clinical response to neoadjuvant chemotherapy in patients with
breast cancer. Cancer Biomark. 22:249–256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang M, Wu WB, Wang ZW and Wang XH:
lncRNA NEAT1 is closely related with progression of breast cancer
via promoting proliferation and EMT. Eur Rev Med Pharmacol Sci.
21:1020–1026. 2017.PubMed/NCBI
|
|
66
|
Li W, Zhang Z, Liu X, Cheng X, Zhang Y,
Han X, Zhang Y, Liu S, Yang J, Xu B, et al: The FOXN3-NEAT1-SIN3A
repressor complex promotes progression of hormonally responsive
breast cancer. J Clin Invest. 127:3421–3440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shin VY, Chen J, Cheuk IW, Siu MT, Ho CW,
Wang X, Jin H and Kwong A: Long non-coding RNA NEAT1 confers
oncogenic role in triple-negative breast cancer through modulating
chemoresistance and cancer stemness. Cell Death Dis. 10:2702019.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Qian K, Liu G, Tang Z, Hu Y, Fang Y, Chen
Z and Xu X: The long non-coding RNA NEAT1 interacted with miR-101
modulates breast cancer growth by targeting EZH2. Arch Biochem
Biophys. 615:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li X, Wang S, Li Z, Long X, Guo Z, Zhang
G, Zu J, Chen Y and Wen L: The lncRNA NEAT1 facilitates cell growth
and invasion via the miR-211/HMGA2 axis in breast cancer. Int J
Biol Macromol. 105:346–353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q,
Su X, Peng L and Jiao B: NEAT1 is required for survival of breast
cancer cells through FUS and miR-548. Gene Regul Syst Bio. 10
(Suppl 1):S11–S17. 2016.
|
|
71
|
Godinho M, Meijer D, Setyono-Han B,
Dorssers LC and van Agthoven T: Characterization of BCAR4, a novel
oncogene causing endocrine resistance in human breast cancer cells.
J Cell Physiol. 226:1741–1749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Godinho MF, Wulfkuhle JD, Look MP,
Sieuwerts AM, Sleijfer S, Foekens JA, Petricoin EF III, Dorssers LC
and van Agthoven T: BCAR4 induces antioestrogen resistance but
sensitises breast cancer to lapatinib. Br J Cancer. 107:947–955.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xing Z, Park PK, Lin C and Yang L: LncRNA
BCAR4 wires up signaling transduction in breast cancer. RNA Biol.
12:681–689. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sun Q, Hao Q and Prasanth KV: Nuclear long
noncoding RNAs: Key regulators of gene expression. Trends Genet.
34:142–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Godinho MF, Sieuwerts AM, Look MP, Meijer
D, Foekens JA, Dorssers LC and van Agthoven T: Relevance of BCAR4
in tamoxifen resistance and tumour aggressiveness of human breast
cancer. Br J Cancer. 103:1284–1291. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Niknafs YS, Han S, Ma T, Speers C, Zhang
C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, et al:
The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1
in breast cancer progression. Nat Commun. 7:127912016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wang Z and Zöller M: Exosomes, metastases,
and the miracle of cancer stem cell markers. Cancer Metastasis Rev.
38:259–295. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ouyang D, Su J, Huang P, Li M, Li Q, Zhao
P, Chen Q, Zou Q, Feng X, Qian K, et al: Identification of lncRNAs
via microarray analysis for predicting HER2-negative breast cancer
response to neoadjuvant chemotherapy. Int J Clin Exp Pathol.
11:2621–2628. 2018.PubMed/NCBI
|
|
79
|
Chen YK and Yen Y: The ambivalent role of
lncRNA Xist in carcinogenesis. Stem Cell Rev Rep. 15:314–323. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Mazor G, Levin L, Picard D, Ahmadov U,
Carén H, Borkhardt A, Reifenberger G, Leprivier G, Remke M and
Rotblat B: The lncRNA TP73-AS1 is linked to aggressiveness in
glioblastoma and promotes temozolomide resistance in glioblastoma
cancer stem cells. Cell Death Dis. 10:2462019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu
Q, Tong X, Yang W, Xu Q, Huang D and Tu K: A novel lncRNA
MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by
targeting miR-194-5p/FOXA1 axis. Mol Cancer. 18:282019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kang CL, Qi B, Cai QQ, Fu LS, Yang Y, Tang
C, Zhu P, Chen QW, Pan J, Chen MH and Wu XZ: LncRNA AY promotes
hepatocellular carcinoma metastasis by stimulating ITGAV
transcription. Theranostics. 9:4421–4436. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang H, Zhu M, Du Y, Zhang H, Zhang Q,
Liu Q, Huang Z, Zhang L, Li H, Xu L, et al: A panel of 12-lncRNA
signature predicts survival of pancreatic adenocarcinoma. J Cancer.
10:1550–1559. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Farooqi AA, Attar R, Qureshi MZ, Fayyaz S,
Sohail MI, Sabitaliyevich UY, Nurmurzayevich SB, Yelekenova A,
Yaylim I and Alaaeddine N: Interplay of long non-coding RNAs and
TGF/SMAD signaling in different cancers. Cell Mol Biol
(Noisy-le-Grand). 64:1–6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Jiang Y, Lin L, Zhong S, Cai Y, Zhang F,
Wang X, Miao R, Zhang B, Gao S and Hu X: Overexpression of novel
lncRNA NLIPMT inhibits metastasis by reducing phosphorylated
glycogen synthase kinase 3β in breast cancer. J Cell Physiol.
234:10698–10708. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu Y, Sharma S and Watabe K: Roles of
lncRNA in breast cancer. Front Biosci (Schol Ed). 7:94–108. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Guan Y, Bhandari A, Xia E, Yang F, Xiang J
and Wang O: lncRNA FOXD3-AS1 is associated with clinical
progression and regulates cell migration and invasion in breast
cancer. Cell Biochem Funct. 37:239–244. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Liu AN, Qu HJ, Gong WJ, Xiang JY, Yang MM
and Zhang W: LncRNA AWPPH and miRNA-21 regulates cancer cell
proliferation and chemosensitivity in triple-negative breast cancer
by interacting with each other. J Cell Biochem. 120:14860–14866.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li Z, Hou P, Fan D, Dong M, Ma M, Li H,
Yao R, Li Y, Wang G, Geng P, et al: The degradation of EZH2
mediated by lncRNA ANCR attenuated the invasion and metastasis of
breast cancer. Cell Death Differ. 24:59–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Guo R, Su Y, Xue J, Si J, Chi Y and Wu J:
Abstract P6-05-01: A novel cleaved cytoplasmic lncRNA LacRNA
interacts with PHB2 and suppresses breast cancer metastasis via
repressing MYC targets. Cancer Res. 792019.doi: 10.1158/1538-7445.
PubMed/NCBI
|
|
94
|
Li W, Jia G, Qu Y, Du Q and Liu B and Liu
B: Long non-coding RNA (LncRNA) HOXA11-AS promotes breast cancer
invasion and metastasis by regulating epithelial-mesenchymal
transition. Med Sci Monit. 23:3393–3403. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Li Z, Dong M, Fan D, Hou P, Li H, Liu L,
Lin C, Liu J, Su L, Wu L, et al: LncRNA ANCR down-regulation
promotes TGF-β-induced EMT and metastasis in breast cancer.
Oncotarget. 8:67329–67343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Naval J, de Miguel D, Gallego-Lleyda A,
Anel A and Martinez-Lostao L: Importance of TRAIL molecular anatomy
in receptor oligomerization and signaling. Implications for Cancer
Therapy. Cancers (Basel). 11:4442019. View Article : Google Scholar
|
|
97
|
Mert U and Sanlioglu AD: Intracellular
localization of DR5 and related regulatory pathways as a mechanism
of resistance to TRAIL in cancer. Cell Mol Life Sci. 74:245–255.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Li T, Liu Y, Xiao H and Xu G: Long
non-coding RNA TUG1 promotes cell proliferation and metastasis in
human breast cancer. Breast Cancer. 24:535–543. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Yang J, Meng X, Yu Y, Pan L, Zheng Q and
Lin W: LncRNA POU3F3 promotes proliferation and inhibits apoptosis
of cancer cells in triple-negative breast cancer by inactivating
caspase 9. Biosci Biotechnol Biochem. 83:1117–1123. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Shan TD, Xu JH, Yu T, Li JY, Zhao LN,
Ouyang H, Luo S, Lu XJ, Huang CZ, Lan QS, et al: Knockdown of
linc-POU3F3 suppresses the proliferation, apoptosis, and migration
resistance of colorectal cancer. Oncotarget. 7:961–975. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Rossi MN and Antonangeli F: LncRNAs: New
players in apoptosis control. Int J Cell Biol. 2014:4738572014.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Qu Y, Wang Y, Wang P, Lin N, Yan X and Li
Y: Overexpression of long noncoding RNA HOXA-AS2 predicts an
adverse prognosis and promotes tumorigenesis via SOX4/PI3K/AKT
pathway in acute myeloid leukemia. Cell Biol Int. May 5–2020.doi:
10.1002/cbin.11370 (Epub ahead of print). View Article : Google Scholar
|
|
103
|
Awasthee N, Rai V, Verma SS, Francis KS,
Nair MS and Gupta SC: Anti-cancer activities of Bharangin against
breast cancer: Evidence for the role of NF-κB and lncRNAs. Biochim
Biophys Acta Gen Subj. 1862:2738–2749. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Dianatpour A, Faramarzi S, Geranpayeh L,
Mirfakhraie R, Motevaseli E and Ghafouri-Fard S: Expression
analysis of AFAP1-AS1 and AFAP1 in breast cancer. Cancer Biomark.
22:49–54. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang H and Lu B: microRNAs as biomarkers
of ovarian cancer. Expert Rev Anticancer Ther. 20:373–385. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Huang YS, Chang CC, Lee SS, Jou YS and
Shih HM: Xist reduction in breast cancer upregulates AKT
phosphorylation via HDAC3-mediated repression of PHLPP1 expression.
Oncotarget. 7:432562016. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Gooding AJ, Zhang B, Jahanbani FK, Gilmore
HL, Chang JC, Valadkhan S and Schiemann WP: The lncRNA BORG drives
breast cancer metastasis and disease recurrence. Sci Rep.
7:126982017. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Gooding AJ, Zhang B, Gunawardane L, Beard
A, Valadkhan S and Schiemann WP: The lncRNA BORG facilitates the
survival and chemoresistance of triple-negative breast cancers.
Oncogene. 38:20202019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Deng R, Liu B, Wang Y, Yan F, Hu S, Wang
H, Wang T, Li B, Deng X, Xiang S, Yang Y and Zhang J: High
expression of the newly found long noncoding RNA Z38 promotes cell
proliferation and oncogenic activity in breast cancer. J Cancer.
7:576–578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang Y, Zheng C, Li T, Zhang R, Wang Y,
Zhang J, He Q, Sun Z and Wang X: Long noncoding RNA Z38 promotes
cell proliferation and metastasis and inhibits cell apoptosis in
human gastric cancer. Oncolo Lett. 16:6051–6058. 2018.
|
|
111
|
Nie ZL, Wang YS, Mei YP, Lin X, Zhang GX,
Sun HL, Wang YL, Xia YX and Wang SK: Prognostic significance of
long noncoding RNA Z38 as a candidate biomarker in breast cancer. J
Clin Lab Anal. 32:e221932018. View Article : Google Scholar
|
|
112
|
Zhang F, Li J, Xiao H, Zou Y, Liu Y and
Huang W: AFAP1-AS1: A novel oncogenic long non-coding RNA in human
cancers. Cell Proliferation. 51:e123972018. View Article : Google Scholar
|
|
113
|
Ma D, Chen C, Wu J, Wang H and Wu D:
Up-regulated lncRNA AFAP1-AS1 indicates a poor prognosis and
promotes carcinogenesis of breast cancer. Breast Cancer. 26:74–83.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Fan S, Yang Z, Ke Z, Huang K, Liu N, Fang
X and Wang K: Downregulation of the long non-coding RNA TUG1 is
associated with cell proliferation, migration, and invasion in
breast cancer. Biomed Pharmacother. 95:1636–1643. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Tang T, Cheng Y, She Q, Jiang Y, Chen Y,
Yang W and Li Y: Long non-coding RNA TUG1 sponges miR-197 to
enhance cisplatin sensitivity in triple negative breast cancer.
Biomed Pharmacother. 107:338–346. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ghavami S, Hashemi M, Ande SR, Yeganeh B,
Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ and
Los M: Apoptosis and cancer: Mutations within caspase genes. J Med
Genet. 46:497–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Rossin A, Miloro G and Hueber AO: TRAIL
and FasL functions in cancer and autoimmune diseases: Towards an
increasing complexity. Cancers. 11:6392019. View Article : Google Scholar
|
|
118
|
Eberle J: Countering TRAIL resistance in
melanoma. Cancers. 11:6562019. View Article : Google Scholar
|
|
119
|
Kolben T, Jeschke U, Reimer T, Karsten N,
Schmoeckel E, Semmlinger A, Mahner S, Harbeck N and Kolben TM:
Induction of apoptosis in breast cancer cells in vitro by Fas
ligand reverse signaling. J Cancer Res Clin Oncol. 144:249–256.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Yang Y, Yang H, Xu M, Zhang H, Sun M, Mu
P, Dong T, Du S and Liu K: Long non-coding RNA (lncRNA) MAGI2-AS3
inhibits breast cancer cell growth by targeting the Fas/FasL
signalling pathway. Hum Cell. 31:232–241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang
E, Shao L, Li A, Yang N, Han X, et al: LncRNA H19 confers
chemoresistance in ERα-positive breast cancer through epigenetic
silencing of the pro-apoptotic gene BIK. Oncotarget. 7:81452–81462.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li
TL, Cai JQ, Zhou HH and Zhu YS: H19 lncRNA mediates
17β-estradiol-induced cell proliferation in MCF-7 breast cancer
cells. Oncol Rep. 33:3045–3052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang K, Luo Z, Zhang Y, Zhang L, Wu L,
Liu L, Yang J, Song X and Liu J: Circulating lncRNA H19 in plasma
as a novel biomarker for breast cancer. Cancer Biomark. 17:187–194.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Lin Y and Tao H: Diagnostic value of
plasma exosomal lncRNA H19 for breast cancer. Chin J Clin
Laboratory Sci. 36:99–101. 2018.
|
|
125
|
Han J, Han B, Wu X, Hao J, Dong X, Shen Q
and Pang H: Knockdown of lncRNA H19 restores chemo-sensitivity in
paclitaxel-resistant triple-negative breast cancer through
triggering apoptosis and regulating Akt signaling pathway. Toxicol
Appl Pharmacol. 359:55–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Li J, Tian H, Yang J and Gong Z: Long
noncoding RNAs regulate cell growth, proliferation, and apoptosis.
DNA Cell Biol. 35:459–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Hung T, Wang Y, Lin MF, Koegel AK, Kotake
Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al:
Extensive and coordinated transcription of noncoding RNAs within
cell-cycle promoters. Nat Genet. 43:621–629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Zhang A, Xu M and Mo YY: Role of the
lncRNA-p53 regulatory network in cancer. J Mol Cell Biol.
6:181–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Pickard MR and Williams GT: The hormone
response element mimic sequence of GAS5 lncRNA is sufficient to
induce apoptosis in breast cancer cells. Oncotarget. 7:101042016.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zong Y, Zhang Y, Sun X, Xu T, Cheng X and
Qin Y: miR-221/222 promote tumor growth and suppress apoptosis by
targeting lncRNA GAS5 in breast cancer. Biosci Rep.
39:BSR201818592019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C,
Xu M, Wu F and Mo YY: Negative regulation of lncRNA GAS5 by miR-21.
Cell Death Differ. 20:1558–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wickramasinghe NS, Manavalan TT, Dougherty
SM, Riggs KA, Li Y and Klinge CM: Estradiol downregulates miR-21
expression and increases miR-21 target gene expression in MCF-7
breast cancer cells. Nucleic Acids Res. 37:2584–2595. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
He X, Chen X, Zhang X, Duan X, Pan T, Hu
Q, Zhang Y, Zhong F, Liu J, Zhang H, et al: An Lnc RNA
(GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by
site-specifically recruiting MLL/COMPASS-like complexes. Nucleic
Acids Res. 43:3712–3725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Wang Y, Chu J, Yi P, Dong W, Saultz J,
Wang Y, Wang H, Scoville S, Zhang J, Wu LC, et al: SMAD4 promotes
TGF-β-independent NK cell homeostasis and maturation and antitumor
immunity. J Clin Invest. 128:5123–5136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Cano-González A and López-Rivas A:
Opposing roles of TGF-β and EGF in the regulation of TRAIL-induced
apoptosis in human breast epithelial cells. Biochim Biophys Acta.
1863:2104–2114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Hou L, Tu J, Cheng F, Yang H, Yu F, Wang
M, Liu J, Fan J and Zhou G: Long noncoding RNA ROR promotes breast
cancer by regulating the TGF-β pathway. Cancer Cell Int.
18:1422018. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Zhang Y, Zhu M, Sun Y, Li W, Wang Y and Yu
W: Upregulation of lncRNA CASC2 suppresses cell proliferation and
metastasis of breast cancer via inactivation of the TGF-β signaling
pathway. Oncol Res. 27:379–387. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Batlle E and Massagué J: Transforming
growth factor-β signaling in immunity and cancer. Immunity.
50:924–940. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Arase M, Horiguchi K, Ehata S, Morikawa M,
Tsutsumi S, Aburatani H, Miyazono K and Koinuma D: Transforming
growth factor-β-induced lnc RNA-Smad7 inhibits apoptosis of mouse
breast cancer JygMC(A) cells. Cancer Sci. 105:974–982. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Hoshino Y, Katsuno Y, Ehata S and Miyazono
K: Autocrine TGF-β protects breast cancer cells from apoptosis
through reduction of BH3-only protein, Bim. J Biochem. 149:55–65.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Xu ST, Xu JH, Zheng ZR, Zhao QQ, Zeng XS,
Cheng SX, Liang YH and Hu QF: Long non-coding RNA ANRIL promotes
carcinogenesis via sponging miR-199a in triple-negative breast
cancer. Biomed Pharmacother. 96:14–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Zhao JJ, Hao S, Wang LL, Hu CY, Zhang S,
Guo LJ, Zhang G, Gao B, Jiang Y, Tian WG and Luo DL: Long
non-coding RNA ANRIL promotes the invasion and metastasis of
thyroid cancer cells through TGF-β/Smad signaling pathway.
Oncotarget. 7:57903–57918. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Chen J, Shin VY, Siu MT, Ho JC, Cheuk I
and Kwong A: miR-199a-5p confers tumor-suppressive role in
triple-negative breast cancer. BMC Cancer. 16:8872016. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Zhang Y, Fan KJ, Sun Q, Chen AZ, Shen WL,
Zhao ZH, Zheng XF and Yang X: Functional screening for miRNAs
targeting Smad4 identified miR-199a as a negative regulator of
TGF-β signalling pathway. Nucleic Acids Res. 40:9286–9297. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X
and Tai S: LncRNA HOXA-AS2 and its molecular mechanisms in human
cancer. Clin Chim Acta. 485:229–233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Fang Y, Wang J, Wu F, Song Y, Zhao S and
Zhang Q: Long non-coding RNA HOXA-AS2 promotes proliferation and
invasion of breast cancer by acting as a miR-520c-3p sponge.
Oncotarget. 8:460902017. View Article : Google Scholar : PubMed/NCBI
|