Introduction
Histone deacetylase inhibitors (HDACIs) have emerged
as novel antitumor agents that are continuously tested for the
treatment of various cancer types due to their significant
antitumor activities, including inhibition of angiogenesis and
induction of cell cycle arrest, differentiation and apoptosis
(1). HDACIs have a promising
therapeutic potential and are approved for lymphoma treatment
(1). Furthermore, HDACIs have been
reported to induce cell cycle arrest, to promote cell
differentiation and to induce apoptosis in a variety of cancer
cells, including lung cancer, colorectal cancer, nasopharyngeal
carcinoma and cervical cancer cells, while having little effect on
healthy cells (1,2). HDACIs can be classified into various
types according to their different structure, such as aliphatic
acid, hydroxamic acids, cyclopeptide and benzamides. Vorinostat
[suberolanilide hydroxamic acid (SAHA)] was the first HDACI
approved by the Food and Drug Administration for the treatment of
cutaneous T-cell lymphoma (3).
Sodium butyrate (NaB) is a structurally similar HDACI that has also
exhibited potent antitumor activity (4).
HDACIs have been reported to exhibit synergistic
effects in combination with a variety of antitumor agents,
including the DNA alkylating agents busulfan (Bu) and melphalan
(5–7). The majority of the antitumor drugs are
more effective when used in combination, and the cytotoxicity
produced from their combined efficacy decreases the development of
chemotherapy-resistant tumor cells (5). However, the combination of antitumor
drugs may also produce antagonistic effects, which highlights the
requirement for the extensive investigation of their
interactions.
In order to investigate the molecular mechanisms of
HDACIs in combination with DNA alkylating agents, it was
hypothesized that HDACIs may affect the expression of drug
transporters, which are involved in the efflux of functionally and
structurally irrelevant antitumor drugs, including DNA alkylating
agents (8). The efflux of antitumor
drugs by ATP binding cassette (ABC) transporters serves an
important role in the development of the multidrug resistance (MDR)
phenotype, which is a main obstacle for successful cancer treatment
(8–10). Drug-resistant-related ABC transporter
genes mainly include ABC subfamily B member 1 (ABCB1), ABCCs
and ABCG2, which encode for MDR1 protein, MDR-associated
proteins (MRPs) and breast cancer resistance protein (BCRP),
respectively (8). Furthermore, a
significant overlap has been reported in the substrate specificity
of the ABC transporters. MDR1 extrudes natural toxins, antitumor
drugs and drug metabolites (11,12),
while MRPs export a variety of structurally diverse glutathione
(GSH)-conjugates or therapeutic drugs (9). It has been revealed that MRP1-3 lead to
resistance to hydrophobic and anionic compounds, including several
natural compounds, whereas MRP4, MRP5 and MRP8 efflux cyclic
nucleotides (13,14). BCRP, a ubiquitous ABC transporter,
has been shown to transport nucleoside drugs and
nucleoside-monophosphate derivatives of clinically relevant
nucleoside drugs (15,16). Moreover, these ABC transporters are
highly expressed in various human cancer types and are closely
associated with poor prognosis (17,18).
The aim of the present study was to investigate the
effects of NaB on the expression levels of MDR1, MRPs and BCRP in
lung cancer and colorectal cancer cells. Since MRP1 and MRP2 export
GSH-conjugated DNA alkylating agents (19), the decrease in their expression
levels may contribute to the synergistic antitumor effect of NaB
and DNA alkylating agents. In contrast to this hypothesis, NaB may
antagonize the anticancer efficacy of drugs that are substrates for
MDR1, MRP7 and MRP9. These differential effects of NaB on the
expression of ABC transporters require detailed investigations in
order to identify its combined antitumor action with other
chemotherapeutic agents.
Materials and methods
Chemicals and reagents
NaB, 5-Carboxyfluorescein diacetate (CFDA) and
3,3′-diethyloxacarbocyanine iodide (DiOC2) were obtained
from Sigma-Aldrich (Merck KGaA). The primary antibody against MDR1
(cat. no. 13978) and acetylate (cat. no. 9441) were purchased from
Cell Signaling Technology, Inc. The primary antibody against
α-tubulin (cat. no. sc-134237) was obtained from Santa Cruz
Biotechnology, Inc. The primary antibodies against MRP2 (cat. no.
24893-1-AP), p21 (cat. no. 10355-1-AP) and p27 (cat. no.
25614-1-AP) were from ProteinTech Group, Inc., while the primary
antibodies against MRP1 (cat. no. BS7474) and BCRP (cat. no.
BS3482) were obtained from Biogot Technology Co., Ltd. The primary
antibodies against MRP3 (cat. no. ab3375), MRP7 (cat. no. ab91451)
and MRP9 (cat. no. ab91453) were purchased from Abcam, and those
against HDAC1 (cat. no. ET1605-35), HDAC2 (cat. no. ET1607-78),
HDAC3 (cat. no. ET1610-5), HDAC4 (cat. no. ET1612-51), were from
Hangzhou HuaAn Biotechnology Co., Ltd. Acetylation at lysine 9 on
histone H3 (AcH3K9) (cat. no. 39917) and methylation at lysine 9 on
histone H3 (2MeH3K9) (cat. no. 39239) were from Active Motif.
Horseradish peroxidase (HRP)-conjugated secondary antibody (cat.
nos. SH001X, SH002X and SH003X) were also purchased from DingGuo
Biotechnology Co., Ltd. PrimeScript® RT reagent kit and
SYBR® Premix Ex Taq™ were obtained from Takara Bio,
Inc.
Cell culture
The human lung cancer cell line A549 and the
colorectal cancer cell line HCT116 were obtained from the Type
Culture Collection of the Chinese Academy of Sciences. HCT116 cells
were maintained in Dulbecco's modified Eagle media (DMEM)/F12
culture medium (Biological Industries) supplemented with 10% fetal
bovine serum (FBS), and A549 cells were cultured in RPMI 1640
culture medium (Biological Industries) supplemented with 10% FBS
under a humidified 5% CO2 atmosphere at 37°C.
Reverse transcription-quantitative
(RT-q)PCR
A549 and HCT116 cells were treated with NaB (2 mM)
at 37°C for 24 h, and total mRNA was extracted using
TRIzol® reagent (Thermo Fisher Scientific Inc.). RNA
concentration was determined using spectrophotometry and 500 ng RNA
was used for cDNA synthesis. RT-qPCR was performed using the Takara
SYBR® Premix Ex Taq™ system in order to quantify the
expression levels of the target genes in an ABI 7500 thermal cycler
(Thermo Fisher Scientific, Inc.). The thermocycling conditions for
RT-qPCR were: Initial denaturation at 95°C for 30 sec, followed by
45 cycles at 95°C for 5 sec and 60°C for 30 sec; and a melt curve
stage at 95°C for 15 sec, 60°C for 60 sec, 95°C for 30 sec and 60°C
for 15 sec. GAPDH was selected as the housekeeping gene.
After normalized to GAPDH gene, each target gene expression
were calculated using the comparative threshold cycle (Cq) method
(20). The ΔCq values were
calculated according to the formula ΔCq=Cq (gene of interest)-Cq
(GAPDH) in correlation analysis, and the 2−ΔΔCq
was calculated according to the formula ΔΔCq=ΔCq (control
group)-ΔCq (experimental group) for determination of relative. The
sequences of the primers used in the RT-qPCR experiments are
presented in Table I.
 | Table I.Primers used in reverse
transcription-quantitative PCR. |
Table I.
Primers used in reverse
transcription-quantitative PCR.
| Gene | Forward primer,
5′→3′ | Reverse primer,
5′→3′ |
|---|
| ABCB1 |
TGCTCAGACAGGATGTGAGTTG |
AATTACAGCAAGCCTGGAACC |
| ABCC1 |
GCCAAGAAGGAGGAGACC |
AGGAAGATGCTGAGGAAGG |
| ABCC2 |
TGGTGGCAACCTGAGCATAGG |
ACTCGTTTTGGATGGTCGTCTG |
| ABCC3 |
GGTTCCCCTTGGAATCATTT |
AATCCTGGTGTGCATCAAACAG |
| ABCC5 |
ACCCGTTGTTGCCATCTTAG |
GCTTTGACCCAGGCATACAT |
| ABCC6 |
GTGGTGTTTGCTGTCCACAC |
ACGACACCAGGGTCAACTTC |
| ABCC10 |
ATTGCCCATAGGCTCAACAC |
AGCAGCCAGCACCTCTGTAT |
| ABCC11 |
GGCTGAGCTACTGGTTGGAG |
TGGTGAAAATCCCTGAGGAG |
| ABCC12 |
GGTGTTCATGCTGGTGTTTGG |
GCTCGTCCATATCCTTGGAA |
| ABCG2 |
TATAGCTCAGATCATTGTCACAGTC |
GTTGGTCGTCAGGAAGAAGAG |
| GAPDH |
GCACCGTCAAGGCTGAGAAC |
TGGTGAAGACGCCAGTGGA |
Western blot analysis
A549 and HCT116 cells were treated with NaB (2 mM)
at 37°C for 24 h. Following washing with ice-cold PBS for three
times, the cells were lysed using western blotting lysis buffer.
The concentration of the total protein was determined using the BCA
reagent. In total, 20 µg proteins were separated by electrophoresis
using 8% SDS-PAGE. The proteins were electrophoretically
transferred onto PVDF membranes. Following blocking with 5% non-fat
milk for 2 h at room temperature, the PVDF membranes were incubated
with primary antibodies at 4°C overnight. The antibodies against
MDR1, MRP1, MRP2, BCRP, α-tubulin, HDAC1, HDAC2, HDAC3, HDAC4, p21,
p27, 2MeH3K9 and AcH3K9 were used at 1:1,000 dilution, while the
antibodies against MRP3, MRP7 and MRP9 were used at 1:50 dilution.
The PVDF membranes were washed three times with PBS-1% Tween-20 and
subsequently incubated with horseradish peroxidase-conjugated
secondary antibodies (1:5,000) for 1.5 h at room temperature.
Specific immune complexes were detected using a chemiluminescence
reagent (Thermo Fisher Scientific, Inc.). Band intensity was
semi-quantified via densitometry analysis using Image-Pro Plus 4.5
software (Media Cybernetics, Inc.).
MTT assay
The cell viability was measured using the MTT assay
(Sigma-Aldrich; Merck KGaA). Cells (1×104) were seeded
on 96-well plates and pretreated with NaB (2 mM) at 37°C for 24 h,
then treated with fluorouracil (5-FU) (2.5, 5, 10, 20 and 40 µg/ml)
or chlorambucil (2, 5, 10, 15 and 20 µM) at 37°C 48 h. The cells
were washed twice with PBS, and 100 µl MTT solution (0.25 mg/ml)
was added to the culture medium. Following incubation for 4 h at
37°C, 100 µl DMSO was added to each well to dissolve the dark blue
crystals. The absorbance was measured at 570 nm using a microplate
reader.
Functional assays for MRP1 and
MDR1
CFDA and DiOC2 were used as fluorescent
substrates to assay the MRP1 and MDR1 transport activity via
fluorescence spectrophotometry. A549 cells were treated with NaB (2
mM) and/or DMSO for 24 h at 37°C, harvested and resuspended in
fresh medium with 1 µM CFDA and/or 0.5 µg/ml DiOC2 at
4°C for 20 min. The cells were centrifuged at 200 × g at 4°C for 1
min, washed with PBS 3 times, resuspended in fresh medium and
incubated at 37°C for 40 min. The mean fluorescence intensities for
CFDA and DiOC2 were determined via fluorescence
spectrophotometer (UV3000; Shanghai Mapada Instruments Co., Ltd.)
using an excitation and emission wavelength of 488 and 525 nm,
respectively.
Immunoprecipitation
A549 cells were treated with NaB (2 mM) or DMSO
(equal volume) at 37°C for 24 h, and washed three times with
ice-cold PBS. The cells were harvested at 4°C in
immunoprecipitation lysis buffer (Beyotime Institute of
Biotechnology) and 20 µg protein was immunoprecipitated using an
anti-MDR1 antibody (1:50) at 4°C overnight. The immune complexes
were bound to protein A/G Sepharose (Beyotime Institute of
Biotechnology) and the beads were washed with lysis buffer. Then,
the protein was subjected to western blotting as aforementioned
with an anti-acetylate antibody (1:1,000).
Statistical analysis
Data are presented as the mean ± SD of three
independent experiments, and were analyzed using a two-tailed
unpaired Student's t-test. The analyses were performed using
GraphPad Prism software version 5.0 (GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effect of NaB on cancer cell
viability
The effect of NaB on the viability of lung cancer
and colorectal cells was investigated. A549 and HCT116 cells were
treated with different concentrations of NaB (0–16 mM) for 24 or 48
h. The viability of cancer cells was measured using the MTT assay,
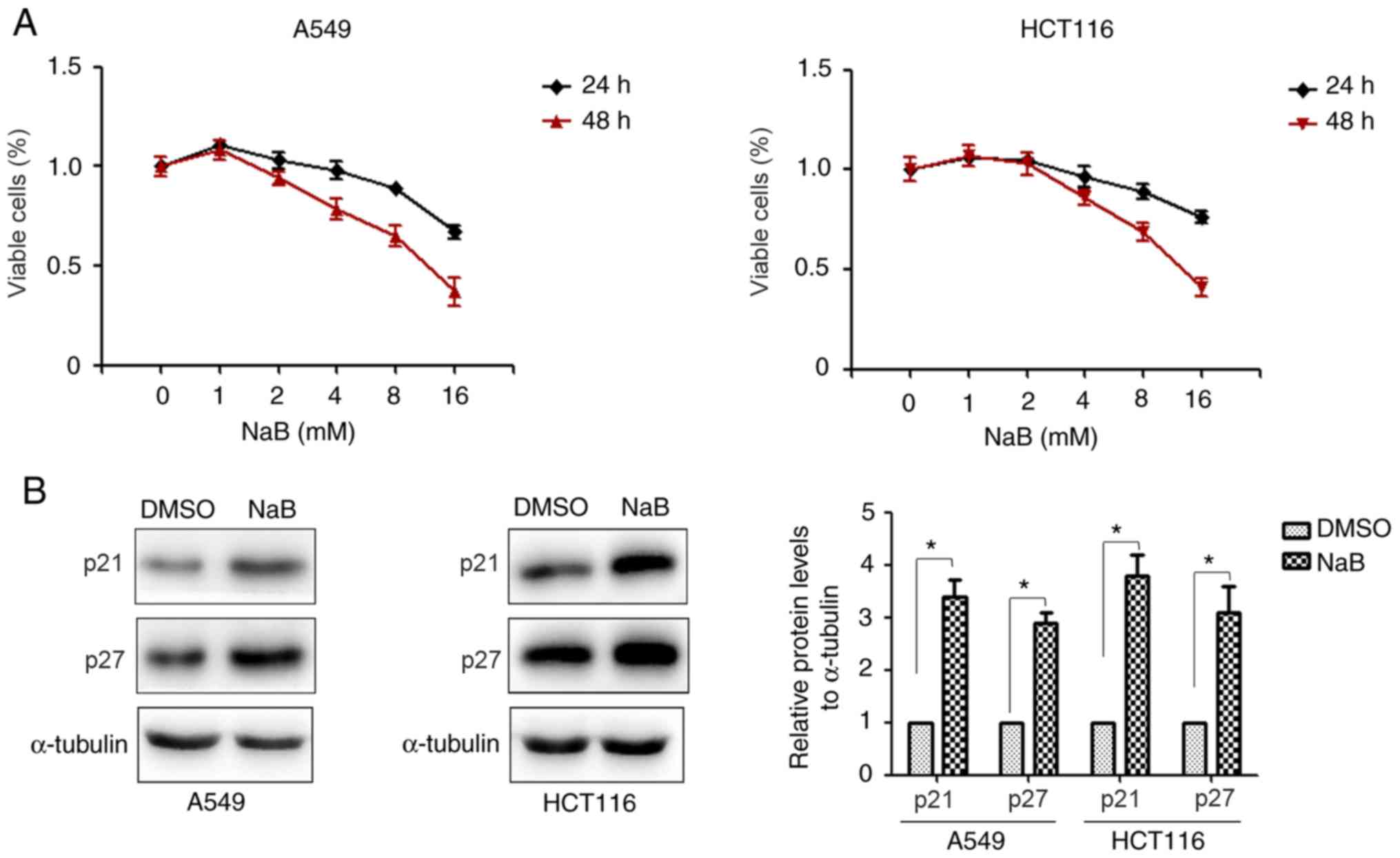
and it was identified that NaB markedly suppressed proliferation of
A549 and HCT116 cells within a 24-h treatment period (Fig. 1A). Furthermore, the effects of NaB on
the expression of the proliferative markers were evaluated. The
results demonstrated that NaB significantly increased the
expression levels of p21 and p27 proteins compared with the DMSO
treatment group (Fig. 1B). These
results suggested that NaB could inhibit the viability of lung
cancer and colorectal cancer cells.
Effect of NaB on the expression of ABC
transporters
Subsequently, the effect of NaB on the expression
levels of the ABC transporters was examined. A549 and HCT116 cells
were treated with 2 mM NaB for 24 h, and the gene and protein
expression levels of the ABC transporters were detected via RT-qPCR
and western blotting, respectively. The results identified that NaB
increased the mRNA expression levels of ABCB1, ABCC10 and
ABCC12, and the protein expression levels of MDR1, MRP7 and
MRP9. However, NaB decreased the expression levels of ABCC1,
ABCC2 and ABCC3 mRNAs, as well as MRP1, MRP2 and MRP3
proteins. In addition, NaB did not alter the expression of
ABCG2 mRNA and BCRP protein in the two cell lines (Fig. 2A and B).
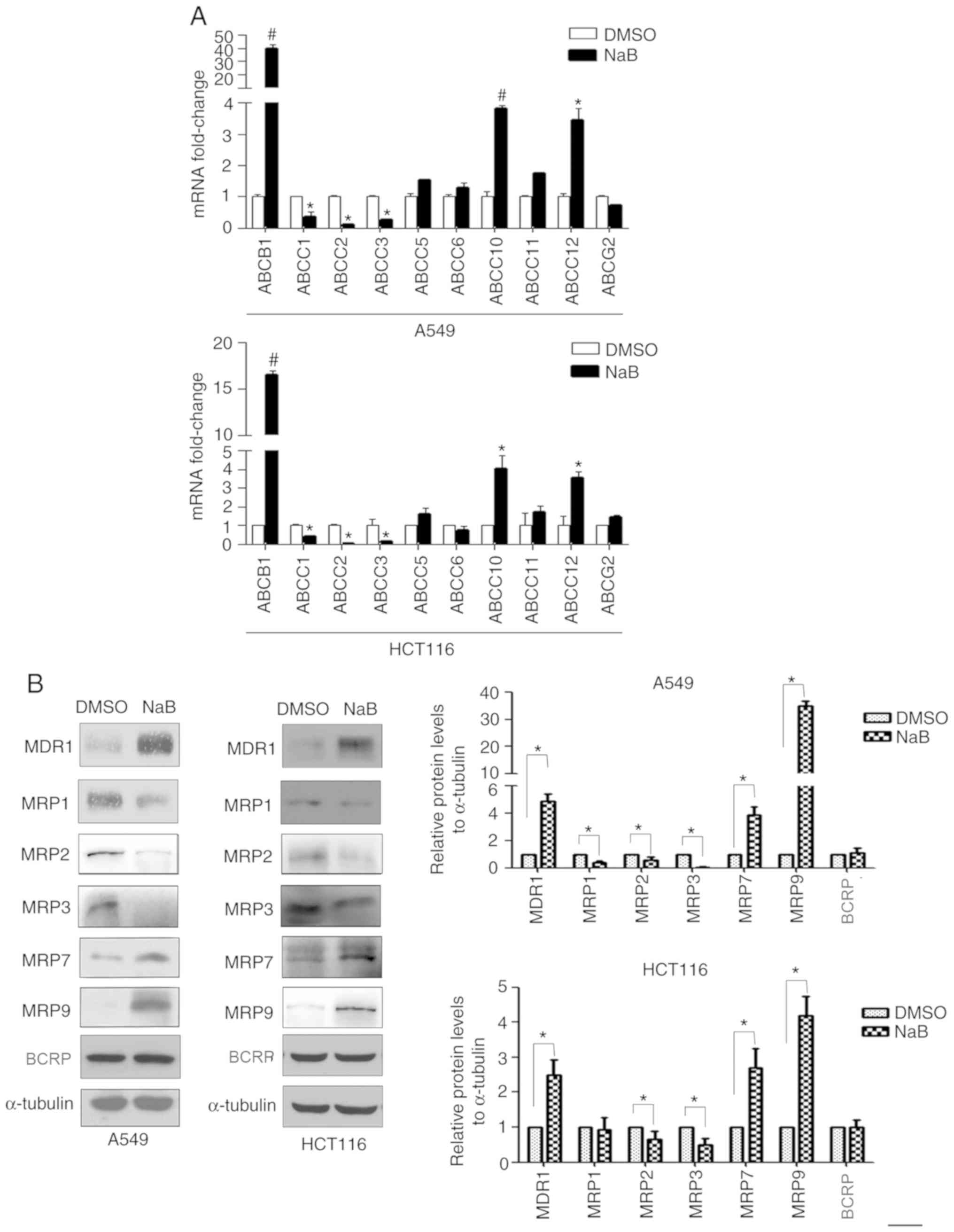 | Figure 2.Effect of NaB on the expression of
ABC transporters. (A) A549 and HCT116 cells were treated with NaB
(2 mM) for 24 h. mRNA expression levels of ABCB1, ABCC1, −2, −3,
−5, −6, −10, −11, −12 and ABCG2 were detected using
reverse transcription-quantitative PCR. (B) A549 and HCT116 cells
were treated with NaB (2 mM) for 24 h, and the protein expression
levels of the MDR1, MRP1, −2, −3, −7, −9 and BCRP were detected via
western blotting. #P<0.01, *P<0.05 compared with
DMSO group. ABC, ATP-binding cassette; NaB, sodium butyrate;
BCRP, breast cancer resistance protein; MDR1, multidrug
resistance-1; MRP, multidrug resistance-associated protein. |
Effect of NaB on the drug sensitivity
of A549 and HCT116 cells
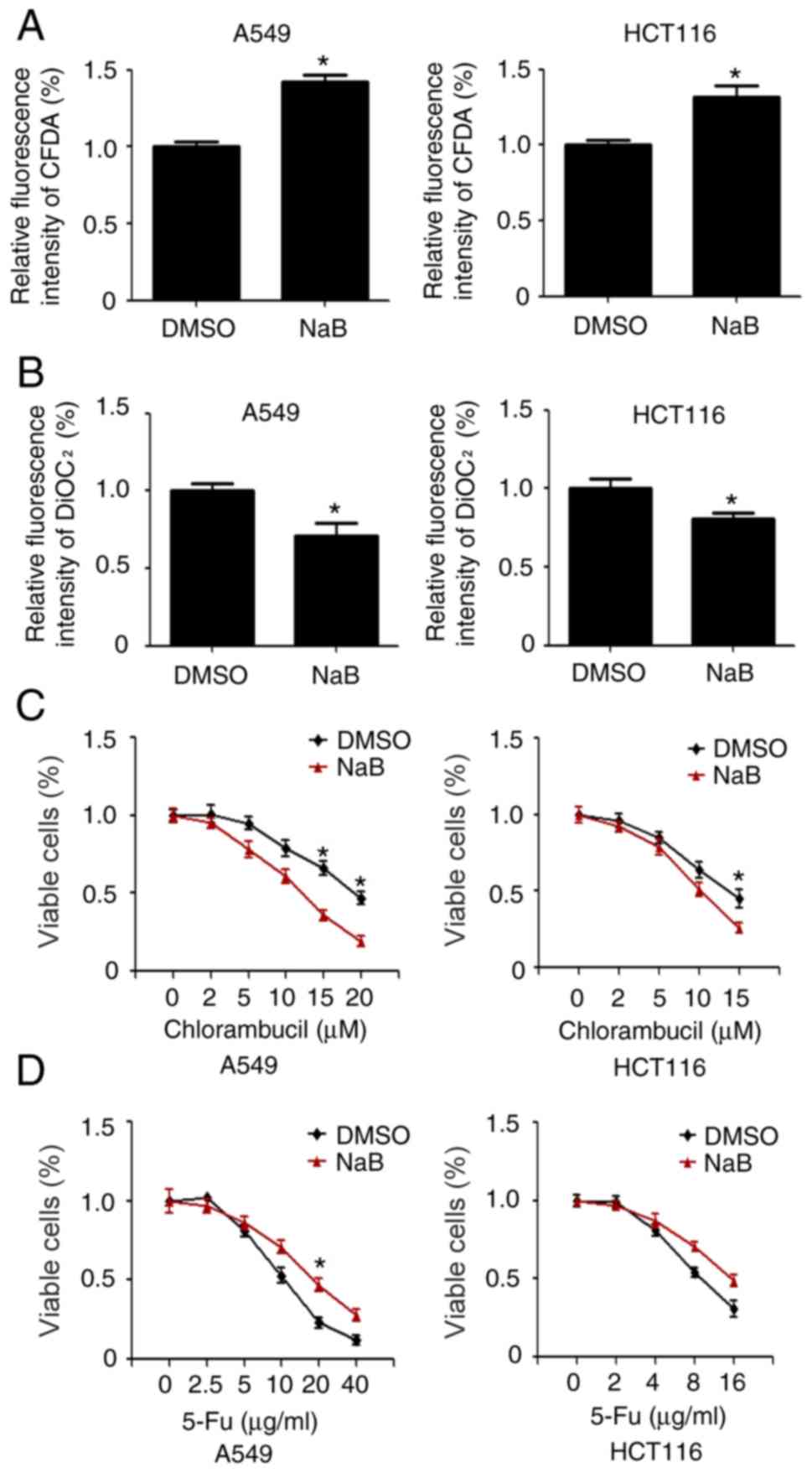
It was further investigated whether cells exposed to
NaB could retain their ability to export specific drugs. CFDA and
DiOC2 are substrates for MRP1 and MDR1, respectively.
A549 and HCT116 cells were stimulated with 2 mM NaB for 24 h. It
was identified that NaB decreased the efflux of CFDA from the cells
(Fig. 3A), while NaB increased
DiOC2 efflux (Fig. 3B).
Moreover, it was evaluated whether NaB treatment could enhance the
drug sensitivity of the cells to chlorambucil, a GSH-conjugated
alkylator exported by MRP1 (21).
A549 and HCT116 cells were pretreated with DMSO and/or NaB for 24 h
and then exposed to increasing concentrations of chlorambucil. The
results demonstrated that NaB significantly promoted the
sensitivity of the cells to chlorambucil (Fig. 3C). Inversely, NaB increased drug
resistance of A549 and HCT116 cells to 5-FU (Fig. 3D).
NaB induces acetylation of MDR1
HDACIs regulate gene transcription mainly by
inhibiting the expression of HDAC enzymes, which serve crucial
roles in tumor progression (22).
The present study detected the effect of NaB on the expression
levels of HDAC1, HDAC2, HDAC3 and HDAC4. The results indicated that
NaB downregulated the expression of HDAC4, but not HDAC1, HDAC2 and
HDAC3 (Fig. 4A). NaB also promoted
AcH3K9 and 2MeH3K9 (Fig. 4A).
Furthermore, NaB significantly increased the acetylation of MDR1
(Fig. 4B).
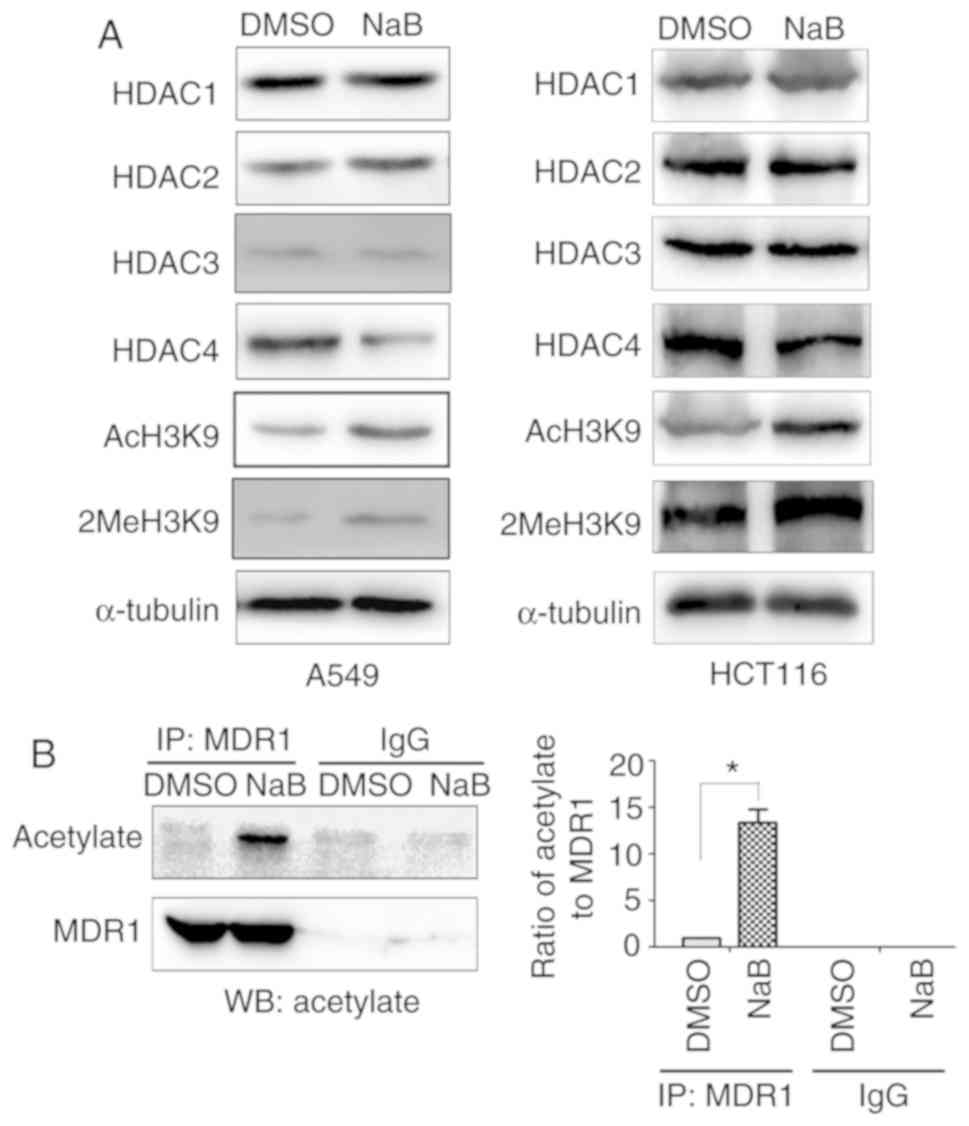 | Figure 4.NaB induces acetylation of MDR1. (A)
A549 and HCT116 cells were treated with NaB (2 mM) or DMSO for 24
h, and the protein expression levels of HDAC1, HDAC2, HDAC3, HDAC4,
AcH3K9 and 2MeH3K9 were detected via WB. (B) A549 cells were
treated with NaB (2 mM) or DMSO for 24 h and MDR1 protein was
immunoprecipitated. The protein expression of MDR1 and the levels
of acetylation were detected using WB. *P<0.05 compared with
DMSO group. NaB, sodium butyrate; HDAC, Histone deacetylase;
AcH3K9, acetylation at lysine 9 on histone H3; 2MeH3K9, methylation
at lysine 9 on histone H3; MDR1, multidrug resistance-1; WB,
western blotting; IP, Immunoprecipitation. |
Discussion
Previous studies have reported the synergistic
cytotoxicity of HDACIs and DNA alkylating agents in a variety of
experimental models, for example HDAC inhibitor panobinostat and
DNA alkylators busulfan showed synergistic cytotoxicity in lymphoma
cells (5–7). In the present study, it was
demonstrated that NaB could downregulate the expression levels of
ABCC1, ABCC2 and ABCC3 mRNAs, and MRP1, MRP2 and MRP3
proteins in lung cancer and colorectal cancer cells. Since MRP1,
MRP2 and MRP3 have been revealed to export GSH-conjugated DNA
alkylating agents from the cytoplasmic regions of cancer cells, the
downregulation of their expression gives rise to cellular
accumulation of these agents, which leads to enhanced cytotoxicity
(9,19). The present findings are consistent
with the inverse association between the sensitivity of DNA
alkylating agents and high expression of MRP1 (23). Cancer cells with high expression of
MRP1 are more resistant to DNA alkylating agents, such as Bu and
chlorambucil (23). A previous study
revealed that SAHA and belinostat downregulated the expression of
MRP1 in T-cell lymphoma and T-cell prolymphocytic leukemia
(21). However, another study
observed that the HDACI FK228 could upregulate MRP1 expression
(14). It has been shown that MRP1
is not induced in all the examined cell lines that are treated with
FK228, suggesting that FK228-induced MRP1 expression is cell
line-dependent (24). Kim et
al (25) also revealed that SAHA
could overcome MDR export of anticancer drugs via the
downregulation of MRP2 in MDR cancer cell lines. The downregulation
of MRP2 that is caused by the combined treatment of paclitaxel and
SAHA leads to an increase in G2/M arrest and apoptosis
(25). In contrast to SAHA, valproic
acid results in upregulation of MRP2 expression (26). Therefore, it was speculated that the
difference in MRP expression that was induced by HDACIs could be
due to the different cancer types and the diverse structures of the
HDACIs used.
Chemotherapy is one of the effective methods for the
treatment of malignant tumors. However, the effectiveness of
chemotherapy is limited by the acquirement of the MDR phenotype
mediated by MDR1, which exports antitumor drugs out of cancer cells
and reduces their intracellular accumulation (8,27).
Although HDACIs are emerging as a novel class of chemotherapeutic
agents, the development of the MDR phenotype is of notable concern
for the efficacy of several antitumor drugs, including HDACIs
(4). In the present study, it was
found that NaB induced 5-FU resistance and increased the expression
of MDR1. Previous studies have reported that HDACIs could promote
MDR1 expression in several cancer types. For example, treatment
with the HDACI apicidine induces paclitaxel resistance and
Rhodamine-123 efflux in HeLa cells (28). Colon and renal cancer cells that are
treated with Trichostatin A (TSA) or depsipeptide also display
increased MDR1 expression (29,30).
Moreover, the same effect was observed for leukocytes isolated from
patients (29,30). MDR1 induction has also been reported
in human and murine cells treated with valproate (31). A recent study demonstrated that SAHA
and TSA induced MDR1 expression via transcriptional activation of
STAT3 and the stabilization of MDR1 mRNA in colorectal cancer
(32). The present study
investigated the molecular mechanism of NaB-induced MDR1
expression. Several HDAC family members are aberrantly expressed in
various types of cancer, and therefore, HDACIs are considered a
promising novel class of anticancer drug targets (22). Previous studies have reported that
some agents can inhibit the expression and function of MDR1, MRP7
and MRP9 (33,34). Lapatinib and erlotinib are potent
reversal agents for MRP7 (34).
Therefore, it was suggested that a drug combination of HDACIs with
these inhibitors may solve the drug resistance mediated by MDR1,
MRP7 and MRP9. Thus, it is worth further evaluating the details of
drug combination in future studies.
Histone acetylation and methylation serve an
important role in gene expression (21). The present study demonstrated that
NaB could inhibit the expression of HDAC4, promote AcH3K9 and
acetylation of MDR1, which may result in the upregulation of its
expression. On the other hand, NaB increased 2MeH3K9, which may
result in the decreased protein expression levels of MRP1, MRP2 and
MRP3. Therefore, it was speculated that different regulation of the
expression levels of the ABC family may due to histone acetylation
and methylation induced by NaB. Although the current study did not
address the reasons for dysregulated expression levels of the
target genes, the present results provide preliminary data for
researchers in this field. In a future study, the molecular
mechanisms underlying the different effects on MRP and HDAC protein
families induced by NaB will be investigated.
In conclusion, the present study demonstrated the
crucial implications for combining NaB with other antitumor drugs.
NaB treatment had different effects on the expression of ABC
transporters, which have various substrates. For example, MRP1 and
MRP2 export GSH-conjugated DNA alkylating agent, and MDR1 exports
anthracyclines (11–14). The combination of NaB with DNA
alkylating agents and/or other MRP1-3 substrates may lead to
synergistic and more efficacious treatments. For instance,
chlorambucil exerts its antitumor activity in variety of cancer
cells, including lymphocytic leukemia, ovarian cancer, colorectal
cancer and lung cancer (35,36). In the current study, the combination
of NaB and chlorambucil exerted a synergistic effect in colorectal
cancer and lung cancer. However, NaB could increase MDR1
expression, which may lead to drug resistance towards MDR1
substrates, such as steroids, vinca alkaloids and anthracyclines.
The present study highlighted the importance of understanding the
mechanism of drug interactions in order to achieve a more
efficacious cytotoxic effect in cancer cells.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Natural Science
Foundation of Anhui Province (grant no. 1608085QH217).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
BS contributed to the study design, data acquisition
and analysis and drafted the manuscript. PF participated in the
study design, data acquisition and revision of the manuscript. FFX,
CPX, RJ, CHY, SQM, NW and AJW performed the statistical analysis.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jiang GM, Wang HS, Zhang F, Zhang KS, Liu
ZC, Fang R, Wang H, Cai SH and Du J: Histone deacetylase inhibitor
induction of epithelial-mesenchymal transitions via up-regulation
of Snail facilitates cancer progression. Biochim Biophys Acta.
1833:663–671. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu W, Liu H, Liu ZG, Wang HS, Zhang F,
Wang H, Zhang J, Chen JJ, Huang HJ, Tan Y, et al: Histone
deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation
and stabilization of Snail to promote metastasis of hepatoma cells.
Cancer Lett. 420:1–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Di Costanzo A, Del Gaudio N, Conte L,
Dell'Aversana C, Vermeulen M, de The H, Migliaccio A, Nebbioso A
and Altucci L: The HDAC inhibitor SAHA regulates CBX2 stability via
a SUMO-triggered ubiquitin-mediated pathway in leukemia. Oncogene.
37:2559–2572. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ni X, Li L and Pan G: HDAC
inhibitor-induced drug resistance involving ATP-binding cassette
transporters (Review). Oncol Lett. 9:515–521. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valdez BC, Nieto Y, Murray D, Li Y, Wang
G, Champlin RE and Andersson BS: Epigenetic modifiers enhance the
synergistic cytotoxicity of combined nucleoside analog-DNA
alkylating agents in lymphoma cell lines. Exp Hematol. 40:800–810.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nieto Y, Valdez BC, Thall PF, Ahmed S,
Jones RB, Hosing C, Popat U, Shpall EJ, Qazilbash M, Gulbis A, et
al: Vorinostat combined with high-dose gemcitabine, busulfan, and
melphalan with autologous stem cell transplantation in patients
with refractory lymphomas. Biol Blood Marrow Transplant.
21:1914–1920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Teo EC, Valdez BC, Ji J, Li Y, Liu Y,
Brammer JE, Hosing C, Nieto Y, Champlin RE and Andersson BS:
Synergistic cytotoxicity of busulfan, melphalan, gemcitabine,
panobinostat, and bortezomib in lymphoma cells. Leuk Lymphoma.
57:2644–2652. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fukuda Y and Schuetz JD: ABC transporters
and their role in nucleoside and nucleotide drug resistance.
Biochem Pharmacol. 83:1073–1083. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen ZS and Tiwari AK: Multidrug
resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic
diseases. FEBS J. 278:3226–3245. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu YR, Liang L, Zhao JM, Zhang Y, Zhang
M, Zhong WL, Zhang Q, Wei JJ, Li M, Yuan J, et al: Twist1 confers
multidrug resistance in colon cancer through upregulation of
ATP-binding cassette transporters. Oncotarget. 8:52901–52912. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maia RC, Vasconcelos FC, Souza PS and
Rumjanek VM: Towards comprehension of the ABCB1/P-glycoprotein role
in chronic myeloid leukemia. Molecules. 23:1192018. View Article : Google Scholar
|
|
12
|
Sharom FJ: The P-glycoprotein multidrug
transporter. Essays Biochem. 50:161–178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Borst P, Evers R, Kool M and Wijnholds J:
A family of drug transporters: The multidrug resistance-associated
proteins. J Natl Cancer Inst. 92:1295–1302. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dean M, Hamon Y and Chimini G: The human
ATP-binding cassette (ABC) transporter superfamily. J Lipid Res.
42:1007–1017. 2001.PubMed/NCBI
|
|
15
|
Wang J, Yunyun Z, Wang L, Chen X and Zhu
Z: ABCG2 confers promotion in gastric cancer through modulating
downstream CRKL in vitro combining with biostatistics mining.
Oncotarget. 8:5256–5267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaehler M, Ruemenapp J, Gonnermann D,
Nagel I, Bruhn O, Haenisch S, Ammerpohl O, Wesch D, Cascorbi I and
Bruckmueller H: MicroRNA-212/ABCG2-axis contributes to development
of Imatinib-resistance in leukemic cells. Oncotarget.
8:92018–92031. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ling V: Multidrug resistance: Molecular
mechanisms and clinical relevance. Cancer Chemother Pharmacol. 40
(Suppl):S3–S8. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cole SP and Deeley RG: Transport of
glutathione and glutathione conjugates by MRP1. Trends Pharmacol
Sci. 27:438–446. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Valdez BC, Li Y, Murray D, Brammer JE, Liu
Y, Hosing C, Nieto Y, Champlin RE and Andersson BS: Differential
effects of histone deacetylase inhibitors on cellular drug
transporters and their implications for using epigenetic modifiers
in combination chemotherapy. Oncotarget. 7:63829–63838. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Witt O, Deubzer HE, Milde T and Oehme I:
HDAC family: What are the cancer relevant targets? Cancer Lett.
277:8–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paumi CM, Ledford BG, Smitherman PK,
Townsend AJ and Morrow CS: Role of multidrug resistance protein 1
(MRP1) and glutathione S-transferase A1-1 in alkylating agent
resistance. Kinetics of glutathione conjugate formation and efflux
govern differential cellular sensitivity to chlorambucil versus
melphalan toxicity. J Biol Chem. 276:7952–7956. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiao JJ, Huang Y, Dai Z, Sadee W, Chen J,
Liu S, Marcucci G, Byrd J, Covey JM, Wright J, et al:
Chemoresistance to depsipeptide FK228 [(E)-(1S,4S,10S,21R)-7-[(Z)-
ethylidene]- 4,21-diisopropyl- 2-oxa-12,13-dithia-5,8,20,23-
tetraazabicyclo[8,7,6]-tricos-16-ene-3,6,9,22-pentanone] is
mediated by reversible MDR1 induction in human cancer cell lines. J
Pharmacol Exp Ther. 314:467–475. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim H, Kim SN, Park YS, Kim NH, Han JW,
Lee HY and Kim YK: HDAC inhibitors downregulate MRP2 expression in
multidrug resistant cancer cells: Implication for
chemosensitization. Int J Oncol. 38:807–812. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fedier A, Dedes KJ, Imesch P, Von Bueren
AO and Fink D: The histone deacetylase inhibitors suberoylanilide
hydroxamic (Vorinostat) and valproic acid induce irreversible and
MDR1-independent resistance in human colon cancer cells. Int J
Oncol. 31:633–641. 2007.PubMed/NCBI
|
|
27
|
Tabe Y, Konopleva M, Contractor R, Munsell
M, Schober WD, Jin L, Tsutsumi-Ishii Y, Nagaoka I, Igari J and
Andreeff M: Up-regulation of MDR1 and induction of doxorubicin
resistance by histone deacetylase inhibitor depsipeptide (FK228)
and ATRA in acute promyelocytic leukemia cells. Blood.
107:1546–1554. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim YK, Kim NH, Hwang JW, Song YJ, Park
YS, Seo DW, Lee HY, Choi WS, Han JW and Kim SN: Histone deacetylase
inhibitor apicidin-mediated drug resistance: Involvement of
P-glycoprotein. Biochem Biophys Res Commun. 368:959–964. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin S and Scotto KW: Transcriptional
regulation of the MDR1 gene by histone acetyltransferase and
deacetylase is mediated by NF-Y. Mol Cell Biol. 18:4377–4384. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Robey RW, Zhan Z, Piekarz RL, Kayastha GL,
Fojo T and Bates SE: Increased MDR1 expression in normal and
malignant peripheral blood mononuclear cells obtained from patients
receiving depsipeptide (FR901228, FK228, NSC630176). Clin Cancer
Res. 12:1547–1555. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eyal S, Lamb JG, Smith-Yockman M, Yagen B,
Fibach E, Altschuler Y, White HS and Bialer M: The antiepileptic
and anticancer agent, valproic acid, induces P-glycoprotein in
human tumour cell lines and in rat liver. Br J Pharmacol.
149:250–260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Huang C, Zhao L, Zhang H, Yang JM,
Luo P, Zhan BX, Pan Q, Li J and Wang BL: Histone deacetylase
inhibitors regulate P-gp expression in colorectal cancer via
transcriptional activation and mRNA stabilization. Oncotarget.
7:49848–49858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karthikeyan S and Hoti SL: Development of
fourth generation ABC inhibitors from natural products: A novel
approach to overcome cancer multidrug resistance. Anticancer Agents
Med Chem. 15:605–615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuang YH, Shen T, Chen X, Sodani K,
Hopper-Borge E, Tiwari AK, Fu LW and Chen ZS: Lapatinib and
erlotinib are potent reversal agents for MRP7 (ABCC10)-mediated
multidrug resistance. Biochem Pharmacol. 79:154–161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kelner MJ, McMorris TC, Rojas RJ, Estes LA
and Suthipinijtham P: Synergy of irofulven in combination with
other DNA damaging agents: Synergistic interaction with
altretamine, alkylating, and platinum-derived agents in the MV522
lung tumor model. Cancer Chemother Pharmacol. 63:19–26. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tacconi EM, Badie S, De Gregoriis G,
Reisländer T, Lai X, Porru M, Folio C, Moore J, Kopp A, Baguña
Torres J, et al: Chlorambucil targets BRCA1/2-deficient tumours and
counteracts PARP inhibitor resistance. EMBO Mol Med. 11:e99822019.
View Article : Google Scholar : PubMed/NCBI
|