Introduction
Oral cancer is one of the most common head and neck
malignancies, while oral squamous cell carcinoma (OSCC) is the most
common (1). It has been reported
that oral cancer accounts for 1.9–3.5% of systemic malignancies. In
addition, it accounts for 4.7–20.3% of head and neck malignancies,
second only to nasopharyngeal carcinoma (2). Today, surgical resection and
radiotherapy are still the two most effective methods for the
treatment of OSCC (3). In addition,
combined treatment with the poly(ADP) ribose polymerase inhibitors
1/2 (PARP1/2) is also a valid therapeutic strategy for OSCC
(4). The overall 5-year survival
rate of OSCC patients is 50–70%. The 5-year survival rate at the
stage I can be as high as 90% or higher, while the 5-year survival
rate at stage IV is only ~10%. The main reason for the failure of
OSCC treatment is local recurrence of the primary tumor (5). Therefore, early diagnosis and treatment
are the keys to the treatment of OSCC.
Long non-coding RNA (lncRNA) is a type of RNA
molecule whose transcript length exceeds 200 nt. lncRNA does not
encode protein, but regulates the expression level of genes in the
form of RNA at multiple levels (epigenetic regulation,
transcription regulation, and post-transcriptional regulation)
(6). In addition, lncRNAs regulate
the tumorigenesis and development of cancers by affecting some
important cellular biological activities, such as cell viability,
differentiation, motility and apoptosis (7). Previous studies have shown that lncRNAs
can serve as tumor suppressors or oncogenes in OSCC, such as lncRNA
MORT and CASC9 (8,9). More specifically, overexpression of
lncRNA MORT restrained cell proliferation in OSCC by downregulating
ROCK1 (8). In addition, the
increased expression of lncRNA CASC9 promoted tumor progression
through the AKT/mTOR pathway in OSCC (9). Recently, the different roles of lncRNA
SNHG5 in malignant tumors have attracted our attention. Low
expression of SNHG5 was found in gastric cancer, and overexpression
of SNHG5 suppressed the progression of gastric cancer (10). However, SNHG5 was upregulated in
glioma, and knockdown of SNHG5 inhibited the malignant cellular
phenotype of glioma (11). These
results prove that SNHG5 may have tissue specificity. However, the
regulatory mechanism of SNHG5 in OSCC is still unknown.
In addition, SNHG5 is predicted to have a binding
site with miR-655-3p. Meanwhile, the abnormal expression and
function of miR-655-3p have been investigated in other cancers.
Downregulation of miR-655-3p has been detected in retinoblastoma
and esophageal squamous cell carcinoma (12,13).
Functionally, miR-655-3p was found to inhibit cell proliferation in
human lip cells (14). In addition,
miR-655-3p restrained the migration and invasion of non-small cell
lung cancer cells by targeting PTTG1 (15). Here, Frizzled-4 (FZD4) is predicted
as the target of miR-655-3p. In addition, abnormal expression of
FZD4 has been identified in human cancers. For example, FZD4 was
abnormally upregulated and acted as an oncogene in lung cancer and
prostate cancer (16,17). In addition, miR-101 has been found to
suppress the migration and invasion of bladder cancer cells by
targeting FZD4 (18). However, the
functional mechanism of miR-655-3p/FZD4 axis in OSCC is unclear.
Besides, lncRNA HOXD-AS1 was found to promote cell proliferation,
migration and invasion in ovarian cancer through the miR-608/FZD4
axis (19). However, it is unclear
how lncRNA SNHG5 regulates OSCC progression by interacting with the
miR-655-3p/FZD4 axis.
Therefore, the functions of lncRNA SNHG5, miR-655-3p
and FZD4 as well as their possible mechanisms were investigated in
OSCC. This research will help us better understand the pathogenesis
of OSCC and provide possible biomarkers for OSCC treatment.
Materials and methods
Clinical tissues
Forty-two OSCC tissues and paired normal tissues
were obtained from Peking Union Medical College Hospital. The
informed consents of OSCC patients were collected before the
experiment. All OSCC patients have not received chemotherapy or
radiotherapy before undergoing surgery. This study was approved by
the Institutional Ethics Committee of Peking Union Medical College
Hospital, approval number was 2017PUMC26. All patients provided
written informed consents.
Cell lines and culture
OSCC cell line SCC-4 (ZKCC-X1937) and Immortalized
Human Oral Mucosal Epithelial Cells hTERT-OME (ATCC®
PCS-200-014™) were obtained from Beijing Zhongke Quality Inspection
Biotechnology Co., Ltd. and American Type Culture Collection
(ATCC). These cells were cultured in Dulbecco's modified Eagle's
medium (DMEM) containing 10% FBS and cultured in an incubator with
5% CO2 at 37°C.
Cell transfection
SNHG5 and FZD4 overexpression vectors or siRNAs, and
miR-655-3p mimics (B01001) or inhibitor (B03001) were obtained from
GenePharma. For transfection, cells were seeded into 6-well plates
at a density of 2×105 cells/well. Cells were transfected
with miR-655-3p mimics or inhibitor (100 pmol), SNHG5 and FZD4
siRNA (100 pmol) or vectors (4 µg) using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) when 60–70%
confluence was achieved, in accordance with the manufacturer's
protocol. Subsequent to transfection for 6–8 h, cell culture medium
was replaced with DMEM without antibiotics and incubated at 37°C
with 5% CO2.
RNA isolation, reverse transcription
and RT-qPCR
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA. RNA was reversely
transcribed to complementary DNA (cDNA) using a Reverse
Transcription Kit (Takara). RT-qPCR assay was performed using
Real-time PCR Mixture assays (Takara) and corresponding primers.
SNHG5 and miR-655-3p expression were normalized to U6, when FZD4
was normalized to GAPDH. Their expressions were quantified by the
2−∆∆cq method. The primers used were: SNHG5 forward
5′-CGAGTAGCCAGTGAAGATAATG-3′; SNHG5 reverse
5′-CACACAACAGTCAAGTAAACC-3′; miR-655-3p forward:
5′-CAATCCTTACTCCAGCCAC-3′ and reverse, 5′-GTGTCTTAAGGCTAGGCCTA-3′;
U6-forward: 5AAAT-3′ and reverse, 5′-TTGCGTGTCAT-3′; FZD4-forward:
5′-GGTGGCTCCCCTCTTTACTT-3′ and reverse, 5′-ATCACACACGTTGCAGAAC-3′;
GAPDH forward: 5′-ACAACTTTGGTATCGTGGAAGG-3′, and reverse,
5′-GCCATCACGCCACAGTTTC-3′.
MTT assay
Prepared transfected SCC-4 cells (2×103
cells/well) were put in a 96-well plate. Next, the SCC-4 cells were
incubated in DMEM medium for 24, 48, 72 or 96 h. Then, the cells
were incubated with 10 µl MTT solution for 4 h. MTT solution was
then aspirated. And Formazan solution was added to completely
dissolve the crystals. The absorbance at 490 nm was examined with a
microplate reader (Olympus Corp.).
Transwell assay
Matrigel matrix (1:8; 50 µl/well; BD Biosciences) in
the upper chamber was used to detect cell invasion. After 30 min,
the SCC-4 cell suspension (2×103 cells/well) was added
to the Transwell upper chamber. The lower chamber was added with
DMEM medium (10% FBS). After 48 h of incubation, crystal violet
(Beyotime Institute of Biotechnology) was used to fix and stain the
cells on the lower surface of the membrane for 30 min. Cell
migration assay was performed without Matrigel. The other steps are
the same as cell invasion. An optical microscope was used to count
the number of stained cells for five randomly selected fields.
Dual luciferase reporter assay
The 3′-UTR of wild-type and mutant SNHG5 (wt-SNHG5
and mut-SNHG5) or FZD4 (wt-FZD4 and mut-FZD4) were inserted into
the pmiR-GLO vectors (Promega Corporation). The above reporter
plasmids and miR-655-3p mimics were transfected into SCC-4 cells.
After 48 h, the activity of firefly and Renilla luciferase was
measured by the dual-luciferase reporter gene system (Promega
Corporation).
Western blot analysis
Protein samples were extracted by RIPA lysis buffer
(Beyotime Institute of Biotechnology). Protein concentration was
measured using Bicinchoninic Acid protein assay kit (Sigma-Aldrich;
Merck KGaA). Next, 25 µg protein was separated by 10% SDS-PAGE and
transferred to PVDF membranes. And the protein was blocked at room
temperature for 2 h with 5% non-fat milk. Then, protein samples
were incubated with FZD4 (ab83042, 1:1,000 dilution, Abcam, rabbit
polyclonal antibodies) and GAPDH (ab8245, 1:1,000 dilution, Abcam,
mouse monoclonal antibody) primary antibodies overnight at 4°C.
After washing with TBST, protein samples were incubated with
corresponding horseradish peroxidase-conjugated secondary
antibodies (ab205719, 1:5,000 dilution; Abcam). Protein bands were
visualized by ECL kit (Beyotime Institute of Biotechnology) and
were quantified with Image Lab Software (Bio-Rad Laboratories,
Inc.).
Statistical analysis
All statistical analyses were conducted by using
SPSS 19.0 or Graphpad Prism 6. All data were presented as the mean
± standard deviation from at least three separate experiments.
Differences between two groups were estimated by unpaired
two-tailed Student t-test. Differences between multiple groups were
compared using one-way analysis of variance followed by Tukey's
post hoc test. The correlations between miR-655-3p expression and
lncRNA SNHG5 or FZD4 expression in OSCC tissues were analyzed using
Spearman's rank test. P<0.05 indicates a statistically
significant difference.
Results
Upregulation of lncRNA SNHG5 promotes
cell proliferation, migration and invasion in OSCC
The role of lncRNA SNHG5 was explored in OSCC.
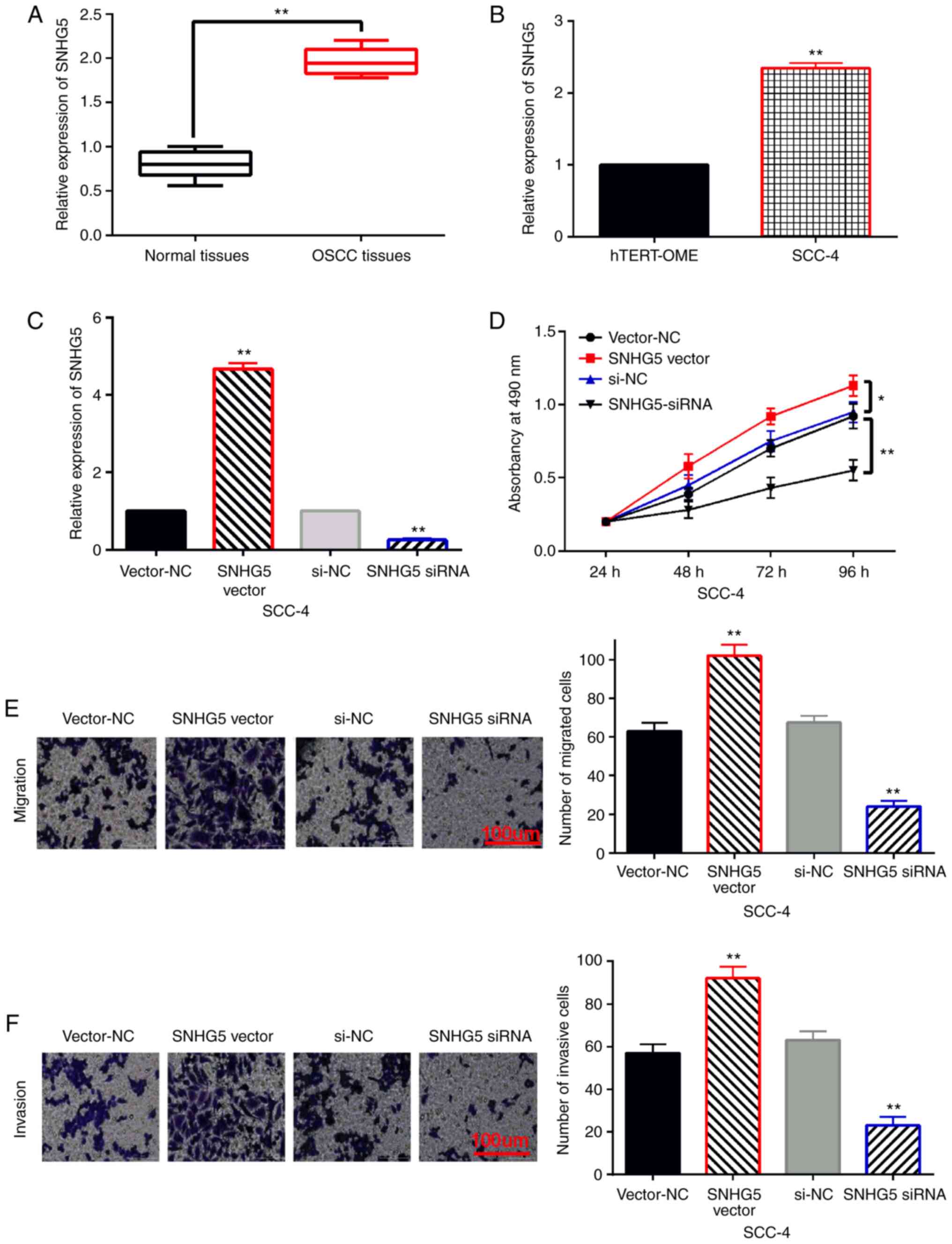
First, SNHG5 expression was detected in OSCC tissues and cells.
Compared to normal tissues, the expression of SNHG5 in OSCC tissues
was increased (Fig. 1A). In
addition, higher expression of SNHG5 was also found in SCC-4 cells
than that in the hTERT-OME cells (Fig.
1B). Next, SNHG5 siRNA or overexpression vector was transfected
into SCC-4 cells. We found that the expression of SNHG5 was reduced
by its siRNA and was enhanced by its overexpression vector in SCC-4
cells (Fig. 1C). In addition,
upregulation of SNHG5 promoted cell proliferation, while knockdown
of SNHG5 restrained cell proliferation in SCC-4 cells (Fig. 1D). Meanwhile, SCC-4 cell migration
was promoted by SNHG5 overexpression and was inhibited by SNHG5
downregulation (Fig. 1E). The same
effect of SNHG5 on cell invasion was also examined in SCC-4 cells
(Fig. 1F). Briefly, upregulation of
lncRNA SNHG5 promotes cell proliferation, migration and invasion in
OSCC.
lncRNA SNHG5 acts as a molecular
sponge of miR-655-3p in OSCC
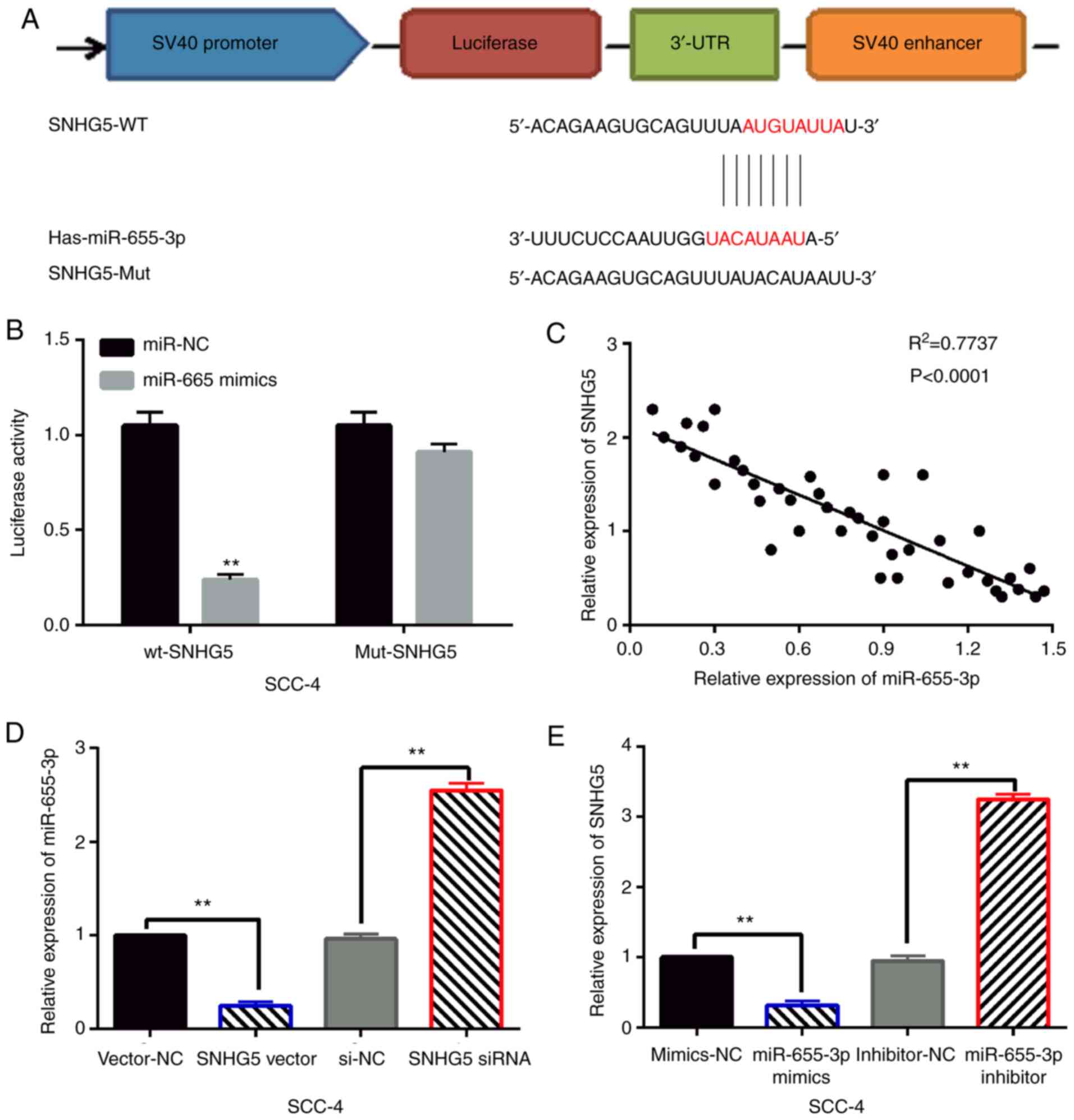
lncRNA SNHG5 was found to have a binding site with
miR-655-3p in the starBase database (http://starbase.sysu.edu.cn/, Fig. 2A). Dual luciferase reporter was used
to verify the relationship between them. It was found that
miR-655-3p mimics reduced the luciferase activity of wt-SNHG5, but
had little effect on mut-SNHG5 luciferase activity in SCC-4 cells
(Fig. 2B). In addition, we found
that SNHG5 was negatively correlated with the expression of
miR-655-3p in OSCC tissues (Fig.
2C). Next, SNHG5 siRNA or vector and miR-655-3p mimics or
inhibitor were transfected into SCC-4 cells, respectively. RT-qPCR
showed that miR-655-3p expression was reduced by SNHG5 upregulation
and enhanced by SNHG5 downregulation in SCC-4 cells (Fig. 2D). Meanwhile, miR-655-3p mimics
reduced SNHG5 expression, while miR-655-3p inhibitor promoted SNHG5
expression in SCC-4 cells (Fig. 2E).
These results indicate that SNHG5 is a competitive miRNA of
miR-655-3p in OSCC.
miR-655-3p is involved in OSCC
progression by mediating lncRNA SNHG5
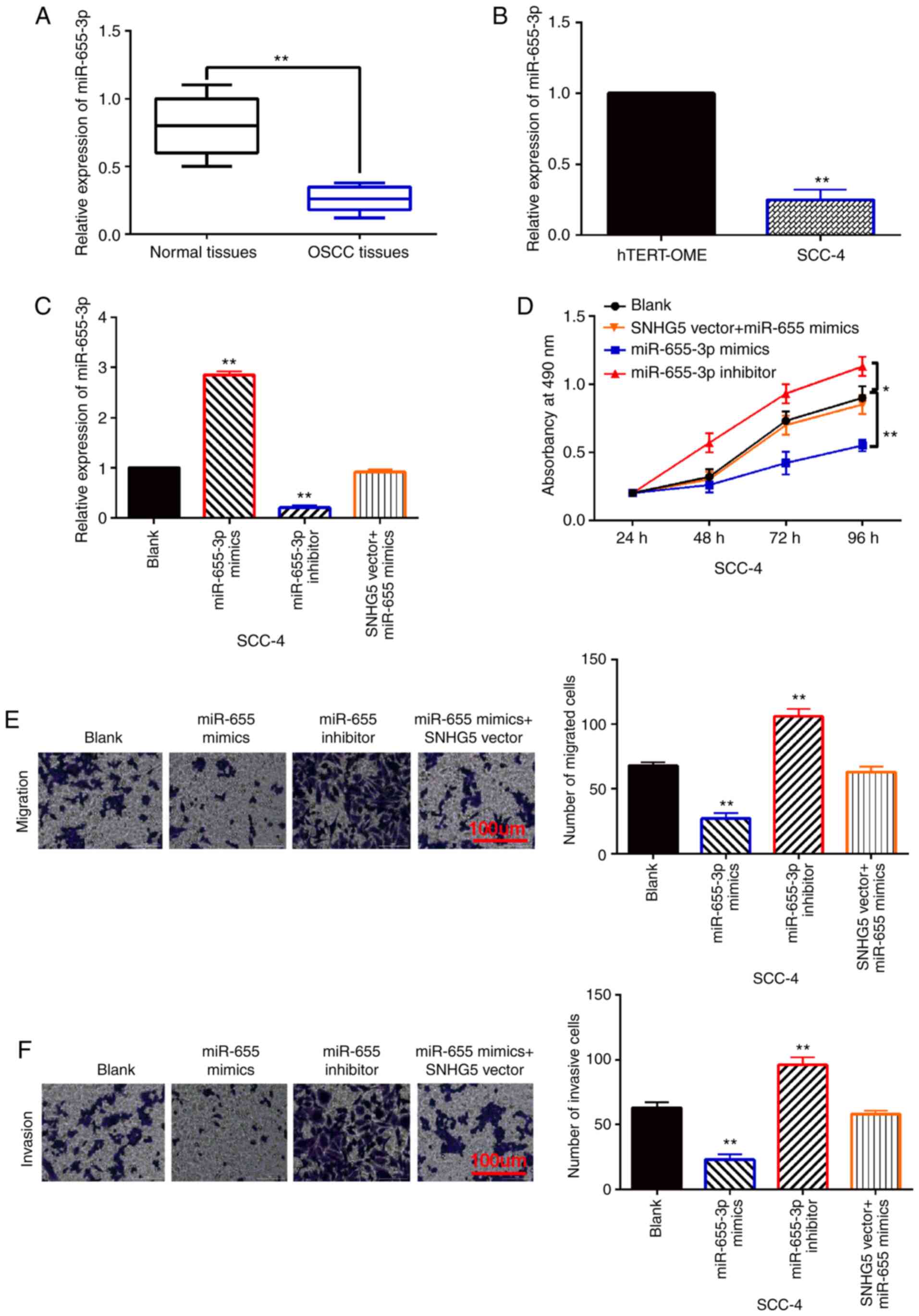
Next, the abnormal expression and function of
miR-655-3p was investigated in OSCC development. Compared to normal
tissues, miR-655-3p expression in OSCC tissues was found to be
downregulated (Fig. 3A). And
compared with hTERT-OME cells, downregulation of miR-655-3p was
detected in SCC-4 cells (Fig. 3B).
To explore the function of miR-655-3p in OSCC, miR-655-3p mimics or
inhibitor were transfected into SCC-4 cells. RT-qPCR showed that
miR-655-3p mimics increased its expression, while miR-655-3p
inhibitor reduced its expression in SCC-4 cells (Fig. 3C). In addition, this increased
expression of miR-655-3p was reduced by SNHG5 upregulation
(Fig. 3C). More importantly, cell
proliferation was restrained by miR-655-3p overexpression and
promoted by miR-655-3p downregulation in SCC-4 cells (Fig. 3D). Similarly, miR-655-3p mimics also
inhibited SCC-4 cell migration and invasion, miR-655-3p inhibitors
also promoted the migration and invasion of SCC-4 cells (Fig. 3E and F). In addition, upregulation of
SNHG5 weakened the inhibitory effect of miR-655-3p on cell
proliferation, migration and invasion in SCC-4 cells (Fig. 3D-F). In conclusion, miR-655-3p acts
as a tumor suppressor in OSCC by competitively binding to
SNHG5.
FZD4 is a direct target of
miR-655-3p
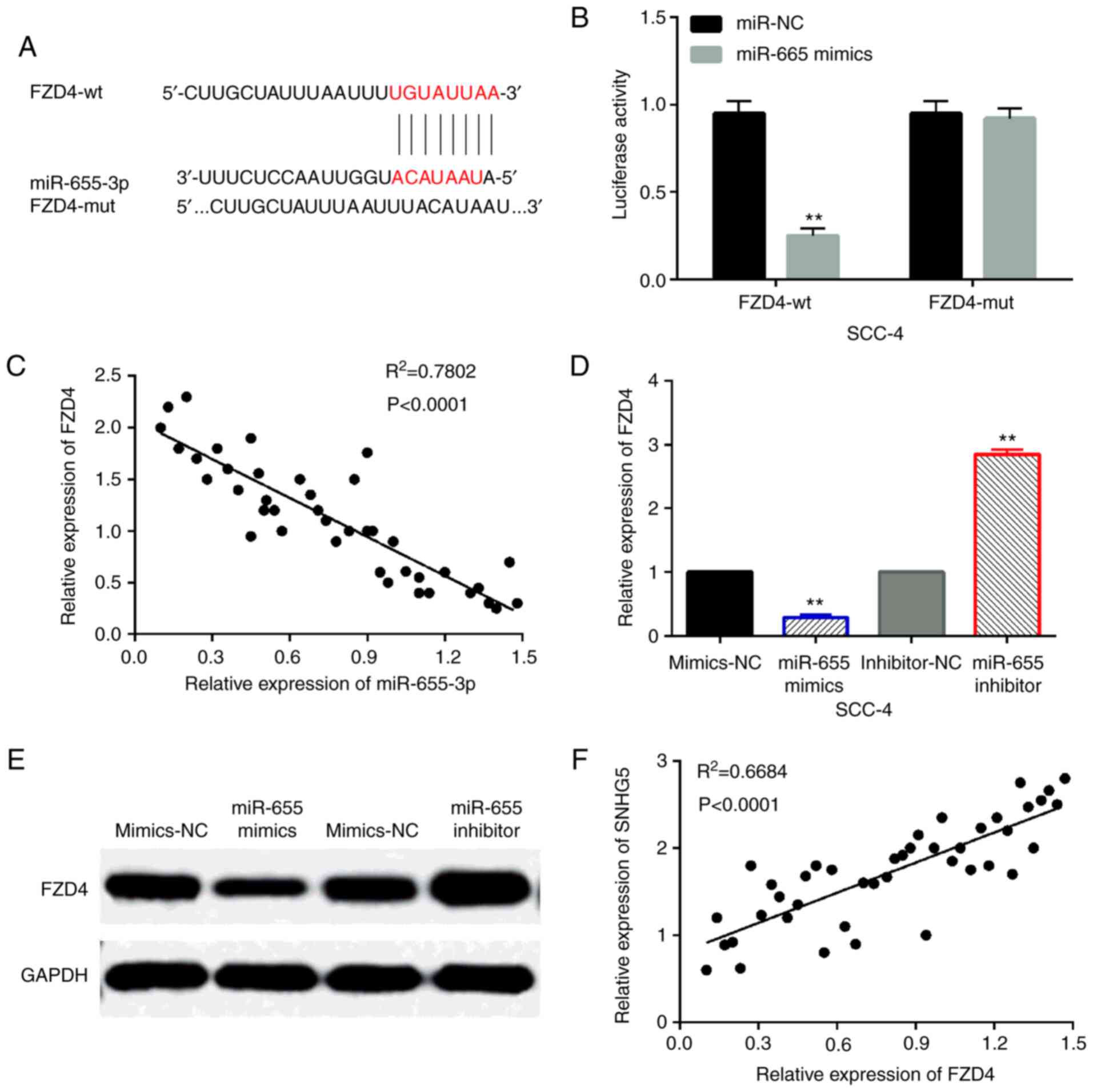
Then, the TargetScan database (http://www.targetscan.org) predicts that FZD4 is the
target of miR-655-3p (Fig. 4A).
Luciferase reporter assay was performed to verify the prediction.
It was found that miR-214-3p mimics reduced the luciferase activity
of wt-FZD4, but had no effect on mut-FZD4 luciferase activity
(Fig. 4B). In addition, FZD4 was
negatively correlated with the expression of miR-655-3p in OSCC
tissues (Fig. 4C). At the same time,
miR-655-3p mimics reduced FZD4 expression, while miR-655-3p
inhibitors promoted FZD4 expression in SCC-4 cells (Fig. 4 and E). Besides that, we also found
that lncRNA SNHG5 was positively correlated with the expression of
FZD4 in OSCC tissues (Fig. 4F).
These results indicate that FZD4 is a direct target of
miR-655-3p.
FZD4 regulates OSCC development by
interacting with SNHG5/miR-655-3p axis
To elucidate the regulatory mechanism of lncRNA
SNHG5, miR-655-3p and FZD4, the SNHG5 vector or miR-655-3p
inhibitor was transfected into SCC-4 cells with FZD4 siRNA
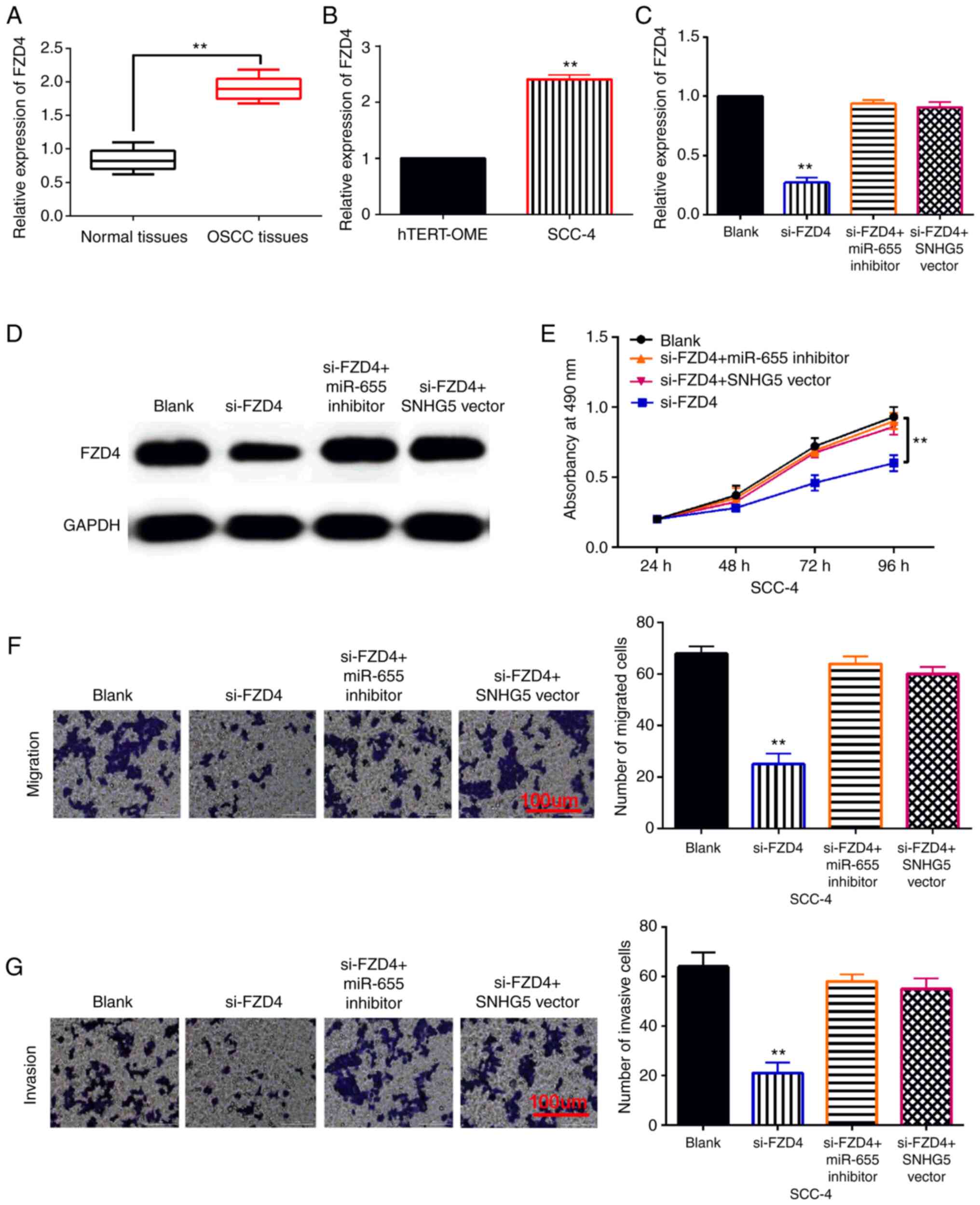
(si-FZD4). First, upregulation of FZD4 was examined in OSCC tissues
and cells (Fig. 5A and B). And we
found that si-FZD4 reduced its expression in SCC-4 cells. However,
upregulation of SNHG5 or downregulation of miR-655-3p restored this
decrease in FZD4 expression (Fig. 5C and
D). Functionally, FZD4 silencing inhibited SCC-4 cell
proliferation. The SNHG5 vector or miR-655-3p inhibitor weakened
the inhibitory effect of si-FZD4 on cell proliferation in SCC-4
cells (Fig. 5E). Moreover, knockdown
of FZD4 suppressed the migration and invasion of SCC-4 cells. And
upregulation of SNHG5 or downregulation of miR-655-3p eliminated
the inhibitory effect of si-FZD4 on SCC-4 cell migration and
invasion (Fig. 5F and G). Therefore,
FZD4 promotes the progression of OSCC by interacting with the
SNHG5/miR-655-3p axis.
Discussion
Recently, the important roles of lncRNAs have been
widely investigated in the progression of OSCC. Previous studies
have indicated shown that lncRNA can play an inhibitory or
carcinogenic effect in OSCC (20,21). In
this study, upregulation of lncRNA SNHG5 was detected in OSCC. And
upregulation of lncRNA SNHG5 promoted cell proliferation, invasion,
and migration in OSCC. Consistent with our results, SNHG5 was also
upregulated in breast cancer and glioma (22,23).
Functionally, SNHG5 was found to promote the survival of colorectal
cancer cells (24). In addition,
SNHG5 promoted cell proliferation, invasion, and migration in human
hepatocellular carcinoma (25). The
same effect of SNHG5 on OSCC progression was also found in this
study, but previous studies have not yet reported. All these
findings suggest that lncRNA SNHG5 acts as a tumor promoter in the
tumorigenesis of OSCC.
As we all know, lncRNA can act as a ‘sponges’ of
miRNA by preventing miRNA from binding to a specific target. lncRNA
SNHG5 has been reported to promote the progression of osteosarcoma
and melanoma through sponging miR-26a-5p and miR-212-3p (26,27).
Here, SNHG5 was identified as a sponge of miR-655-3p. In addition,
downregulation of miR-655-3p was found in OSCC. And overexpression
of miR-655-3p restrained the proliferation, invasion, and migration
of OSCC cells. Reduced expression of miR-655-3p was also found in
esophageal squamous cell carcinoma and triple-negative breast
cancer (13,28). In addition, miR-655-3p inhibited the
proliferation and migration of ovarian cancer cells (29). It was also found that overexpression
of miR-655 suppressed cell invasion (30). These findings are similar to our
results, indicating that miR-655 plays an inhibitory role in
OSCC.
Previous studies have shown that miRNAs are involved
in tumor development by regulating gene expression. It has been
reported that ADAM10, ZEB1 and TGFBR2 are direct targets of
miR-655-3p (31,32). In our study, FZD4 was confirmed as a
direct target of miR-655-3p. Moreover, FZD4 was upregulated in
OSCC, and knockdown of FZD4 inhibited OSCC tumorigenesis. The same
effect of FZD4 has also been found in non-small-cell lung cancer
(33). Besides, it has been found
that miR-505 acted as a tumor suppressor in cervical carcinoma by
inversely regulating FZD4 (34).
Here, we also found that miR-655-3p restrained the development of
OSCC by downregulating FZD4. And lncRNA SNHG5 was found to promote
the occurrence of OSCC by upregulating FZD4. Similarly, lncRNA
DlX6-AS1 has been reported to promote the tumorigenesis of
pancreatic cancer by regulating the miR-497-5p/FZD4 pathway
(35). In conclusion, lncRNA SNHG5
promotes OSCC cell proliferation and metastasis by mediating the
miR-655-3p/FZD4 axis. However, there are some limitations in this
study, such as the absence of in vivo experiment. This study
only explored the regulatory mechanism of lncRNA SNHG5 in
vitro. The verification of our conclusion in vivo is
still needed to be done in the future.
In conclusion, lncRNA SNHG5 promotes cell
proliferation, migration and invasion in OSCC. In addition, lncRNA
SNHG5 accelerates the progression of OSCC by downregulating
miR-655-3p and upregulating FZD4. However, there is still an
unknown about the regulatory mechanism of mechanism in OSCC.
Therefore, further research will be conducted to clarify the
functional mechanism of lncRNA SNHG5 in OSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
LY and LH were responsible for the conception or
design of the work. LY, XS and JZ contributed to the acquisition,
analysis, or interpretation of data for the work. LY and XS
provided the tissue samples. LH helped in the follow-up of the
patients. JZ helped in reviewing the histopathology slides. JZ is
the guarantor of the article. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study was approved by Ethical Committee of
Peking Union Medical College Hospital and conducted in accordance
with the ethical standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma-an update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang SK, Zheng R, Chen Q, Zhang S, Sun X
and Chen W: Oral cancer incidence and mortality in China, 2011.
Chin J Cancer Res. 27:44–51. 2015.PubMed/NCBI
|
|
3
|
Brands MT, Brennan PA, Verbeek ALM, Merkx
MAW and Geurts SME: Follow-up after curative treatment for oral
squamous cell carcinoma. A critical appraisal of the guidelines and
a review of the literature. Eur J Surg Oncol. 44:559–565. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morra F, Merolla F, Picardi I, Russo D,
Ilardi G, Varricchio S, Liotti F, Pacelli R, Palazzo L, Mascolo M,
et al: CAF-1 subunits levels suggest combined treatments with
PARP-inhibitors and ionizing radiation in advanced HNSCC. Cancers
(Basel). 11:15822019. View Article : Google Scholar
|
|
5
|
Sasahira T and Kirita T: Hallmarks of
cancer-related newly prognostic factors of oral squamous cell
carcinoma. Int J Mol Sci. 19:24132018. View Article : Google Scholar
|
|
6
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peng Z, Zhang C and Duan C: Functions and
mechanisms of long noncoding RNAs in lung cancer. Onco Targets
Ther. 9:4411–4424. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin Z, Jiang S, Jian S and Shang Z: Long
noncoding RNA MORT overexpression inhibits cancer cell
proliferation in oral squamous cell carcinoma by downregulating
ROCK1. J Cell Biochem. Feb 25–2019.(Epub ahead of print). doi:
10.1002/jcb.28449. View Article : Google Scholar
|
|
9
|
Yang Y, Chen D, Liu H and Yang K:
Increased expression of lncRNA CASC9 promotes tumor progression by
suppressing autophagy-mediated cell apoptosis via the AKT/mTOR
pathway in oral squamous cell carcinoma. Cell Death Dis. 10:412019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao L, Guo H, Zhou B, Feng J, Li Y, Han
T, Liu L, Li L, Zhang S, Liu Y, et al: Long non-coding RNA SNHG5
suppresses gastric cancer progression by trapping MTA2 in the
cytosol. Oncogene. 35:5770–5780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu X, Hong Y and Shang C: Knockdown of
long non-coding RNA SNHG5 inhibits malignant cellular phenotypes of
glioma via Wnt/CTNNB1 signaling pathway. J Cancer. 10:1333–1340.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang M, Li Q, Pan Y, Wang H, Liu G and
Yin H: MicroRNA-655 attenuates the malignant biological behaviours
of retinoblastoma cells by directly targeting PAX6 and suppressing
the ERK and p38 MAPK signalling pathways. Oncol Rep. 39:2040–2050.
2018.PubMed/NCBI
|
|
13
|
Kiuchi J, Komatsu S, Imamura T, Nishibeppu
K, Shoda K, Arita T, Kosuga T, Konishi H, Shiozaki A, Okamoto K, et
al: Low levels of tumour suppressor miR-655 in plasma contribute to
lymphatic progression and poor outcomes in oesophageal squamous
cell carcinoma. Mol Cancer. 18:22019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gajera M, Desai N, Suzuki A, Li A, Zhang
M, Jun G, Jia P, Zhao Z and Iwata J: MicroRNA-655-3p and
microRNA-497-5p inhibit cell proliferation in cultured human lip
cells through the regulation of genes related to human cleft lip.
BMC Med Genomics. 12:702019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang W, Cao R, Su W, Li Y and Yan H:
miR-655-3p inhibits cell migration and invasion by targeting
pituitary tumor-transforming 1 in non-small cell lung cancer.
Biosci Biotechnol Biochem. 83:1703–1708. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin J, Zandi R, Shao R, Gu J, Ye Y, Wang
J, Zhao Y, Pertsemlidis A, Wistuba II, Wu X, et al: A miR-SNP
biomarker linked to an increased lung cancer survival by
miRNA-mediated down-regulation of FZD4 expression and Wnt
signaling. Sci Rep. 7:90292017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gupta S, Iljin K, Sara H, Mpindi JP,
Mirtti T, Vainio P, Rantala J, Alanen K, Nees M and Kallioniemi O:
FZD4 as a mediator of ERG oncogene-induced WNT signaling and
epithelial-to-mesenchymal transition in human prostate cancer
cells. Cancer Res. 70:6735–6745. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen L, Long Y, Han Z, Yuan Z, Liu W, Yang
F, Li T, Shu L and Zhong Y: MicroRNA-101 inhibits cell migration
and invasion in bladder cancer via targeting FZD4. Exp Ther Med.
17:1476–1485. 2019.PubMed/NCBI
|
|
19
|
Wang Y, Zhang W, Wang Y and Wang S:
HOXD-AS1 promotes cell proliferation, migration and invasion
through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res.
8:170–182. 2018.PubMed/NCBI
|
|
20
|
Tan J, Xiang L and Xu G: lncRNA MEG3
suppresses migration and promotes apoptosis by sponging miR-548d-3p
to modulate JAK-STAT pathway in oral squamous cell carcinoma. IUBMB
Life. 71:882–890. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li B, Wang W, Miao S, Li G, Lv Y, Xiang C
and Pei R: HOXA11-AS promotes the progression of oral squamous cell
carcinoma by targeting the miR-518a-3p/PDK1 axis. Cancer Cell Int.
19:1402019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chi JR, Yu ZH, Liu BW, Zhang D, Ge J, Yu Y
and Cao XC: SNHG5 promotes breast cancer proliferation by sponging
the miR-154-5p/PCNA axis. Mol Ther Nucleic Acids. 17:138–149. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, Liu L, Luo Y, Cui S, Chen W, Zeng A,
Shi Y and Luo L: Long non-coding RNA SNHG5 promotes glioma
progression via miR-205/E2F3 axis. Biosci Rep. 39:BSR201906682019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Damas ND, Marcatti M, Come C, Christensen
LL, Nielsen MM, Baumgartner R, Gylling HM, Maglieri G, Rundsten CF,
Seemann SE, et al: SNHG5 promotes colorectal cancer cell survival
by counteracting STAU1-mediated mRNA destabilization. Nat Commun.
7:138752016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y,
Zhang J, Mi C, He S and Lu X: Long non-coding RNA SNHG5 promotes
human hepatocellular carcinoma progression by regulating
miR-26a-5p/GSK3β signal pathway. Cell Death Dis. 9:8882018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ju C, Zhou R, Sun J, Zhang F, Tang X, Chen
KK, Zhao J, Lan X, Lin S, Zhang Z and Lv XB: lncRNA SNHG5 promotes
the progression of osteosarcoma by sponging the miR-212-3p/SGK3
axis. Cancer Cell Int. 18:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao J, Zeng K, Liu Y, Gao L and Liu L:
lncRNA SNHG5 promotes growth and invasion in melanoma by regulating
the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. 12:169–179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lv ZD, Kong B, Liu XP, Jin LY, Dong Q, Li
FN and Wang HB: miR-655 suppresses epithelial-to-mesenchymal
transition by targeting Prrx1 in triple-negative breast cancer. J
Cell Mol Med. 20:864–873. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zha JF and Chen DX: miR-655-3p inhibited
proliferation and migration of ovarian cancer cells by targeting
RAB1A. Eur Rev Med Pharmacol Sci. 23:3627–3634. 2019.PubMed/NCBI
|
|
30
|
Wang Y, Zang W, Du Y, Ma Y, Li M, Li P,
Chen X, Wang T, Dong Z and Zhao G: Mir-655 up-regulation suppresses
cell invasion by targeting pituitary tumor-transforming gene-1 in
esophageal squamous cell carcinoma. J Transl Med. 11:3012013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu G, Zheng K, Xia S, Wang Y, Meng X, Qin
X and Cheng Y: MicroRNA-655-3p functions as a tumor suppressor by
regulating ADAM10 and β-catenin pathway in hepatocellular
carcinoma. J Exp Clin Cancer Res. 35:892016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Harazono Y, Muramatsu T, Endo H, Uzawa N,
Kawano T, Harada K, Inazawa J and Kozaki K: miR-655 is an
EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One.
8:e627572013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang Y, Sun Y, Wu Y, Tang D, Ding X, Xu W,
Su B and Gao W: Downregulation of miR-3127-5p promotes
epithelial-mesenchymal transition via FZD4 regulation of
Wnt/β-catenin signaling in non-small-cell lung cancer. Mol
Carcinog. 57:842–853. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma C, Xu B, Husaiyin S, Wang L,
Wusainahong K, Ma J, Zhu K and Niyazi M: MicroRNA-505 predicts
prognosis and acts as tumor inhibitor in cervical carcinoma with
inverse association with FZD4. Biomed Pharmacother. 92:586–594.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang J, Ye Z, Mei D, Gu H and Zhang J:
Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating
miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer.
Cancer Manag Res. 11:4209–4221. 2019. View Article : Google Scholar : PubMed/NCBI
|