Introduction
Mitochondria are the bioenergetic and metabolic hubs
of the cell, and are harbor sites of free radical generation
(1). Free radicals can regulate
cellular signaling pathways and contribute to cellular
proliferation and death mechanisms (2). Changes in mitochondria, such as
respiration impairments, oxidative stress and metabolic
alterations, affect these signaling pathways and are associated
with various diseases, including cancer (3). In cancer, mitochondrial contribution is
variable and dependent on the type of cancer, genetic factors,
tissue origin and other unknown factors (4). In mitochondria, respiratory chain (RC)
complexes are large multi-subunit inner membrane structures that
facilitate electron transfer, ATP production via oxidative
phosphorylation (OXPHOS) and reactive oxygen species (ROS)
generation. Since ROS levels act as signaling molecules to activate
cellular pathways, excessive ROS levels can cause oxidative damage
and contribute to oncogenic signaling for cancer development
(5). Previous studies have reported
that oxidative stress is a mitigating factor in metastasis, and the
threshold of oxidative stress is crucial in enhancing or decreasing
metastatic potential (6,7). From the metabolic perspective, while
cancer cells are highly glycolytic (Warburg effect), it is now
evident that reprogramming of metabolic pathways serves a critical
role in proliferation of cancer cells (8). However, the contribution and role of RC
in metabolic reprogramming in cancer progression are yet to be
fully elucidated.
Among the five types of mitochondrial RC complexes,
mitochondrial complex I (C-I; NADH: Ubiquinone oxidoreductase) is
the largest and most prolific ROS producing site, and has been
implicated in cancer progression (9–14). C-I
alterations are involved in mitochondrial dysfunction in different
cancer types, including lung, liver, prostrate breast and colon
cancer (14). While the specific
role of C-I in promoting or suppressing tumors remains unknown, it
largely depends on cancer types and the experimental system
(15). Since metastatic processes
involve cell migration, invasion and proliferation at distal sites,
C-I-mediated changes could serve an important role in regulating
metastatic signaling (16). Hence,
it is imperative to understand how different RC complexes regulate
proliferative pathways and contribute to these pathways at
different stages of neoplastic transformations. Such studies are
clinically relevant in targeting specific RC complexes, as well as
combining therapeutic approaches to specific metastatic conditions
and tumor types.
Colorectal cancer (CRC) is the fourth most common
type of cancer with high incidence (6.1%) and mortality (5.8%)
rates across all cancers worldwide according to global cancer
statistics 2018 (17). Although
mitochondrial alterations have been associated with CRC metastasis,
the functionality of different RC complexes, contribution of
oxidative stress and the role of metastatic signaling in CRC remain
unknown. Therefore, it is clinically relevant to identify
therapeutic targets.
The current study aimed to identify specific changes
in RC complexes, as well as their contribution to mitochondrial
dysfunctions and metabolism in metastatic CRC cells. Using CRC
cells with low and high metastatic potentials, the present study
aimed to investigate the effect of RC inhibition on cell survival,
mitochondrial functions and contribution to metastatic signaling
pathways.
Materials and methods
Cell culture
Established CRC cell lines (low metastatic, HT-29
and HCT-15; high metastatic, HCT-116 and COLO-205) were used in the
current study. These authenticated cell lines were purchased from
the national repository at National Centre for Cell Sciences Pune,
and early passage cultures were used for performing all the
experiments. Cell culture reagents, including growth media, FBS and
antibiotics were purchased from HiMedia Laboratories LLC. HT-29,
HCT-15 and COLO-205 cells were maintained in RPMI-1640 medium,
while HCT-116 cells were maintained in McCoy's growth medium. The
medium was supplemented with 10% FBS and 1% antibiotic-antimycotic
solution, and cells were grown at 37°C with 95% humidity in
incubator maintaining 5% CO2.
In vitro tumorigenesis assay
Tumorigenic potential was determined using soft agar
colony-forming assay (10). Briefly,
a bottom layer containing 0.4% agar, 1X growth media and 10% FBS
was prepared in 60-mm culture dishes. An overlay media, containing
1,000 cells in 0.3% agarose, 1X growth media and 10% FBS, was added
to each plate in triplicate. The cells were incubated for 3–4 weeks
at 37°C in a humidified CO2 incubator and given fresh
media every 4th day. Colony formation was observed after 3 weeks of
plating; bright field images (at 10× magnification) were captured
using the CMOS camera application in ChemiDoc Imaging system
(Bio-Rad Laboratories, Inc.). Clones were counted and quantified as
relative colony-forming units between CRC cells.
Transwell cell migration assay
The migratory capacity of metastatic cells was
measured by their ability to invade through extracellular matrix
(ECM), following a previously described protocol (12). Cells (~5,000) were plated in
triplicates in 8-µm pore-sized cell culture inserts (BD Labware; BD
Biosciences) and supplemented with media without serum. These cell
inserts were positioned in 12-well plates containing media with 10%
FBS, followed by incubation for 12–16 h at 37°C in a 5%
CO2 incubator. Migration of cells was visualized by
staining the cells with Hema 3 staining following manufacturer's
instructions (Thermo Fisher Scientific, Inc.) and images were
captured at 100× magnification using a microscope (Nikon Eclipse
TE2000; Nikon Corporation). Total number of stained and migrated
cells were counted, and calculated as relative migration units
between CRC cells.
Cell viability measurements
A total of ~5×105 cells/ml were seeded in
triplicates in 12-well plates in culture medium and incubated at
37°C for 24 h. Cells were treated with different agents at multiple
concentrations [rotenone, 0.05–200 µM; paraquat, 0.025–20 mM;
antimycin, 0.25–20 µM; oligomycin, 5–100 µM;
H2O2, 0.025–2 mM; N-acetyl cysteine (NAC), 5
mM; all purchased from Sigma-Aldrich) followed by 24 h incubation
at 37°C. Cell viability assay was performed by staining the cells
with trypan blue (0.4%) for 3 min at room temperature and counting
by haemocytometer. Bright field images of cells were captured at
×100 magnification.
Blue native gel electrophoresis and
C-I activity assay
Blue native poly acrylamide gel electrophoresis
(BN-PAGE) was performed following optimized protocol as published
previously (18). Cultured cells
were harvested and centrifuged (at 100 × g for 5 min at room
temperature), and the pellet was re-suspended in 0.5 ml ice-cold HB
buffer (50 mM KPO4; pH: 7.4; 1 mM EDTA; 2.5% glycerol; 250 mM
sucrose) containing protease inhibitor cocktail (Sigma-Aldrich).
Cells were disrupted using a dounce homogenizer at 4°C for 5 min
and enriched for mitochondria via differential centrifugation at
4°C (initial spin at 500 × g for 5 min where the supernatant was
removed, and respun to pellet the mitochondria at 10,000 × g for 5
min). The mitochondrial pellet was washed twice and re-suspended in
a final protein concentration of 2–5 mg/ml in HB buffer. Optimal
solubility of mitochondrial super-complexes was achieved using
optimized concentration of 8 mg digitonin/mg of protein in HB
buffer without EDTA. The samples were incubated on ice for 20 min
in a coomassie blue solution (5% coomassie blue G-250 in 750 mM
6-aminocaproic acid) to a ratio of 1:30 v/v. The supernatant (total
of 80 µg protein) was then run on a 3–12% Native PAGE (Novex
Bis-Tris gel) for 4 h at 80 V and 4°C in the buffer provided by the
supplier (Invitrogen; Thermo Fisher Scientific, Inc.), to resolve
the mitochondrial complexes, and the resulting gels were stained
with Bio-safe coomassie R-250 (Bio-Rad Laboratories, Inc.) for 30
min at room temperature.
In parallel, a similarly run gel without staining
was used for the gel activity assay for C-I following a previously
published protocol (19). Both the
stained and activity gels were scanned and analyzed using ImageJ
software (version 1.8.0; National Institutes of Health), to
determine the relative band intensities, which were normalized with
total mitochondrial protein levels and calculated as fold change
relative to HT-29 bands. Equal loading (80 µg) of mitochondrial
protein was confirmed by running a similar aliquot of mitochondrial
extract on a separate 12% denaturing gel and western blotting with
mitochondrial voltage-dependent anion channel protein (cat. no
4661, Cell Signaling Technology, Inc.).
Western blotting
Protein samples were extracted using RIPA buffer
(Cell Signaling Technology, Inc.) and the concentration of protein
was determined using a BCA Protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). The samples (30 µg) were electrophoresed on 12%
SDS-PAGE at 120 V for 2 h. The resolved proteins were transferred
on PVDF membrane and blocked with 5% non-fat dry milk in TBS-0.5%
Tween-20 (TBS-T) for 1 h at room temperature. The membrane was
incubated with primary antibodies at dilution of 1:1,000, overnight
at 4°C following manufacturer's instructions (Cell Signaling
Technology, Inc.). The antibodies against pAKT-Thr (308) (cat. no.
13038), p-AKT-Ser (473) (cat. no. 4060), total AKT (cat. no. 4691),
Actin (cat. no. 4970), HIF1-α (cat. no. 36169), cMyc (cat. no.
18583), GAPDH (cat. no. 5174), SOD1 (cat. no. 37385), Beclin-1
(cat. no. 3495) and ATG5 (cat. no. 12994) were purchased from Cell
Signaling Technology, Inc. The membrane was washed thrice with
TBS-T and then incubated with anti-rabbit IgG horseradish
peroxidase-conjugated secondary antibody (cat. no. 7074; Cell
Signaling Technology, Inc.) at 1:5,000 dilution. Specific bands
were detected using super signal west pico-chemi-luminescent
substrate kit (Thermo Fisher Scientific, Inc.) and imaged on a
ChemiDoc Imaging system (Bio-Rad Laboratories, Inc.). Experiments
were performed twice or thrice, and one of the representative
images was analyzed for densitometry using ImageJ (version 1.8.0;
National Institutes of Health).
Mitochondrial functional analysis
Reactive oxygen species
measurement
To measure cellular ROS levels, cell-permeant
2′,7′-dichlorodihydrofluorescein diacetate (H2-DCFDA)
dye was used as per manufacturer's instructions (Invitrogen; Thermo
Fisher Scientific, Inc.). Briefly, 1×106 live cells were
re-suspended in Hank's buffered salt solution (HBSS) with
H2DCFDA (10 µM) and Hoechst-33342 (10 µg/ml), and
incubated at 37°C for 15 min. Cells were then washed, centrifuged
at 100 × g for 5 min at room temperature and the resulting pellet
was re-suspended in PBS. The fluorescence intensity was measured
using HT-BioTek fluorescence plate reader at Excitation/Emission
(Ex/Em): 495/529 nm for H2DCFDA and 350/497 nm for
Hoechst-33342 at 37°C.
To measure mitochondrial superoxide levels,
mitochondrial specific superoxide indicator dye MitoSOX™ Red
(Invitrogen; Thermo Fisher Scientific, Inc.) was used. Equal number
of cells (1×106) were re-suspended in HBSS buffer with
MitoSOX™ Red (5 µM) and Hoechst-33342 (10 µg/ml), and incubated at
37°C for 15 min. Cells were washed, and the fluorescence (MitoSOX™
Red; Ex/Em, 510/580 nm) was measured using HT-BioTek fluorescence
plate reader. Relative values for both measurements were calculated
after normalizing with Hoechst-33342 fluorescence. All the
experiments were performed in triplicates, and the results are
presented as relative mean fluorescence intensity.
ATP measurement
Total ATP content was measured using
luciferase-based ATP determination kit according to manufacturer's
instructions (Invitrogen; Thermo Fisher Scientific, Inc.). For ATP
measurement, 1×106 cells were harvested, centrifuged at
100 × g for 5 min at room temperature and re-suspended in 100 µl
buffer (25 mM Tris; pH 7.4; 150 mM EDTA). The re-suspended cells
were boiled at 100°C for 5 min, followed by centrifugation at
10,000 × g for 5 min at room temperature. The supernatant was
collected, added to the luciferin-luciferase mixture and
luminescence was measured in BioTek Synergy HT Multi-detection
Microplate reader. The ATP concentration was determined using a
standard ATP/luminescence curve ranging from 0.001–10 mM ATP,
normalized to total protein and calculated as relative fold change
between CRC cells.
Mitochondrial Membrane Potential (MMP)
measurement
Similar to ATP measurement, 1×106 cells
were incubated with 200 nM MMP indicator dye tetra-methyl-rhodamine
methyl ester perchlorate (TMRM) for 15 min at 37°C. Cells were
washed and counterstained with Hoechst-33342 (10 µg/ml) for 5 min
at 37°C. Fluorescence was recorded for TMRM (Ex/Em, 540/575 nm)
using a fluorescence plate reader. Relative values for both
measurements were calculated after normalizing to Hoechst-33342
fluorescence. All experiments were performed in triplicates, and
the results are presented as relative mean fluorescence
intensity.
Mitochondrial DNA (mtDNA) copy number
measurement
Total DNA was isolated from 1×106 CRC
cells using DNeasy kit (Qiagen China Co., Ltd.). A total of 20 ng
of DNA was used for quantitative PCR (qPCR) for mtDNA copy number
determination using the Fast SYBR Green Master mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.) on a 7900HT Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.. The steps were as follows: 95°C For 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min. Quantitation of mtDNA
copy number relative to nuclear DNA was done by amplifying mtDNA
(human tRNA leucine1, transcription terminator and 5S-like
sequence), and nuclear reference gene (18s ribosomal DNA) as
previously described (20). The
primer sequences were: mtDNA (Human-tRNA leucine 1, transcription
terminator and 5S-like sequence), forward:
5′-CACCCAAGAACAGGGTTTGT-3′ and reverse:
5′-TGGCCATGGGTATGTTGTTAA-3′; nDNA (18s ribosomal DNA), forward:
5′-TAGAGGGACAAGTGGCGTTC-3′ and reverse: 5′-CGCTGAGCCAGTCAGTGT-3′.
Relative mtDNA copy number was calculated with normalization to the
nuclear DNA (18s ribosomal DNA) copy number using 2−ΔΔCq
method (21).
Reverse transcription-quantitative PCR
(RT-qPCR)
For gene expression studies, RNA was isolated from
1×106 non-treated cells or treated with rotenone (100
µM)/paraquat (10 mM) for different time points (0, 3, 6 and 12 h),
using RNeasy kit (Qiagen China Co., Ltd.). A total of 1 µg RNA was
reversed transcribed into cDNA using the QuantiTect Reverse
Transcription kit (Qiagen China Co., Ltd.) at 42°C for 15 min, and
95°C for 3 min. Subsequently qPCR was performed using Fast SYBR
Green Master mix following similar PCR conditions as
aforementioned. The list of primers used for RT-qPCR of the various
genes (PGC1-α, TFAM, β-actin, AKT-1, HIF-1α, cMyc, Survivine,
SLC2A1, CA-9, VEGF) is presented in Table SI with relevant references (22–25). In
addition, qPCR of C-I subunit genes was performed using mtDNA
encoded ND-1,-2,-4, −4L and −6 gene primers as reported by Salehi
et al (26). Relative gene
expression of target genes was normalized to β-actin expression
(reference gene) using 2−ΔΔCq method (21).
Statistical analysis
Graphs were prepared and analyzed using GraphPad
Prism 5 software (GraphPad Software, Inc.). Data in graphs are
presented as the mean ± SEM. Experiments were performed at least
thrice with ≥3 replicates for each condition. Morphological images
were representative of ≥3 independent experiments with similar
results. Significant statistical differences were measured using
unpaired Student's t-test or one-way ANOVA followed by Dunnett's
post hoc test for comparisons between treatment and control groups
or by Tukey's test for comparisons among multiple groups. P<0.05
was considered to indicate a statistically significant
difference.
Results
Properties of cell lines
To study the role of mitochondrial functions in the
metastatic potential of CRC cells, low metastatic HT-29 and high
metastatic HCT-116 CRC lines were used. To confirm whether these
cells demonstrate their respective cancer properties, the
tumorigenic and metastatic potentials were examined using soft agar
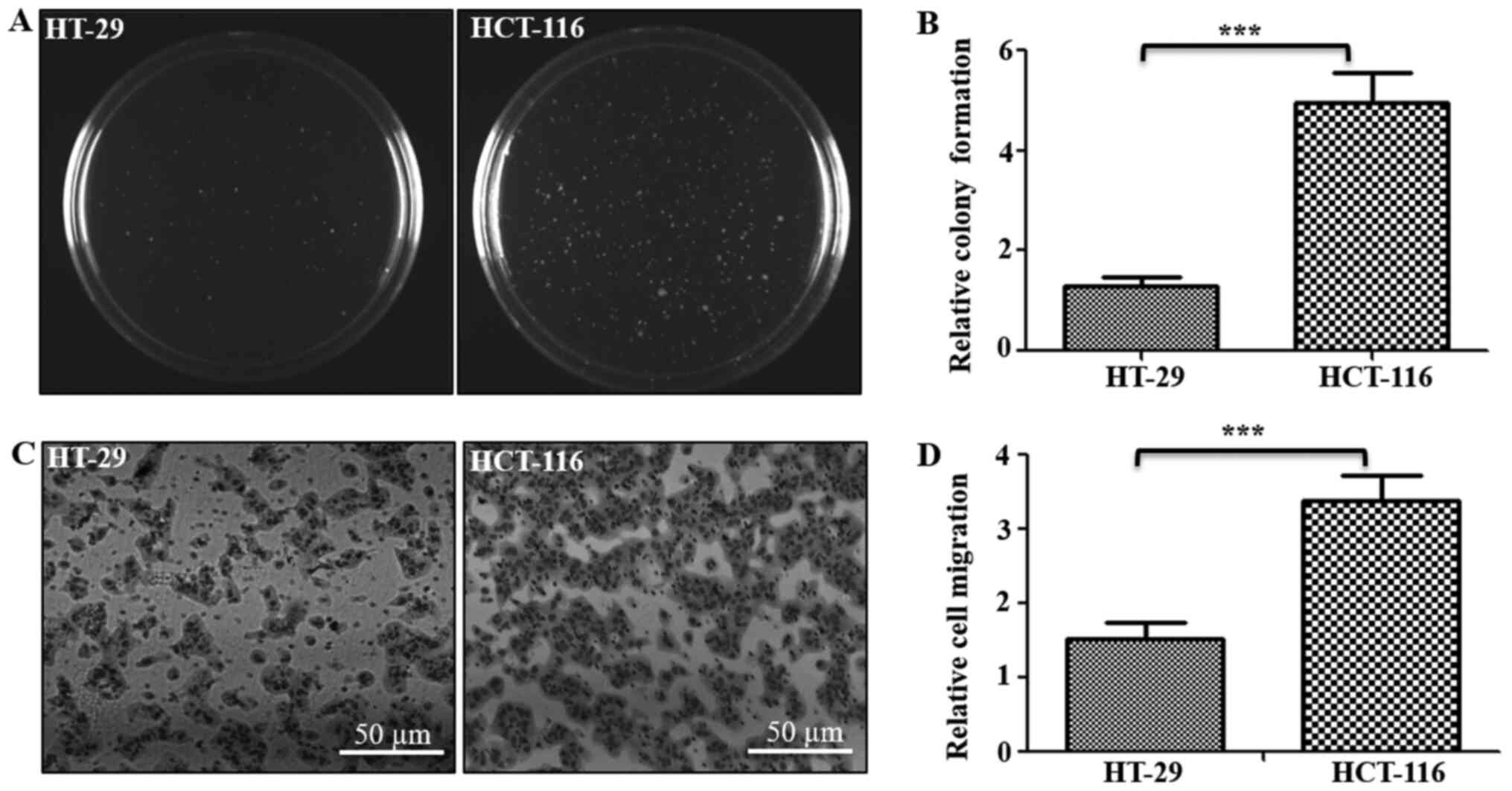
and Transwell assays, respectively (Fig.
1). Results of soft agar assay indicated that HCT-116 cells
formed ~3.8-fold higher numbers of clones on soft agar compared
with HT-29 cells (Fig. 1A and B).
Similarly, Transwell assay results identified that the number of
cells that migrated through the ECM matrix were ~2.3-fold higher in
HCT-116 cells compared with HT-29 cells (Fig. 1C and D). Thus, these assays confirmed
the tumorigenic and metastatic potentials of both cells, indicating
HT-29 cells as low tumorigenic and metastatic, with HCT-116 cells
as highly tumorigenic and metastatic in nature.
Resistance to C-I inhibition in low
metastatic cells
In mitochondria, C-I and Complex III (C-III) are
considered as the major producers of superoxide anions among RC
complexes, and inhibition of these complexes results in an
increased mitochondrial oxidative stress (27–29). The
present study investigated the effect of mitochondrial oxidative
stress via pharmacological inhibition of these complexes by
measuring cellular viability of metastatic cells. Rotenone is a C-I
inhibitor that acts by blocking the transfer of electrons from
iron-sulfur centers in C-I to ubiquinone, which results in the
inhibition of OXPHOS, limited ATP production and increased free
radical production (30). Similarly,
antimycin-A is a C-III inhibitor that binds to the quinone
reduction site of C-III, leading to increased superoxide production
(31). Cells were treated with
different concentrations of rotenone and antimycin-A to measure the
effect of C-I and C-III inhibition on cellular viability,
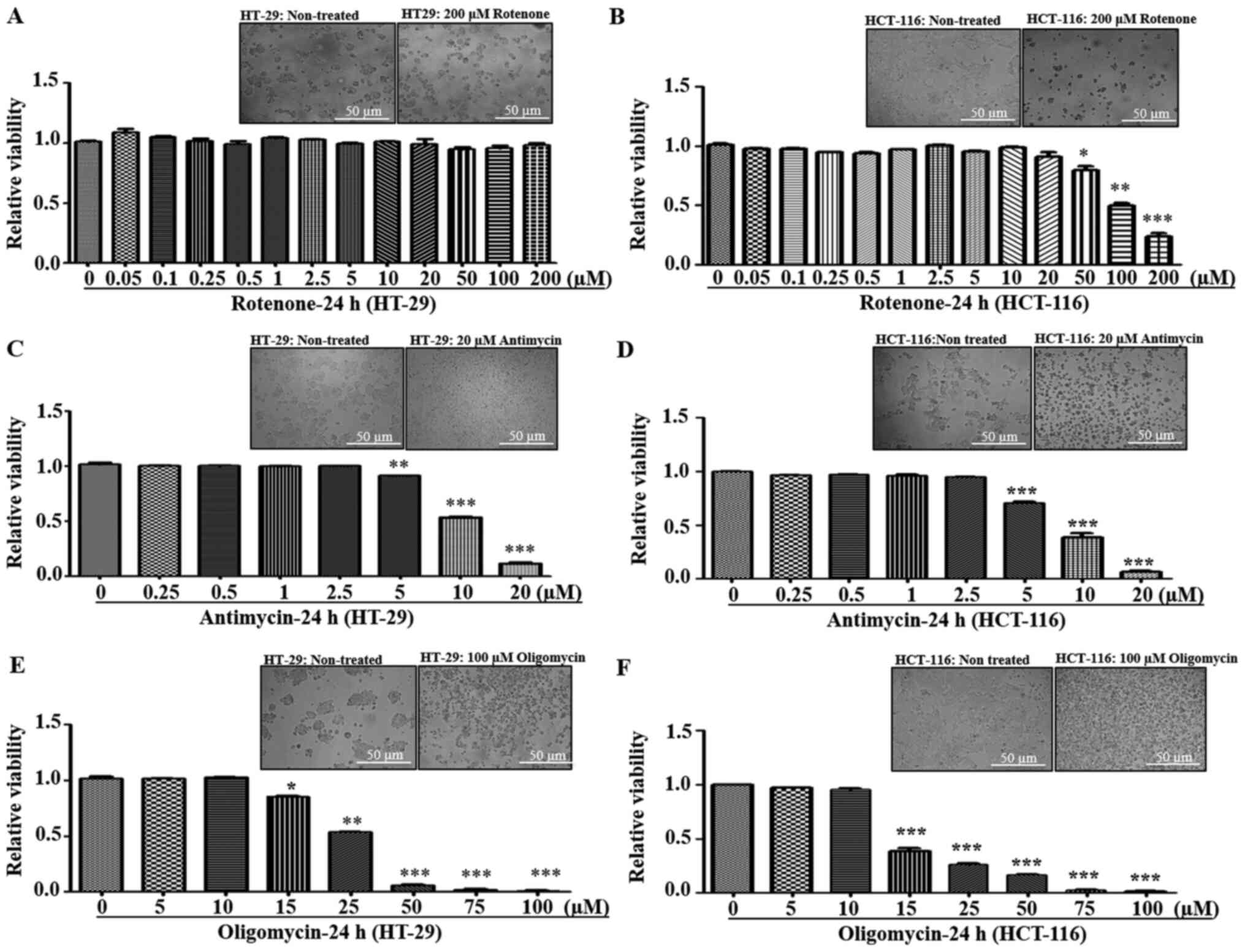
respectively. It was found that both HT-29 and HCT-116 cells were
tolerant to lower concentrations of rotenone (0–20 µM). However, at
>20 µM concentration, HCT-116 cells demonstrated increased
sensitivity and cell death, while HT-29 cells had resistance up to
200 mM (Fig. 2A and B). With regards
to antimycin, both cell lines exhibited a similar trend of declined
viability at ≥5 µM (Fig. 2C and
D).
Since electron transfer via RC complexes is
associated with ATP production, the effect of ATP synthase
[Complex-V (C-V)] inhibition using oligomycin was examined.
Oligomycin is an inhibitor of C-V that functions by inducing
conformational change in the F0 subunit and impairs
binding with substrate at the catalytic sites, leading to ATP
depletion (32). Both HT-29 and
HCT-116 cell lines demonstrated similar sensitivity to C-V
inhibition, with decreased viabilities at ≥15 µM oligomycin
concentrations (Fig. 2E and F).
Overall, in response to different RC inhibitors, low metastatic
HT-29 cells had significant resistance towards rotenone treatment
compared with high metastatic cells, indicating possible C-I
abnormalities.
Resistance of low metastatic cells to C-I inhibition
was assessed via repeatedly measuring cell viability after paraquat
treatment, another C-I inhibitor. Paraquat is reduced by C-I to
form paraquat cation radicals, which react with oxygen to form
superoxide (28). C-I inhibition was
similar in both the inhibitors (rotenone and paraquat); HT-29 cells
were resistant to higher concentration of paraquat compared with
HCT-116 cells, and could tolerate concentrations up to 20 mM
without decreasing cell viability (Fig.
S1). Thus, the investigation of RC inhibitors on cell viability
suggested that low metastatic HT-29 cells were tolerant to higher
concentrations of C-I inhibitors (Rotenone and paraquat) compared
with high metastatic cells, which indicated a compromised or
non-functional C-I in HT-29 cells.
Upregulated C-I and mitochondrial
functions in high metastatic cells
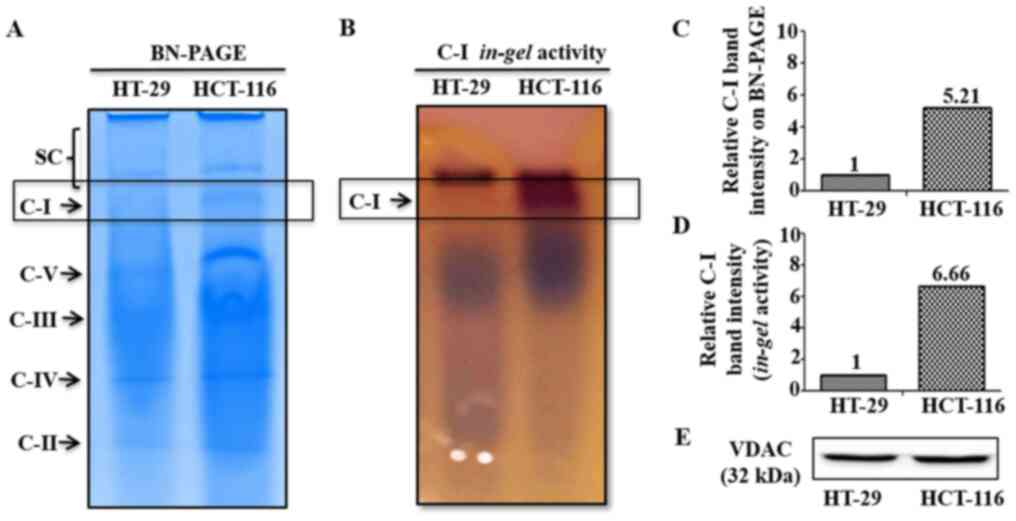
In order to investigate potential differences in C-I
functionality between low and high metastatic cells that may
contribute to their sensitivity to C-I inhibition, C-I assembly and
activity were analyzed using BN-PAGE and C-I in-gel activity
assay, respectively. These measurements were performed in the
isolated mitochondria from low and high metastatic cells. BN-PAGE
results demonstrated an enhanced C-I assembly via increased
expression of C-I specific band in HCT-116 cells, while its
corresponding band was almost absent in HT-29 cells (Fig. 3A and C). Similarly, functional
activity of assembled C-I was higher in HCT-116 compared with HT-29
cells as indicated by increased staining of C-I band in C-I
specific in-gel activity assay (Fig. 3B and D). Equal loading of
mitochondrial preparation for BN-PAGE/in-gel assay from
these cells was confirmed by western blotting of similar aliquot
with mitochondrial marker protein VDAC as loading control (Fig. 3E).
To assess whether this upregulation of C-I was a
common feature in other high metastatic cells, two different CRC
cells, HCT-15 and COLO-205, with low and high metastatic potential,
respectively (33), were used. C-I
functionality was determined by measuring the gene expression
profile of mtDNA encoded C-I genes. mtDNA encodes seven C-I subunit
genes (ND-1, −2, −3, −4, −4L, −5 and −6) (13), and RT-qPCR analysis was performed to
detect the mRNA expression of 5 of these subunit genes (except ND-3
and ND-5 genes) in metastatic cells. Significantly higher
expression levels of C-I genes were identified in high metastatic
COLO-205 cells, compared with low metastatic HCT-15 cells (Fig. S2A), which confirmed C-I upregulation
in high metastatic cells.
C-I is the major entry point for electrons in
electron transfer chain, and its inhibition may cause changes in
free radicals and mitochondrial functions (10,12).
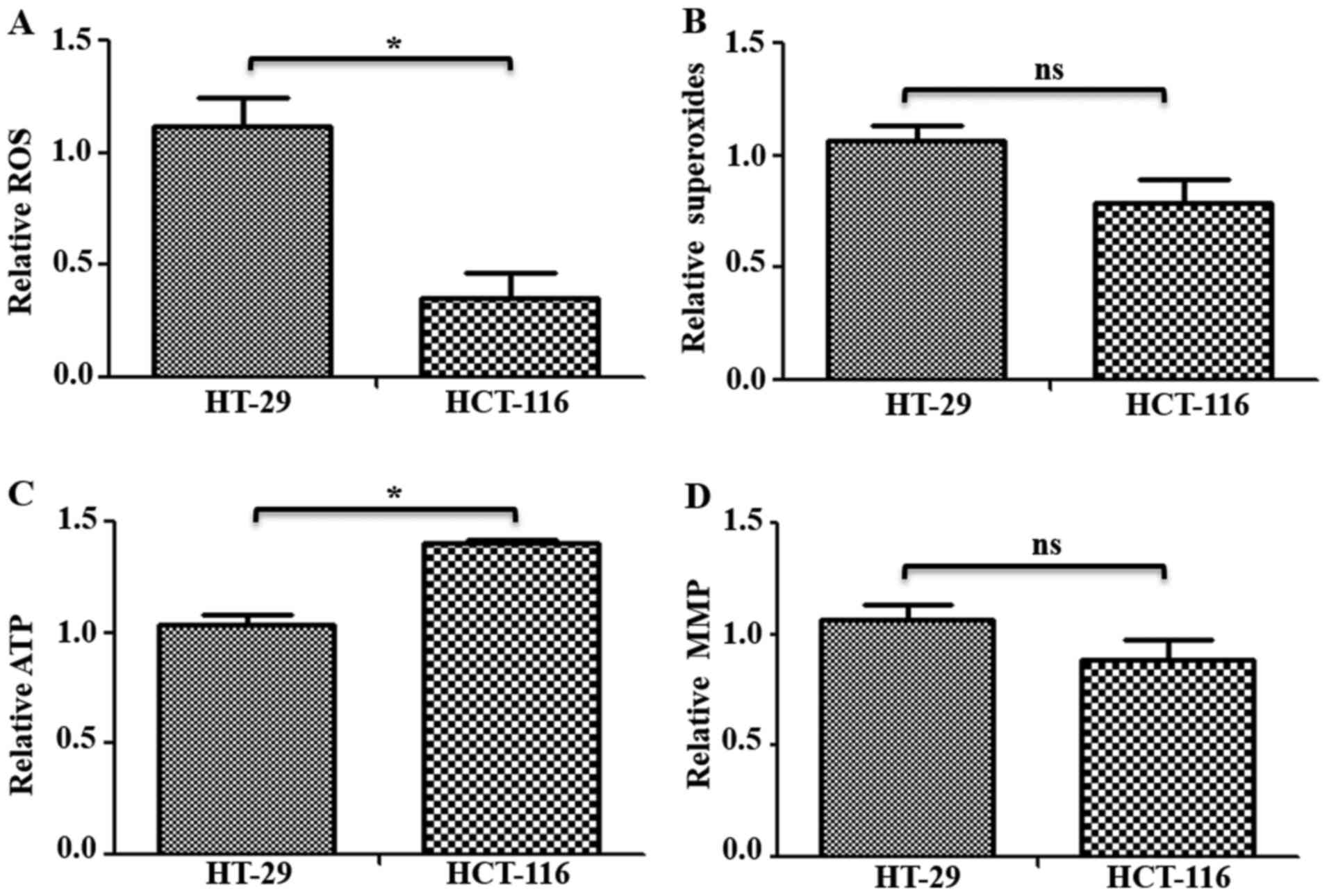
Therefore, to investigate the status of mitochondrial functions in
these cells, ROS, mitochondrial superoxides, ATP and MMP levels
were measured in HT-29 and HCT-116 cells. Total ROS levels were
measured using H2DCFDA, which remains non-fluorescent
until oxidized to the highly fluorescent 2′,7′-dichlorofluorescein
radicals. Compared with HT-29 cells, HCT-116 cells had
significantly lower levels (~0.76-fold lower) of total ROS
(Fig. 4A). Furthermore,
mitochondrial superoxide levels were measured using indicator dye
MitoSOX™ Red, which is oxidized by mitochondrial superoxides. While
a decrease in mitochondrial superoxide levels was observed in
HCT-116 cells, it was not significantly different compared with
HT-29 cells (Fig. 4B). Measurement
of total ATP in these cells identified a ~0.36-fold significantly
higher ATP content in HCT-116 compared with HT-29 cells (Fig. 4C). However, there was no significant
difference in MMP levels (Fig. 4D).
Similar changes in mitochondrial functions were observed when
examined in additional CRC cells (Fig.
S2B).
Therefore, the results suggested that C-I assembly
and activity were worse in low metastatic cells, while C-I was
upregulated in high metastatic cells, which may contribute to
improved mitochondrial functions, such as decreased oxidative
stress, increased ATP levels and enhanced cellular
proliferation.
Increased sensitivity to oxidative
stress in high metastatic cells
Various chemotherapeutic agents can induce cell
death in tumor cells via generation of oxidative stress. To further
evaluate whether the metastatic potential of CRC cells depends upon
changes in C-I and mitochondrial functionality via their response
to higher oxidative stress, their susceptibility towards a general
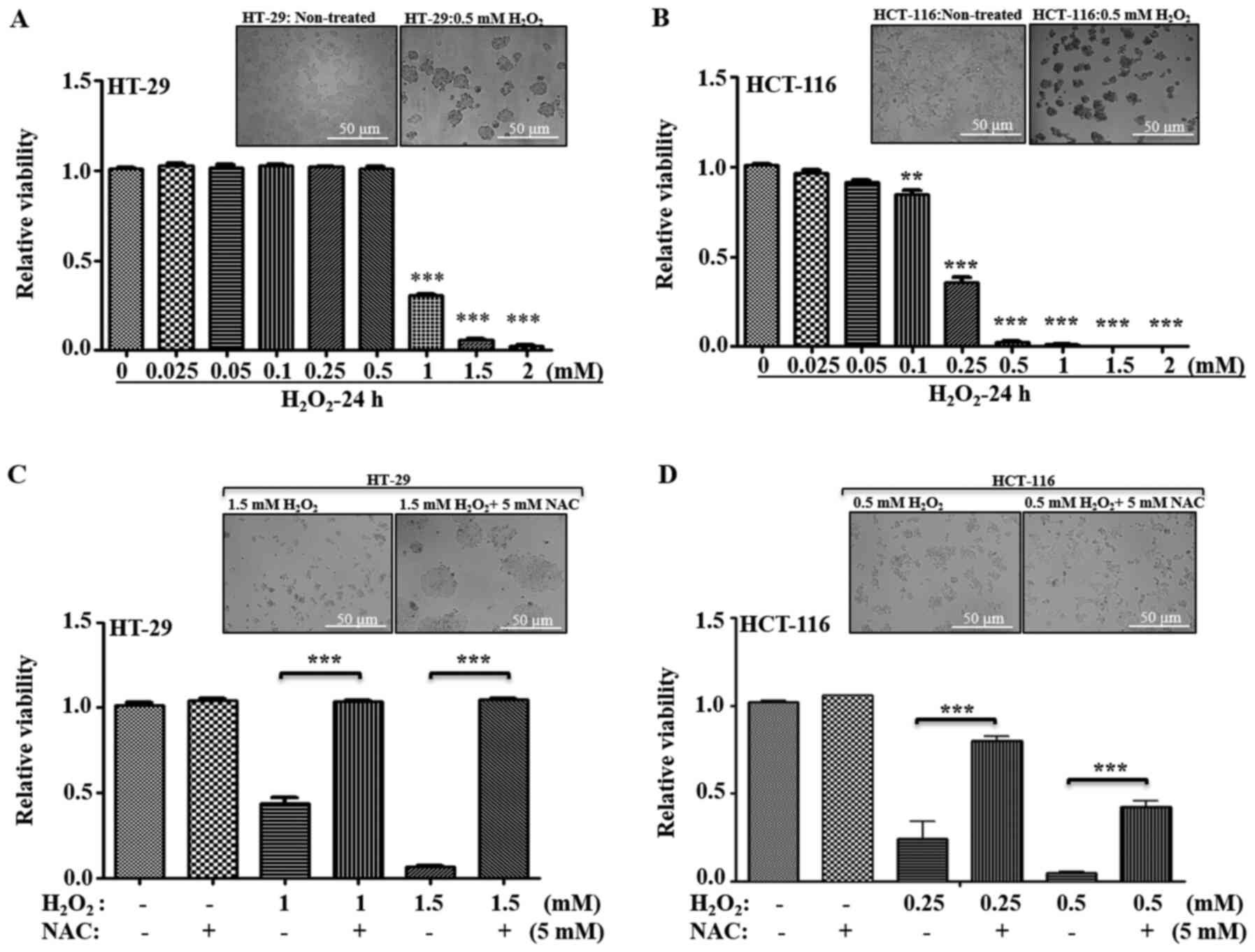
oxidative stress agent, H2O2, was examined.
H2O2 is a well-studied oxidative
stress-inducing agent, where transient exposure triggers apoptosis
in a variety of mammalian cells (34–37).
Thus, low and high metastatic cells were treated with varied
concentration of H2O2, and their viability
was measured (Fig. 5A and B). In
HT-29 cells, no significant cell death was observed from 0.025–0.5
mM H2O2 concentrations; a significant
decrease in viability was observed at ≥1 mM (Fig. 5B). Compared with HT-29, HCT-116 cells
demonstrated a higher sensitivity to
H2O2-induced cell death at ≥0.1 mM; with a
~0.40-fold viability at 0.25 mM, and <0.05-fold viability at
≥0.5 mM concentrations of H2O2 (Fig. 5A and B).
The general antioxidant NAC was used in combination
with H2O2, and cellular viability was
measured to examine whether the cells differed in their response to
recovery after oxidative stress. Inhibitory concentrations of
H2O2 for HT-29 cells (1–1.5 mM
H2O2) and for HCT-116 cells (0.25–0.5 mM
H2O2) were used with or without 5 mM NAC, and
cellular viability was measured. It was identified that, with
regards to HT-29 cells, antioxidant treatment significantly
restored the viability, while HCT-116 cells demonstrated only
partial recovery at the given inhibitory concentrations (Fig. 5C and D). Therefore, these results
suggested that low metastatic cells were more tolerant to higher
oxidative stress, and high metastatic cells were more sensitive to
oxidative stress-induced cell death.
Elevated mitochondrial biogenesis in
high metastatic cells
HCT-116 cells had increased C-I function and
relatively improved mitochondrial functionality, such as lower
levels of ROS and higher ATP levels, compared with HT-29 cells
(Figs. 3 and 4A), suggesting an association between
effective C-I activity and proliferative signaling pathways, which
may contribute to tumor aggressiveness. To understand how high
metastatic cells maintain a functional C-I, it was evaluated
whether high metastatic cells upregulate the compensatory pathway
of mitochondrial biogenesis.
Measurement of mtDNA copy number is an important
aspect of mitochondrial biogenesis and reflects the mitochondrial
requirements for cellular function (38). Peroxisome proliferator-activated
receptor γ coactivator α (PGC1-α), is a major regulator of the
mitochondrial biogenesis, which stimulates transcription of
numerous mitochondrial genes via mitochondrial transcription factor
A (TFAM), a key regulator in mtDNA replication and transcription
(39). The present study measured
mtDNA copy number and mitochondrial biogenesis markers to determine
mitochondrial requirements during metastasis (Fig. 6A). It was found that HCT-116 cells
had higher mtDNA copy numbers compared with HT-29 cells (Fig. 6A-a). RT-qPCR analysis also identified
significantly higher expression levels of mitochondrial biogenesis
markers (PGC1-α and TFAM) in HCT-116 cells compared with HT-29
cells (Fig. 6A-b and c), indicating
an increase in mitochondrial numbers and biogenesis. Similarly,
enhanced mitochondrial copy number, biogenesis and transcription
were observed in other high metastatic cell lines (Fig. S2C).
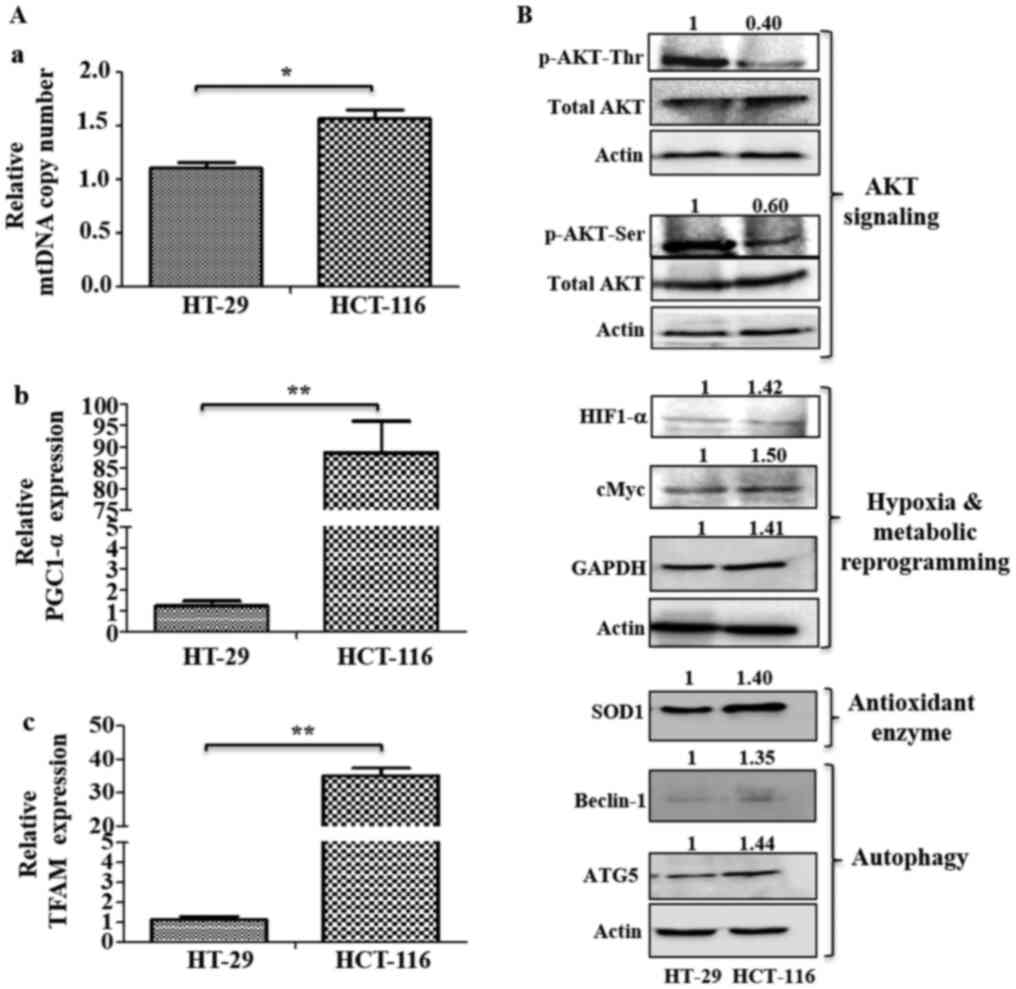 | Figure 6.Analysis of mitochondrial biogenesis
and signaling pathways. (A) Mitochondrial biogenesis was measured
via mtDNA copy number and mRNA expression of mitochondrial
biogenesis markers. (A-a) Changes in mtDNA copy number were
determined using the SYBR green qPCR method. mRNA expression levels
of (A-b) mitochondrial biogenesis marker PGC1-α and (A-c) TFAM were
analyzed using reverse transcription-quantitative PCR. (B)
Immunoblotting was performed to analyze the expression levels of
p-AKT (Ser 478 and Thr 308), HIF1-α, cMyc, GAPDH, SOD1, Beclin-1
and ATG5. Values represent relative band intensities of protein
that were measured by densitometry, normalized with actin loading
control and presented as relative to HT-29. p-AKT (Ser 478 and Thr
308) proteins were normalized with total AKT as well as actin, and
other proteins were normalized with β-actin as loading control.
*P<0.05 and **P<0.01. p-, phosphorylated; HIF1-α, hypoxia
inducible factor 1 subunit α; SOD1, superoxide dismutase 1; ATG5,
autophagy related 5; PGC1-α, Peroxisome proliferator-activated
receptor γ coactivator α; TFAM, mitochondrial transcription factor
A; mtDNA, mitochondrial DNA. |
Investigation of metastatic signaling
based on metastatic potential
HT-29 cells had relatively higher levels of p-AKT at
both Ser473 and Thr308 residues compared with HCT-116 cells
(Fig. 6B). However, HCT-116 cells
had upregulated expression levels of HIF1-α, cMyc, GAPDH, the
antioxidant enzyme SOD1 and autophagy markers (Beclin-1 and ATG5),
compared with HT-29 cells (Fig.
6B).
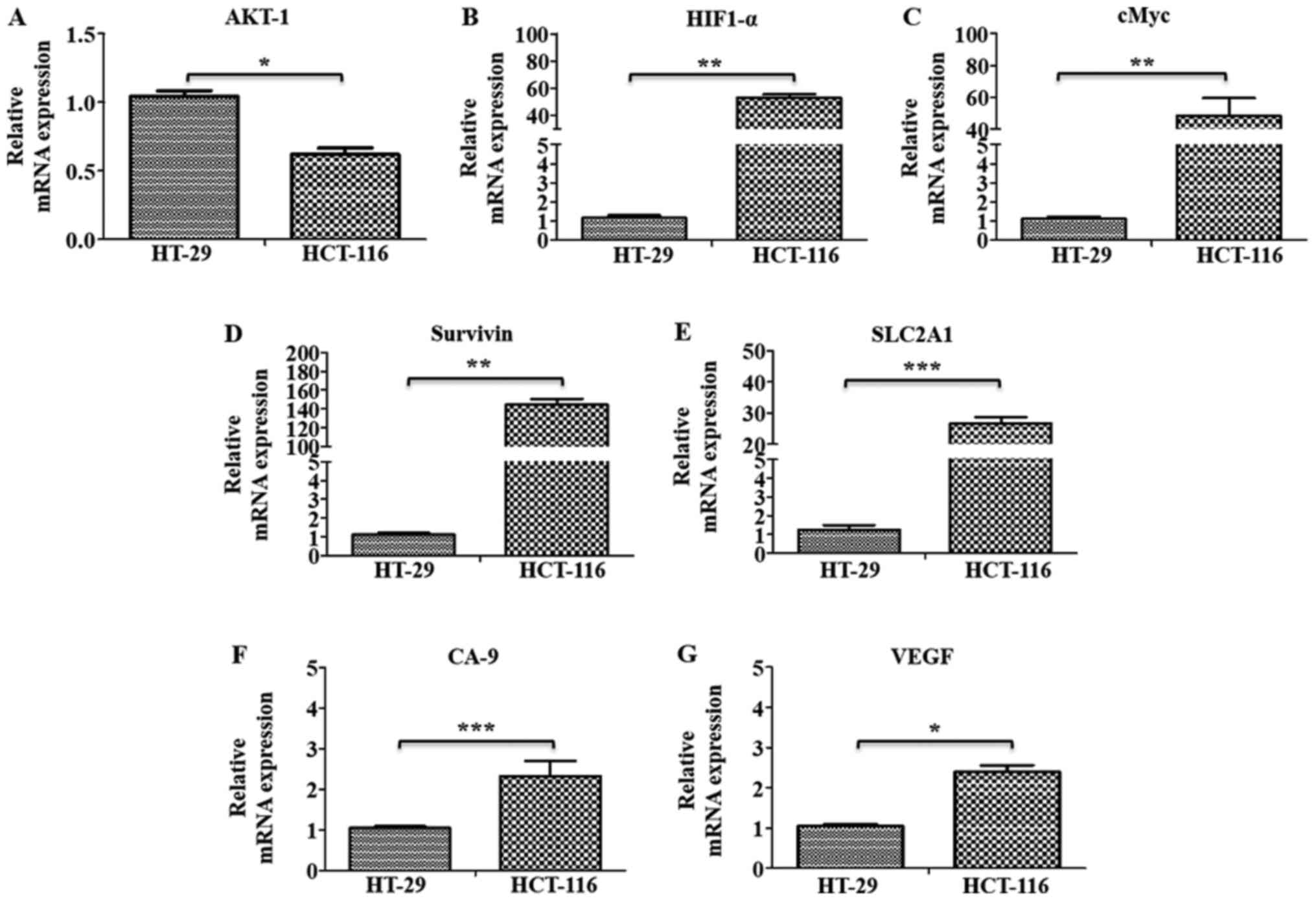
The gene expression levels of marker proteins of
major pathways, such as oncogenic signaling (AKT-1), hypoxia
(HIF1-α) and metabolic reprogramming (cMyc and GAPDH), were further
examined using RT-qPCR analysis, along with the HIF1-α target genes
SLC2A1, Survivin, CA-9 and VEGF in these cells. All these markers,
(except AKT-1), were found to be significantly upregulated in
HCT-116 cells compared with HT-29 cells, indicating their
involvement in metastatic progression in these cells (Fig. 7A-G).
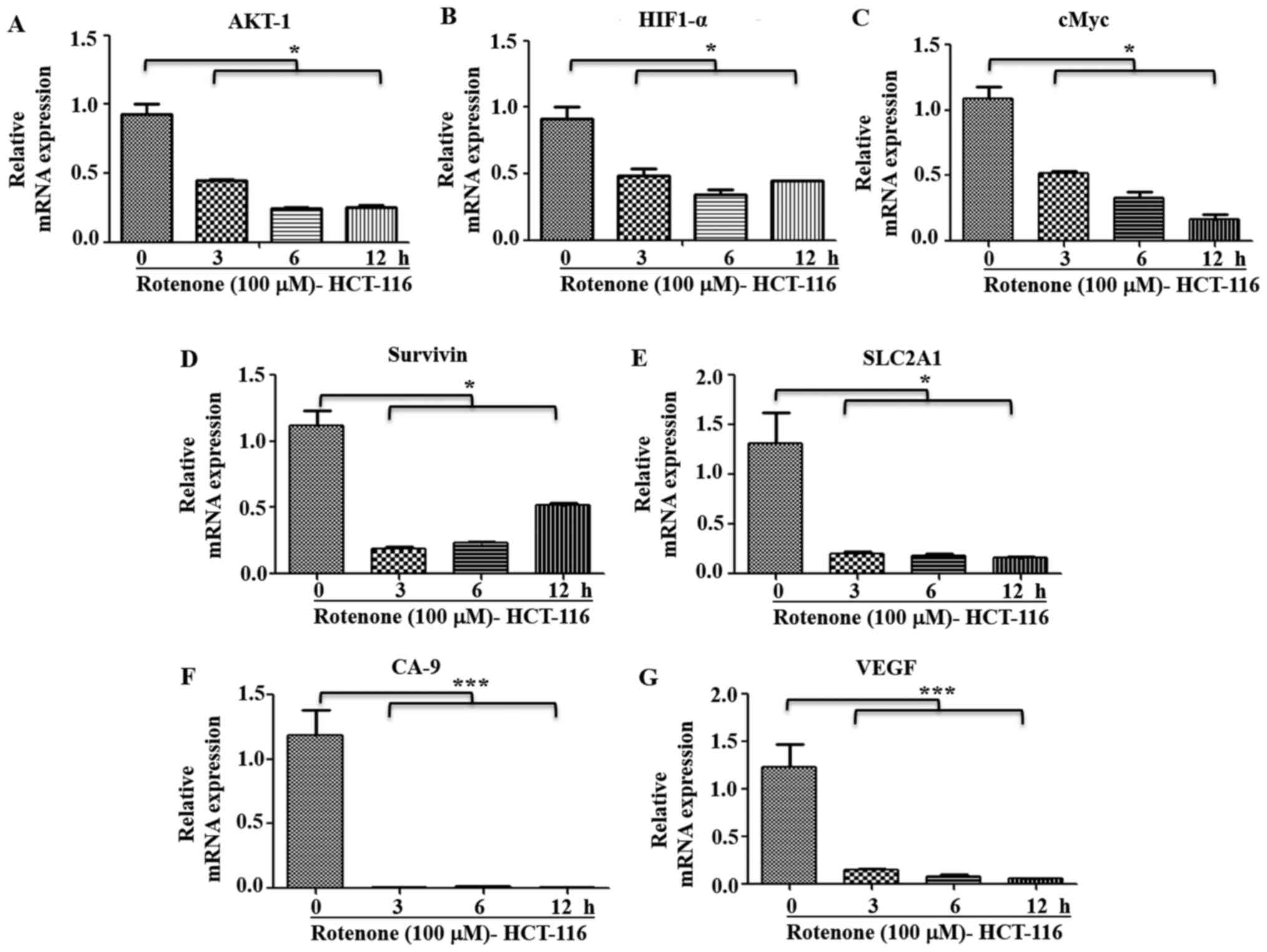
Pharmacological inhibition of C-I in
high metastatic cells blocks metastatic signaling
It was observed that metastatic HCT-116 cells
demonstrated a functional C-I and pharmacological inhibition of
C-I, and these cells had higher sensitivity to C-I-induced cell
death compared with HT-29 cells (Figs.
2B and S1B). It was
hypothesized that C-I recovery may serve an important role in the
transition to the metastatic stage by enhancing metastatic
signaling. Thus, the effect of C-I inhibition on metastatic
signaling in high metastatic cells was also studied. HCT-116 cells
were treated with inhibitory concentration of rotenone (100 µM),
and early time points (0, 3, 6 and 12 h) were selected for
measuring gene expression profile of metastatic pathways. It was
identified that rotenone treatment significantly decreased the mRNA
expression levels of AKT-1, HIF1-α, cMyc, SLC2A1, Survivin, GAPDH,
CA-9 and VEGF compared with non-treated controls (Fig. 8A-G). In addition, paraquat treatment
in these cells with similar conditions resulted in a similar
pattern of decreased gene expression levels of these markers, as
compared with the control group cells (Fig. S3A-G).
These results suggested that the functional C-I was
restored or re-activated in high metastatic cells to maintain or
activate oncogenic and metastatic signaling. Moreover, it was
indicated that pharmacological inhibition of C-I in high metastatic
cells resulted in a decrease in these signaling pathways leading to
cell death, thus implicating C-I as a therapeutic target for highly
metastatic conditions.
Discussion
Cancer metastasis includes the migration, invasion
and survival of cancer cells in the circulation, followed by their
proliferation at distal sites (40).
Moreover, this is a highly complex process in which cancer cells
modify their metabolic requirements for growth and proliferation to
achieve metastasis (41). It has
been proposed that mitochondria are implicated in the malignant
transformation of healthy cells mainly by three major mechanisms:
i) By generating ROS or oxidative stress, which may cause oncogenic
mutations and activation of oncogenic signaling (42); ii) reprogramming of mitochondrial
metabolic pathways, leading to accumulation of onco-metabolites
(43); and iii) resistance to
mitochondrial permeability transition-driven cell death (44). Among these mechanisms, oxidative
stress has been examined extensively, and several studies have
reported that oxidative stress serves a major role in malignant
transformation of primary tumors and enhances their metastatic
potential (6,7,16).
However, the precise role of ROS is debatable as the threshold
levels of ROS vary between cancer cell types, cancer cell responses
and signaling mechanisms to ROS insults.
Mitochondrial RC complexes, mainly C-I and C-III,
are involved in generating the majority of free radicals from
mitochondria (27,29). Thus, it is important to understand
the specific changes in these RC complexes, as well as their
contribution in altering the mitochondrial functions and associated
signaling pathways. Since CRC rapidly progresses to metastatic
phase, the present study used CRC cells with different metastatic
potential as in vitro model systems. The current report
investigated the effect of mitochondrial stress on respiratory
complexes, to understand the functional changes in these complexes
and their contribution in metastatic signaling.
The metastatic properties of CRC cells were
validated in vitro, and it was demonstrated that HCT-116
cells were more aggressive and metastatic in nature compared with
HT-29 cells. Although healthy colon cells would be a more
appropriate choice for comparison with low and high metastasis, due
to non-availability of these cells and the present focus on
comparing low and high metastasis, a known pair of low (HT-29) and
high metastatic cells (HCT-116) were selected for the current
study. The response of these cells towards mitochondrial RC
inhibition and their adaptability of higher oxidative stress were
investigated.
Since ROS are generated by leakage of electrons from
RC complexes, the present study examined the effect of RC
inhibition primarily via C-I and C-III, which are major
contributors of mitochondrial oxidative stress-induced cell death
in mammalian cells (27,29). Low metastatic HT-29 cells were found
to be more resistant to C-I inhibition compared with HCT-116 cells,
suggesting a higher threshold for C-I mediated oxidative stress in
low metastatic conditions. These findings were further corroborated
by increased tolerance and improved adaptation to additional
oxidative stress by low metastatic cells, as evidenced by their
resistance to higher concentrations of H2O2
and significant recovery after antioxidant treatment compared with
other cells. In general, increasing oxidative stress beyond the
threshold in cancer cells has been key for current cancer
therapeutics to induce targeted cancer cell death (45). However, in multiple types of tumors,
including prostate, melanoma and breast cancer, the increased
metastatic ability of tumor cells is positively associated with
their intracellular ROS levels (46), and exogenous treatment of ROS can
enhance certain stages of metastasis (47). The present results suggested a higher
tolerance to oxidative stress in low metastatic conditions compared
with in high metastatic conditions. Furthermore, inducing apoptosis
by enhancing oxidative stress may not be an effective strategy in
early stages, as it may significantly enhance cytotoxicity in
healthy cells and contribute to detrimental outcomes.
Previous studies have reported that partial
impairment of C-I due to heteroplasmic mtDNA mutation in C-I
subunit gene increases tumorigenic potential via ROS-mediated
oncogenic activation (10,12). An inhibitory effect on tumorigenesis
is observed when these C-I defects are severe during conditions of
homoplasmic mtDNA mutations in the same C-I subunit gene (10). Moreover, other studies on nuclear
encoded C-I subunits or assembly proteins revealed that these C-I
associated proteins act as tumor suppressors (48–50).
However, there is no consensus on the role of C-I subunits, as
several findings observed the upregulation of these proteins in
tumor samples (51,52), which may be due to their differential
involvement during metastatic process (15). The present study demonstrated that
C-I sassembly and activity were inhibited in low metastatic cells
compared with high metastatic cells, indicating that inhibited C-I
may limit the cellular ability for rapid metastatic
transformations. Analysis of mitochondrial functions demonstrated
that low metastatic cells with C-I inhibition had increased ROS
levels and decreased ATP levels.
The metastatic process involves cell proliferation
at distal sites, adaptation to low oxygen environment, metabolic
reprogramming and activation of cellular recycling machinery; thus,
the contribution of different component of these signaling pathways
in metastatic cells was investigated. Mitochondrial alteration,
specifically activated via C-I defects, is associated with
activation of AKT, which inhibits apoptotic proteins, leading to
cell survival in stressed conditions (10,53). The
current study further identified that increased phosphorylation of
AKT in low metastatic conditions may provide a survival advantage,
specifically to oxidatively-stressed and energy-deprived cells,
acting as an adaptive response. This finding was in line with
previous reports, which suggested that AKT upregulation is an early
event in colon carcinogenesis and is more common in sporadic cases
compared with microsatellite instability-high colon cancer cases
(54,55). AKT is differentially regulated in
CRC, and specifically, the AKT-1 isoform has opposite and
inhibitory effect on metastasis compared with other isoforms
(56). In the current study, the
mRNA expression profiling of AKT-1 and immunoblotting results
indicated that low metastatic cells demonstrated increased
oncogenic AKT signaling compared with high metastatic cells. Thus,
it was hypothesized that while this enhanced AKT signaling may
contribute to local cellular proliferation and cellular adaptation
of low metastatic cells to high oxidative stress, it may be
insufficient for metastatic progression. In addition, this process
may involve metabolic alterations and activation of angiogenic
pathways for adaptation to microenvironment at distal sites. High
metastatic cells had increased mitochondrial biogenesis to restore
C-I and mitochondrial functions, and C-I and mitochondrial
functions were compromised during early metastasis, as observed in
low metastatic cells.
Similarly, during metastatic transformation, cells
experience low oxygen levels, and therefore, stabilize hypoxia
inducible transcription factors, such as hypoxia inducible factor 1
subunit α (HIF1-α), which activates several downstream
targets, including the glucose transporter solute carrier family 2
member 1 (SLC2A1) (57), Survivin
for anti-apoptosis (58) and
angiogenic factors, such as carbonic anhydrase-9 (CA-9) (59) and VEGF (60), which are associated with tumor
invasion and metastasis. The cMyc oncogene, an important
transcription factor, is a key regulator of mitochondrial
biogenesis and targets 100s of mitochondrial genes (61). Oncogenic activation of cMyc is
reported to increase biosynthetic and respiratory capacity, and
contribute to upregulating glycolytic and mitochondrial metabolism
for enhanced metastatic potential (61). Although mitochondrial biogenesis and
HIF1-α expression are inversely related in certain cancer types, a
positive correlation between these factors has also been revealed
during cMyc activation and confers metabolic advantages to tumor
cells, which tend to exist in a hypoxic microenvironment (62). The present study observed a
synergistic role of mitochondrial biogenesis, cMyc and HIF1-α in
high metastatic cells that may explain the restoration of C-I
expression and activity, decreasing ROS levels and partially
improving ATP levels, thus indicating their role in enhancing
metastatic activity. Furthermore, gene expression analysis
identified the upregulation of HIF1-α targeted genes, such as the
glucose transporter SLC2A1, the anti-apoptotic protein Survivin and
the metastatic markers, VEGF and CA-9, suggesting a metabolic
reprogramming mechanism in high metastatic cells that may
contribute to their aggressiveness.
The current study demonstrated a direct association
of C-I functions in metastatic signaling, as inhibition of C-I in
high metastatic cells resulted in a decrease in oncogenic and
metastatic signaling, leading to a decline in cellular viability.
Therefore, the results highlighted the role of functional C-I in
the survival of high metastatic cells. Highly metastatic cells
demonstrate aggressive features and can survive under harsh
environments, including oxidative stress (63), and the current study observed
elevated levels of the antioxidant enzyme SOD1, which is known to
scavenge cytoplasmic free radicals (64). This partly explains the lower levels
of ROS in high metastatic cells compared with low metastatic cells.
In addition, the upregulation of the autophagy-indicator proteins
Beclin-1 and ATG5 was found in high metastatic cells. Autophagy has
reported to serve an important role in different stages of
metastasis (65). Specifically in
CRC, autophagy exerts a pro-active effect as revealed by increased
expression of Beclin-1 (66) and
inhibition of ATG5, which results in an inhibitory effect on
tumorigenesis both in vitro and in vivo (67). Therefore, upregulation of these
proteins in high metastatic cells indicates a positive contribution
of autophagy in metastasis, possibly by enhanced clearance of
damaged mitochondria via mitophagy, but this requires further
investigation.
There are certain limitations to the present study,
including the lack of investigation into the specific components of
these signaling pathways associated with C-I functionality and the
absence of in vivo experiments. The effect of complex IV
(C-IV) inhibition, could not be determined due to potential hazard
and regulatory restriction on the use of C-IV inhibitor potassium
cyanide. The upregulation of autophagy proteins indicated the role
of autophagy in metastasis. However, the role of selective
clearance of mitochondria via mitophagy requires further
investigation. Therefore, additional studies are required to
identify molecular mechanism of C-I targeting molecules and their
potential use in developing effective therapies for highly fatal
metastatic cancer types.
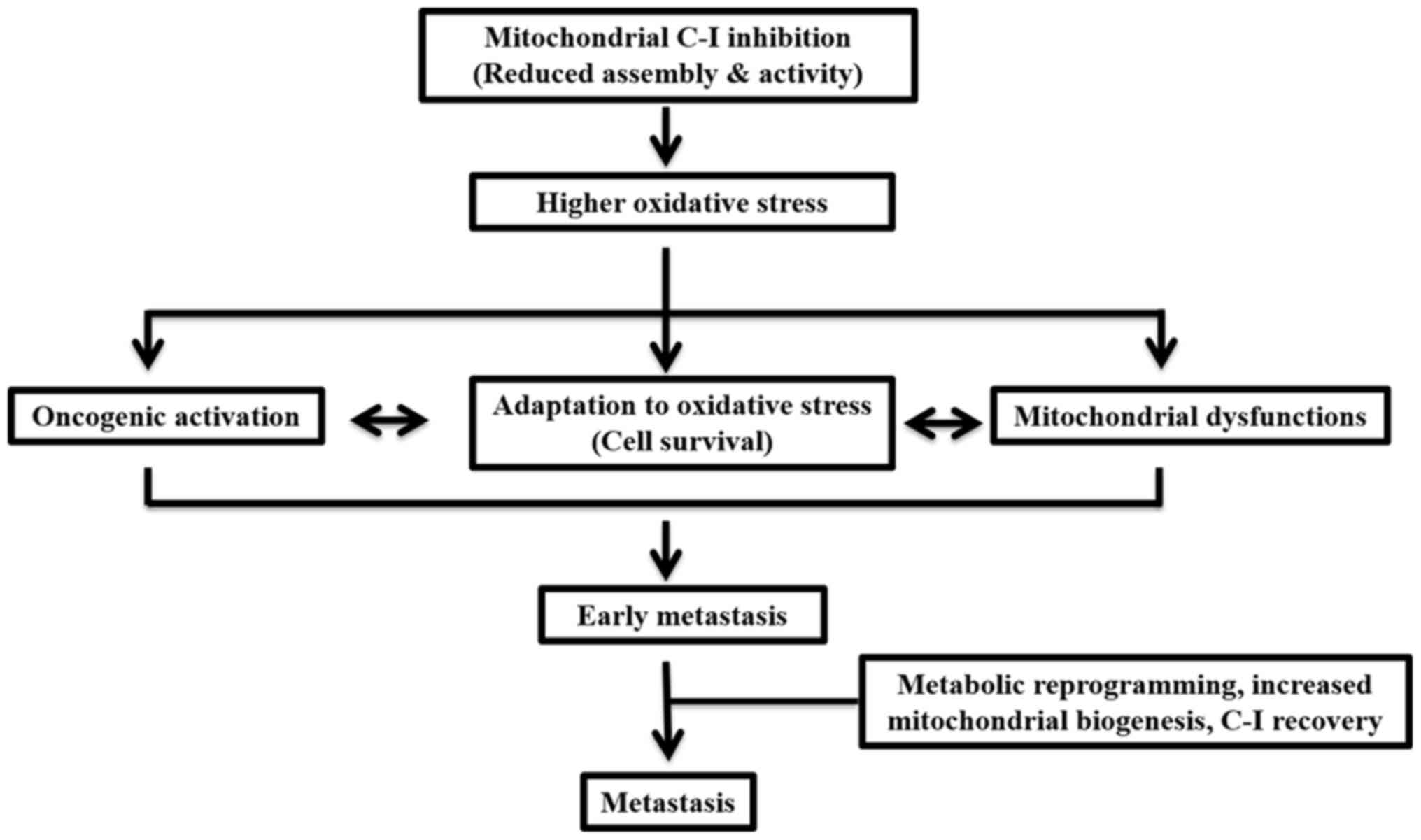
In conclusion, the present study demonstrated that,
during early metastasis, impairments in C-I may contribute to
enhanced ROS levels, which may lead to cellular adaptation via
activation of cell survival pathways (Fig. 9). In a state of high metastasis,
cells may be reprogrammed via a coordinated upregulation of
mitochondrial biogenesis and cMyc to restore C-I and the overall
mitochondrial functions required for their aggressive features. The
current results also suggested that threshold levels of ROS depend
on C-I activity and the level of metastasis. Therefore, these
should be considered when selecting therapies for cancer, as the
threshold and adaptation for oxidative stress are different in
early and late phases of metastasis, and can change the outcome of
the disease after pro- or anti-oxidant therapies. Moreover, a
functional role of C-I was identified, and suggested C-I as a
potential therapeutic target for highly metastatic cancer types
that are otherwise resistant to chemotherapy.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Lia R. Edmunds
(University of Pittsburgh) for proofreading the manuscript.
Funding
Funding was received from Science and Engineering
Research Board (grant nos. SB/YS/LS-95/2013 and CRG/2018/001559)
and DBT Bio-CARe grant (grant no. BT/P19357/BIC/101/927/2016). NKR
was supported by junior research fellowship [grant no. 16-6
(Dec.2017)/2018] from CSIR-UGC.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NKR, SM and SKS planned and performed the
experiments, complied and analyzed the data. MT, ST and LKS were
involved in study design, planning of experiments, data analysis
and interpretation and writing the manuscript. RH helped in
performing experiments on additional cell lines to validate the
findings. VKS was involved in statistical analysis and revising the
manuscript critically for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zorov DB, Juhaszova M and Sollott SJ:
Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS
release. Physiol Rev. 94:909–950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Simon HU, Haj-Yehia A and Levi-Schaffer F:
Role of reactive oxygen species (ROS) in apoptosis induction.
Apoptosis. 5:415–418. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wallace DC: Mitochondria as chi. Genetics.
179:727–735. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vyas S, Zaganjor E and Haigis MC:
Mitochondria and cancer. Cell. 166:555–566. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schieber M and Chandel NS: ROS function in
redox signaling and oxidative stress. Curr Biol. 24:R453–R462.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Le Gal K, Ibrahim MX, Wiel C, Sayin VI,
Akula MK, Karlsson C, Dalin MG, Akyürek LM, Lindahl P, Nilsson J
and Bergo MO: Antioxidants can increase melanoma metastasis in
mice. Sci Transl Med. 7:308re82015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Piskounova E, Agathocleous M, Murphy MM,
Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ
and Morrison SJ: Oxidative stress inhibits distant metastasis by
human melanoma cells. Nature. 527:186–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ward PS and Thompson CB: Metabolic
reprogramming: A cancer hallmark even warburg did not anticipate.
Cancer Cell. 21:297–308. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Sharma LK and Bai Y: Implications of
mitochondrial DNA mutations and mitochondrial dysfunction in
tumorigenesis. Cell Res. 19:802–815. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park JS, Sharma LK, Li H, Xiang R,
Holstein D, Wu J, Lechleiter J, Naylor SL, Deng JJ, Lu J and Bai Y:
A heteroplasmic, not homoplasmic, mitochondrial DNA mutation
promotes tumorigenesis via alteration in reactive oxygen species
generation and apoptosis. Hum Mol Genet. 18:1578–1589. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Santidrian AF, Matsuno-Yagi A, Ritland M,
Seo BB, LeBoeuf SE, Gay LJ, Yagi T and Felding-Habermann B:
Mitochondrial complex I activity and NAD+/NADH balance
regulate breast cancer progression. J Clin Invest. 123:1068–1081.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sharma LK, Fang H, Liu J, Vartak R, Deng J
and Bai Y: Mitochondrial respiratory complex I dysfunction promotes
tumorigenesis through ROS alteration and AKT activation. Hum Mol
Genet. 20:4605–4616. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sharma LK, Lu J and Bai Y: Mitochondrial
respiratory complex I: Structure, function and implication in human
diseases. Curr Med Chem. 16:1266–1277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Urra FA, Munoz F, Lovy A and Cardenas C:
The mitochondrial complex(I)ty of cancer. Front Oncol. 7:1182017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leone G, Abla H, Gasparre G, Porcelli AM
and Iommarini L: The oncojanus paradigm of respiratory complex I.
Genes (Basel). 9:2432018. View Article : Google Scholar
|
|
16
|
Gill JG, Piskounova E and Morrison SJ:
Cancer, oxidative stress, and metastasis. Cold Spring Harb Symp
Quant Biol. 81:163–175. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edmunds LR, Sharma L, Wang H, Kang A,
d'Souza S, Lu J, McLaughlin M, Dolezal JM, Gao X, Weintraub ST, et
al: c-Myc and AMPK control cellular energy levels by cooperatively
regulating mitochondrial structure and function. PLoS One.
10:e01340492015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wittig I, Karas M and Schagger H: High
resolution clear native electrophoresis for in-gel functional
assays and fluorescence studies of membrane protein complexes. Mol
Cell Proteomics. 6:1215–1225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang H, Liu X, Shen L, Li F, Liu Y, Chi H,
Miao H, Lu J and Bai Y: Role of mtDNA haplogroups in the prevalence
of knee osteoarthritis in a southern Chinese population. Int J Mol
Sci. 15:2646–2659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li QF, Wang XR, Yang YW and Lin H: Hypoxia
upregulates hypoxia inducible factor (HIF)-3alpha expression in
lung epithelial cells: Characterization and comparison with
HIF-1alpha. Cell Res. 16:548–558. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Onishi Y, Ueha T, Kawamoto T, Hara H, Toda
M, Harada R, Minoda M, Kurosaka M and Akisue T: Regulation of
mitochondrial proliferation by PGC-1α induces cellular apoptosis in
musculoskeletal malignancies. Sci Rep. 4:39162014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Spandidos A, Wang X, Wang H and Seed B:
PrimerBank: A resource of human and mouse PCR primer pairs for gene
expression detection and quantification. Nucleic Acids Res.
38:D792–D799. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wheaton WW, Weinberg SE, Hamanaka RB,
Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM,
Budigner GS and Chandel NS: Metformin inhibits mitochondrial
complex I of cancer cells to reduce tumorigenesis. Elife.
3:e022422014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salehi MH, Kamalidehghan B, Houshmand M,
Meng GY, Sadeghizadeh M, Aryani O and Nafissi S: Gene expression
profiling of mitochondrial oxidative phosphorylation (OXPHOS)
complex I in Friedreich ataxia (FRDA) patients. PLoS One.
9:e940692014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL
and Lesnefsky EJ: Production of reactive oxygen species by
mitochondria: Central role of complex III. J Biol Chem.
278:36027–36031. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cocheme HM and Murphy MP: Complex I is the
major site of mitochondrial superoxide production by paraquat. J
Biol Chem. 283:1786–1798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dröse S and Brandt U: Molecular mechanisms
of superoxide production by the mitochondrial respiratory chain.
Adv Exp Med Biol. 748:145–169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Palmer G, Horgan DJ, Tisdale H, Singer TP
and Beinert H: Studies on the respiratory chain-linked reduced
nicotinamide adenine dinucleotide dehydrogenase. XIV. Location of
the sites of inhibition of rotenone, barbiturates, and piericidin
by means of electron paramagnetic resonance spectroscopy. J Biol
Chem. 243:844–847. 1968.PubMed/NCBI
|
|
31
|
Huang LS, Cobessi D, Tung EY and Berry EA:
Binding of the respiratory chain inhibitor antimycin to the
mitochondrial bc1 complex: A new crystal structure reveals an
altered intramolecular hydrogen-bonding pattern. J Mol Biol.
351:573–597. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Penefsky HS: Mechanism of inhibition of
mitochondrial adenosine triphosphatase by dicyclohexylcarbodiimide
and oligomycin: Relationship to ATP synthesis. Proc Natl Acad Sci
USA. 82:1589–1593. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Trainer DL, Kline T, McCabe FL, Faucette
LF, Field J, Chaikin M, Anzano M, Rieman D, Hoffstein S, Li DJ, et
al: Biological characterization and oncogene expression in human
colorectal carcinoma cell lines. Int J Cancer. 41:287–296. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singh M, Sharma H and Singh N: Hydrogen
peroxide induces apoptosis in HeLa cells through mitochondrial
pathway. Mitochondrion. 7:367–373. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Viola HM, Arthur PG and Hool LC: Transient
exposure to hydrogen peroxide causes an increase in
mitochondria-derived superoxide as a result of sustained alteration
in L-type Ca2+ channel function in the absence of
apoptosis in ventricular myocytes. Circ Res. 100:1036–1044. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Whittemore ER, Loo DT, Watt JA and Cotman
CW: A detailed analysis of hydrogen peroxide-induced cell death in
primary neuronal culture. Neuroscience. 67:921–932. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiang J, Wan C, Guo R and Guo D: Is
hydrogen peroxide a suitable apoptosis inducer for all cell types?
Biomed Res Int. 2016:73439652016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Clay Montier LL, Deng JJ and Bai Y: Number
matters: Control of mammalian mitochondrial DNA copy number. J
Genet Genomics. 36:125–131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Virbasius JV and Scarpulla RC: Activation
of the human mitochondrial transcription factor A gene by nuclear
respiratory factors: A potential regulatory link between nuclear
and mitochondrial gene expression in organelle biogenesis. Proc
Natl Acad Sci USA. 91:1309–1313. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
van Zijl F, Krupitza G and Mikulits W:
Initial steps of metastasis: Cell invasion and endothelial
transmigration. Mutat Res. 728:23–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Phan LM, Yeung SC and Lee MH: Cancer
metabolic reprogramming: Importance, main features, and potentials
for precise targeted anti-cancer therapies. Cancer Biol Med.
11:1–19. 2014.PubMed/NCBI
|
|
42
|
Sabharwal SS and Schumacker PT:
Mitochondrial ROS in cancer: Initiators, amplifiers or an Achilles'
heel? Nat Rev Cancer. 14:709–721. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sullivan LB, Gui DY and Vander Heiden MG:
Altered metabolite levels in cancer: Implications for tumour
biology and cancer therapy. Nat Rev Cancer. 16:680–693. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Izzo V, Bravo-San Pedro JM, Sica V,
Kroemer G and Galluzzi L: Mitochondrial permeability transition:
New findings and persisting uncertainties. Trends Cell Biol.
26:655–667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang J and Yi J: Cancer cell killing via
ROS: To increase or decrease, that is the question. Cancer Biol
Ther. 7:1875–1884. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lim SD, Sun C, Lambeth JD, Marshall F,
Amin M, Chung L, Petros JA and Arnold RS: Increased Nox1 and
hydrogen peroxide in prostate cancer. Prostate. 62:200–207. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jing X, Ueki N, Cheng J, Imanishi H and
Hada T: Induction of apoptosis in hepatocellular carcinoma cell
lines by emodin. Jpn J Cancer Res. 93:874–882. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
He X, Zhou A, Lu H, Chen Y, Huang G, Yue
X, Zhao P and Wu Y: Suppression of mitochondrial complex I
influences cell metastatic properties. PLoS One. 8:e616772013.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kalakonda S, Nallar SC, Jaber S, Keay SK,
Rorke E, Munivenkatappa R, Lindner DJ, Fiskum GM and Kalvakolanu
DV: Monoallelic loss of tumor suppressor GRIM-19 promotes
tumorigenesis in mice. Proc Natl Acad Sci USA. 110:E4213–E4222.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li LD, Sun HF, Liu XX, Gao SP, Jiang HL,
Hu X and Jin W: Down-regulation of NDUFB9 promotes breast cancer
cell proliferation, metastasis by mediating mitochondrial
metabolism. PLoS One. 10:e01444412015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Su CY, Chang YC, Yang CJ, Huang MS and
Hsiao M: The opposite prognostic effect of NDUFS1 and NDUFS8 in
lung cancer reflects the oncojanus role of mitochondrial complex I.
Sci Rep. 6:313572016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Suhane S, Berel D and Ramanujan VK:
Biomarker signatures of mitochondrial NDUFS3 in invasive breast
carcinoma. Biochem Biophys Res Commun. 412:590–595. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Pelicano H, Xu RH, Du M, Feng L, Sasaki R,
Carew JS, Hu Y, Ramdas L, Hu L, Keating MJ, et al: Mitochondrial
respiration defects in cancer cells cause activation of Akt
survival pathway through a redox-mediated mechanism. J Cell Biol.
175:913–923. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Agarwal E, Brattain MG and Chowdhury S:
Cell survival and metastasis regulation by Akt signaling in
colorectal cancer. Cell Signal. 25:1711–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Roy HK, Olusola BF, Clemens DL, Karolski
WJ, Ratashak A, Lynch HT and Smyrk TC: AKT proto-oncogene
overexpression is an early event during sporadic colon
carcinogenesis. Carcinogenesis. 23:201–205. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ericson K, Gan C, Cheong I, Rago C,
Samuels Y, Velculescu VE, Kinzler KW, Huso DL, Vogelstein B and
Papadopoulos N: Genetic inactivation of AKT1, AKT2, and PDPK1 in
human colorectal cancer cells clarifies their roles in tumor growth
regulation. Proc Natl Acad Sci USA. 107:2598–2603. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen C, Pore N, Behrooz A, Ismail-Beigi F
and Maity A: Regulation of glut1 mRNA by hypoxia-inducible
factor-1. Interaction between H-ras and hypoxia. J Biol Chem.
276:9519–9525. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen P, Zhu J, Liu DY, Li HY, Xu N and Hou
M: Over-expression of survivin and VEGF in small-cell lung cancer
may predict the poorer prognosis. Med Oncol. 31:7752014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Loncaster JA, Harris AL, Davidson SE,
Logue JP, Hunter RD, Wycoff CC, Pastorek J, Ratcliffe PJ, Stratford
IJ and West CM: Carbonic anhydrase (CA IX) expression, a potential
new intrinsic marker of hypoxia: Correlations with tumor oxygen
measurements and prognosis in locally advanced carcinoma of the
cervix. Cancer Res. 61:6394–6399. 2001.PubMed/NCBI
|
|
60
|
Shweiki D, Itin A, Soffer D and Keshet E:
Vascular endothelial growth factor induced by hypoxia may mediate
hypoxia-initiated angiogenesis. Nature. 359:843–845. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li F, Wang Y, Zeller KI, Potter JJ, Wonsey
DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA and Dang CV: Myc
stimulates nuclearly encoded mitochondrial genes and mitochondrial
biogenesis. Mol Cell Biol. 25:6225–6234. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dang CV, Kim JW, Gao P and Yustein J: The
interplay between MYC and HIF in cancer. Nat Rev Cancer. 8:51–56.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kumari S, Badana AK, Gavara MM, Gugalavath
S and Malla R: Reactive oxygen species: A key constituent in cancer
survival. Biomark Insights. 13:11772719187553912018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Che M, Wang R, Li X, Wang HY and Zheng
XFS: Expanding roles of superoxide dismutases in cell regulation
and cancer. Drug Discov Today. 21:143–149. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mowers EE, Sharifi MN and Macleod KF:
Autophagy in cancer metastasis. Oncogene. 36:1619–1630. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH,
Kim SS, Yoo NJ and Lee SH: Expression of beclin-1, an
autophagy-related protein, in gastric and colorectal cancers.
APMIS. 115:1344–1349. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sakitani K, Hirata Y, Hikiba Y, Hayakawa
Y, Ihara S, Suzuki H, Suzuki N, Serizawa T, Kinoshita H, Sakamoto
K, et al: Inhibition of autophagy exerts anti-colon cancer effects
via apoptosis induced by p53 activation and ER stress. BMC Cancer.
15:7952015. View Article : Google Scholar : PubMed/NCBI
|