Introduction
Multiple myeloma (MM) is one of the most common
hematologic malignancies and accounts for ~1% of all cancer cases
(1). Although MM has been studied
for 150 years, it still remains an incurable disease (2). In recent years, immunomodulatory agents
such as lenalidomide and proteasome inhibitors (PIs), including
bortezomib, have substantially improved the survival of patients
with MM. However, patients with MM may acquire drug resistance and
thus exhibit recurrence and progression (3). Therefore, investigating the pathogenic
mechanisms underlying the development and progression of MM is
vital, particularly in regard to proliferation and apoptosis, to
identify novel treatment strategies.
Interleukin-6 (IL-6) is a pivotal growth factor that
mediates the proliferation of MM via autocrine mechanisms when
released from MM cells or via paracrine mechanisms when released
from stromal cells (4). IL-6 exerts
its biological activities by binding to its receptor (IL-6R). The
levels of both IL-6 and IL-6R are upregulated in patients with MM
(5). IL-6R leads to activation of
the JAK-STAT signaling pathway to regulate the proliferation and
apoptosis of myeloma cells (6).
However, the mechanism by which IL-6R is upregulated in MM is still
unknown.
MicroRNAs (miRNAs/miRs) bind to the 3′-untranslated
regions (3′-UTRs) of specific target mRNAs, thereby inhibiting the
expression of the target genes post-transcriptionally. miR-451a was
discovered in 2005 and is located on chromosome 17qll.2 (7). miR-451a acts as a tumor suppressor and
is associated with several types of cancer, including lung cancer
(8), hepatocellular carcinoma
(9), glioma (10) and osteosarcoma (11), and inhibits the proliferation,
migration and invasion of tumors (12). miR-451a targets IL-6R in several
types of solid tumors and inhibits tumor proliferation, migration
and angiogenesis through the IL-6R signaling pathway (13). However, whether miR-451a also
inhibits tumor growth in MM and targets IL-6R remains unknown.
Additionally, the clinical significance of miR-451a in MM has not
been established. The aim of the present study was to address these
gaps in our knowledge.
Materials and methods
Ethics statement
The present study was approved by the Institutional
Review Board of Sichuan Provincial Peoples' Hospital of China.
Written informed consent was obtained from each eligible patient,
and the study was performed in accordance with the guidelines
stated in the Declaration of Helsinki.
Study subjects
The diagnostic criteria for active MM were based on
the International myeloma working group (IMWG) guidelines (14). Because the study did not involve any
interventions, a sample meeting the diagnostic criteria was
considered acceptable. The sub-typing criteria were based on
subtypes of abnormal proliferative immunoglobulin (14). The staging criteria were based on the
Revised International Staging System (R-ISS) (14). In total, 10 unrelated control
subjects from among donors were recruited. Disease responses were
assessed according to the IMWG guidelines (15). A total of 66 patients with MM (37 men
and 29 women; age range, 38–88 years; median age, 63.0 years) and
10 healthy controls (six men and four women; age range, 31–83
years; median age, 59 years) were enrolled in the present study
between January 2017 and December 2018.
Sampling
Bone marrow (BM) aspirates were collected from
normal controls and patients after obtaining informed consent and
were subjected to red blood cell lysis using Versalyse Lysing
Solution (cat. no. A09777; Beckman Coulter, Inc.) and mononuclear
cell isolation. Mononuclear cells were isolated using a Ficoll
gradient (density 1.077 g/ml) with Lymphoprep (cat. no. 07801;
STEMCELL Technologies, http://www.stemcell.com/products/lymphoprep.html).
Subsequently, plasma cells (PCs) were incubated and separated by
positive selection using CD138-coated magnetic beads (Miltenyi
Biotec, GmbH) according to the manufacturer's protocol. RNA from
PCs was extracted to analyze miR-451a levels, and total protein was
extracted to assess IL-6R. The remaining BM was used for
multiparameter flow cytometry (MFC). The patients were sampled at
initial diagnosis and at the indicated time points during the
follow-up.
Cell culture
U266 and INA-6 cells were purchased from the
National Infrastructure of Cell Line resource and kindly provided
by Dr. Gramatzki (Laboratory of Hematology, GIGA-I3, University of
Liège, Belgium), respectively, and were maintained in RPMI 1640
medium with GlutaMAX™ supplemented with 10% fetal calf serum and 1%
penicillin/streptomycin. All these reagents were purchased from
Thermo Fisher Scientific, Inc.
miR-451a transfection
For miR-451a mimic transfection, an miR-451a mimic
(5′-AAACCGUUACCAUUACUGAGUU-3′; 50 nM; Guangzhou RiboBio Co., Ltd.)
and the corresponding negative control (50 nM; Guangzhou RiboBio
Co., Ltd.) were separately transfected into 293T cells (purchased
from China Center for Type Culture Collection) in the absence of
any other treatments. Transfection was performed using
Lipofectamine 3000 (Thermo Fisher Scientific, Inc.). miR-451a
expression was measured via reverse transcription-quantitative
(RT-q)PCR analysis to confirm successful transfection, and the
subsequent effect was determined only in cells expressing miR-451a
48 h post-transfection.
RT-qPCR
Total RNA was isolated from the indicated cells
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Then, miR-451a was
reverse transcribed with a Bulge-Loop™ miRNA RT-qPCR Starter kit
(Guangzhou RiboBio Co., Ltd.). The reaction conditions were as
follows: 42°C for 60 min and 70°C for 10 min. Relative
quantification was performed via qPCR using the SYBR Green PCR kit
(Guangzhou RiboBio Co., Ltd.). The following thermocycling
conditions were used: Initial denaturation at 95°C for 10 min,
followed by 40 cycles at 95°C for 2 sec, 60°C for 20 sec and 70°C
for 10 sec. The following primer sequences were used: miR-451a
forward, 5′-ACCGTTACCATTACT-3′ and reverse, 5′-CTCACACGACTCACGA-3′.
cDNA was synthesized using EasyScript One-Step gDNA Removal and
cDNA Synthesis SuperMix (TransGen Biotech). The RT protocol was as
follows: 45°C for 15 min, 85°C for 5 sec, and end at 4°C. qPCR was
performed to analyze IL-6R expression using SYBR Premix Ex Taq
(Takara Bio, Inc.). The qPCR was performed with an initial
denaturation step at 95°C for 5 min; 39 cycles at 95°C for 10 sec
and 60°C for 30 sec; and a final stage at 95°C for 15 sec, 60°C
elongation for 1 min and 95°C for 15 sec. The following primer
sequences were used: IL-6R forward, 5′-CCTCTGCATTGCCATTGTTC-3′ and
reverse, 5′-GAGATGAGAGGAACAAGCAC-3′; BAX forward,
5′-ACCATCATGGGCTGGACATTG-3′ and reverse,
5′-CTGGAGACAGGGACATCAGTCG-3′; and Bcl-2 forward,
5′-ACTTCGCCGAGATGTCCAG-3′ and reverse, 5′-CCACAATCCTCCCCCAGTTCA-3′.
Quantification was performed using the 2−∆∆Cq method
(16), and the expression values
were normalized to the expression of GAPDH as the loading control.
Each sample was run in triplicate. The expression levels of the
target miRNAs and a control gene, U6, were measured simultaneously.
All reactions, including the no-template controls, were performed
in triplicate.
Western blotting
To assess changes in cellular protein levels, MM
cells and PCs from patients were harvested after 72 h, washed with
ice-cold PBS and resuspended in 100 µl lysis buffer (99 µl RPMI and
1 µl PMSF). After 2 h on ice, the solution was centrifuged at
12,000 × g at 4°C for 20 min. The supernatants were stored at −80°C
until required. Proteins (20 µg/lane) were subjected to
electrophoresis on a 10% SDS-gel, resolved using SDS-PAGE and
transferred to nitrocellulose membranes (Bio-Rad Laboratories,
Inc.). The membranes were blocked with 5% nonfat dried milk in
Tris-buffered saline with 0.05% Tween-20 and incubated with the
following primary antibodies overnight at 4°C: Rabbit anti-IL-6R
(1:400; cat. no. ab128008; Abcam), rabbit anti-BAX (1:1,000; cat.
no. ab32503; Abcam), rabbit anti-Bcl-2 (1:1,000; cat. no. ab185002;
Abcam), rabbit anti-JAK2 (1:5,000; cat. no. ab108596; Abcam),
rabbit anti-p-JAK2 (1:1,000; cat. no. ab32101; Abcam), mouse
anti-STAT3 (1:5,000; cat. no. ab119352; Abcam) and rabbit
anti-p-STAT3 (1:1,000, ab76315, Abcam). Subsequently, the membranes
were incubated with horseradish peroxidase-conjugated secondary
antibodies (1:2,000; cat. nos. ab6789 and ab6721; Abcam) for 2 h at
room temperature and visualized using enhanced chemiluminescence
reagent (Amersham; Cytiva).
Measurement of serum IL-6 levels
The levels of IL-6 in serum were measured using an
IL-6 ELISA kit (cat. no. ab46027; Abcam) according to the
manufacturer's protocol. After color development was stopped, the
absorbance was measured using a computer-connected microtiter plate
reader at 450 nm. The sensitivity for IL-6 detection was 2
pg/ml.
Flow cytometry analysis
A fixative-free erythrocyte lysis (Beckman Coulter,
Inc.) method was used for phenotypic characterization of most
participants. Intraprep permeabilization reagent (cat. no. A07803;
Beckman Coulter, Inc.) was used to assess intracellular
immunoglobulin levels at room temperature. The number and viability
of cells obtained were assessed using trypan blue (cat. no. T8070;
Solarbio) for 10 min at room temperature and observed under a light
microscope (Olympus Corporation; magnification, 10×10) by counting
the number of unstained cells. If the viability of recovered cells
was ≥90%, 2×106 cells were stained with two independent
6-color panels (1×106 cells each) at room temperature
for 30 min: One tube contained CD19-FITC, CD20-PE, CD56-ECD,
CD38-PECY5.5, CD138-APC and CD45-PECy7, and the other tube
contained CD19-FITC, CD117-PE, CD56-ECD, CD38-PECY5.5, CD138-APC
and CD45-PECy7. The intracytoplasmic panel consisted of the markers
Cyκ-APC, Cyλ-APC750, CD19-PE, CD38-FITC and CD138-APC (all
antibodies were purchased from Beckman Coulter, Inc.). A total of
1×106 cells were suspended in a final volume of 200
µl/tube and labeled. A minimum of 5×106 events were
recorded per tube in a Navios cytometer (Beckman Coulter, Inc.)
with a set forward scatter (FSC) threshold of 10,000 within a
maximum of 1 h after the final washing step of the preparation
process. Data analysis was performed using Kaluza (version no. 2.1;
Beckman Coulter, Inc.). After exclusion of cell doublets and
debris, PCs from the remaining BM cells were mainly selected using
CD138/CD38, CD45/CD38 and side scatter/FSC bivariate dot plots.
CD19 and CD56 were predicted to be applicable to ≥90% of patients,
and the markers CD20 and CD117 were considered likely to increase
the percentage to ≥95% of patients. Assessment of cytoplasmic κ/λ
expression by flow cytometry is important to demonstrate clonality
(with a value >3 or <0.33 representing the monoclonal light
chain) (15,17). Abnormal PCs were identified as CD45
negative/weakly positive, CD19 negative, strongly CD56 positive,
CD117 positive, CD20 positive and cytoplasmic κ or λ monoclonal
positive.
IL-6R expression in PCs from patients and normal
controls was detected by MFC according to the manufacturer's
protocol (cat. no. ab27321; Abcam). To analyze the proliferative
capacity, Ki-67 expression was detected in U266 cells using MFC
according to the manufacturer's protocol (cat. no. B27259;
BioLegend, Inc.).
U266 cells were cultured in 60-mm petri dishes for
48 h at 37°C with 5% CO2 and transfected with mimics or
inhibitors for another 48 h. The pro-apoptotic effects of miR-451a
on U266 cells were examined by MFC following double-staining with
Annexin V/FITC and propidium iodide using an Annexin V Apoptosis
Detection kit (eBioscience; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
Immunohistochemistry
Paraffin-embedded sections of patients' BM biopsies
in the department of pathology were deparaffinized using xylene and
ethanol. Then, the sections were blocked by 2% bovine serum protein
(Sigma-Aldrich) for 30 min at room temperature.
Immunohistochemistry was performed on BM sections using the labeled
Streptavidin-Horseradish Peroxidase (HRP) Conjugate (Invitrogen,
SA10001). Antigen retrieval techniques were applied as needed for
each specific antibody using IHC Antigen Retrieval Solution
(Invitrogen, 00-4956-58). The following antibodies were used: IL-6R
(1:4,000; cat. no. ab128008; Abcam) and CD138 (1:8,000; cat. no.
ab128936; Abcam) at room temperature for 1 h. Following the primary
incubation, tissue sections were incubated with a goat anti-rabbit
IgG (H&L) secondary antibody (1:500; ab97051; Abcam) at room
temperature for 1 h. DAB was used as a substrate, and the positive
signal was dark brown in color (Invitrogen, TA-060-QHDX). Samples
were observed under a light microscope (Olympus Corporation;
magnifications, 20×10, 10×10).
Cell viability assay
A Cell Counting Kit-8 assay was used to determine
cell viability (Dojindo Molecular Technologies, Inc.) according to
the manufacturer's protocol. Cell proliferation was assessed using
BrdU staining (Roche Diagnostics, GmbH) at room temperature for
approximately 2.5 h. The absorbance was assessed using a microplate
reader (BioTek Instruments, Inc.) at a wavelength of 370 nm.
Luciferase reporter assay
The online software TargetScan (TargetScan Human
7.0, http://www.targetscan.org/vert_70) was used to
determine the association between miR-451a and IL-6R. A human IL-6R
3′-UTR with a mutation in the miR-451a seed sequence was amplified
and inserted into the firefly and Renilla luciferase reporter
vector pmiR-RB-REPORT (Guangzhou RiboBio Co., Ltd.) to form a
mutant (Mut) vector. Cells were transfected with the luciferase
reporter vectors (30 ng). Firefly and Renilla luciferase activity
levels were consecutively measured according to the manufacturer's
protocol (Promega Corporation) 24 h after transfection. The Renilla
luciferase signal was normalized to the respective firefly
luciferase signal.
Statistical analysis
Statistical analysis was performed using SPSS
version 19.0 (IBM Corp.). The χ2 test was used to
determine whether sex and age ratios significantly differed between
the experimental group and the controls. Data are presented as the
means ± standard deviations of three independent repeats. A one-way
ANOVA with post hoc Tukey's test was used to compare differences
between multiple groups. Correlation coefficients were calculated
using Spearman's correlation test to analyze correlations between
two variables. The χ2 test was used to compare
frequencies between two groups. A two-sided P<0.05 was
considered indicative of a statistically significant
difference.
Results
miR-451a expression in the BM of
patients with MM at diagnosis
Among the patients, 15 (22.7%) had R-ISS stage I MM,
17 (25.8%) had R-ISS stage II MM, and 34 (51.5%) had R-ISS stage
III MM. These patients received immunomodulatory drugs or PIs for
induction therapy without autologous stem cell transplantation.
miR-451a expression was low in MM patients at 0.39±0.13-times that
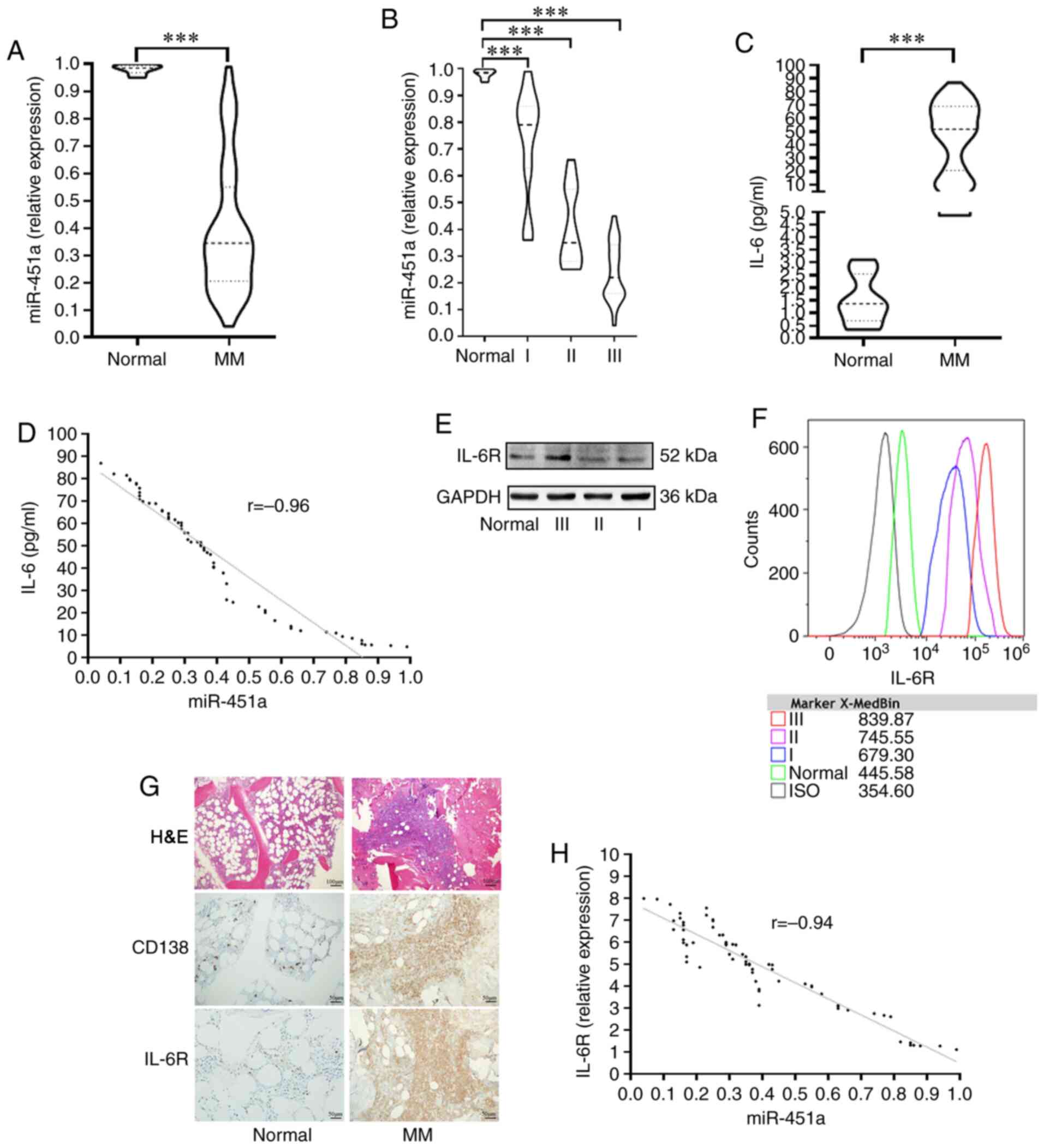
in normal subjects (t=7.66, P<0.0001) (Fig. 1A). Among the 66 patients with MM, the
median level of miR-451a in patients with R-ISS stage I MM was
0.72±0.05-times that in normal subjects. The median level in
patients with R-ISS stage II MM was 0.40±0.03-times that in normal
subjects. R-ISS stage III patients showed the lowest level
(0.24±0.01-times that in normal subjects; F=101.14, P<0.0001;
Fig. 1B). The concentration of IL-6
in MM patients was significantly higher than that in the normal
controls (t=5.62, P<0.0001; Fig.
1C) and was inversely related to the levels of miR-451a
(r=−0.96, P<0.0001; Fig. 1D).
Compared with those from the controls, PCs from MM
patients, particularly R-ISS stage III patients, showed upregulated
expression of IL-6R as indicated by western blot analysis (Fig. 1E). Furthermore, the MFC results
showed differences in the expression intensity of IL-6R among the
different R-ISS stages, with stage III patients exhibiting the
highest intensity (Fig. 1F). BM
histopathology showed that the tumor cells formed foci and were
strongly positive for both CD138 (plasma positive) and IL-6R
(membrane positive) (Fig. 1G). IL-6R
expression was negatively correlated with the level of miR-451a
(r=−0.94, P<0.0001; Fig. 1H).
The mechanism underlying miR-451a
regulation of myeloma cell proliferation and apoptosis
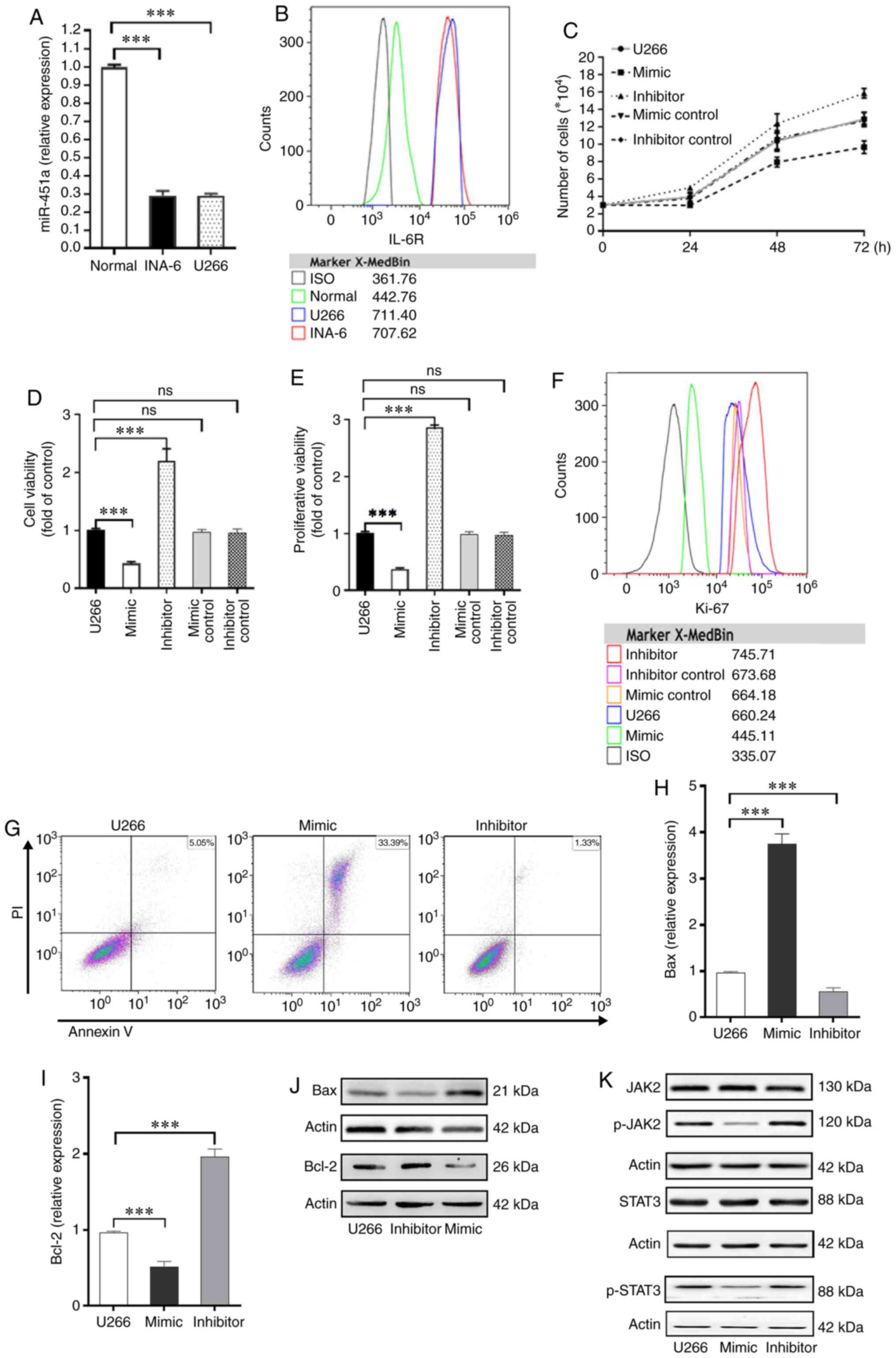
The levels of miR-451a among INA-6 cells, U266 cells
and PCs from normal controls were compared. The expression of
miR-451a was 0.28±0.02-times normal levels in INA-6 cells and
0.29±0.03-times normal levels in U266 cells (F=478.13, P<0.0001;
Fig. 2A). The expression of IL-6R
was analyzed by MFC. IL-6R expression on the surfaces of U266 and
INA-6 cells was significantly higher than on the surfaces of normal
cells. The median fluorescence intensity (MFI) of U266 cells was
711.4, the MFI of INA-6 cells was 707.62, and the MFI of normal
cells was only 442.76 (Fig. 2B).
In U266 cells transfected with the miR-451a mimic or
miR-451a inhibitor, miR-451a expression was increased or reduced,
respectively. Furthermore, the levels of miR-451a in cells
transfected with the mimic control and inhibitor control did not
differ from those in the untransfected U266 cells (Fig. S1). Among the groups, U266 cells
transfected with the miR-451a inhibitor exhibited the highest
proliferation rate after 72 h, whereas the miR-451a
mimic-transfected cells exhibited the lowest levels of
proliferation. Cells treated with the mimic control and inhibitor
control did not significantly differ from the untransfected U266
cells with regard to proliferation (Fig.
2C). Treatment with the miR-451 mimic decreased the viability
of U266 cells by 57.3%±3.1% and decreased the proliferative ability
of U266 cells by 62.6%±2.6%. The miR-451a inhibitor showed the
opposite effects (F=54.628, P<0.0001, and F=2749.244,
P<0.0001, respectively). The groups treated with the mimic
control and inhibitor control did not significantly differ from the
untransfected U266 cells (P>0.05; Fig. 2D and E). Using MFC, the miR-451a
mimic was found to significantly reduce the intensity of Ki-67
expression in U266 cells, whereas transfection with the miR-451a
inhibitor significantly increased Ki-67 expression. The mimic
control and inhibitor control did not affect the intensity of Ki-67
expression in U266 cells compared with the untransfected U266 cells
(Fig. 2F).
The miR-451a mimic also significantly increased the
rate of apoptosis in U266 cells from 5.05% (control) to 33.39%
(transfected U266 cells) (Fig. 2G).
The expression of the apoptotic protein Bax was significantly
upregulated in the mimic-transfected cells, and the expression of
the antiapoptotic protein, Bcl-2, was significantly decreased. When
miR-451a was inhibited, the opposite trend was observed in the mRNA
(Bcl-2, F=172.577, P<0.0001; Bax, F=112.922, P<0.0001;
Fig. 2H and I) and protein
expression levels of Bcl-2 and Bax (Fig.
2J). The JAK2/STAT3 pathway is the classical pathway stimulated
by IL-6, and its cancer-promoting effects are well established
(18,19). Thus, the expression levels of JAK2
and STAT3 and their phosphorylated forms were assessed. Total JAK2
and STAT3 levels among the three groups did not differ. However,
the levels of phospho-(p-)JAK2 and p-STAT3 were significantly lower
in U266 cells treated with the miR-451a mimic than in the control
cells (Fig. 2K).
miR-451a target verification and
assessment of the target gene IL-6R
3′-UTR luciferase reporter assays were used to
assess the binding of miR-451a to its targets. Luciferase activity
in cells containing the wild-type (WT) vector was reduced in cells
transfected with miR-451a and the 3′UTR of IL-6R by 0.36±0.07-fold.
Cells transfected with a miR-451a Mutant (Mut) vector did not
exhibit a decrease in luciferase activity (Fig. 3A). Furthermore, the miR-451a mimic
control had no effect on the luciferase activity of the WT vector
or Mut vector. These results showed that miR-451a directly
repressed IL-6R by binding to the 3′-UTR. Additionally, in cells
transfected with the miR-451a mimic, IL-6R mRNA (Fig. 3B) and protein expression levels
(Fig. 3C) were significantly
decreased.
Discussion
IL-6 plays a crucial role in the pathogenesis of MM,
and the discovery of the involvement of IL-6 in MM is of notable
significance (20). IL-6 is critical
for the growth and survival of malignant PCs, and several factors
and mechanisms can increase IL-6 levels (21). However, IL-6 must bind to its
receptor to elicit its effects as IL-6R transduces the IL-6 signal
(5). The molecular mechanisms
regulating IL-6R expression have not yet been fully elucidated
(22). In the present study,
miR-451a was shown to target IL-6R to offset the effects induced by
IL-6. To the best of our knowledge, the present study is the first
to highlight miR-451a as a vital antitumor factor in MM.
Abnormal expression of miR-451a is negatively
correlated with the degree of malignancy in several types of
malignant tumors (23). Recently,
miR-451a was shown to be significantly downregulated in patients
with MM and was correlated with a poor clinical prognosis (24), which is consistent with the results
of the present study. However, the mechanisms by which miR-451a
expression is downregulated remain unclear. In addition, long
non-coding (lnc)RNAs have been shown to sponge miRNAs, resulting in
a decrease in miRNA expression, and LINC00657, AC084082.3 and
LINC00657 have been found to interact with miR-451a in BM (25). However, in MM, the expression levels
of the lncRNAs that sponge miR-451a are unknown. IL-6R was
confirmed to be a target of miR-451a in the present study,
indicating that miR-451a was associated with the IL-6R/JAK2/STAT3
pathway. Whether lncRNAs that sponge miR-451a are also associated
with this pathway, which exhibits a high level of activity in MM,
will be addressed in future studies.
The downstream mechanism by which downregulation of
miR-451a influences MM is unknown. In osteosarcoma, miR-451a
inhibits the proliferation, migration and angiogenesis of cancer
cells by silencing IL-6R (26). In
the present study, miR-451a levels were negatively correlated with
the R-ISS stage and IL-6 and IL-6R levels. The dual-luciferase
reporter assays confirmed the relationship between miR-451a and
IL-6R. Therefore, miR-451a may function through the
IL-6R/JAK2/STAT3 pathway. Further analyses verified that miR-451a
altered the levels of pJAK2 and pSTAT3 (active forms) to induce
apoptosis of myeloma cells with no changes in the levels of total
JAK2 and STAT3; this mechanism may apply to clinical
transformation. Thus, miR-451a may serve as a potential biomarker
for the prediction of patient outcomes. In addition to signaling
via the JAK2/STAT3 pathway, IL-6 can also promote the proliferation
of myeloma cells through the Ras/MAPK pathway (27) and reduce apoptosis of MM cells by
regulating the PI3K/AKT pathway (28). Furthermore, IL-6 interacts with VEGF
to promote angiogenesis, migration and invasion (29). Several pathways activated by IL-6
result in activation of NF-кB (30).
Therefore, miR-451a may exert its effects through other mechanisms
and signaling pathways, which may be tumor-type dependent. However,
these hypotheses must be confirmed in future studies.
In conclusion, the present study showed that
miR-451a specifically silences IL-6R-induced myeloma cell apoptosis
and promotes JAK2/STAT3 pathway inactivation to exert its antitumor
effects. Thus, miR-451a may be an attractive alternative target for
the mapping of tumor loads in real time and for MM treatment via
novel strategies.
Supplementary Material
Supporting Data
Acknowledgements
The authors of the present study would like to thank
Mr. Yi Shi (Sichuan Academy of Medical Sciences and Sichuan
Provincial People's Hospital, School of Medicine, University of
Electronic Science and Technology of China), for his ongoing advice
and assistance in analyzing the data. The authors would also like
to thank Dr Gramatzki (Laboratory of Hematology, University of
Liège, Belgium), for kindly providing the cell line used in the
present study.
Funding
The present study was supported by the Health
Commission of Sichuan Province (grant no. 150193), the Commission
of the Cardre Health Care in Sichuan Province (grant no. 2017-228),
the Science & Technology Department of Sichuan Province (grant
no. 19YJ0593), the Sichuan Provincial People's Hospital (grant no.
2018LY03), the Chengdu Science and Technology Bureau (grant no.
2015-HM01-00470-SF) and the Science & Technology Department of
Sichuan Province (grant no. 2020YFS0433).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
LZ, ZYX, XJ, YH, JBZ, TJ and JC participated in the
design and interpretation of the studies, the analysis of the data
and in editing the manuscript. LZ, ZYX and XJ performed the
experiments. YH and JBZ performed the statistical analysis and the
follow-up of the patients. LZ, TJ, and JC wrote the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of Sichuan Provincial Peoples' Hospital of China
(Chengdu, Sichuan; approval no. 2018-49). Written informed consent
was obtained from each eligible patient, and the entire study was
performed in accordance with the guidelines stated in the
Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chang SH, Luo S, Thomas TS, O'Brian KK,
Colditz GA, Carlsson NP and Carson KR: Obesity and the
transformation of monoclonal gammopathy of undetermined
significance to multiple myeloma: A population-based cohort study.
J Natl Cancer Inst. 109:djw2642016. View Article : Google Scholar
|
|
2
|
Popovic M, Lao N, Bedard G, Zeng L, Zhang
L, Cella D, Beaumont JL, Chiu N, Chiu L, Lam H, et al: Quality of
life in patients with advanced cancer using the functional
assessment of cancer therapy-general assessment tool: A literature
review. World J Oncol. 4:8–17. 2013.PubMed/NCBI
|
|
3
|
Tandon N, Rajkumar SV, LaPlant B,
Pettinger A, Lacy MQ, Dispenzieri A, Buadi FK, Gertz MA, Hayman SR,
Leung N, et al: Clinical utility of the revised international
staging system in unselected patients with newly diagnosed and
relapsed multiple myeloma. Blood Cancer J. 7:e5282017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Furukawa M, Ohkawara H, Ogawa K, Ikeda K,
Ueda K, Shichishima-Nakamura A, Ito E, Imai JI, Yanagisawa Y, Honma
R, et al: Autocrine and paracrine interactions between multiple
myeloma cells and bone marrow stromal cells by growth
arrest-specific gene 6 cross-talk with interleukin-6. J Biol Chem.
292:4280–4292. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mishra AK and Dingli D: Metformin inhibits
IL-6 signaling by decreasing IL-6R expression on multiple myeloma
cells. Leukemia. 33:2695–2709. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Oliveira MB, Fook-Alves VL, Eugenio
AIP, Fernando RC, Sanson LFG, de Carvalho MF, Braga WMT, Davies FE
and Colleoni GWB: Anti-myeloma effects of ruxolitinib combined with
bortezomib and lenalidomide: A rationale for JAK/STAT pathway
inhibition in myeloma patients. Cancer Lett. 403:206–215. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Altuvia Y, Landgraf P, Lithwick G, Elefant
N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T and Margalit H:
Clustering and conservation patterns of human microRNAs. Nucleic
Acids Res. 33:2697–2706. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang R, Wang ZX, Yang JS, Pan X, De W and
Chen LB: MicroRNA-451 functions as a tumor suppressor in human
non-small cell lung cancer by targeting ras-related protein 14
(RAB14). Oncogene. 30:2644–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang JY, Zhang K, Chen DQ, Chen J, Feng
B, Song H, Chen Y, Zhu Z, Lu L, De W, et al: MicroRNA-451:
Epithelial-mesenchymal transition inhibitor and prognostic
biomarker of hepatocellular carcinoma. Oncotarget. 6:18613–18630.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim Y, Powathil G, Kang H, Trucu D, Kim H,
Lawler S and Chaplain M: Strategies of eradicating glioma cells: A
multi-scale mathematical model with MiR-451-AMPK-mTOR control. PLoS
One. 10:e1143702015.
|
|
11
|
Yuan J, Lang J, Liu C, Zhou K, Chen L and
Liu Y: The expression and function of miRNA-451 in osteosarcoma.
Med Oncol. 32:3242015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gits CM, van Kuijk PF, Jonkers MB, Boersma
AWM, Smid M, van Ijcken WF, Coindre JM, Chibon F, Verhoef C,
Mathijssen RHJ, et al: MicroRNA expression profiles distinguish
liposarcoma subtypes and implicate miR-145 and miR-451 as tumor
suppressors. Int J Cancer. 135:348–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu X, Zhang A, Xiang J, Lv Y and Zhang X:
miR-451 acts as a suppressor of angiogenesis in hepatocellular
carcinoma by targeting the IL-6R-STAT3 pathway. Oncol Rep.
36:1385–1392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chng WJ, Dispenzieri A, Chim CS, Fonseca
R, Goldschmidt H, Lentzsch S, Munshi N, Palumbo A, Miguel JS,
Sonneveld P, et al: IMWG consensus on risk stratification in
multiple myeloma. Leukemia. 28:269–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kumar S, Paiva B, Anderson KC, Durie B,
Landgren O, Moreau P, Munshi N, Lonial S, Bladé J, Mateos MV, et
al: International myeloma working group consensus criteria for
response and minimal residual disease assessment in multiple
myeloma. Lancet Oncol. 17:e328–e346. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Flores-Montero J, Sanoja-Flores L, Paiva
B, Puig N, García-Sánchez O, Böttcher S, van der Velden VHJ,
Pérez-Morán JJ, Vidriales MB, García-Sanz R, et al: Next generation
flow for highly sensitive and standardized detection of minimal
residual disease in multiple myeloma. Leukemia. 31:2094–2103. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kolosenko I, Grander D and Tamm KP: IL-6
activated JAK/STAT3 pathway and sensitivity to Hsp90 inhibitors in
multiple myeloma. Curr Med Chem. 21:3042–3047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Wang J, Wang H, Yin G, Liu Y, Lei X
and Xiang M: REG3A accelerates pancreatic cancer cell growth under
IL-6-associated inflammatory condition: Involvement of a
REG3A-JAK2/STAT3 positive feedback loop. Cancer Lett. 362:45–60.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suematsu S, Hibi M, Sugita T, Saito M,
Murakami M, Matsusaka T, Matsuda T, Hirano T, Taga T and Kishimoto
T: Interleukin 6 (IL-6) and its receptor (IL-6R) in
myeloma/plasmacytoma. Curr Top Microbiol Immunol. 166:13–22.
1990.PubMed/NCBI
|
|
21
|
Piddock RE, Marlein CR, Abdul-Aziz A,
Shafat MS, Auger MJ, Bowles KM and Rushworth SA: Myeloma-derived
macrophage inhibitory factor regulates bone marrow stromal
cell-derived IL-6 via c-MYC. J Hematol Oncol. 11:66–69. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Birmann BM, Neuhouser ML, Rosner B,
Albanes D, Buring JE, Giles GG, Lan Q, Lee IM, Purdue MP, Rothman
N, et al: Prediagnosis biomarkers of insulin-like growth factor-1,
insulin, and interleukin-6 dysregulation and multiple myeloma risk
in the multiple myeloma cohort consortium. Blood. 120:4929–4937.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao S, Li J, Zhang G, Wang Q, Wu C, Zhang
Q, Wang H, Sun P, Xiang R and Yang S: Exosomal miR-451a functions
as a tumor suppressor in hepatocellular carcinoma by targeting
LPIN1. Cell Physiol Biochem. 53:19–35. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Meng YB, He X, Huang YF, Wu QN, Zhou YC
and Hao DJ: Long noncoding RNA CRNDE promotes multiple myeloma cell
growth by suppressing miR-451. Oncol Res. 25:1207–1214. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Balakrishnan I, Yang X, Brown J,
Ramakrishnan A, Torok-Storb B, Kabos P, Hesselberth JR and Pillai
MM: Genome-wide analysis of miRNA-mRNA interactions in marrow
stromal cells. Stem Cells. 32:662–673. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu SY, Deng SY, He YB and Ni GX: miR-451
inhibits cell growth, migration and angiogenesis in human
osteosarcoma via down-regulating IL-6R. Biochem Biophys Res Commun.
482:987–993. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gocke CB, McMillan R, Wang Q, Begum A,
Penchev VR, Ali SA, Borrello I, Huff CA and Matsui W: IQGAP1
scaffold-MAP kinase interactions enhance multiple myeloma
clonogenic growth and self-renewal. Mol Cancer Ther. 15:2733–2739.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mimura N, Hideshima T, Shimomura T, Suzuki
R, Ohguchi H, Rizq O, Kikuchi S, Yoshida Y, Cottini F, Jakubikova
J, et al: Selective and potent Akt inhibition triggers anti-myeloma
activities and enhances fatal endoplasmic reticulum stress induced
by proteasome inhibition. Cancer Res. 74:4458–4469. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Berenstein R, Nogai A, Waechter M, Blau O,
Kuehnel A, Schmidt-Hieber M, Kunitz A, Pezzutto A, Dörken B and
Blau IW: Multiple myeloma cells modify VEGF/IL-6 levels and
osteogenic potential of bone marrow stromal cells via
Notch/miR-223. Mol Carcinog. 55:1927–1939. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matthews GM, de Matos Simoes R, Dhimolea
E, Sheffer M, Gandolfi S, Dashevsky O, Sorrell JD and Mitsiades CS:
NF-κB dysregulation in multiple myeloma. Semin Cancer Biol.
39:68–76. 2016. View Article : Google Scholar : PubMed/NCBI
|