Introduction
Colorectal cancer (CRC) was the third most common
malignant type of tumor worldwide in 2017 (1–3), and
mainly results from metastasis (4,5). For
advanced CRC, 5-year survival rates have been reported to be
<50% in 2019 (6). Accumulating
evidence has suggested that aging and rapid population growth
contribute to high morbidity and mortality rates both in
high-income countries and, especially, in low- and middle-income
countries (7,8). According to a previous report, ~1.5
million individuals are diagnosed with CRC each year (9). Furthermore, the 5-year survival rate is
<44% after resection of hepatic colorectal metastases (10,11).
Although progress has been achieved in understanding the potential
biochemical mechanism of CRC (12,13),
current clinical diagnosis and gene-targeted or pharmacologic
therapies present numerous limitations in decreasing the mortality
rate of patients with CRC. Therefore, it is urgent to develop novel
and effective therapeutic strategies for CRC.
Neurensin-2 (NRSN2) is a small neuronal membrane
protein located in small vesicles of neural cells (14). A previous study has revealed that
NRSN2 is highly expressed in osteosarcoma tissues and promotes
osteosarcoma cell proliferation via dysregulation of the
PI3K/Akt/mTOR and Wnt/β-catenin signaling pathways (14). Furthermore, Tang et al
(15) reported that NRSN2 is closely
associated with the malignant phenotype of ovarian cancer. However,
contradictory roles of NRSN2 have been observed in non-small cell
lung cancer (NSCLC) and hepatocellular carcinoma (HCC). For
example, a previous study suggested that NRSN2 is upregulated in
malignant tissues and promotes NSCLC cell proliferation via the
PI3K/Akt/mTOR pathway (16). By
contrast, Wang et al (17)
demonstrated that NRSN2 is downregulated in HCC tissues and exerts
a suppressive role in HCC tumorigenesis. These findings indicate
that NRSN2 may serve a pivotal role in tumorigenesis, but its role
in CRC is yet to be elucidated.
To further investigate the potential value of NRSN2
in CRC, the present study selected the CRC SW620 cell line. The aim
of present study was to investigate the association between NRSN2
and CRC and further investigate the underlying mechanism of its
effect on CRC metastasis.
Materials and methods
Cell culture and transfection
The human CRC SW480, SW620 and HCT116 cell lines,
and the human normal colon epithelial FHC cell line were obtained
from the American Type Culture Collection (ATCC). Cells were
cultured in DMEM containing 10% FBS (both HyClone; Cytiva), 100
µg/ml streptomycin and 100 U/ml penicillin with a 5% CO2
atmosphere at 37°C.
SW620 cells were transfected in 6-well plates with
short hairpin (sh) RNA-NRSN2 (100 pmol), shRNA-NC (100 pmol) or
overexpression-NRSN2 (Ov-NRSN2; 100 pmol) and Ov-NC (100 pmol) for
modulating NRSN2 expression. shRNA-NRSN2 causes downregulation of
NRSN2 expression, while Ov-NRSN2 is a plasmid that specifically
elevates NRSN2 expression. The sequences are as follows:
shRNA-NRSN2-1 (5′-GCACCTCTTCTATGAGGACTG-3′); shRNA-NRSN2-2
(5′-GCAATCTTCTGTGCAGACTAT-3′); shRNA-NC (scrambled sequence,
5′-TTCTCCGAACGTGTCACGT-3′). Ov-NRSN2 plasmid (pRK5-NRSN2, forward,
5′-CTAGCTAGCATGATGCCGAGCTGCAATC-3′ and reverse,
5′-CGCGGATCCTCAGGAGTCCCTCTTGGG-3′) and Ov-NC (empty vector,
pRK5-control plasmid) were constructed by Shanghai GenePharma, Co.,
Ltd. After SW620 cells were grown to ~80% confluence, transfection
was performed using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C according to the
manufacturer's protocol. At 24 h post-transfection, cells were
harvested for subsequent experimentation and RT-qPCR was used to
detect the efficiency of transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. RT was performed to synthesize cDNA
with a Reverse Transcription kit (Thermo Fisher Scientific, Inc.),
and qPCR was performed using SYBR Green Real-Time PCR Master Mix
(Bio-Rad Laboratories, Inc.)and primers for NRSN2 and GAPDH (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The expression levels of NRSN2 were normalized to
those of GAPDH from the same RT sample and were detected using an
Applied Biosystems 7500 Real Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with a SYBR Green QPCR Master mix
(Thermo Fisher Scientific, Inc.). The relative expression levels
were calculated using the 2−ΔΔCq method (18). qPCR amplification parameters were:
annealing at 25°C for 10 min, extension at 42°C for 30 min;
inactivation at 85°C for 10 min, after extension of the detection
of fluorescence signals, a total of 40 cycles. Primer sequences
were as follows: NRSN2 forward, 5′-GATGGCAAGTGGTATGGGGTC-3′ and
reverse, 5′-CGAGGACAGGCTGATCTTCC-3′; and GAPDH forward,
5′-CCAGGTGGTCTCCTCTGA-3′ and reverse,
5′-GCTGTAGCCAAATCGTTGT-3′.
Western blot analysis
SW620 cells were harvested and lysed using RIPA
lysis buffer containing 1 mM PMSF, 1 µM Ben, 1 mM
Na3VO4, 10 µg/ml leupetin and 10 µg/ml
Aprotinin. Protein concentration was measured using a BCA protein
assay (Thermo Fisher Scientific, Inc.). Total protein (25 µg/lane)
was separated via 12% SDS-PAGE and transferred to PVDF membranes.
Membranes were blocked with 5% skimmed milk at room temperature for
2 h. Subsequently, appropriate primary antibody dilutions (1:1,000)
were prepared and incubated with membranes overnight at 4°C. The
antibodies included rabbit anti-NRSN2 (1:1,000; cat. no. ab237739;
Abcam), rabbit anti-MMP2 (1:1,000; cat. no. 4022; Cell Signaling
Technology, Inc.), rabbit anti-MMP9 (1:1,000; cat. no. 3852; Cell
Signaling Technology, Inc.), rabbit anti-SOX12 (1:1,000; cat. no.
SAB4502835; Sigma-Aldrich; Merck KGaA) and rabbit anti-GAPDH
(1:1,000; cat. no. 5014; Cell Signaling Technology, Inc.). After
washing with PBS three times, the membranes were incubated with
HRP-conjugated goat anti-rabbit IgG secondary antibodies (1:1,000;
cat. no. 21537; EMD Millipore) for 2 h at room temperature. The
membranes were visualized using Tanon-5200 Chemiluminescence Imager
(Tanon Science and Technology Co., Ltd.) with ECL western blotting
substrate (EMD Millipore). Band intensity was quantified using
ImageJ v1.8.0 (National Institutes of Health) and relative protein
levels were normalized to GAPDH.
Immunoprecipitation (IP)
The target proteins of NRSN2 were predicted using
the STRING database (https://string-db.org/cgi/network). For IP, cells
lysed with lysis buffer (20 mM Tris-HCl (pH 8.0), 300 mM NaCl, 1 mM
EDTA and 0.5% Nonidet P-40). After centrifugation (12,000 × g for
10 min at 4°C), 2 mg cell lysate supernatants were incubated with
Anti-FLAGM2 Magnetic beads (cat. no. M8823, Sigma-Aldrich; Merck
KGaA) or protein A/G Sepharose beads (20 µl, cat. no. sc-2003,
Santa Cruz Biotechnology, Inc.) according to manufacturer's
protocol and then with primary antibodies for 12 h at 4°C, and the
collected beads were washed four times with washing buffer (0.05%
Triton X-100 immunoprecipitation buffer without a protease
inhibitor cocktail), using IgG as a blank control.
Immunoprecipitates were boiled in sample loading buffer with 1X SDS
loading buffer (50 µl) for 5 min. Captured proteins on Sepharose
beads were then separated via 8% SDS-PAGE and western blot analysis
was performed as previously mentioned.
Cell viability
A Cell Counting Kit-8 assay (CCK-8; Thermo Fisher
Scientific, Inc.) was performed to detect the cell viability. In
brief, SW620 cells were seeded into a 96-well plate at a density of
5,000 cells/well. All experiments were performed ≥3 times in
triplicate. After transfection, cell viability at 24, 48 and 72 h
was determined using a CCK-8 assay. CCK-8 reagent (10 µl) was added
per well and incubated with the cells for another 2 h according to
the manufacturer's protocol.
Colony formation assay
SW620 cells (5×103 cells/ml) were seeded
in 6-well plates overnight at 37°C. After transfection and culture
for 14 days at 37°C, cell colonies were formed in the dish and were
fixed using 4% paraformaldehyde at room temperature for 15 min and
stained using 0.5% crystal violet solution at room temperature for
10 min to manually count the numbers of colonies containing >50
cells. A cell group containing >50 cells was identified as a
colony.
Wound-healing assay
Wound-healing assays were performed to detect the
migratory ability of SW620 cells. The transfected SW620 cells were
seeded in 6-well plates, allowed to reach ~80% confluence and
cultivated in serum-free medium for 24 h. The cell monolayers were
scratched vertically with a 200-µl RNase-free pipette tip. The
width of the wound was captured at the indicated time points (0 and
24 h) using an inverted fluorescence microscope (Olympus
Corporation) (magnification, ×100) to measure the percentage of the
area covered by the migrated cells relative to the control.
Transwell assay
Cell invasive ability was assessed using Transwell
assays. A Transwell chamber was precoated with Matrigel®
(BD Biosciences) at 37°C for 30 min. After transfection, SW620
cells (1×105 cells/ml in DMEM containing 1% bovine serum
albumin) were seeded into the upper chamber and cultured at 37°C
for 24 h. DMEM containing 20% FBS was added into the lower chamber
as a chemoattractant. Subsequently, the non-invasive cells were
removed using a cotton-tipped swab and the cells in the lower
chamber were fixed with 4% formaldehyde solution at room
temperature for 15 min and stained with 0.1% crystal violet at 25°C
for 20 min. The stained cells were observed and counted manually
under an inverted fluorescence microscope (Olympus Corporation)
with three random fields at a magnification of ×100.
Statistical analysis
Data are presented as the mean ± SEM. Statistical
analysis was performed using GraphPad Prism 5.0 (GraphPad Software,
Inc.) using a two-sided unpaired Student's t-test for comparison
between two independent groups or one-way ANOVA followed by Tukey's
post hoc test to analyze differences among multiple groups. A
Pearson's correlation analysis was used to analyze the relationship
between NRSN2 expression level and cell growth in SW620 cells.
P<0.05 was considered to indicate a statistically significant
difference.
Results
NRSN2 is highly expressed in CRC
cells
To investigate the role of NRSN2, RT-qPCR and
western blotting were performed to detect the expression levels of
NRSN2 in CRC cells. The human CRC SW480, SW620 and HCT116 cell
lines, and the human normal colon epithelial FHC cell line were
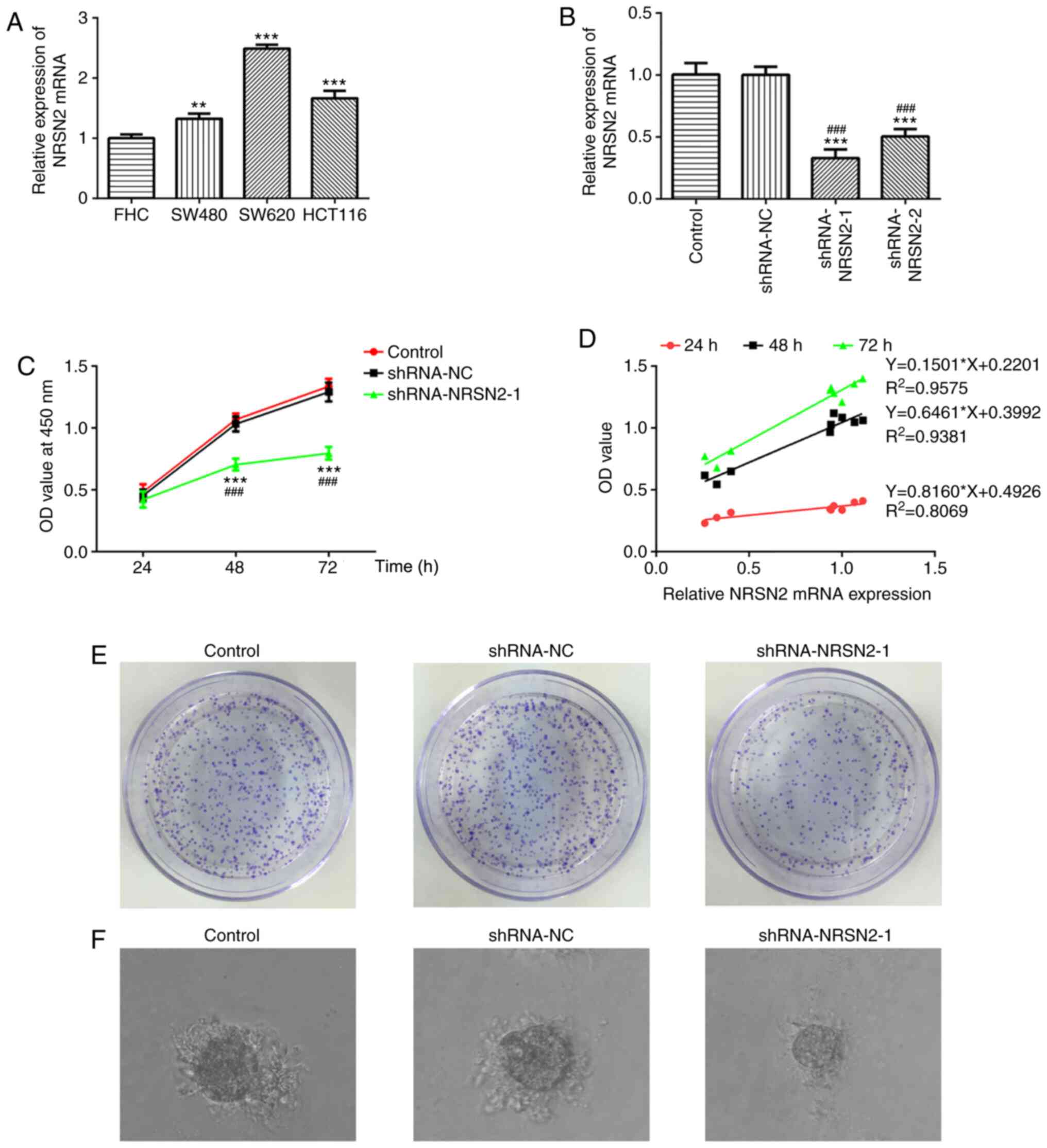
used. RT-qPCR results demonstrated that NRSN2 expression was
significantly increased in CRC cells, especially in SW620 cells,
compared with FHC cells (Fig. 1A).
Therefore, SW620 cells were selected for subsequent experiments.
The results suggested that dysregulated NRSN2 expression may be
closely associated with CRC pathogenesis.
Knockdown of NRSN2 inhibits
proliferation, invasion and migration of CRC cells
To study the effect of NRSN2 on proliferation,
invasion and migration of SW620 cells, shRNA-NRSN2 was constructed
to knock down NRSN2 expression. According to RT-qPCR analysis, both
shRNA-NRSN2-1 and shRNA-NRSN2-2 resulted in a significant decrease
in NRSN2 expression, compared with control and shRNA-NC groups
(Fig. 1B). Since shRNA-NRSN2-1
appeared to decrease NRSN2 expression to a greater extent compared
with shRNA-NRSN2-2, shRNA-NRSN2-1 was selected for further
experiments.
CCK-8 and colony formation assays were performed to
determine the viability and proliferative ability of CRC cells. The
correlation analysis was applied to analyze the relationship
between NRSN2 expression level and cell growth in SW620 cells
(Fig. 1D). Transfection with
shRNA-NRSN2-1 significantly decreased the viability (Fig. 1C) and proliferative ability (Fig. 1E) of SW620 cells at 48 and 72 h
compared with the control and shRNA-NC groups, as well as colony
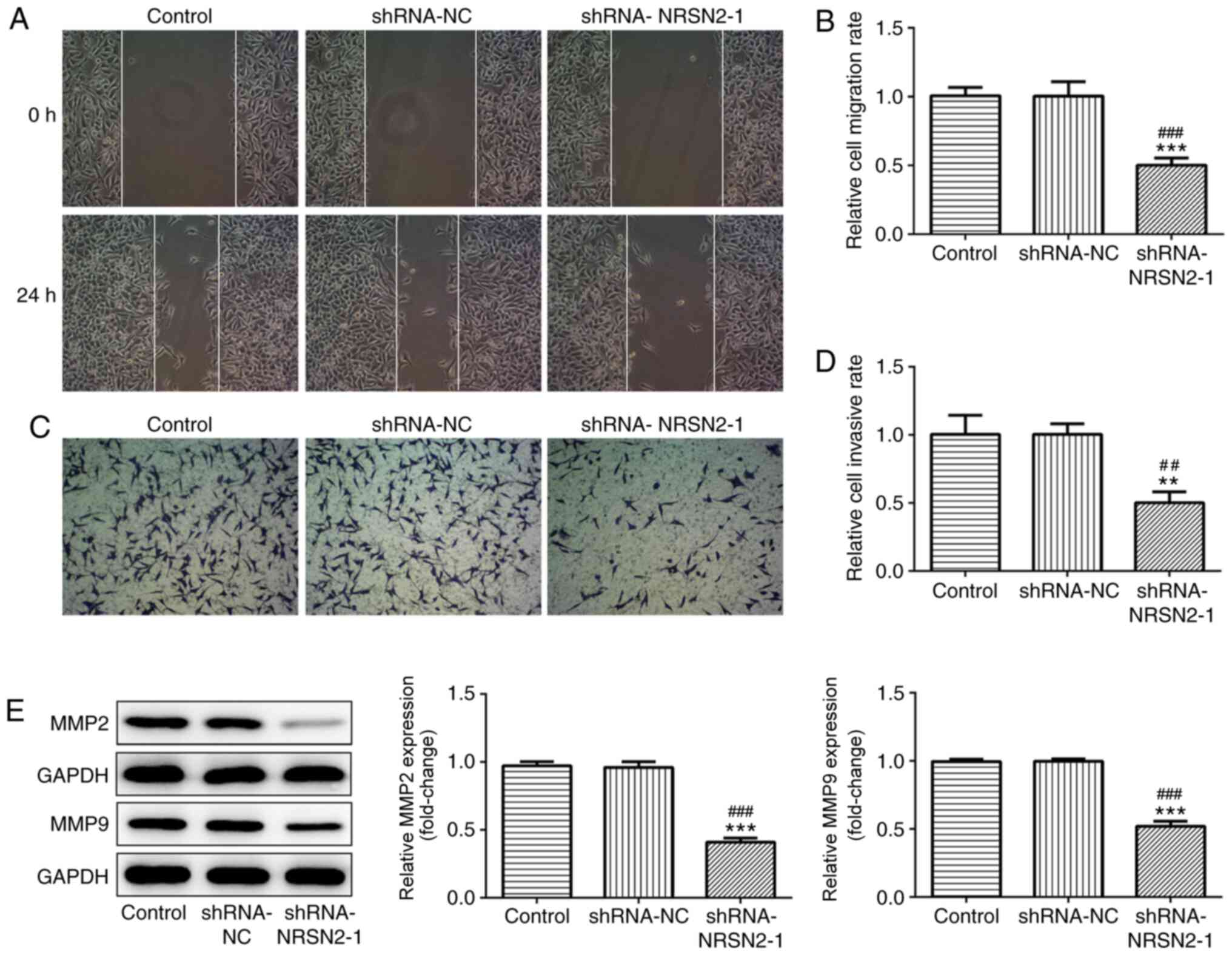
formation ability from a single cell (Fig. 1F). Wound-healing and Transwell assays
were used to analyze the migratory and invasive abilities of SW620
cells, respectively. It was revealed that SW620 cells had a
decreased migratory capacity after transfection with shRNA-NRSN2-1
(Fig. 2A and B). Furthermore, the
invasive ability of SW620 cells was significantly suppressed by
transfection with shRNA-NRSN2-1 (Fig. 2C
and D).
MMP2 and MMP9 expression levels are associated with
tumor invasion and metastasis of malignant tumors (19). As measured via western blot analysis,
shRNA-NRSN2-1 transfection led to a significant decrease in MMP2
and MMP9 expression (Fig. 2E).
Overall, these findings indicated that knockdown of NRSN2 inhibited
the proliferation, invasion and migration of SW620 cells.
Overexpression of NRSN2 promotes
proliferation, invasion and migration of CRC cells
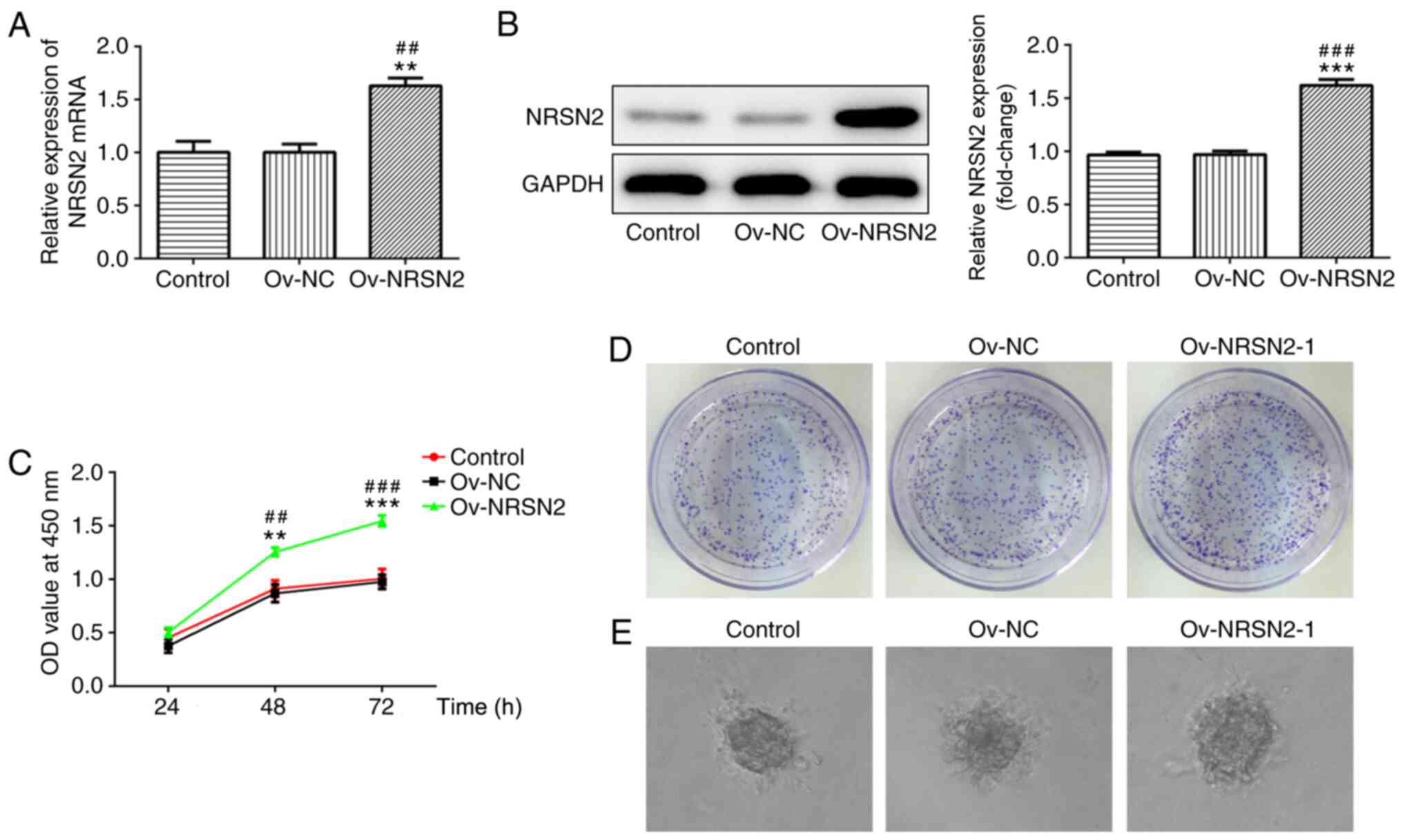
To evaluate the effect of NRSN2 on the
proliferation, invasion and migration of SW620 cells, an NRSN2
plasmid was constructed to induce the overexpression of NRSN2.
According to RT-qPCR and western blot analysis results, the NRSN2
plasmid significantly upregulated NRSN2 expression, indicating that
the NRSN2 plasmid was successfully constructed (Fig. 3A and B).
Subsequently, CCK-8 and colony formation assays were
performed to analyze the viability (Fig.
3C) and proliferative ability (Fig.
3D and E) of SW620 cells, which were significantly promoted by
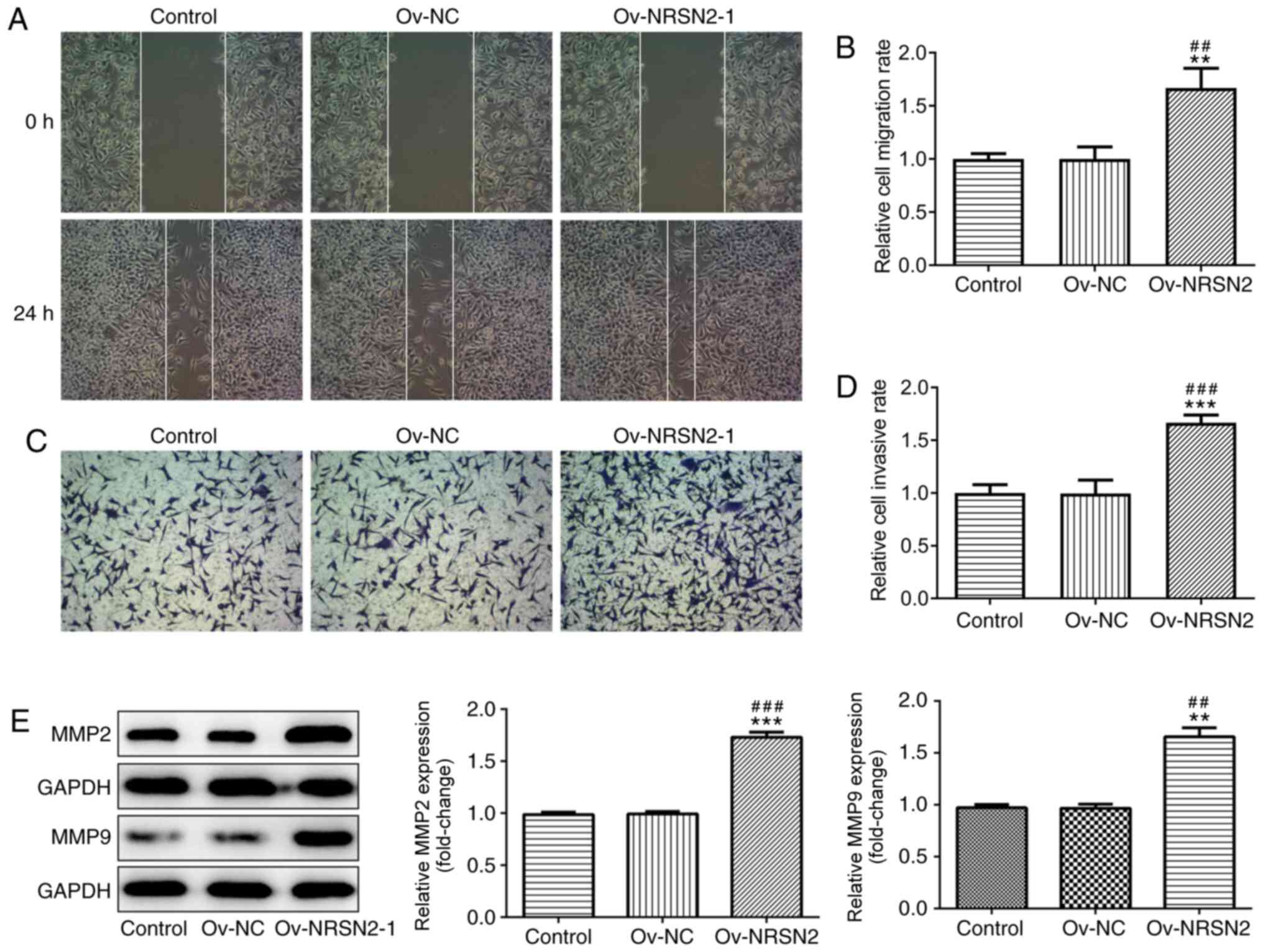
overexpression of NRSN2 at 48 and 72 h. In addition, wound-healing
and Transwell assay results suggested that overexpression of NRSN2
significantly increased the migratory (Fig. 4A and B) and invasive (Fig. 4C and D) ability of SW620 cells,
respectively. The expression levels of MMP2 and MMP9 were evaluated
using western blotting, revealing that they were significantly
upregulated in SW620 cells transfected with Ov-NRSN2 (Fig. 4E). Therefore, the data suggested that
NRSN2 overexpression promoted the proliferation, invasion and
migration of SW620 cells.
SOX12 may be involved in the
suppressive role of NRSN2 in CRC
To investigate the mechanism underlying the effect
of NRSN2 promotion of the proliferation, invasion and migration of
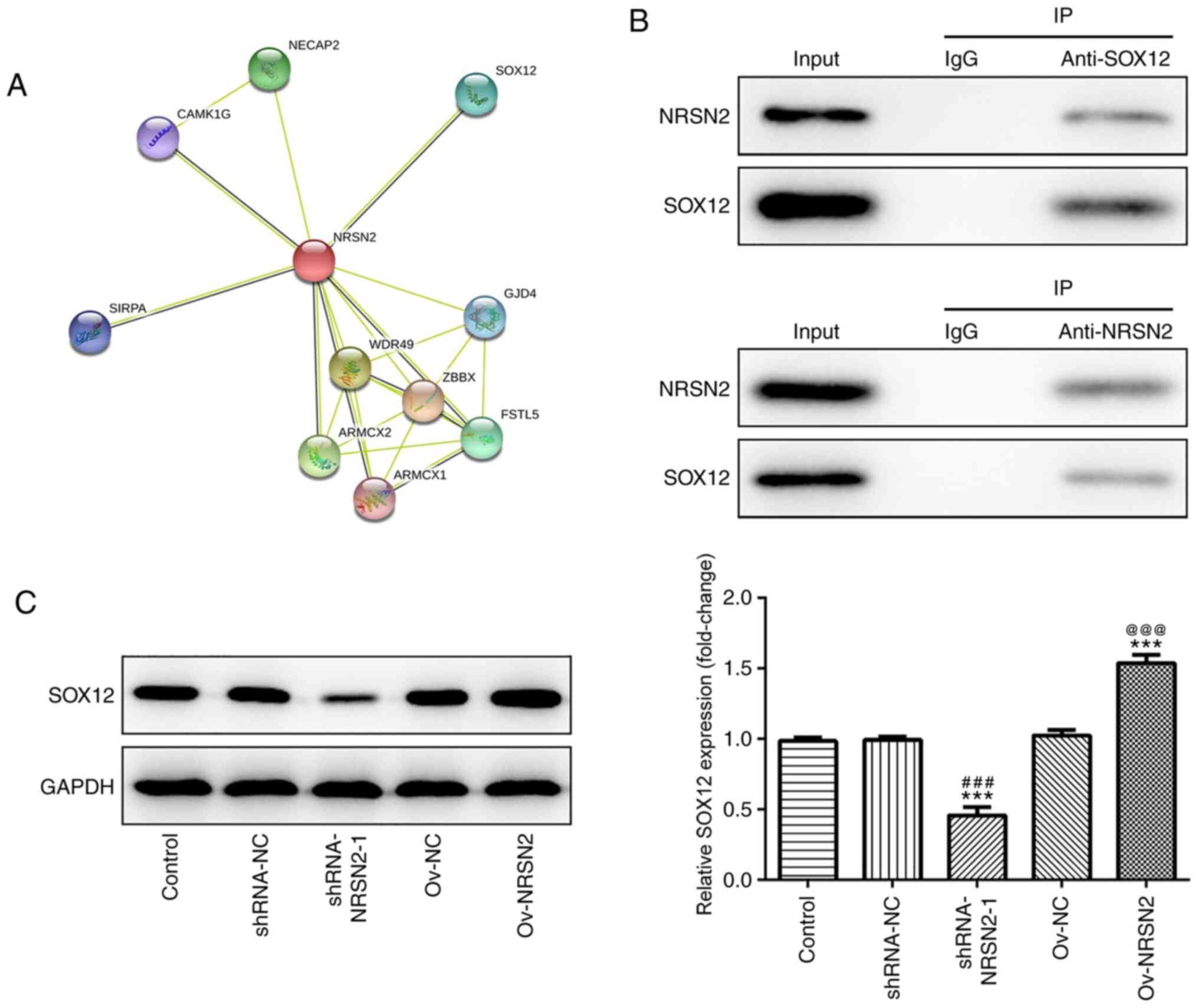
SW620 cells, the target proteins of NRSN2 were predicted using the
STRING database (https://string-db.org/cgi/network), revealing that
SOX12 may be recruited by NRSN2 (Fig.
5A). SOX12 expression is increased in CRC and promotes the
proliferation, invasion and migration of CRC cells. Hence, SOX12
was chosen for investigation in the present study. Additionally,
the physical association between NRSN2 and SOX12 was confirmed via
IP in SW620 cells (Fig. 5B). The
protein expression levels of SOX12 were determined after
transfection with shRNA-NRSN2-1 and Ov-NRSN2 plasmid. As detected
via western blot analysis, the results demonstrated that
NRSN2-knockdown significantly decreased SOX12 expression, while
NRSN2 overexpression significantly upregulated SOX12 expression,
compared to control group. Overall, these results indicated that
NRSN2 exerted a promoting effect on CRC tumorigenesis by recruiting
SOX12.
Discussion
CRC has been identified as the fifth leading cause
of cancer-associated mortality, with increasing incidence and
mortality rates (20). For instance,
it is estimated that there will be >1.1 million mortalities and
2.2 million new cases globally by 2030 due to CRC (21).
Previous studies have reported that various
biomarkers can predict cancer progression and are associated with
prognosis in patients with cancer (13,22). In
the present study, NRSN2 was observed to be associated with CRC
tumorigenesis, and the current findings provided evidence to
demonstrate that NRSN2 may serve a pivotal role in CRC progression.
In addition, it is well-established that excessive proliferation of
medial colorectal epithelial cells is a critical risk factor for
CRC pathogenesis (23). According to
the ATCC, the human colon cancer SW620 cell line isolated from the
tissue of a Caucasian male was highly tumorigenic in nude mice
(24), and has been employed in
numerous genetic, biochemical and pharmacological studies (25,26).
Therefore, SW620 cells were used in the current study to determine
the biological functions of NRSN2 in the development of CRC.
NRSN2, a novel gene that may act as a tumor
promoter, encodes a small neuronal membrane protein (16). Tang et al (15) reported that NRSN2 expression is
upregulated in ovarian cancer and serves an important role in tumor
formation, anchorage-independent colony formation, cell invasion
and chemoresistance. Additionally, Selga et al (27) demonstrated that NRSN2 is highly
expressed in a methotrexate-resistant colon cancer cell line by
searching the Gene Expression Omnibus database. Considering that
NRSN2 exerts crucial roles in cancer progression, the present study
analyzed NRSN2 expression in human CRC cells, revealing that NRSN2
was highly expressed in these cells, especially in SW620 cells.
Subsequently, shRNA-NRSN2-1 was used for NRSN2 gene silencing and
the Ov-NRSN2 plasmid was used for NRSN2 overexpression.
NRSN2-knockdown suppressed the proliferation, invasion and
migration of SW620 cells, while NRSN2 overexpression promoted these
processes in SW620 cells, consistent with the aforementioned
studies. Both loss- and gain-of-function assays indicated that
NRSN2 promoted CRC cell proliferation and colony formation to
facilitate cancer progression.
As predicted using the STRING database, SOX12 was a
potential target protein of NRSN2. Furthermore, IP results
demonstrated the interaction between NRSN2 and SOX12 in SW620
cells, and western blot analysis further confirmed an association
between NRSN2 and SOX12. Chen et al (28) reported that the circular RNA
hsa_circ_001895 sponges microRNA-296-5p to promote clear cell renal
cell carcinoma progression by regulating SOX12. In addition, Du
et al (29) demonstrated that
SOX12 may represent a novel prognostic biomarker and regulator of
gastric cancer metastasis via upregulating MMP7 and insulin-like
growth factor 1. According to a previous study, SOX12 may serve as
a prognostic biomarker of CRC, and promotes CRC cell proliferation
and metastasis via regulating asparagine synthesis (30). In the present study, western blot
analysis demonstrated that NRSN2-knockdown decreased SOX12
expression, while NRSN2 overexpression upregulated SOX12
expression, indicating that NRSN2 may exert accelerative effects on
CRC progression by recruiting SOX12. Overall, the present findings
suggested that NRSN2 may be a potential biomarker and may provide a
novel therapeutic strategy for the treatment of CRC.
In conclusion, the present study demonstrated that
NRSN2 promoted the proliferation, invasion and migration of CRC
cells. SW620 cells were used to investigate the role of NRSN2 in
CRC and to identify its mechanism, and the results suggested that
NRSN2 exerted accelerative effects on CRC progression by recruiting
SOX12. Therefore, NRSN2 may be used as a potential biomarker and
promising therapeutic target for CRC. However, the interaction
between NRSN2 and SOX12 has not been fully elucidated, and future
experiments, such as invitro binding or co-localization
assays, should be performed to understand the role of SOX12
underlying the effect of NRSN2.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GW was responsible for the writing of the
manuscript, performed the experiments, analyzed and interpreted the
data. KY reviewed the manuscript and designed the experiments. Both
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
González-Llorente L, Santacatterina F,
García-Aguilar A, Nuevo-Tapioles C, González-García S, Tirpakova Z,
Toribio ML and Cuezva JM: Overexpression of Mitochondrial IF1
Prevents Metastatic Disease of Colorectal Cancer by Enhancing
Anoikis and Tumor Infiltration of NK Cells. Cancers (Basel).
12:122019. View Article : Google Scholar
|
|
2
|
Khelwatty SA, Essapen S, Bagwan I, Green
M, Seddon AM and Modjtahedi H: Co-expression and prognostic
significance of putative CSC markers CD44, CD133, wild-type EGFR
and EGFRvIII in metastatic colorectal cancer. Oncotarget.
10:1704–1715. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goldstein DA, Zeichner SB, Bartnik CM,
Neustadter E and Flowers CR: Metastatic Colorectal Cancer: A
Systematic Review of the Value of Current Therapies. Clin
Colorectal Cancer. 15:1–6. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu X, Zhang C, Xia Y and Yu J: Over
expression of METRN predicts poor clinical prognosis in colorectal
cancer. Mol Genet Genomic Med. 8:e11022020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Issa IA and Noureddine M: Colorectal
cancer screening: An updated review of the available options. World
J Gastroenterol. 23:5086–5096. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al CONCORD Working Group, : Global surveillance of
trends in cancer survival 2000–14 (CONCORD-3): Analysis of
individual records for 37 513 025 patients diagnosed with one of 18
cancers from 322 population-based registries in 71 countries.
Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pessaux P, Chenard MP, Bachellier P and
Jaeck D: Consequences of chemotherapy on resection of colorectal
liver metastases. J Visc Surg. 147:e193–e201. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lima PMA, Torres LC, Martins MR, da Matta
MC, Lima JTO, de Mello MJG, da Silva LM, Cintra EB Jr, Lira CCR, da
Fonte EJA, et al: Soluble levels of sCD40L and s4-1BB are
associated with a poor prognosis in elderly patients with
colorectal cancer. J Surg Oncol. 121:901–905. 2020.PubMed/NCBI
|
|
13
|
Leiphrakpam PD, Lazenby AJ, Chowdhury S,
Smith LM, Mathiesen M, Brattain MG, Wang J, Black JD and Are C:
Prognostic and therapeutic implications of NHERF1 expression and
regulation in colorectal cancer. J Surg Oncol. 121:547–560. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Keremu A, Maimaiti X, Aimaiti A, Yushan M,
Alike Y, Yilihamu Y and Yusufu A: NRSN2 promotes osteosarcoma cell
proliferation and growth through PI3K/Akt/MTOR and Wnt/β-catenin
signaling. Am J Cancer Res. 7:565–573. 2017.PubMed/NCBI
|
|
15
|
Tang W, Ren A, Xiao H, Sun H and Li B:
Highly expressed NRSN2 is related to malignant phenotype in ovarian
cancer. Biomed Pharmacother. 85:248–255. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang XY, Kuang JL, Yan CS, Tu XY, Zhao
JH, Cheng XS and Ye XQ: NRSN2 promotes non-small cell lung cancer
cell growth through PI3K/Akt/mTOR pathway. Int J Clin Exp Pathol.
8:2574–2581. 2015.PubMed/NCBI
|
|
17
|
Wang X, Han L, Zhang J and Xia Q:
Down-Regulated NRSN2 Promotes Cell Proliferation and Survival
Through PI3K/Akt/mTOR Pathway in Hepatocellular Carcinoma. Dig Dis
Sci. 60:3011–3018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Björklund M and Koivunen E:
Gelatinase-mediated migration and invasion of cancer cells. Biochim
Biophys Acta. 1755:37–69. 2005.PubMed/NCBI
|
|
20
|
Li W, Xu Y, Wang X, Cao G, Bu W, Wang X,
Fang Z, Xu Y, Dong M and Tao Q: circCCT3 Modulates Vascular
Endothelial Growth Factor A and Wnt Signaling to Enhance Colorectal
Cancer Metastasis Through Sponging miR-613. DNA Cell Biol.
39:118–125. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu G, Lin C, Cheng Z, Wang Q, Hoffman RM,
Singh SR, Huang Y, Zheng W, Yang S and Ye J: TRAF6-Mediated
Inflammatory Cytokines Secretion in LPS-induced Colorectal Cancer
Cells Is Regulated by miR-140. Cancer Genomics Proteomics.
17:23–33. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang XJ, Yu Q, Chi P, Lin HM, Lu XR, Huang
Y, Xu ZB, Huang SH, Sun YW and Ye DX: Identification of gene
biomarkers to predict responses to neoadjuvant chemoradiotherapy in
patients with rectal cancer and pathways enrichment analysis.
Zhonghua Wei Chang Wai Ke Za Zhi. 22:1183–1187. 2019.(In Chinese).
PubMed/NCBI
|
|
23
|
Dziki Ł, Puła A, Stawiski K, Mudza B,
Włodarczyk M and Dziki A: Patients' Awareness Of The Prevention And
Treatment Of Colorectal Cancer. Pol Przegl Chir. 87:459–463. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huerta S, Harris DM, Jazirehi A, Bonavida
B, Elashoff D, Livingston EH and Heber D: Gene expression profile
of metastatic colon cancer cells resistant to cisplatin-induced
apoptosis. Int J Oncol. 22:663–670. 2003.PubMed/NCBI
|
|
25
|
26. Melcher R, Steinlein C, Feichtinger W,
Müller CR, Menzel T, Lührs H, Scheppach W and Schmid M: Spectral
karyotyping of the human colon cancer cell lines SW480 and SW620.
Cytogenet Cell Genet. 88:145–152. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Broznić D, Ratkaj I, Malenica Staver M,
Kraljević Pavelić S, Žurga P, Bubalo D and Gobin I: Evaluation of
the Antioxidant Capacity, Antimicrobial and Antiproliferative
Potential of Fir (Abies alba Mill.) Honeydew Honey Collected
from Gorski kotar (Croatia). Food Technol Biotechnol. 56:533–545.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Selga E, Noé V and Ciudad CJ:
Transcriptional regulation of aldo-keto reductase 1C1 in HT29 human
colon cancer cells resistant to methotrexate: Role in the cell
cycle and apoptosis. Biochem Pharmacol. 75:414–426. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Z, Xiao K, Chen S, Huang Z, Ye Y and
Chen T: CircRNA hsa_circ_001895 serves as a sponge of miR-296-5p to
promote cell carcinoma progression via regulating SOX12. Cancer
Sci. 111:713–726. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du F, Feng W, Chen S, Wu S, Cao T, Yuan T,
Tian D, Nie Y, Wu K, Fan D, et al: Sex determining region Y-box 12
(SOX12) promotes gastric cancer metastasis by upregulating MMP7 and
IGF1. Cancer Lett. 452:103–118. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du F, Chen J, Liu H, Cai Y, Cao T, Han W,
Yi X, Qian M, Tian D, Nie Y, et al: SOX12 promotes colorectal
cancer cell proliferation and metastasis by regulating asparagine
synthesis. Cell Death Dis. 10:2392019. View Article : Google Scholar : PubMed/NCBI
|