Introduction
Melanoma, originating from melanocytes that produce
melanin, is the most common type of skin cancer with a poor
prognosis (1). Epidemiological
studies demonstrated that ~55,500 people die of melanoma annually
worldwide (2,3), and its incidence rate was the highest
among Caucasians, particularly in New Zealand (0.0358%) and
Australia (0.0349%) in 2012 (4).
Clinically, melanoma lesions are classified according to their
location and progression, ranging from benign nevi to metastatic
melanoma (5). The exact cause of
melanoma is unknown; however, exposure to ultraviolet radiation
from sunlight increases the risk of developing melanoma (6). Additionally, genetic background and
pigmentation status contribute to cancer predisposition (7–9).
Melanoma can be cured at the early stage; however, advanced
melanoma easily metastasizes to lymph glands without cell adhesion
between lymph nodes and other metastases, leading to treatment
resistance and a poor prognosis (5).
Before the approval of selective kinase inhibitors, such as
dabrafenib, vemurafenib and trametinib, for metastatic melanoma in
2010 (10), the 5-year survival rate
for patients with advanced melanoma was only 16% (11,12).
It is estimated that ~40% of melanoma cases contain
oncogenic B-Raf oncogene serine/threonine-kinase (BRAF) somatic
mutations, which lead to the constitutive activation of MAPK
signaling and resistance to chemotherapy. The most common BRAF
mutation is V600E, which accounts for ~5-90% of the BRAF mutations
in melanoma (13). Since 2010,
vemurafenib and dabrafenib targeting BRAF V600E have been approved
for the treatment of patients with advanced melanoma in numerous
countries around the world (10).
Both progression-free survival and overall survival times are now
extended in patients with metastatic melanoma with BRAF V600E
mutation (14). However, the 5-year
survival rates of melanoma remain unfavorable, due to resistance to
treatment in most patients within ~12 months (12). Therefore, identification of novel
therapeutic targets and understanding their molecular mechanisms
are urgently required for melanoma therapy.
The precursor of vitamin D3,
7-dehydrocholesterol (7-DHC), is stored under human skin and serves
an important role in regulating calcium and phosphorus metabolism
(13,15). 7-DHC has been demonstrated to be
associated with numerous processes, such as the maintenance of the
epidermal barrier and the treatment of common skin diseases
(16). Recently, researchers
revealed that 7-DHC treatment could specifically promote the
phosphorylation of interferon regulatory factor 3 and enhance type
I interferon production in macrophages, suggesting 7-DHC as a
potential therapeutic agent against emerging or highly pathogenic
viruses (17). In addition, in
ovarian cancer, the replacement of cholesterol by 7-DHC efficiently
enhances the anticancer activity of photosensitizer-encapsulated
liposomes upon irradiation (18),
and 7-DHC peroxide exhibits improved anticancer activity and
selectivity over ergosterol peroxide (19). Although the mechanism has not been
fully elucidated yet, 7-DHC has been demonstrated to elicit
cytotoxic and apoptosis-promoting effects in melanoma cells
(20).
In the present study, the transcriptome analysis of
A375 cells with 7-DHC treatment and mutational hotspots of
kinase-coding genes recorded in The Cancer Genome Atlas (TCGA)
database were integrated to determine the molecular mechanism
underlying the anticancer property of 7-DHC and provide a
theoretical basis for its clinical application.
Materials and methods
Cell lines and cell culture
A375 malignant melanoma and 293T embryonic kidney
cells were purchased from the China Infrastructure of Cell Line
Resources (Institute of Basic Medical Sciences; Chinese Academy of
Medical Sciences). A2058 melanoma cells were kindly provided by Dr
Qiong Yang from the Fang Lab of the Beijing Institute of Genomics
of the Chinese Academy of Sciences. All cells were maintained in
high-glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin (Beijing Solarbio Science &
Technology Co., Ltd.) at 37°C, 95% humidity and 5%
CO2.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
Total RNA was extracted from A375 and A2058 melanoma
transfected cells or cells treated with the AKT1 inhibitor MK-2206
(1 µM; cat. no. HY-10358; MedChemExpress) or the AKT inhibitor III
(1 µM; cat. no. HY-10355; MedChemExpress) using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA was
produced using a reverse transcription kit (TransScript All-in-One
First-Strand cDNA Synthesis SuperMix for qPCR; TransGen Biotech
Co., Ltd.) at 42°C for 15 min according to the manufacturer's
protocol. qPCR was performed using SYBR PCR mix (TransGen Biotech
Co., Ltd.) and GAPDH was used as the housekeeping gene for internal
reference. The following primers were used to amplify genes at a
concentration of 5 µM: RELA proto-oncogene NF-κB subunit (RELA;
gene ID, ENSG00000173039) forward, 5′-TTCCGACCGGGAGCTCAGTG-3′ [20
bp; melting temperature (Tm), 61°C] and reverse,
5′-TGCGGGAAGGCACAGCAAT-3′ (19 bp; Tm, 62°C); long intergenic
non-protein coding RNA 552 (LINC00552; gene ID, ENSG00000279770)
forward, 5′-ACACAGACATGCACACACAGG−3′ (21 bp; Tm, 60°C) and reverse,
5′-GGGGCCAGTGGATTTTCATG-3′ (20 bp; Tm, 58°C); ADAM metallopeptidase
domain 19 (ADAM19; gene ID, ENSG00000135074) forward,
5′-CGAGAAGGTGAATGTGGCAGGA-3′ (22 bp; Tm, 61°C) and reverse,
5′-AGCTCTGACACTGGATCTTCCC-3′ (22 bp; Tm, 60°C); C-X-C motif
chemokine ligand 3 (CXCL3; gene ID, ENSG00000163734) forward,
5′-TCCGTGGTCACTGAACTGCG-3′ (20 bp; Tm, 61°C) and reverse,
5′-AGTTGGTGCTCCCCTTGTTCA−3′ (21 bp; Tm, 60°C); insulin-like growth
factor binding protein 5 (IGFBP5; gene ID, ENSG00000115461)
forward, 5′-ACCTGAGATGAGACAGGAGTC−3′ (21 bp; Tm, 56°C) and reverse,
5′-GTAGAATCCTTTGCGGTCACAA-3′ (22 bp; Tm, 57°C); MMP9 (gene ID,
ENSG00000100985) forward, 5′-AACTACGACCGGGACAAGCTC−3′ (21 bp; Tm,
60°C) and reverse, 5′-ACAGGTCGAGTACTCCTTACCCAG−3′ (24 bp; Tm,
60°C); α-methylacyl-CoA racemase (AMACR; gene ID, ENSG00000242110)
forward, 5′-AGAACCCCAGTTCTACGAGCTG-3′ (22 bp; Tm, 60°C) and
reverse, 5′-ACTCTGCCTTCGTCTTCTCTGC-3′ (22 bp; Tm, 61°C); heme
oxygenase 1 (HMOX1; gene ID, ENSG00000100292) forward,
5′-ACCCAGGCAGAGAATGCTGAG−3′ (21 bp; Tm, 61°C) and reverse,
5′-ACATAGATGTGGTACAGGGAGGC-3′ (23 bp; Tm, 60°C); IL11 (gene ID,
ENSG00000095752) forward, 5′-TCCTGACCCGCTCTCTCCTG-3′ (20 bp; Tm,
62°C) and reverse, 5′-AGGGAATCCAGGTTGTGGTCC-3′ (21 bp; Tm, 60°C);
lipocalin 2 (LCN2; gene ID, ENSG00000148346) forward,
5′-AGAGCTACAATGTCACCTCCGTC-3′ (23 bp; Tm, 60°C) and reverse,
5′-TCTTAATGTTGCCCAGCGTGAACT-3′ (24 bp; Tm, 61°C);
prostaglandin-endoperoxide synthase 2 (PTGS2; gene ID,
ENSG00000073756) forward, 5′-TACCCACTTCAAGGGATTTTGGAACG-3′ (26 bp;
Tm, 61°C) and reverse, 5′-AGTCAGCATTGTAAGTTGGTGGACT-3′ (25 bp; Tm,
61°C); and GAPDH (gene ID, ENSG00000111640) forward,
5′-AGCCACATCGCTCAGACAC-3′ (19 bp; Tm, 59°C) and reverse,
5′-ACCAGGCGCCCAATACGA−3′ (18 bp; Tm, 60°C). The qPCR thermocycling
conditions were as follows: 94°C for 30 sec, followed by 40 cycles
of 5 sec at 94°C for denaturation and 60°C for 30 sec for
annealing/extension using the two-step method. The expression fold
changes were calculated using the 2−ΔΔCq method
(21).
Lentivirus packaging and
infection
Three short hairpin RNAs (shRNAs) of RELA, namely
shRNA-1
(5′-CGGATTGAGGAGAAACGTAAATTCAAGAGATTTACGTTTCTCCTCAATCCGTTTTTTG-3′),
shRNA-2
(5′-GCCTTAATAGTAGGGTAAGTTTTCAAGAGAAACTTACCCTACTATTAAGGCTTTTTTG-3′)
and shRNA-3
(5′-GGATTCATTACAGCTTAATTCAAGAGATTAAGCTGTAATGAATCCATTTTTTG-3′), and
a scrambled control shRNA
(5′-CGGTCCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGGTTTTTG-3′),
were synthesized by Genewiz, Inc., and were then cloned into
PLKO.1-EGFP-T2A-Puro (Addgene, Inc.) via AgeI/EcoRI
enzyme digestion sites. High titer production of lentivector was
achieved by transiently co-transfecting 293T cells with lentiviral
packaging plasmids, 4 µg pCMVR8.2, 1 µg pCMV–VSVG (Addgene, Inc.)
and 3 µg RELA-shRNA-expressing constructs via
Lipofectamine® 3000 (1 µl Lipofectamine 3000 per µg
plasmid; Invitrogen; Thermo Fisher Scientific, Inc.). At 70–90%
confluence, A375 melanoma cells were infected with the
aforementioned shRNA lentiviruses with the help of Polybrene with a
final concentration of 6 µg/ml (Sigma-Aldrich; Merck KGaA),
according to the manufacturer's protocol. RNA was harvested 48 h
post-infection to determine the knockdown efficiency using RT-qPCR,
as aforementioned.
Cell proliferation assay
A375 cells treated with multiple concentrations (0,
1, 2.5, 5, 10 and 25 µM) of 7-DHC or infected with RELA-shRNA
lentivirus, were cultured in a 96-well plate at a density of 5,000
cells/well at 37°C for 48 h. Subsequently, 10 µl Cell Counting
Kit-8 (APeXBIO Technology LLC) solution was added to each well and
incubated at 37°C with 5% CO2 for another 4 h, according
to the manufacturer's protocol. Absorbance was measured at a
wavelength of 450 nm using an ELISA plate reader (Thermo Fisher
Scientific, Inc.).
Apoptosis and cell cycle analysis
using flow cytometry
A375 and A2058 cells were treated with ethanol
(negative control) and different concentrations (1, 5 and 10 µM) of
7-DHC at 37°C for 48 h and were then harvested at 500 g at 20°C for
5 min and washed twice with PBS. Apoptosis was detected using the
TransDetect Annexin V-FITC/PI Cell Apoptosis Detection kit
(TransGen Biotech Co., Ltd.) according to the manufacturer's
protocol; briefly, cells were resuspended with 100 µl Annexin V
binding buffer and stained with 5 µl Annexin V-FITC and 5 µl PI for
20 min at room temperature. For cell cycles analysis, the cells
were fixed with 70% ethanol at 4°C overnight. On the next day,
using the DNA Content Quantitation Assay kit (Beijing Solarbio
Science & Technology Co., Ltd.), the cells were stained with 20
µl PI (50 µg/ml) and 20 µl RNase A (50 µg/ml) for 30 min at 4°C.
All samples were detected using a flow cytometer (NovoCyte 2040R;
ACEA Bioscience, Inc.; Agilent Technologies) and analyzed using the
NovoExpress 1.4.1 software (ACEA Bioscience, Inc.; Agilent
Technologies).
Hoechst-PI double staining
A total of 5×105 A375 and A2058 melanoma
cells were cultured in 6-well plates at 37°C for 48 h. During this
period, some cells were treated with 25 µM 7-DHC, while negative
control cells were treated with ethanol. Subsequently, 5 µl Hoechst
and 5 µl PI staining solution from a Hoechst/PI double-staining kit
(Beijing Solarbio Science & Technology Co., Ltd.) were added to
the cells at 37°C for 1 h. Finally, the plates were observed under
a fluorescence microscope (eye lens × objective lens, 10×10; Leica
S/N 443077; Leica Microsystems, Inc.).
Real-time cellular analysis (RTCA)
assay
The migration rates of A375 and A2058 cells were
analyzed using RTCA. A375 and A2058 cells were seeded into the
upper chamber of individual CIM-Plate-16 (5,000 cells/well; ACEA
Bioscience, Inc.; Agilent Technologies) in 130 µl DMEM without FBS.
The upper chamber was then placed on the lower chamber of the
CIM-Plate-16, which contained DMEM supplemented with 10% FCS as an
attractant. Changes in impedance resulting from cells that had
migrated to the bottom side of the membrane were recorded every 5
min and were monitored for a total of 36 h. Finally, the cell index
values representing the migration rates were calculated using the
xCELLigence 1.1 software (ACEA Bioscience, Inc.; Agilent
Technologies, Inc.).
Chromatin immunoprecipitation
(ChIP)-qPCR
ChIP experiments were performed using a SimpleChIP
Plus Sonication Chromatin IP kit (Cell Signaling Technology, Inc.),
according to the manufacturer's protocol. Normal rabbit IgG
antibody was used as a control. Antibodies were as follows: Normal
rabbit IgG (1:5,000; cat. no. 2729S; Cell Signaling Technology,
Inc.) and anti-RELA (1:1,000; cat. no. ab19870; Abcam). Antibodies
were incubated at 4°C overnight. Subsequently, qPCR was performed
using SYBR PCR mix kit (TransGen Biotech Co., Ltd.), and GAPDH was
used as the reference gene. D130D1 was used as the negative
control. The qPCR thermocycling conditions were as follows: 94°C
for 30 sec, followed by 40 cycles of 5 sec at 94°C for denaturation
and 60°C for 30 sec for annealing/extension using the two-step
method. The expression fold changes were calculated using the
2−ΔΔCq method (21).
Primers for RELA target genes promoter regions were as follows:
LINC00552-P-1 forward, 5′-TCCATTATGAGCCCTGGGAC-3′, and reverse,
5′-TCCCGGAGCCTCATTGATAC-3′; LINC00552-P-2 forward,
5′-GCTCTCTCACCATGCGATTG-3′, and reverse,
5′-AGGCTGAGAAGTCCAAGGTC-3′; ADAM19-P-1 forward,
5′-GGCTTTGTTCCCACGTTCTG-3′, and reverse,
5′-GAGGAAGGAGAAGGCGAGAA-3′; ADAM19-P-2 forward,
5′-TGGAAGAACACACGTCTGGT-3′, and reverse, 5′-CTTTCCCCACACACCAAGC-3′;
ADAM19-P-3 forward, 5′-TTTGTCGCCCAGGAGCAATA-3′, and reverse,
5′-AGGGGATTTTGTCATGGGAAC−3′; CXCL1-P-1 forward,
5′-ATCCCAAAGTCCCAGAGTGC-3′, and reverse,
5′-CAAGATCGGCGAACCCTTTT-3′; MMP9-P-1 forward,
5′-CAGTACCGAGAGAAAGCCTATT-3′, and reverse,
5′-CAGGATGTCATAGGTCACGTAG-3′; and GAPDH forward,
5′-AGCCACATCGCTCAGACAC-3′, and reverse,
5′-ACCAGGCGCCCAATACGA−3′.
Wound-healing assay
The A375 cell line is a typical malignant melanoma
cell line, and therefore, wound-healing assays were performed in
A375 cells to illustrate the inhibition of migration by 7-DHC. A375
melanoma cells were cultured in six-well plates in DMEM with 10%
FBS at 37°C, 95% humidity and 5% CO2 for 24 h until the
cells reached 100% confluence. Subsequently, 200-µl sterile pipette
tips were used to scratch the cells in the middle of each well. The
medium of each well was discarded, PBS was added to wash cells and
remove floating cells, and serum-free DMEM was added into each
well. Different concentrations of 7-DHC were added into each well,
with the final 7-DHC concentrations being 0, 1, 5, 10 and 25 µM.
Wound-healing was observed and captured using a light inverted
microscope at a magnification of 10×10 (eye lens × objective lens)
at 0, 24 and 48 h. Lastly, the migration rate was evaluated by
detecting the width of the cell-free region using GraphPad Prism 5
(GraphPad Software, Inc.) for the quantification and plotting.
Western blotting
Total protein lysates isolated from A375 and A2058
melanoma cells treated with different concentrations of 7-DHC (0,
1, 2.5, 5, 10 and 25 µM) or 1 µM insulin-like growth factor 1
(IGF1; Beijing Solarbio Science & Technology Co., Ltd.) at 37°C
for 48 h, were collected using RIPA buffer with a protease
inhibitor cocktail (Beijing Solarbio Science & Technology Co.,
Ltd.). In addition, the nuclear and cytoplasmic proteins of A375
and A2058 cells were collected using the NE-PER Nuclear and
Cytoplasmic Extraction Reagents kit (cat. no. 78835; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. For
western blotting, protein concentration was determined using a BCA
assay and proteins (20 µg/lane) were separated via 12% SDS-PAGE and
transferred onto a nitrocellulose membrane (EMD Millipore). The
membrane was blocked with 5% skimmed milk in TBS-Tween (0.1% Tween
20) at room temperature for 1 h and then incubated at 4°C overnight
with primary antibodies against Akt1 (1:1,000; cat. no. 33224;
Signalway Antibody LLC), phosphorylated (p)Akt1-Ser473 (1:1,000;
cat. no. 13357; Signalway Antibody LLC), pAkt1-Thr308/309 (1:1,000;
cat. no. 13311; Signalway Antibody LLC), MEK1 (1:1,000; cat. no.
21428; Signalway Antibody LLC), pMEK1-ser217/ser221 (1:1,000; cat.
no. 11205; Signalway Antibody LLC), histone H3 (H3; 1:1,000; cat.
no. ab1791; Abcam), RELA (1:1,000; cat. no. ab19870; Abcam),
TUBULIN (1:5,000; cat. no. RM2007L; Beijing Ray antibody Biotech)
and GAPDH (1:5,000; cat. no. ab181602; Abcam). After washing, the
membrane was incubated with anti-mouse (1:5,000; cat. no. ab97040;
Abcam) or anti-rabbit (1:5,000; cat. no. ab7090; Abcam)
HRP-conjugated secondary antibodies at room temperature for 1 h.
Bands recognized by antibodies were revealed using an ECL reagent
(Thermo Fisher Scientific, Inc.) on Exposure meter Azure Imager 300
(Azure Biosystems, Inc.). Finally, ImageJ 1.52 software (National
Institutes of Health) was used for quantification of protein
expression.
Dual-luciferase assay
A375 melanoma cells were co-transfected with vectors
of firefly luciferase and Renilla luciferase (Addgene, Inc.)
using Lipofectamine 3000 (1 µl Lipofectamine 3000/µg plasmid;
Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h.
Notably, the transcription of firefly luciferase was regulated by a
mini-promoter constructed with 4 repeating RELA motifs
(5′-GGGAATTTCC-3′). The growth medium from the cultured cells was
removed, and a sufficient volume of PBS was gently added to wash
the surface of the culture vessel. The vessel was briefly swirled
to remove the detached cells and residual growth medium, and PBS
was removed. Subsequently, 1X passive lysis buffer from the
dual-luciferase reporter assay system kit (cat. no. E1960; Promega
Corporation) was added to each culture well, and the culture plates
were rocked at room temperature for 15 min. A total of 100 µl
Luciferase Assay Reagent II from the dual-luciferase reporter assay
system kit (cat. no. E1960; Promega Corporation) was added to
luminometer tubes including 20 µl lysate of samples. The absorbance
was measured at a wavelength of 560 nm to detect the firefly
luciferase activity. Stop & Glo Reagent (100 µl; Promega
Corporation) was added to each tube, and the absorbance at a
wavelength of 470 nm was measured to detect the Renilla
luciferase activity. The luciferase results were determined via
dividing the absorbance value of 560 nm by the absorbance value of
470 nm to normalize the firefly luciferase activity by the
Renilla luciferase activity.
Bioinformatics analysis
R 3.5.1 software (https://www.r-project.org/) was used to analyze the
RNA sequencing (RNA-seq) data of 7-DHC-treated (10 and 25 µM) A375
melanoma cells and the bioinformatics data from TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-enomics/tcga)
and Gene-Tissue Expression (GTEX; http://genome.ucsc.edu/gtex.html) databases. Firstly,
the limma 3.44.3 R package (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
was used to analyze the gene expression pattern of A375 cells
treated with 7-DHC vs. ethanol to determine the differentially
expressed genes [log fold change >1, adjusted (adj-)P<0.05].
ClusterProfiler 3.16.1 R package (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
was used for the Gene Set Enrichment Analysis (GSEA) (22), which used the gene set
c2.all.v6.2.entrez.gmt downloaded from the GSEA website (http://software.broadinstitute.org/gsea/downloads.jsp)
for enrichment pathway, based on the aforementioned differential
expression results.
Secondly, for the public data from TCGA and GTEX
databases, the transcriptome data from 471 samples of patients with
melanoma were downloaded from TCGA skin cutaneous melanoma
(TCGA-SKCM) dataset and data from 813 normal skin samples were
downloaded from the GTEX (http://genome.ucsc.edu/gtex.html) database. In order
to compare the gene expression in normal and tumor samples, the
RNA-seq data from TCGA and GTEX databases were normalized by log2
(fragments per kilobase of exon per million value) and the limma R
package was used to analyze the gene expression pattern to screen
for significantly differential genes (log fold change >1,
adj-P<0.05).
Statistical analysis
Univariate and multivariate Cox proportional hazards
regression analyses were performed, based on 19 genes of
intersection between upregulated genes in melanoma tissues from
TCGA-SKCM dataset and downregulated genes in melanoma cells treated
with 7-DHC. Furthermore, a 5-gene prognosis predictive model was
constructed, and patients with melanoma from TCGA-SKCM dataset were
classified into high- and low-risk groups according to the mean
value of the risk score calculated using the survival 2.43 R
package (https://CRAN.R-project.org/package=survivalAnalysis).
The survivalROC 1.0.3 R package (https://cran.r-project.org/web/packages/survivalROC/index.html)
was used to construct the time-dependent receiver operating
characteristic (ROC) curve, in order to assess the sensitivity and
specificity of the 5-gene signature in predicting survival. The
Kaplan-Meier method and log-rank test were used to estimate the
overall survival (OS). In addition, GraphPad Prism 5 (GraphPad
Software, Inc.) was used for statistical analysis, and unpaired
Student's t-test was used to compare the difference between two
groups, while one-way ANOVA was used to compare differences among
≥3 groups, followed by Tukey's post hoc test. Data were presented
as the mean ± standard deviation, and the experiments were repeated
≥3 times in each group. P<0.05 was considered to indicate a
statistically significant difference.
Results
7-DHC increases apoptosis and inhibits
proliferation and migration of melanoma cells
To confirm the anticancer property of 7-DHC on
melanoma cells, proliferation assays were performed. Human
embryonic kidney 293T cells and A375 and A2058 melanoma cells were
treated with increasing concentrations of 7-DHC (0, 1, 2.5, 5, 10
and 25 µM) for 48 h. The results revealed that 10 and 25 µM 7-DHC
significantly inhibited the proliferation of melanoma cells
(Fig. 1A and B), but not that of
293T cells (Fig. S1). Subsequently,
Annexin V-FITC and PI staining was used to evaluate the apoptosis
rate of A375 and A2058 melanoma cells treated with different
concentrations (1, 5 and 10 µM) of 7-DHC. The results of the flow
cytometer assay revealed that the proportion of late apoptotic A375
and A2058 cells was markedly increased with the increasing doses of
7-DHC compared with the ethanol control (Fig. 1C and D). In addition, Hoechst/PI
double staining assay revealed that 7-DHC induced necrosis in
melanoma cells (Fig. S2A). Tumor
metastasis is one of the hallmarks of malignant melanoma and is
closely associated with a poor prognosis (23). In order to investigate the migration
capability of melanoma cells treated with 7-DHC, RTCA assay was
used to continuously monitor the cell index value for 36 h, and
wound-healing assay was used to detect the cell migration rate. The
results of RTCA demonstrated that the cell index of A375 and A2058
cells treated with 25 µM 7-DHC was lower compared with that of
cells treated with ethanol or with low concentrations of 7-DHC at
36 h, which demonstrated that only a high concentration (25 µM) of
7-DHC had the ability to inhibit the migration rate of melanoma
cells (Fig. 1E and F). Similarly,
the wound-healing assay revealed that the migration rate of
melanoma cells treated with 7-DHC was lower compared with that of
the ethanol control (Fig. S2B),
suggesting that the migration of melanoma cells was negatively
affected by 7-DHC.
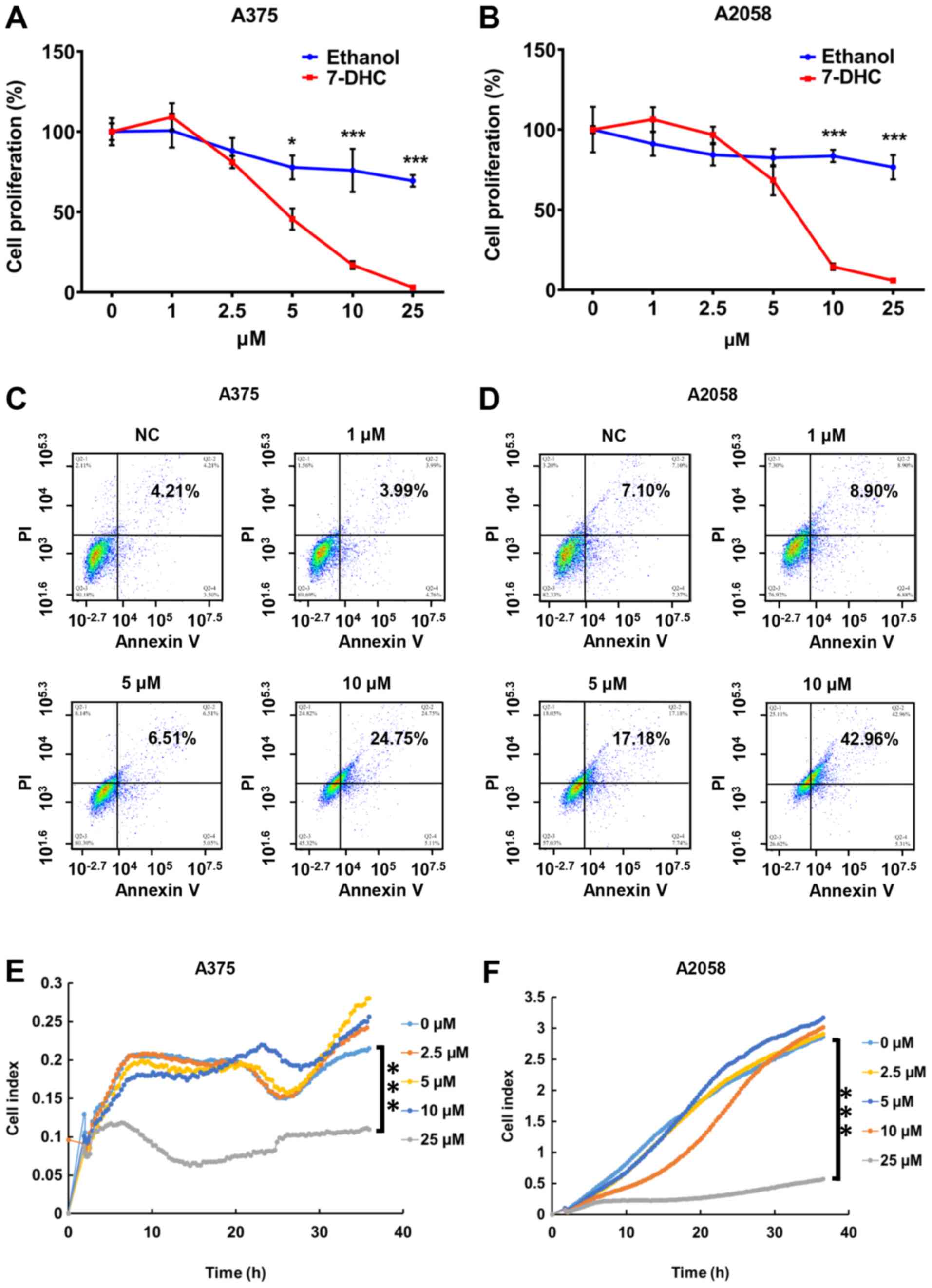 | Figure 1.7-DHC increases apoptosis and
inhibits proliferation and migration of melanoma cells. Cell
proliferation in (A) A375 and (B) A2058 melanoma cells treated with
multiple concentrations (0, 1, 2.5, 5, 10 and 25 µM) of 7-DHC and
ethanol as a control for 48 h. Apoptosis of (C) A375 and (D) A2058
melanoma cells treated with 7-DHC (1, 5 and 10 µM) for 48 h was
examined using flow cytometry, with ethanol used as a control.
Migration of (E) A375 and (F) A2058 melanoma cells treated with
7-DHC (0, 2.5, 5, 10 and 25 µM) was measured via real-time cellular
analysis assay. *P<0.05, ***P<0.001. 7-DHC,
7-dehydrocholesterol; NC, negative control. |
Considering that cell proliferation is regulated by
the cell cycle, flow cytometry was used to evaluate any changes in
the cell cycle that may lead to apoptosis and low proliferation of
melanoma cells. Flow cytometry revealed that the cell cycle of A375
and A2058 melanoma cells treated with 7-DHC was blocked at the S
phase in a dose-dependent manner (Fig.
S3A-D), which may result in apoptosis of melanoma cells and may
limit their proliferation rate.
RNA-seq analysis of melanoma cells
treated with 7-DHC
In order to further investigate the mechanism of
7-DHC on inhibiting melanoma, whole transcriptome sequencing of
A375 melanoma cells treated with 7-DHC (10 and 25 µM) and ethanol
control was performed three times. Significant differential
expression of genes in A375 cells treated with 7-DHC versus ethanol
control were screened for, revealing an intersection of
differentially expressed genes, including 65 downregulated genes
and 14 upregulated genes, shown using a Venn diagram and a heatmap
in Fig. 2A. Subsequently, RNA-seq of
A375 cells treated with 7-DHC was analyzed via GSEA to screen for
altered signaling pathways. GSEA results revealed that genes
associated with positive regulation of NF-ĸB import into nucleus
and NF-ĸB signaling were significantly downregulated in A375 cells
treated with 7-DHC compared with the ethanol negative control
(Fig. 2B). Due to the abnormality of
NF-ĸB signaling, it was postulated that 7-DHC may repress melanoma
via NF-ĸB signaling, and therefore the association between 7-DHC
and RELA, an important transcription factor of NF-ĸB signaling
(24), was analyzed. Firstly,
combined with the aforementioned RNA-seq results, three RELA-shRNA
lentiviruses were infected into A375 cells, revealing that RELA
downregulation also decreased the expression levels of target genes
(LINC00552, ADAM19 and IGFBP5) of 7-DHC compared with scrambled
shRNA (Fig. 2C). Subsequently,
ChIP-qPCR of 7-DHC target genes revealed that RELA could bind to
the promoters of the aforementioned genes, especially ADAM19 and
LINC00522 (Fig. 2D).
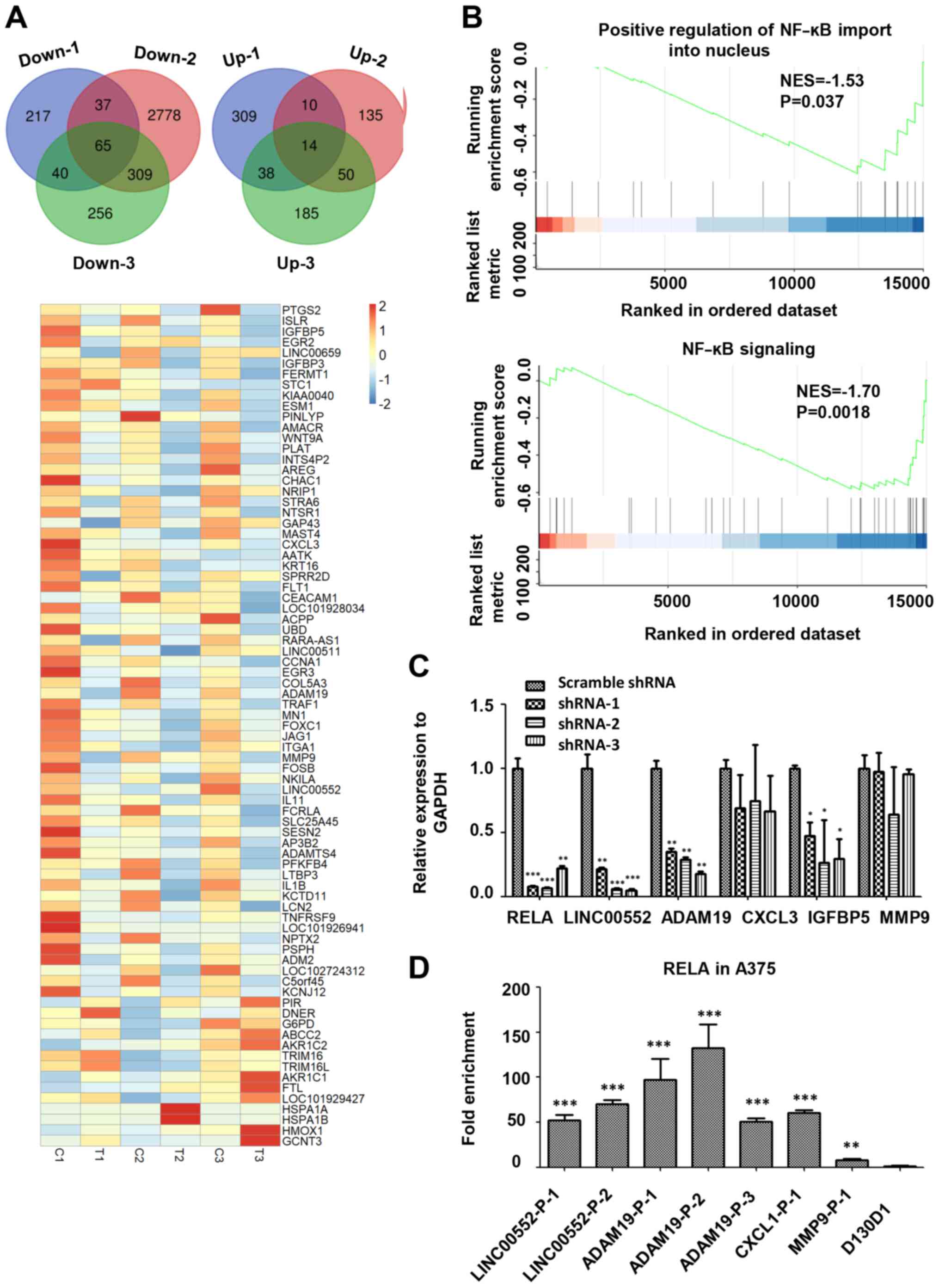 | Figure 2.RNA-seq analysis of A375 melanoma
cells treated with 7-DHC. (A) Venn diagrams show the intersection
of significant differentially expressed genes (upregulated and
downregulated) in A375 cells treated with 7-DHC (compared with
ethanol) in RNA sequencing performed three times. Heatmap of 79
significant differentially expressed genes in A375 melanoma cells
treated with 7-DHC (T1, T2, T3) compared with ethanol control (C1,
C2, C3). (B) Gene Set Enrichment Analysis demonstrated the
downregulated signaling pathways associated with NF-κB signaling in
A375 melanoma cells treated with 7-DHC. (C) Relative expression of
7-DHC target genes (RELA, LINC00552, ADAM19, CXCL3, IGFBP5 and
MMP9) in A375 cells infected with RELA-shRNA lentiviruses compared
with scrambled shRNA lentiviruses. *P<0.05, **P<0.01,
***P<0.001 vs. scramble shRNA. (D) Chromatin
immunoprecipitation-quantitative PCR of RELA bound to different
promoter regions of 7-DHC target genes (LINC00552-P-1,
LINC00552-P-2, ADAM19-P-1, ADAM19-P-2, ADAM19-P-3, CXCL1-P-1 and
MMP9-P-1) and negative control D130D1. D130D1 (negative group)
cannot bind to RELA. **P<0.01, ***P<0.001 vs. D130D1. 7-DHC,
7-dehydrocholesterol; shRNA, short hairpin RNA; NES, normalized
enrichment score; RELA, RELA proto-oncogene NF-κB subunit;
LINC00552, long intergenic non-protein coding RNA 552; ADAM19, ADAM
metallopeptidase domain 19; CXCL3, C-X-C motif chemokine ligand 3;
IGFBP5, insulin-like growth factor binding protein 5; P-1/2,
promoter region 1/2. |
7-DHC inhibits melanoma via the
Akt1/NF-κB signaling pathway
The abnormal activation of signal transduction
caused by somatic mutations is very common in patients with cancer,
particularly in those with advanced melanoma (25–28). In
order to determine the underlying molecular mechanism of 7-DHC in
melanoma, the somatic mutations of kinases, such as Akt1, Akt2 and
Akt3, involved in signal transduction were analyzed in 471 patients
with melanoma from TCGA database. As shown in Fig. 3A, the mutation rates of Akt1, Akt2
and Akt3, which belong to the PI3K/AKT signaling pathway, and BRAF,
upstream of MAPK signaling, were higher compared with kinases from
other signaling pathways. The PI3K/AKT and MAPK signaling pathways
were chosen to determine the specific effect of 7-DHC via western
blotting. Compared with the ethanol negative control, treatment
with 7-DHC decreased the protein expression levels of pAkt1-Ser473
rather than those of pAkt-Thr308 or pMEK1 (Figs. 3B and S4A). Furthermore, 7-DHC increased the
expression levels of RELA in cytoplasm in melanoma cells, which
demonstrated that the inhibition of free RELA translocation into
the nucleus (Figs. 3C and S4B) suggested that 7-DHC may promote tumor
regression by decreasing NF-κB signaling. To confirm whether 7-DHC
also inhibited RELA expression, the luciferase activity in A375
cells treated with various concentrations of 7-DHC was detected,
revealing that 7-DHC could inhibit RELA expression in a
dose-dependent manner (Fig. 3D). In
addition, although the PI3K/AKT signaling pathway is downstream of
the NF-κB signaling pathway, the association between 7-DHC and the
two aforementioned signaling pathways was investigated. Therefore,
gradient concentrations (0, 1, 5 and 10 µM) of 7-DHC were added to
A375 melanoma cells together with 1 µM IGF1, an Akt1 activator, at
37°C for 48 h, which demonstrated that 7-DHC inhibited the protein
expression levels of pAkt1-Ser473, while addition of IGF1 partially
rescued this inhibition (Fig. S4C).
Additionally, the Akt1 inhibitors MK-2206 (1 µM at 37°C for 48 h)
and AKT inhibitor III (1 µM at 37°C for 48 h) were added to A375
and A2058 melanoma cells, and reported RELA target genes were
detected via RT-qPCR. The results demonstrated that inhibiting Akt1
could restrain the expression levels of RELA target genes (Fig. S4D and E). In summary, 7-DHC may
inhibit melanoma via the Akt1/NF-κB signaling pathway.
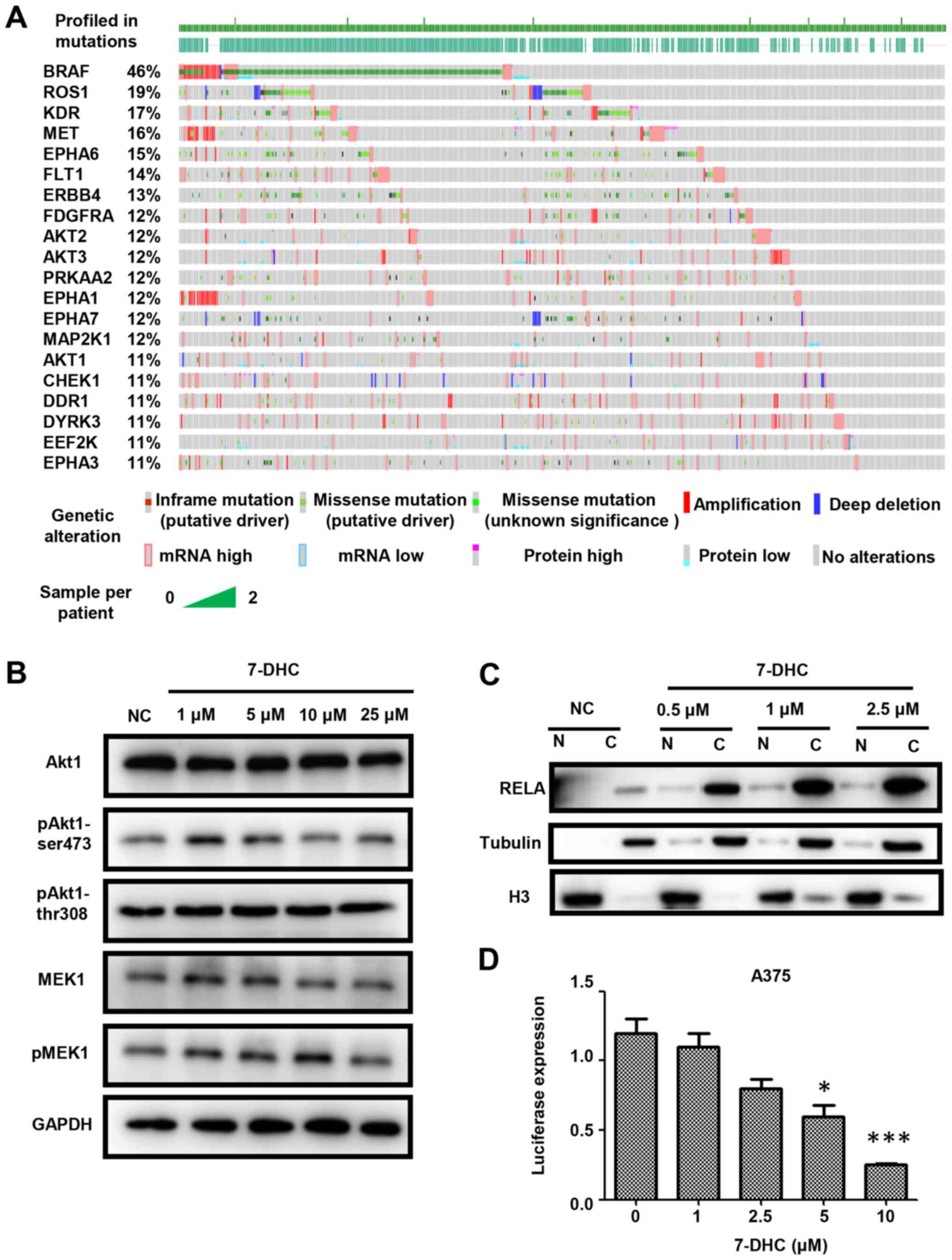 | Figure 3.7-DHC inhibits melanoma via the
Akt1/NF-κB signaling pathway. (A) Top 20 somatic mutations of
kinases via analyzing The Cancer Genome Atlas-skin cutaneous
melanoma dataset. (B) Western blot analysis of A375 cells treated
with several concentrations (1, 5, 10 and 25 µM) of 7-DHC were
compared with cells treated with ethanol as the NC for the analysis
of the levels of pAkt1-Ser473, pAkt1-Thr308/309 and pMEK1
normalized to GAPDH. (C) Western blot analysis of A375 cells
treated with several concentrations (0, 0.5, 1 and 2.5 µM) of 7-DHC
were compared with cells treated with ethanol as the NC to analyze
the entry of RELA into the nucleus. (D) Luciferase expression of
A375 cells transfected with 4 repeating motifs of RELA after adding
various concentrations (1, 2.5, 5, and 10 µM) of 7-DHC compared
with the ethanol negative control (0 µM 7-DHC). *P<0.05,
***P<0.001. 7-DHC, 7-dehydrocholesterol; RELA, RELA
proto-oncogene NF-κB subunit; p, phosphorylated; NC, negative
control; H3, histone H3; N, nuclear; C, cytoplasmic. |
7-DHC gene signature is associated
with a poor prognosis in patients with melanoma
In order to determine the core target genes of 7-DHC
in melanoma combined with prognosis, the clinical data and
transcriptome sequencing data of 471 patients with melanoma from
TCGA database (TCGA-SKCM) were analyzed. Firstly, 19 genes were
obtained by intersecting the downregulated genes in A375 cells
treated with 7-DHC and the upregulated genes in patients with
melanoma (Fig. 4A). Subsequently, a
7-DHC gene signature associated with prognosis, including STRA6,
UBD, SESN2, CHAC1, TRAF1, KIAA0040 and MMP9, was identified based
on the prognosis coefficient of the 19 aforementioned genes via
univariate Cox proportional hazards regression analysis, which
revealed that the 7 genes of the 7-DHC signature may be used as
preliminary prognostic factors in patients with melanoma (Fig. 4B). According to the median value of
the risk score calculated using the survival R package, patients
with melanoma were divided into high- and low-risk groups. The
distribution of the risk scores, along with the corresponding OS
data and the expression levels of five genes in the 7-DHC gene
signature were plotted and shown in Fig.
4D. Patients with higher risk scores tended to experience a
shorter OS time and a higher death rate compared with those with
lower risk scores. The 5-gene signature, including UBD, CHAC1,
KIAA0040, SESN2 and MMP9, was constructed to provide the guideline
for prognosis, which demonstrated that the higher the expression
levels of CHAC1, KIAA0040, SESN2 and MMP9, the higher the risk
score, while the lower the expression levels of UBD, the lower the
risk score in patients with melanoma (Fig. 4D). In addition, ROC analysis was used
to estimate the significance of the five genes in the gene
signature, and the area under the curve value of the ROC analysis
for the prognostic signature was 0.643 (Fig. 4C). To assess the overall prognosis of
the five genes in the prognosis model, Kaplan-Meier analysis
revealed that a higher risk score was associated with a higher
mortality risk (Fig. 4E).
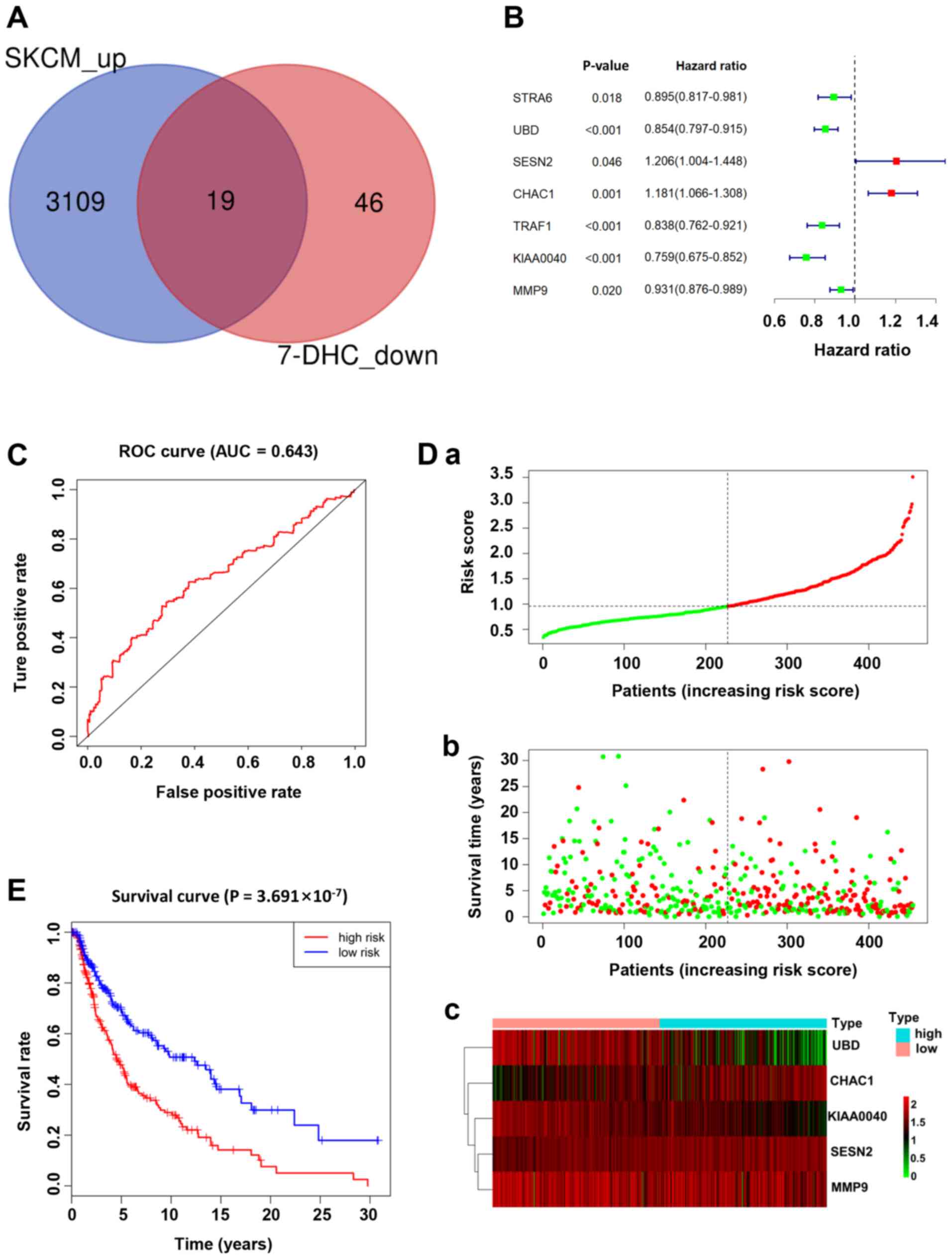 | Figure 4.7-DHC gene signature is associated
with a poor prognosis in patients with melanoma. (A) Venn diagram
showing the interaction of upregulated genes in The Cancer Genome
Atlas-skin cutaneous melanoma dataset and downregulated genes in
A375 melanoma cells treated with 7-DHC. (B) Multivariate Cox
regression analysis of seven significant genes (STRA6, UBD, SESN2,
CHAC1, TRAF1, KIAA0040 and MMP9). (C) ROC curve demonstrating the
specificity and selectivity of the gene signature. (D) Distribution
of (a) risk scores, (b) survival status and (c) expression levels
of five genes of the gene signature with significant upregulated
expression. (green and red lines/dots represent low and high risk,
respectively). (E) Survival curve of five significant differential
genes. 7-DHC, 7-dehydrocholesterol; ROC, receiver operating
characteristic; AUC, area under the curve. |
Discussion
Mammalian cholesterol synthesis is one of the most
complicated biological processes that includes 21 enzymatic steps
that generate numerous cholesterol metabolites (29). The dysregulation of cholesterol
synthesis is implicated in the progression of hypocholesterolemia,
hypercholesteremia and diabetes (30–32).
Furthermore, metabolites in cholesterol metabolism, both in
synthesis and in metabolism and transport, have been demonstrated
to promote or delay tumorigenesis and metastasis (33,34). For
example, it has been demonstrated that glucocorticoids and
Dendrogenin A function as tumor suppressors to slow tumor growth
and increase cell apoptosis in breast cancer (33). Additionally, a previous study
revealed that 7-DHC had a cytotoxic effect on melanoma cell lines
(20), but the mechanism of 7-DHC in
melanoma cells is poorly understood.
The present data confirmed the antitumor property of
7-DHC in inducing apoptosis and inhibiting proliferation and
metastasis in A375 and A2058 melanoma cells. To uncover the
molecular mechanism underlying the anticancer property of 7-DHC,
according to RNA-seq data, GSEA demonstrated that the downregulated
genes were associated with the PI3K-AKT and NF-κB signaling
pathways. Consistently, the top 20 most frequent somatic mutations
of kinase-coding genes were analyzed, revealing that AKT1, AKT2 and
AKT3 in the PI3K/AKT signaling pathway, and BRAF in the MAPK
signaling pathway were the most frequent hotspots of mutations in
patients with melanoma. Subsequent validation demonstrated that
7-DHC restrained melanoma via decreasing the phosphorylation levels
of Akt1-Ser473 and further blocked the translocation into the
nucleus of free RELA. Furthermore, 7-DHC decreased the expression
levels of RELA as assessed via the luciferase assay. Therefore, it
can be concluded that 7-DHC may repress melanoma via the Akt1/NF-κB
signaling pathway. Subsequently, a 7-DHC gene signature was
established via univariate and multivariate Cox proportional
hazards regression analysis, including five prognosis-dependent
genes identified by combining RNA-seq data from melanoma cell lines
with expression data from patients in the TCGA-SKCM dataset.
In conclusion, compared with a previous study
demonstrating the inhibitory effect of 7-DHC in melanoma cells
(20), the molecular mechanism of
the antitumor effect of 7-DHC was further investigated in the
present study. The present findings provide a molecular basis for
7-DHC as a potential novel anti-neoplastic drug. The anticancer
properties of 7-DHC were analyzed in A375 and A2058 melanoma cells,
demonstrating that 7-DHC promoted the regression of melanoma by
decreasing the phosphorylation levels of Akt1 rather than those of
MEK1, and by inhibiting the nuclear translocation of RELA.
Furthermore, the 7-DHC gene signature of genes that were downstream
of 7-DHC was identified, which was negatively associated with the
survival of patients with melanoma. The present findings shed light
on the molecular mechanism of 7-DHC in melanoma and provide a
theoretical basis for its clinical application. Although the
mechanism of 7-DHC inhibiting melanoma was elucidated in the
present study, future studies should perform further in vivo
and clinical experiments of the effect of 7-DHC in treating
melanoma.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Tianjin City (grant no. 18JCQNJC13300) and
the National Natural Science Foundation of China (grant no.
81700153).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL, CL and YY conceived and supervised the study. BL
and JL designed the experiments. JL, FZ, RZ, LC and JQ performed
the experiments. JL, LY, TC, YH, YW, MY and WX analyzed the data.
BL, CL and YY wrote the manuscript. All authors read and approved
the final version of manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schadendorf D, van Akkooi ACJ, Berking C,
Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A and Ugurel
S: Melanoma. Lancet. 392:971–984. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leong SP, Mihm MC Jr, Murphy GF, Hoon DS,
Kashani-Sabet M, Agarwala SS, Zager JS, Hauschild A, Sondak VK,
Guild V and Kirkwood JM: Progression of cutaneous melanoma:
Implications for treatment. Clin Exp Metastas. 29:775–796. 2012.
View Article : Google Scholar
|
|
4
|
Crocetti E, Mallone S, Robsahm TE, Gavin
A, Agius D, Ardanaz E, Lopez MC, Innos K, Minicozzi P, Borgognoni
L, et al: Survival of patients with skin melanoma in Europe
increases further: Results of the EUROCARE-5 study. Eur J Cancer.
51:2179–2190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Whiteman DC, Pavan WJ and Bastian BC: The
melanomas: A synthesis of epidemiological, clinical,
histopathological, genetic, and biological aspects, supporting
distinct subtypes, causal pathways, and cells of origin. Pigment
Cell Melanoma Res. 24:879–897. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pennello G, Devesa S and Gail M:
Association of surface ultraviolet B radiation levels with melanoma
and nonmelanoma skin cancer in United States blacks. Cancer
Epidemiol Biomarkers Prev. 9:291–297. 2000.PubMed/NCBI
|
|
7
|
Rodriguez-Cerdeira C, Gregorio MC,
López-Barcenas A, Sánchez-Blanco E, Sánchez-Blanco B, Fabbrocini G,
Bardhi B, Sinani A and Guzman RA: Advances in immunotherapy for
melanoma: A comprehensive review. Mediators Inflamm.
2017:32642172017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shain AH, Yeh I, Kovalyshyn I, Sriharan A,
Talevich E, Gagnon A, Dummer R, North J, Pincus L, Ruben B, et al:
The genetic evolution of melanoma from precursor lesions. N Engl J
Med. 373:1926–1936. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo D, Lui GYL, Lai SL, Wilmott JS, Tikoo
S, Jackett LA, Quek C, Brown DL, Sharp DM, Kwan RYQ, et al: RAB27A
promotes melanoma cell invasion and metastasis via regulation of
pro-invasive exosomes. Int J Cancer. 144:3070–3085. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Michielin O, van Akkooi ACJ, Ascierto PA,
Dummer R and Keilholz U; ESMO Guidelines Committee. Electronic
address, : clinicalguidelines@esmo.org: Cutaneous melanoma: ESMO
clinical practice guidelines for diagnosis, treatment and
follow-up†. Ann Oncol. 30:1884–1901. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Duggan MA, Anderson WF, Altekruse S,
Penberthy L and Sherman ME: The surveillance, epidemiology, and end
results (SEER) program and pathology: Toward strengthening the
critical relationship. Am J Surg Pathol. 40:e94–e102. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Flaherty KT: Narrative review: BRAF opens
the door for therapeutic advances in melanoma. Ann Intern Med.
153:587–591. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Menzies AM and Long GV: Systemic treatment
for BRAF-mutant melanoma: Where do we go next? Lancet Oncol.
15:E371–E381. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Korman JB and Fisher DE: Developing
melanoma therapeutics: Overview and update. Wiley Interdiscip Rev
Syst Biol Med. 5:257–271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prabhu AV, Luu W, Sharpe LJ and Brown AJ:
Cholesterol-mediated degradation of 7-dehydrocholesterol reductase
switches the balance from cholesterol to vitamin D synthesis. J
Biol Chem. 291:8363–8373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Piotrowska A, Wierzbicka J and Żmijewski
MA: Vitamin D in the skin physiology and pathology. Acta Biochim
Pol. 63:17–29. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao J, Li W, Zheng X, Qi L, Wang H, Zhang
C, Wan X, Zheng Y, Zhong R, Zhou X, et al: Targeting
7-dehydrocholesterol reductase integrates cholesterol metabolism
and IRF3 activation to eliminate infection. Immunity.
52:109–122.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Tian N, Li C, Hou Y, Wang X and
Zhou Q: Incorporation of 7-dehydrocholesterol into liposomes as a
simple, universal and efficient way to enhance anticancer activity
by combining PDT and photoactivated chemotherapy. Chem Commun
(Camb). 55:14081–14084. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tian NN, Li C, Tian N, Zhou QX, Hou YJ,
Zhang BW and Wang XS: Syntheses of 7-dehydrocholesterol peroxides
and their improved anticancer activity and selectivity over
ergosterol peroxide†. New J Chem. 41:14843–14846. 2017.
View Article : Google Scholar
|
|
20
|
Gelzo M, Granato G, Albano F, Arcucci A,
Dello Russo A, De Vendittis E, Ruocco MR and Corso G: Evaluation of
cytotoxic effects of 7-dehydrocholesterol on melanoma cells. Free
Radical Bio Med. 70:129–140. 2014. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chistiakov DA and Chekhonin VP:
Circulating tumor cells and their advances to promote cancer
metastasis and relapse, with focus on glioblastoma multiforme. Exp
Mol Pathol. 105:166–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaltschmidt B, Greiner JFW, Kadhim HM and
Kaltschmidt C: Subunit-specific role of NF-κB in cancer.
Biomedicines. 6:442018. View Article : Google Scholar
|
|
25
|
Hinz N and Jücker M: Distinct functions of
AKT isoforms in breast cancer: A comprehensive review. Cell Commun
Signal. 17:1542019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wise HM, Hermida MA and Leslie NR:
Prostate cancer, PI3K, PTEN and prognosis. Clin Sci (Lond).
131:197–210. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tilborghs S, Corthouts J, Verhoeven Y,
Arias D, Rolfo C, Trinh XB and van Dam PA: The role of nuclear
factor-kappa B signaling in human cervical cancer. Crit Rev Oncol
Hematol. 120:141–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Atefi M, Avramis E, Lassen A, Wong DJ,
Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, et
al: Effects of MAPK and PI3K pathways on PD-L1 expression in
melanoma. Clin Cancer Res. 20:3446–3457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Silvente-Poirot S and Poirot M: Cancer.
Cholesterol and cancer, in the balance. Science. 343:1445–1446.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moutzouri E, Elisaf M and Liberopoulos EN:
Hypocholesterolemia. Curr Vasc Pharmacol. 9:200–212. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bentz MH and Magnette J:
Hypocholesterolemia in the acute phase of inflammation during
sepsis. Rev Med Int. 19:168–172. 1998. View Article : Google Scholar
|
|
32
|
Mokdad AH, Bowman BA, Ford ES, Vinicor F,
Marks JS and Koplan JP: The continuing epidemics of obesity and
diabetes in the United States. JAMA. 286:1195–1200. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
de Medina P, Paillasse MR, Segala G,
Voisin M, Mhamdi L, Dalenc F, Lacroix-Triki M, Filleron T, Pont F,
Saati TA, et al: Dendrogenin A arises from cholesterol and
histamine metabolism and shows cell differentiation and anti-tumour
properties. Nat Commun. 4:18402013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Q, Ishikawa T, Sirianni R, Tang H,
McDonald JG, Yuhanna IS, Thompson B, Girard L, Mineo C, Brekken RA,
et al: 27-Hydroxycholesterol promotes cell-autonomous, ER-positive
breast cancer growth. Cell Rep. 5:637–645. 2013. View Article : Google Scholar : PubMed/NCBI
|


















