Introduction
Tripartite motif 59 (TRIM59) is a member of the TRIM
family, which is characterized by an N-terminal really interesting
new gene (RING)-finger domain, followed by two zinc-finger domains,
B2 box and coiled-coil (1). TRIM59
protein has a domain at the C-terminal trans-membrane region
(2). Ubiquitination serves a vital
role in the degradation of several types of proteins that function
in the intracellular signaling pathway, cell cycle, DNA repair and
transcriptional regulation (2). E3
ubiquitin ligases directly recognize substrates on the basis of
domain structures. There are two families of E3 ubiquitin ligases:
The homologous to E6-AP COOH terminal family and the
RING-finger-containing protein family (2). Due to the RING-finger domain, TRIM59
protein functions as a E3 ubiquitin ligase and can selectively
target ubiquitin-modified proteins for proteasomes or degradation
(2). In addition, TRIM proteins can
positively or negatively promote carcinogenesis (3). The present study focused on the TRIM59
protein and its oncogenic activity in the promotion of human
esophageal cancer (hESC) cell proliferation and metastasis. Matrix
metalloproteinases (MMPs) play a key role in tumorigenesis,
including early carcinogenesis events, tumor metastasis and
invasion (4). Given the ability of
tumor cells to infiltrate and disseminate, the role of MMP in cell
invasion is important (5). Certain
MMPs, such as MMP2 and MMP9 have special mechanism to help the
invasion of cancer cells (5). MMP2
and MMP9 are family members of zinc-dependent endoproteases,
responsible for degrading the extracellular matrix (ECM) by
destroyingthe structure of various proteins (5). Degradation of the basement membrane and
subsequently of the ECM is critical for invasion, and MMP2/9 are
vital factors in providing the invasive and metastatic
characteristics of malignant tumor cells by enabling their
infiltration and migration in the process of EMT (5).
The p53 protein, the tumor suppressor gene tumor
protein p53 (TP53), is mutated in ~50% of human cancer and serves a
vital role in the response of malignant and non-transformed cells
to several anticancer therapeutics, apoptosis, cell cycle arrest
and DNA repair (6). A number of
studies have demonstrated that mutant p53 expression promotes
cancer cell proliferation, and wild-type p53 overexpression
decreases transformed cancer cell proliferation, which is evidence
of p53 serving as a tumor suppressor (7,8). Several
types of p53 mutations in cancer are point mutations in the
DNA-binding domain; however, patients with Li-Fraumeni syndrome
carry heterozygous germline p53 mutations and present with
different types of cancer throughout their lifetime (7,8). The
mutant p53 proteins are not considered as functional proteins that
can regulate the transcription of wild-type p53 target genes
(6). Gene expression studies have
reported that the wild-type p53 protein level was decreased in hESC
compared with adjacent healthy tissues (7,8). The
reactivation of the p53 signaling pathway in response to several
types of cancer may become an attractive means to understand hESC
growth and mobility. However, why the wild-type p53 protein level
is downregulated in hESC tissues and cell lines is not completely
understood.
hESC is the most common type of digestive tumor,
with high incidence (572,000 new cases in 2018) and mortality
(509,000 mortalities in 2018) rates worldwide (9). Esophageal squamous cell carcinoma
(ESCA) is the most common in the Taihang Mountain Area of China,
with patients usually diagnosed at an advanced stage, due to the
lack of no obvious symptoms at the early stage (10). Chemoradiation therapy is the most
effective treatment strategy for hESC after surgery (10). Cisplatin is an effective chemotherapy
drug, but cisplatin resistance frequently occurs in the
cisplatin-based treatment of ESCA (11). Therefore, identifying the mechanism
underlying cisplatin resistance in hESC requires further
investigation.
Materials and methods
Surgical hESC samples
A total of 112 hESC and paired adjacent healthy
tissues (5 cm away from tumor margin) were obtained by surgical
removal from patients with hESC at the Forth Affiliated Hospital of
Hebei Medical University (Shijiazhuang, China) between January 2015
and December 2017 (Table I). The
clinicopathological characteristics of the patients are presented
in Table I. A total of 85 men and 27
women were included in the present study. The median age was 60
years (age range, 50–80 years). All tissues were immediately frozen
in liquid nitrogen and stored at −80°C. Written informed consent
was provided by all patients or their relatives. There were several
informed consents signed by patients' relatives because these
patients had limited capacity, resulting in them being unable to
sign the contents. The present study was approved by the Research
Ethics Committees of the Forth Hospital of Hebei Medical University
(Shijiazhuang, China).
 | Table I.Characteristics of patients with
human esophageal cancer. |
Table I.
Characteristics of patients with
human esophageal cancer.
|
Characteristics | n | TRIM59 negative/low
expression (n=42) | TRIM59 high
expression (n=70) | P-value | Statistical
method |
|---|
| Age (years) |
|
|
| 0.658 |
|
|
<60 | 84 | 29 | 55 |
| Mann-Whitney U
test |
|
≥60 | 28 | 13 | 15 |
|
|
| Gender |
|
|
| 0.284 |
|
|
Male | 85 | 26 | 59 |
| Mann-Whitney U
test |
|
Female | 27 | 16 | 11 |
|
|
| Histological
type |
|
|
| 0.987 |
|
|
Squamous cell carcinoma | 110 | 40 | 70 |
| Mann-Whitney U
test |
|
Adenocarcinomas | 2 | 2 | 0 |
|
|
| Localization |
|
|
| 0.692 |
|
|
Upper | 20 | 8 | 12 |
| Kruskal-Wallis
test |
|
Middle | 56 | 14 | 42 |
|
|
|
Lower | 36 | 20 | 16 |
|
|
| Invasive depth |
|
|
| 0.000a |
|
|
T1-2 | 20 | 11 | 9 |
| Kruskal-Wallis
test |
| T3 | 50 | 25 | 25 |
|
|
| T4 | 42 | 6 | 36 |
|
|
| pTNM stage |
|
|
| 0.000a |
|
| I | 20 | 8 | 12 |
| Kruskal-Wallis
test |
| II | 30 | 18 | 12 |
|
|
|
III | 56 | 16 | 40 |
|
|
| IV | 6 | 0 | 6 |
|
|
| Lymph node
metastasis |
|
|
| 0.003a |
|
|
Absence | 55 | 14 | 41 |
| Mann-Whitney U
test |
|
Presence | 57 | 28 | 29 |
|
|
Cell lines and antibodies
hESC cell lines (Eca109, KYSE150, KYSE30, KYSE510
and EC9706) and human esophageal epithelial cells (HEEC) were
purchased from the American Type Culture Collection. hESC cells
were maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.),
while HEEC cells were maintained in RPMI-1640 (Corning Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C with 5% CO2. The Eca109 cell line was a
stable cisplatin-resistance cell line, as determined by continuous
screening. The HEK293T cell line was purchased from the Cell Bank
of the Institute for Biological Science, Chinese Academic of
Science and cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS and 1% penicillin-streptomycin (all
purchased from Gibco; Thermo Fisher Scientific, Inc.) at 37°C with
5% CO2.
The following antibodies were used for western
blotting, immunofluorescence and immunoprecipitation. Primary
antibodies targ-eted against: TRIM59 (cat. no. ab69639; 1:500 for
western blotting, 1:300 for immunofluorescence and 10 µg/ml for
immunoprecipitation; Abcam), p53 (cat. no. ab26; 10 µg/ml for
western blotting and 5 µg/ml for immunoprecipitation; Abcam),
matrix metallopeptidase (MMP)9 (cat. no. ab38898; 1:1,000; Abcam),
MMP2 (cat. no. ab97779; 1:1,000; Abcam), Bcl-2 (cat. no. ab32124;
1:1,000; Abcam), Tublin (cat. no. 801213; 1:5,000; BioLegend, Inc.)
and GAPDH (cat. no. 649201; 1:5,000; BioLegend, Inc.). The
secondary antibody (anti-Rabbit: Cat. no. 926-32211; anti-Mouse:
Cat. no. 925-32210; anti-Human: Cat. no. 926-32232; 1:5,000;
LI-COR) was purchased from LI-COR Biosciences. The FITC Annexin V
Apoptosis Detection kit with PI (cat. no. 640914) was purchased
from BioLegend, Inc.
Chemotherapy drug
Cisplatin was purchased from Nanjing Pharmaceutical
Factory Co., Ltd.
Gene expression profiling of
esophageal cancer
The Cancer Genome Atlas database (https://portal.gdc.cancer.gov) and Gene Expression
Omnibus (GEO) database (dataset no. GEO30480, http://www.ncbi.nlm.nih.gov/geo) were used to
analyze the gene expression profile of TRIM59 protein (12).
Immunoblotting, immunofluorescence,
co-immunoprecipitation
For immunoblotting, cells were collected and total
protein was isolated using high RIPA lysis buffer (cat. no.
Ab156034; 1:10; Abcam) containing 1 mM PMSF. Cell lysates were
centrifuged at 12,000 × g for 15 min at 4°C, and total protein was
quantified using the BCA method (cat. no. Ab102536; Abcam).
Proteins (40 µg) were added to 5X loading buffer for 5 min at 95°C,
separated via 12% SDS-PAGE and transferred to PVDF membranes
(Immobilon-FL; EMD Millipore). The membranes were blocked with 5%
skimmed milk powder in PBS for 2 h at room temperature. The
membranes were incubated with primary antibodies overnight at 4°C.
The second antibodies were used to incubate the membrane for 2 h at
25°C. TRIM59, p53, MMP9 and MMP2 protein bands were visualized
using the Odyssey Imaging Systems (ODYSSEY®, LI-COR
Biosciences) and Application software (version 2.1.12; LI-COR
Biosciences).
For immunofluorescence, following fixation with 4%
paraformaldehyde for 2 h at 4°C, frozen sections embedded with
tissue freezing medium (cat. no. 03812525; Leica Biosystem
Richmond, Inc.) were permeabilized with 0.3% Triton X-100/PBS for 1
h at 37°C and blocked with 10% normal goat serum (cat. no. Ab7481;
1:1,000; Abcam) in PBS for 30 min at 37°C. Tissue slides were
incubated with primary antibodies overnight at 4°C. Subsequently,
tissues were incubated with a fluorescent secondary antibody (cat.
no. 111-165-003; 1:400; Jackson ImmunoResearch Inc.) for 1 h at
room temperature, followed by washing three times with PBS (10 min
per time). Nuclear staining was performed using DAPI for 10 min in
the dark at room temperature. Following washing one time with PBS
and drying, SlowFade™ Diamond Antifade Mountant solution (cat. no.
S36963; Thermo Fisher Scientific, Inc.) was added to the tissues
and cover glasses were placed on the slides. Stained slides were
observed under a fluorescence microscope (magnification, ×20) and
analyzed using LAS AF software (version 2.5.2 build 6939; Leica
Microsystems).
For immunoprecipitation assays, at 36 h
post-transfection (Lipofectamine™ 2000 Transfection Reagent; cat.
no. 11668019; Thermo Fisher Scientific, Inc.), cells were rinsed
with PBS and lysed in RIPA buffer (cat. no. Ab156034; 1:10; Abcam)
supplemented with a protease and phosphatase inhibitor cocktail
(cat. no. 78427; 1:100; Thermo Fisher Scientific, Inc.) for 30 min
on ice. Whole cell extract was incubated with indicated primary
antibodies or with anti-IgG (cat. no. Ab181236; 1:10,000; Abcam) in
a vertical rocker overnight at 4°C. Protein A/G beads (50 µl; cat.
no. 20421; Thermo Fisher Scientific, Inc.) were added and incubated
for another 4 h at 4°C to pull down the TRIM59 or p53 protein.
Following incubation, the agarose gels were washed five times with
lysis buffer and boiled for 5 min at 98°C with 4X SDS loading
buffer, and then subjected to 12% SDS-PAGE. Proteins were
transferred onto PVDF membranes (Immobilon-FL; EMD Millipore).
Membranes were blocked with 5% BSA (cat. no. V900933;
Sigma-Aldrich; Merck KGaA) at 25°C for 2 h and incubated with
primary antibodies overnight at 4°C. Following the primary
incubation, membranes were incubated with anti-rabbit second
antibody (cat. no. 926-32211; 1:5,000) or anti-mouse second
antibody (cat. no. 925-32210; 1:5,000) at 25°C for 1.5 h. The
membranes were detected using an Odyssey infrared scanner (LiI-COR
Biosciences) and assayed using Application software (version
2.1.12; LI-COR Biosciences).
Wound healing and clone formation
assays
Eca109 cells were plated in 6-well plates. At
90–100% confluence, cells were cultured in medium containing 0.5%
FBS (cat. no. 10099141; Gibco; Thermo Fisher Scientific, Inc.) for
24 h. Subsequently, cells were treated with 20 µg/ml mitomycin C
(cat. no. 4287; Merck KGaA) for 4 h at 37°C. Subsequently, a wound
was scratched using sterile pipette tips in vertical and horizontal
directions. Cells were gently washed three times with PBS and
cultured with complete medium containing 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) (12).
Notably, cells in 10% FBS-supplemented medium exhibited a
significantly enhanced proliferative ability after the
aforementioned treatments. Thus, the present study investigated
whether 10% FBS-supplemented medium can improve cell viability
while maintaining effective propagation. The width of the scratched
area was recorded and photographed using an inverted light-field
microscope (magnification, ×10) every 12 h. The quantitative
analysis of the scratch was computed and the differentiated images
were presented when cells were cultured for 48 h following
transfection.
For clone formation assay, cells were seeded
(0.5–1×104 cells/well) into 6-well plates and cultured
for ~2 weeks. When clones grew to >50 µm in diameter, clones
were defined and subsequently fixed with 4% paraformaldehyde for 30
min at 25°C and stained with 0.5% crystal violet staining solution
for 30 min at 25°C. Cells were observed under an inverted
light-field microscope (magnification, ×10).
Cell Counting Kit-8 (CCK-8)
A CCK-8 assay (cat. no. HY-K0301; MedChemExpress)
was performed to evaluate cell proliferation according to the
manufacturer's instructions. Cells were seeded (1×105)
into 96-well plates and incubated for 0, 24, 48 and 72 h at 37°C.
Subsequently, 10 µl CCK-8 reagent was added into each well and
incubated at 4 h at 37°C. Absorbance was measured at a wavelength
of 450 nm using a FLUOstar Omega microplate reader (BMG Labtech
GmbH).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from tissues and cells using
the RNeasy mini kit (Qiagen AB) according to the manufacturer's
instructions. The remaining DNA was removed by adding DNase I.
Total RNA was reverse transcribed into cDNA using the
Advantage® RT-for-PCR kit (cat. no. 639505; Clontech
Laboratories, Inc.). The reaction conditions for reverse
transcription were 37°C for 30 min and 85°C for 10 sec.
Subsequently, qPCR was performed using the Real-Time PCR Detection
system (Bio-Rad Laboratories, Inc.) with TB Green®
Premix Ex Taq™ (cat. no. RR420Q; Takara Bio Inc.). The following
thermocycling conditions were used for qPCR: 95°C for 50 sec; 40
cycles at 95°C for 5 sec and 60°C for 50 sec. The following primers
were used for qPCR: TRIM59 forward,
5′-CCTGTGTTTGAGATAGATTTAAGAGC-3′ and reverse,
5′-GCAACAAGGTGAGACCCAGT-3′; GAPDH forward,
5′-TTGATGGCAACAATCTCCAC-3′ and reverse, 5′-CGTCCCGTAGACAAAATGGT-3′;
Bcl-2 forward, 5′-ACTGAGTACCTGAACCGGCA-3′ and reverse,
5′-GAAATCAAACAGAGGCCGCAT-3′; MMP2 forward,
5′-ACAAAGGGATTGCCAGGACC-3′ and reverse, 5′-CGGTGGCCTGGGGTTTG-3′;
MMP9 forward, 5′-CCTGGGCAGATTCCAAACCT-3′ and reverse,
5′-CAAAGGCGTCGTCAATCACC-3′; p53 forward, 5′-AAGTCTAGAGCCACCGTCCA-3′
and reverse, 5′-CAATCCAGGGAAGCGTGTCA-3′. mRNA expression levels
were quantified using the 2−ΔΔCq method (12) and normalized to the internal
reference gene GAPDH.
RNA interference
TRIM59 siRNA, and scrambled NC were synthesized by
Sangon Biotech Co., Ltd. Eca109 cells (1×105/well) were
seeded and cultured for 24 h. Subsequently, Eca109 cells were
transfected with 2 µg TRIM59 siRNA or NC using
Lipofectamine® 3000 (cat. no. L3000015; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. At 48 h post-transfection, transfection efficiency
was assessed via RT-qPCR, fluorescence microscopy and western
blotting. The method of siTP53transfection performed according to
the same protocol described for TRIM59 siRNA transfection. The
sequences of the siRNAs were as follows: siTRIM59a,
5-CCCUGAACAUUACAGGCAATT-3′; siTRIM59b, CCAGCATGTACAGATCTTGAAA; NC
forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′; and siTP53, 5′-CUACUUCCUGAAAACAACG-3′;
and NC for TP53, 5′-UUCUCCGAACGUGUCACGUTT-3′.
Statistical analysis
All experiments were performed in triplicate and
data are presented as the mean ± standard deviation. Statistical
analyses were performed using GraphPad Prism (version 6; GraphPad
Software, Inc.) and SPSS (version 21.0; IBM Corp.). A χ2
test was used to examine the association between TRIM59 expression
and clinicopathological features. The Kaplan-Meier method with
log-rank tests was used to estimate overall patient survival.
Comparisons between two groups were analyzed using the paired
Student's t-test. Comparisons among multiple groups were analyzed
using one-way analysis of variance, followed by Tukey's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Upregulation of TRIM59 in hESC tissues
and cell lines, and its association with poor prognosis
In a previous study, TRIM59 promoted the growth of
several types of cancer, including non-small cell lung cancer
(12); however, its function in hESC
cells is not completely understood. To investigate the significance
of TRIM59 in hESC cells, the GSE30480 dataset was downloaded from
the GEO database and used to analyze the multiple microarray
dataset from Tan et al (12).
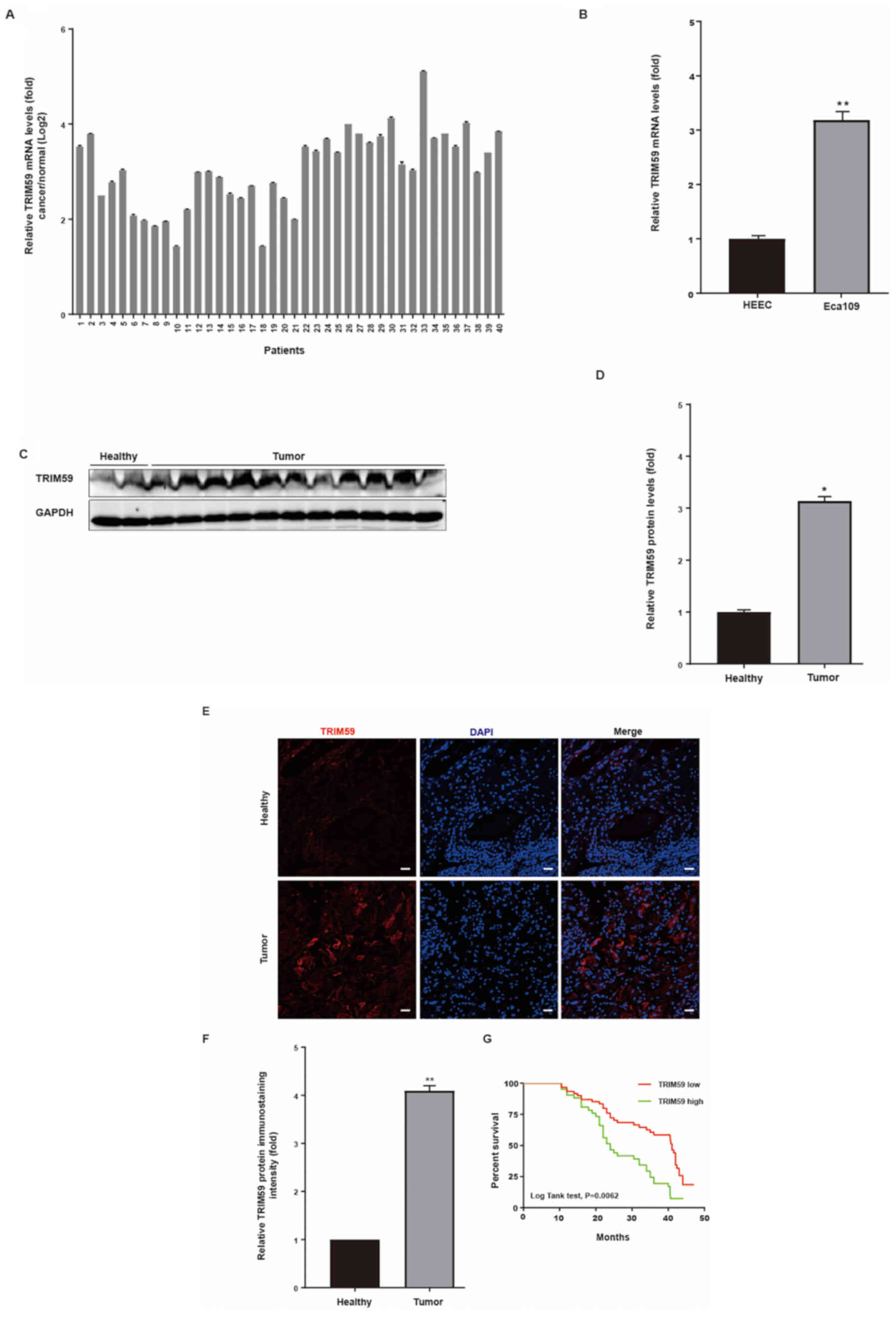
The results indicated that TRIM59 mRNA levels were enhanced in hESC
cells derived from tumor tissues compared with hESC cells derived
from healthy adjacent tissues (Fig.
1A). Moreover, by analyzing the relevant clinical data, the
results indicated that increased TRIM59 expression was associated
with decreased survival in patients with hESC (Fig. 1A). Collectively, the results
suggested that the high expression of TRIM59 in patients with hESC
was positively associated with poor prognosis.
To verify the results of the microarray analysis,
the mRNA and protein expression levels of TRIM59 were detected in
hESC tissues and matched adjacent healthy tissues via RT-qPCR. The
RT-qPCR results indicated that TRIM59 was significantly increased
in hESC tissues compared with the adjacent healthy esophageal
tissues (Fig. 1D). The mRNA
expression levels of TRIM59 were detected in a hESC cell line
(Eca109) using HEEC as a non-malignant control. The results
indicated that TRIM59 expression was significantly increased in
Eca109 cells compared with HEEC cells (Fig. 1B).
The protein expression levels of TRIM59 in hESC
tissues were further verified via western blotting. Among the 112
patients, TRIM59 protein expression was detected in 112 biopsy
samples compared with the matched adjacent healthy tissues, and the
representational figure using the tissues from 11 cancer tissues
and 2 adjacent normal tissues (Fig. 1C
and D). Therefore, the high protein expression level of TRIM59
in hESC may originate from the upregulation of TRIM59
transcription.
To further investigate the association between
TRIM59 and hESC, immunofluorescence staining of TRIM59 was
performed on primary tumors and matched adjacent healthy tissues.
Among the 112 specimens, 112 biopsy samples contained tumor and
matched adjacent tissues. TRIM59 expression was significantly
increased in hESC tissues compared with adjacent healthy tissues,
as determined by immunofluorescence staining (Fig. 1E and F). In addition, Kaplan-Meier
analysis was conducted to assess the association between high
TRIM59 expression and patient survival. High TRIM59 expression was
associated with decreased patient survival (Fig. 1G).
TRIM59 promotes hESC cell migration
and invasion in vitro
To determine the clinical characteristics and
prognostic value of TRIM59 expression in hESC, its potential
biological function was further explored. First, the protein
expression levels of TRIM59 were detected in Eca109, EC9706, HYSE30
and HYSE150 cell lines. The results indicated that TRIM59
expression levels were significantly higher in Eca109 cells
compared with the HEEC cell line (Fig.
2A and B), which was observed in all four cell lines. Higher
TRIM59 expression levels were observed in Eca109 cells, whereas
lower TRIM59 expression levels were observed in EC9706, HYSE30 and
HYSE150 cells. Increased TRIM59 protein expression levels in Eca109
cells were consistent with the trends of increased TRIM59
expression in tumor specimens compared with healthy tissues
identified via western blotting and RT-qPCR.
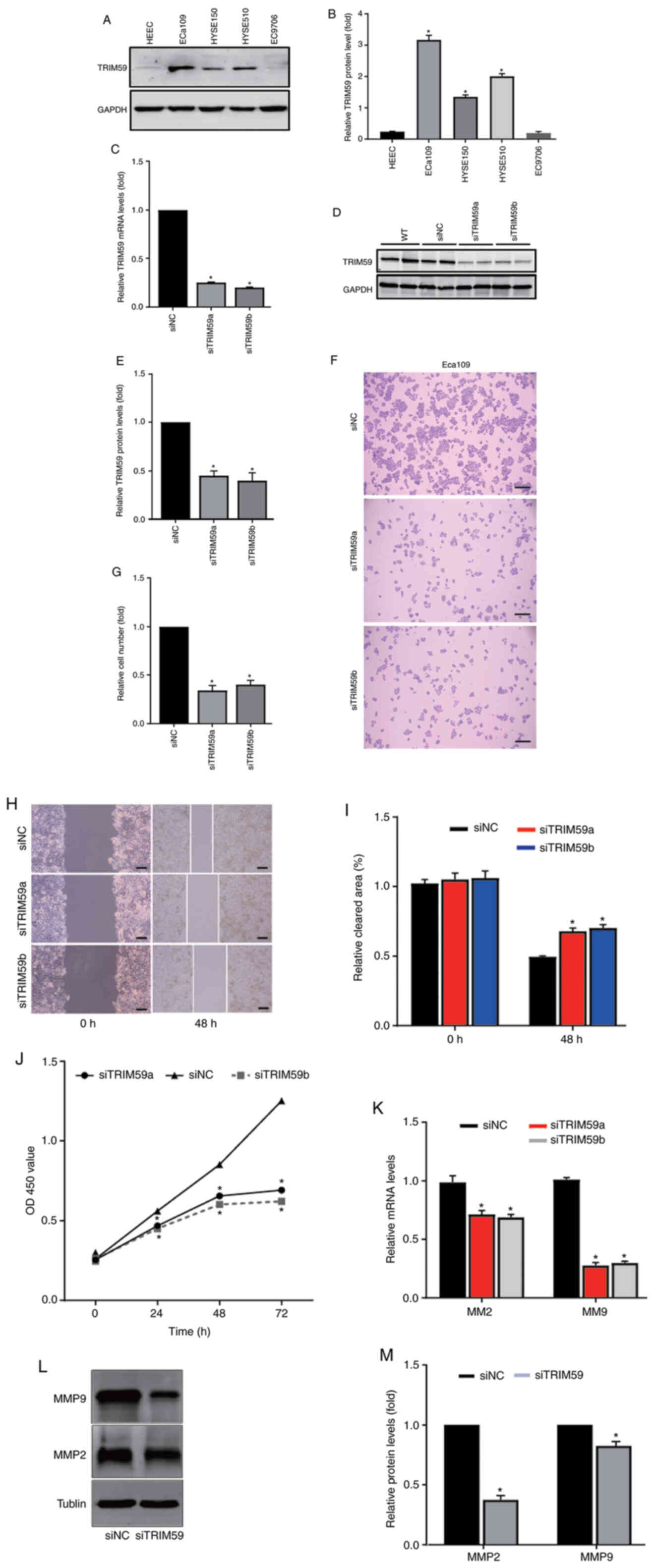 | Figure 2.TRIM59 knockdown inhibits hESC cell
migration and invasion. TRIM59 protein expression levels were (A)
determined via western blotting and (B) semi-quantified. (C) At 48
h post-transfection, TRIM59 mRNA expression levels were measured
via reverse transcription-quantitative PCR. At 48 h
post-transfection, TRIM59 protein expression levels were (D)
determined via western blotting and (E) semi-quantified. The effect
of TRIM59 knockdown on hESC cell proliferation was (F) determined
by performing a colony formation assay (scale bar, 200 µm) and (G)
quantified. The effect of TRIM59 knockdown on hESC cell migration
was (H) assessed by performing a wound healing assay (scale bar,
200 µm) and (I) quantified. (J) Eca109 cell viability was measured
at different time points using a Cell Counting Kit-8 assay. (K) The
effect of TRIM59 knockdown on MMP2 and MMP9 mRNA expression levels.
The effect of TRIM59 knockdown on MMP2 and MMP9 protein expression
levels was (L) assessed via western blotting and (M)
semi-quantified. *P<0.05 vs. Eca109 or siNC. TRIM59, tripartite
motif-containing 59; hESC, human esophageal cancer; MMP, matrix
metallopeptidase; si, small interfering RNA; NC, negative control;
WT (function as an ineffective proof of siNC), wild-type; OD,
optical density. |
To explore the effect of TRIM59 knockdown on cell
proliferation, two TRIM59-targeting siRNAs (siRNAa and siRNAb) or
scramble siRNA were used to examine the biological function of
TRIM59 in hESC cell lines. The results suggested that both
TRIM59-targeting siRNAs significantly decreased TRIM59 protein
expression levels in the Eca109 cell line compared with the
scramble siRNA. Therefore, both siRNAs were used in subsequent
experiments due to their knockdown effect. The RT-qPCR and western
blotting results suggested that TRIM59 siRNAs significantly
decreased the mRNA and protein expression levels of TRIM59 in the
Eca109 cell line compared with siNC (Fig. 2C-E).
Following TRIM59 knockdown, alterations in cell
numbers, migration and viability were investigated using a hESC
cell line. To assess cell proliferation, colony formation assays
were performed. The results indicatedthat TRIM59 knockdown
significantly decreased the colony formation ability of the Eca109
cell line compared with the siNC group (Fig. 2F and G).
The clinical investigation and analysis of hESC
indicated that patients with higher TRIM59 expression displayed
poor survival (Fig. 1G). First, to
examine the biological function of TRIM59 in tumor migration, wound
healing assays were performed. The results suggested that TRIM59
knockdown significantly inhibited Eca109 cell migration by 50–60%
compared with the siNC group (Fig. 2H
and I). Subsequently, the effects of TRIM59 knockdown on Eca109
cell viability were assessed by performing a CCK-8 assay. The
results indicated that TRIM59-knockdown Eca109 cells displayed
significantly reduced cell viability compared with the siNC group
at 24, 36 and 48 h (Fig. 2J).
Based on the association between cell proliferation
and migration-related proteins, the association between matrix
metalloproteinases, including MMP2 and MMP9, and TRIM59 was
examined in Eca109 cells. RT-qPCR and western blotting were
performed to quantify MMP2 and MMP9 expression levels in
TRIM59-knockdown Eca109 cells.
MMP2 and MMP9 mRNA expression levels were
significantly decreased in TRIM59-knockdown cells compared with the
siNC group (Fig. 2K). The results
also indicated that TRIM59 knockdown significantly reduced MMP2 and
MMP9 protein expression levels compared with the siNC group
(Fig. 2L and M). The results
suggested that TRIM59 promoted hESC cell proliferation, migration
and invasion.
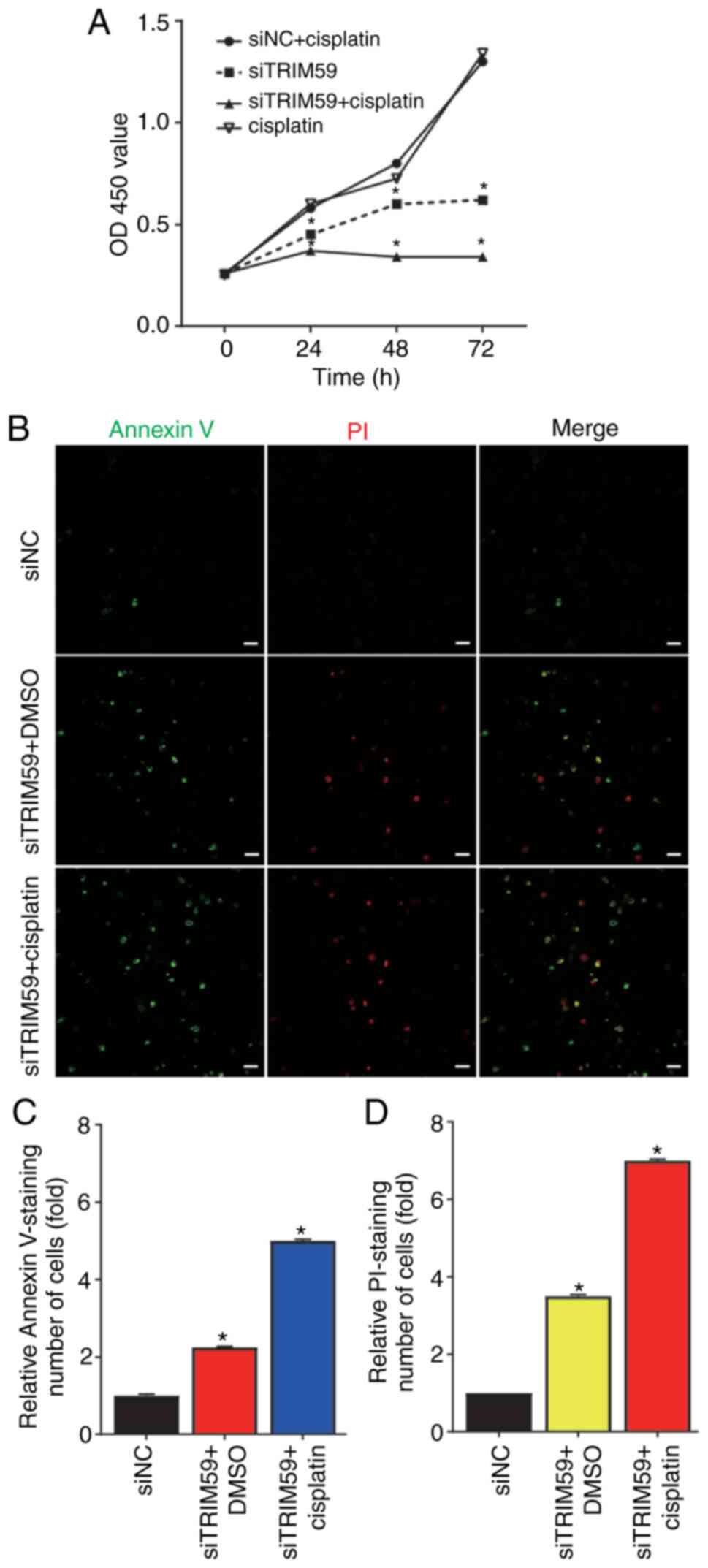
TRIM59 knockdown enhances Eca109
chemosensitivity to cisplatin
hESC progression and chemoresistance serve important
roles in patient survival and prognosis (11). It was predicted that cisplatin, the
most prevalent first-line treatment for hESC, would become
non-sensitive to tumor cells, resulting in treatment failure.
Therefore, elucidating the mechanism underlying cisplatin in hESC
is important. The results indicated that TRIM59 expression was
significantly increased in hESC tissues compared with healthy
esophageal epithelial tissues (Fig. 1C
and D). Compared with the siNC group, TRIM59 knockdown
inhibited esophageal cancer proliferation and migration, which
indicated that the chemosensitivity effects of TRIM59 in hESC
should be investigated further. Therefore, it was hypothesized that
enhancing TRIM59 promoted hESC progression and contributed to
chemoresistance to cisplatin. Eca109 cells were selected as the
cisplatin-resistant hESC cell line. Subsequently, drug sensitivity
was measured to assess TRIM59 knockdown-mediated alterations in
cisplatin resistance. Of note, cisplatin-induced cell viability in
the siTRIM59-knockdown group was significantly lower compared with
the siNC + cisplatin group. Theresults also suggested that
cisplatin enhanced hESC cell death in siTRIM59-transfected cells
(Fig. 3A). To examine whether
cisplatin treatment induced Eca109 cell apoptosis, cells were
transfected with siTRIM59 or siNC. Following incubation for 48 h at
37°C, cells were treated with cisplatin for 48 h. Cell apoptosis
was analyzed via immunostaining with Annexin V and PI. The number
of apoptotic cells was significantly increased in the siTRIM59 +
cisplatin group compared with the siTRIM59 + DMSO and si-NC groups
(Fig. 3B-D). Collectively, the
results indicated that Eca109 cells pretreated with siTRIM59
displayed increased cisplatin-related cell apoptosis.
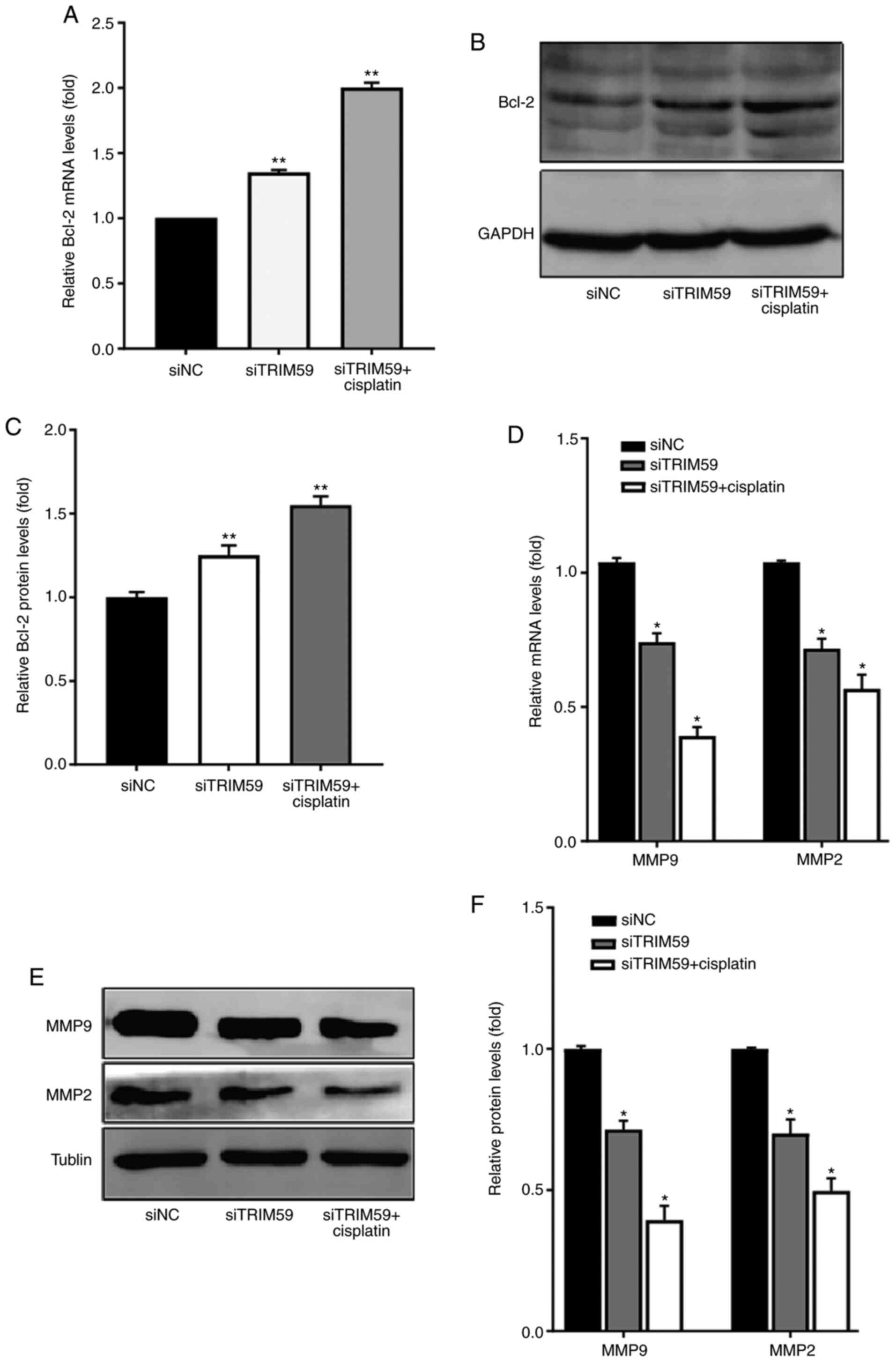
TRIM59 knockdown regulates the
expression levels of antiapoptotic and migration-related proteins
in response to cisplatin
To investigate whether the combined treatment of
siTRIM59 and cisplatin inhibited Eca109 cell migration, the
expression levels of antiapoptotic and migration-related proteins
were compared among the combined, cisplatin, siTRM59 and siNC
groups. The protein expression level of Bcl-2 was determined via
western blotting to explore the molecular mechanism underlying
apoptotic upregulation in Eca109 cells. The results indicated that
the antiapoptotic protein Bcl-2 was significantly upregulated in
the siTRIM59 group compared with the siNC group. Furthermore, a
slightly higher protein expression level of Bcl-2 was observed in
the siTRIM59 + cisplatin group compared with the siNC group
(Fig. 4A-C). To better understand
the observed synergy between TRIM59 knockdown and cisplatin,
expressive difference of Bcl-2 between siTRIM59 + cisplatin group
and siTRIM59 was assessed, and the results demonstrated that the
union group exhibited higher Bcl-2 levels. The expression levels of
the migration-related proteins MMP2 and MMP9 were also measured via
RT-qPCR and western blotting in the siTRIM59, siNC and combined
treatment groups. The results suggested that MMP2 and MMP9
expression levels were significantly decreased in the siTRIM59
group compared with the siNC group (Fig.
4D-F). Moreover, the protein expression levels of MMP2 and MMP9
were further downregulated in the siTRIM59 + cisplatin group
compared with the siNC group. To determine the synergy function of
decreased invasion in union group of siTRIM59 and cisplatin, the
expressive difference of MMP2 and MMP9 between the union and
siTRIM59 groups was compared, and the results demonstrated that the
union group exhibited lower MMP2 and MMP9 levels. Collectively, the
results indicated that TRIM59 knockdown increased the protein
expression levels of the antiapoptotic protein Bcl-2 and restrained
tumor cell migration by decreasing MMP2 and MMP9 expression
levels.
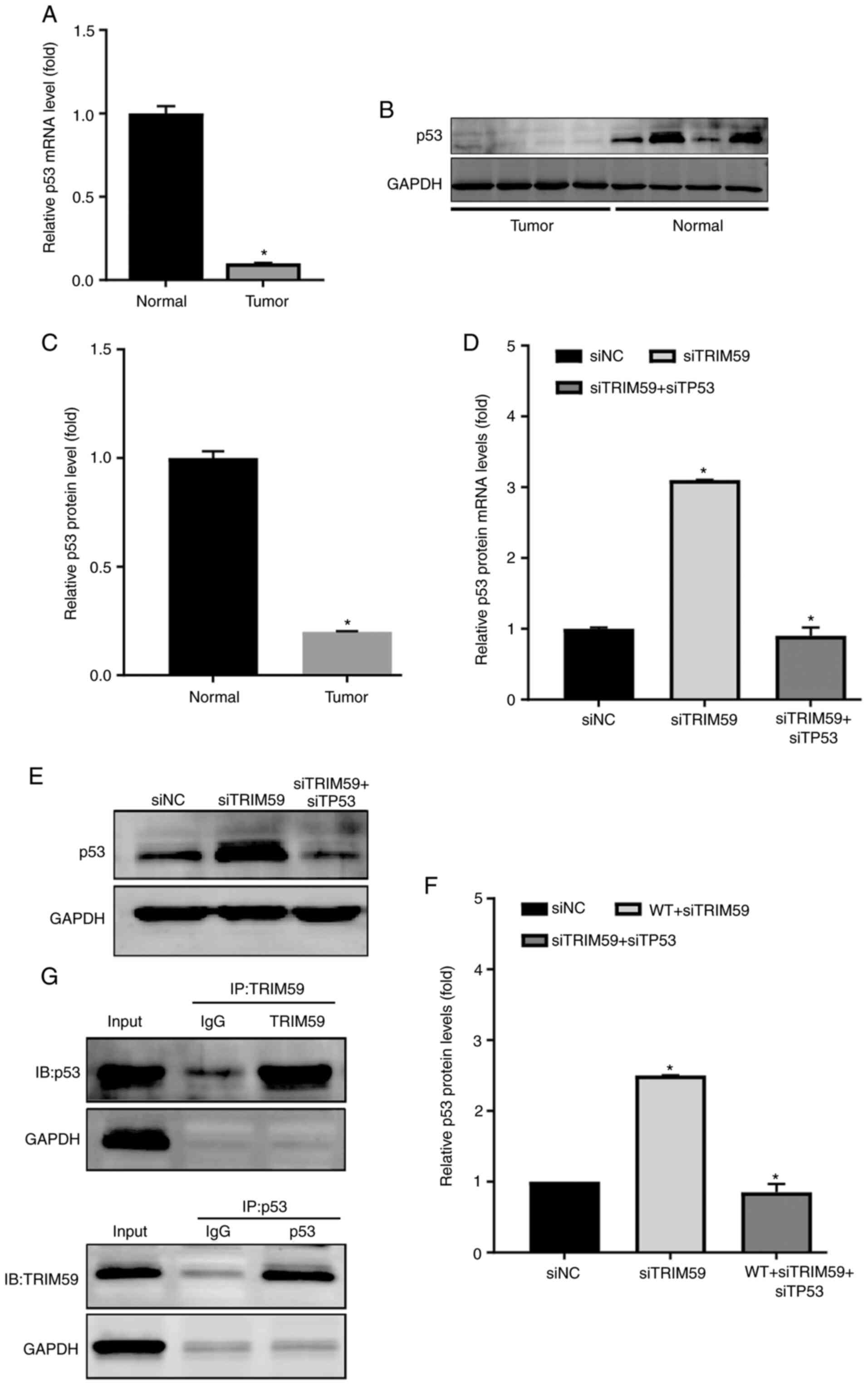
TRIM59 expression is negatively
associated with p53 in hESC samples and Eca109 cells
To understand the molecular mechanism underlying
TRIM59-mediated promotion of hESC, several proteins previously
reported to serve a critical role in esophageal tumorigenesis and
possibly be regulated by TRIM59 were examined (12). Since several studies have identified
that p53 is lowly expressed in hESC (13,14), it
was hypothesized that p53 expression was correlated with TRIM59
expression. Therefore, p53 expression was detected in hESC tissues
and Eca109 cells to explore the mechanism underlying
TRIM59-mediated promotion of hESC proliferation. The RT-qPCR
results indicated that p53 mRNA expression levels were
significantly decreased in patients with higher TRIM59 expression
levels compared with the control group (Fig. 5A). In addition, the protein
expression level of p53 was analyzed by performing western blotting
on surgically dissected hESC tissues. Consistently, p53 protein
expression levels in hESC tissues were significantly lower compared
with adjacent healthy tissues (Fig. 5B
and C). Based on measurements in tissues isolated from
patients, a strong negative association was identified between
TRIM59 and p53 protein levels (Figs.
1C and 5B). To determine whether
TRIM59 knockdown affected p53 expression, RT-qPCR was performed to
detect the mRNA expression levels of TP53 in the different
treatment groups, including siTRIM59 and siTP53 double-knockdown
cells. The transfection efficiency of siTP53 was confirmed via
RT-qPCR and western blotting (Fig.
S1). The results from the Eca109 cell line suggested that
TRIM59 knockdown significantly increased TP53 mRNA expression
levels compared with the siNC group, which was inhibited by
co-transfection with siTP53 (Fig.
5D).
The western blotting results further suggested the
negative role of TRIM59 in the regulation of p53; therefore, the
impact of TRIM59 expression on p53 was investigated in Ec109 cells
(Fig. 5E and F). In combination, the
results suggested that TRIM59 was highly expressed in patients with
hESC, resulting in accelerated tumor growth and migration by
inhibiting the p53 tumor-suppressor signaling pathway.
Subsequently, the molecular mechanism underlying
TRIM59-mediated promotion of p53 degradation required further
investigation. Therefore, whether TRIM59 interacted with p53 was
investigated. A co-immunoprecipitation assay was designed to detect
the physical association between TRIM59 and p53 using 293T cells
transfected with TRIM59 and p53 plasmids. The results indicated
that TRIM59 bound to p53 (Fig. 5G).
Collectively, the results indicated that TRIM59 was a negative
regulator of p53, which enhanced p53 degradation.
Discussion
hESC is the leading cause of cancer-related
mortality worldwide, particularly in men, and is the fifth most
common type of cancer, with >400,000 cases of mortality in the
first 5 years from diagnosis worldwide (15,16).
In-depth studies are required to explore and clarify the
pathogenesis of hESC, in order to provide the tools for a more
accurate diagnosis and effective treatment strategy. TRIM59, an
important member of the TRIM protein family, is upregulated in hESC
tissues and serves as an oncogene by promoting proliferation and
migration (12). In addition, TRIM59
overexpression significantly increased cisplatin resistance in hESC
cells. Therefore, TRIM59 was identified as a prognostic or
predictive biomarker of hESC (12).
However, identifying the mechanism underlying TRIM59 to develop
advanced treatment strategies for hESC is important.
TRIM59, a member of the TRIM family of proteins,
serves a crucial role in the proliferation and metabolism of
several types of cancer, such as lung (17,18) and
gastric cancer (19). TRIM59 protein
functions as an E3 ubiquitin ligase, and is comprised of an
N-terminal really interesting RING-finger domain (1). A previous study analyzed TRIM59
expression profiles in different types of cancer, and demonstrated
that TRIM59 was markedly upregulated across 12 cancer types
(12). A correlation between higher
expression levels of TRIM59 in cancer tissues, and particularly its
involvement in advanced stages of malignant transformation, and
poor patient prognosis was identified. Previous studies have
reported that TRIM59-silencing could decrease the growth rate of
lung cancer, prostate cancer (20),
breast cancer (21,22), colorectal cancer (23), cholangiocarcinoma (24), human cervical cancer (25), gliomagenesis (26,27),
medulloblastoma (28), ovarian
cancer (29,30) and gastric cancer (19). The aforementioned studies indicated
that TRIM59 may function as an oncogene in the majority of cancer
types (31).
However, whether the TRIM59 protein promotes hESC
proliferation and migration, and whether the increased protein
expression of TRIM59 is correlated with poor prognosis in patients
with hESC is not completely understood. In the present study,
TRIM59 was upregulated in hESC tissues and cell lines compared with
adjacent healthy tissues and cell lines. Following TRIM59 knockdown
in Eca109 cells, cells displayed significantly lower viability and
migration, as well as reduced colony formation ability compared
with the siNC group. Moreover, higher protein expression of TRIM59
in patients with hESC was associated with a significantly poorer
prognosis, as determined by Kaplan-Meier analysis. The results
indicated that TRIM59 was associated with hESC tumorigenesis and
migration, and may function as an oncogene.
Cisplatin is the most common anticancer drug used to
inhibit cancer proliferation and migration (11). However, due to the long time period
required for clinical chemotherapeutic treatment with cisplatin,
cancer cells develop chemoresistance to the drug, and its efficacy
diminishes, resulting in cancer treatment failure (32,33). In
the present study, siTRIM59 was used to knock down the protein
expression of TRIM59 in Eca109 esophageal cancer cells, which were
then exposed to cisplatin. Treatment with siTRIM59 and cisplatin
decreased cell viability and increased apoptosis to a greater level
compared with cells treated with siTRIM59 or cisplatin alone. To
study the effectiveness of the combined treatment of siTRIM59 and
cisplatin, the protein expression levels of MMP2 and MMP9 was
detected. The results suggested that MMP2 and MMP9 expression
levels were downregulated in the si-TRIM59 + cisplatin group
compared with the siTRIM59 group. To the best of our knowledge, the
present study was the first to report the effectiveness of the
combination of siTRIM59 and cisplatin in hESC.
In the present study, TP53 protein expression was
decreased in hESC tissues compared with adjacent healthy tissues.
TP53 is one of the most important tumor suppressors and the first
line of defense that keeps the genome of several types of cells
stable by preventing genome changes (6). In humans, activation of p53 is a vital
regulator of post-target resistance to chemotherapy and can induce
cell apoptosis in the presence of strong survival signals (6,14). Cells
with decreased p53 protein expression are unable to respond in a
timely manner to cellular stress and are more prone to a decreased
response to chemotherapeutic drugs and radiotherapy, resulting in
cancer migration (6). In the present
study, TRIM59 knockdown increased p53 protein expression and
enhanced the effective anticancer functions in Eca109 cells
compared with the siNC group. Co-immunoprecipitation was used to
detect the direct action by transfecting 293T cells with TRIM59 and
p53. The results suggested that TRIM59 promoted p53 degradation,
resulting in hESC cell proliferation and metastasis via direct
bonding. Therefore, the results indicated that the combination of
TRIM59 inhibitor and cisplatin might serve as a novel therapeutic
strategy for hESC.
In conclusion, the present study suggested that
upregulation of TRIM59 in hESC tissues was associated with poor
patient prognosis, and TRIM59 may serve as an oncogene to promote
the proliferation and metastasis of hESC. The present study also
indicated that TRIM59 knockdown inhibited Eca109 cancer cell
proliferation and migration, and enhanced chemosensitivity to
cisplatin by increasing p53 expression. Collectively, the results
indicated that the combination of TRIM59 knockdown and cisplatin
treatment may serve as a promising therapeutic strategy for
patients with hESC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Key Research Program of Hebei Science and Technology committee in
China (grant no. 182777206).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
JL and ZW conceived and designed the study. RL
performed the experiments and wrote the manuscript. HL performed
statistical analysis and revised the manuscript for intellectual
content. YX and XG followed up the patients and performed the
immunofluorescence experiments. YC and XL performed bioinformatics
analysis. JS contributed to the design and analysis of
immunoprecipitation experiments. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Forth Hospital of Hebei Medical University (Hebei,
China; approval no. Science Research-2014-Oncology-99). Written
informed consent was provided by all patients prior to the study
start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Esposito D, Koliopoulos MG and Rittinger
K: Structural determinants of TRIM protein function. Biochem Soc
Trans. 45:183–191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hatakeyama S: TRIM family proteins: Roles
in autophagy, immunity, and carcinogenesis. Trends Biochem Sci.
42:297–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hatakeyama S: TRIM proteins and cancer.
Nat Rev Cancer. 11:792–804. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nabeshima K, Inoue T, Shimao Y and
Sameshima T: Matrix metalloproteinases in tumor invasion: Role for
cell migration. Pathol Int. 52:255–264. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Scheau C, Badarau IA, Costache R, Caruntu
C, Mihai GL, Didilescu AC, Constantin C and Neagu M: The role of
matrix metalloproteinases in the epithelial-mesenchymal transition
of hepatocellular carcinoma. Anal Cell Pathol (Amst).
94:239072019.
|
|
6
|
Yanqing L, Omid T and Wei G: p53
modifications: Exquisite decorations of the powerful guardian. J
Mol Cell Biol. 11:564–577. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malkin D: Li-fraumeni syndrome. Genes
Cancer. 2:475–484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guha T and Malkin D: Inherited TP53
mutations and the Li-Fraumeni syndrome. Cold Spring Harb Perspect
Med. 7:a0261872017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freddie B, Jacques F, Isabelle S, Rebecca
SL, Lindey TA and Ahmedin J: Global cancer statistics 2018:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 68:394–424. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jackie Oh S, Han S, Lee W and Lockhart AC:
Emerging immunotherapy for the treatment of esophageal cancer.
Expert Opin Investig Drugs. 25:667–677. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang F, Wang Y, Wang ZQ, Sun P, Wang DS,
Jiang YX, Zhang DS, Wang FH, Xu RH and Li YH: Efficacy and safety
of cisplatin-based versus nedaplatin-based regimens for the
treatment of metastatic/recurrent and advanced esophageal squamous
cell carcinoma: A systematic review and meta-analysis. Dis
Esophagus. 30:1–8. 2017. View Article : Google Scholar
|
|
12
|
Tan P, Ye Y, He L, Xie J, Jing J, Ma G,
Pan H, Han L, Han W and Zhou Y: TRIM59 promotes breast cancer
motility by suppressing p62-selective autophagic degradation of
PDCD10. PLoS Biol. 16:e30000512018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z and Li C, Li Y, Guo X, Yan Z, Gao F
and Li C: DpdtbA-induced growth inhibition in human esophageal
cancer cells involved inactivation of the p53/EGFR/AKT pathway.
Oxid Med Cell Longev. 2019:54146702019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valletti A, Marzano F, Pesole G, Sbisa E
and Tullo A: Targeting chemoresistant tumors: Could trim
proteins-p53 axis be a possible answer? Int J Mol Sci. 20:17762019.
View Article : Google Scholar
|
|
15
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rustgi A and El-Serag HB: Esophageal
carcinoma. N Engl J Med. 372:1472–1473. 2015.PubMed/NCBI
|
|
17
|
Hao L, Du B and Xi X: TRIM59 is a novel
potential prognostic biomarker in patients with non-small cell lung
cancer: A research based on bioinformatics analysis. Oncol Lett.
14:2153–2164. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui Z, Liu Z, Zeng J, Zhang S, Chen L,
Zhang G, Xu W, Song L and Guo X: TRIM59 promotes gefitinib
resistance in EGFR mutant lung adenocarcinoma cells. Life Sci.
224:23–32. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo D, Wang Y, Huan X, Huang C, Yang C,
Fan H, Xu Z and Yang L: Identification of a synonymous variant in
TRIM59 gene for gastric cancer risk in a Chinese population.
Oncotarget. 8:11507–11516. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin WY, Wang H, Song X, Zhang SX, Zhou PS,
Sun JM and Li JS: Knockdown of tripartite motif 59 (TRIM59)
inhibits tumor growth in prostate cancer. Eur Rev Med Pharmacol
Sci. 20:4864–4873. 2016.PubMed/NCBI
|
|
21
|
Zhang Y and Yang W: Down-regulation of
tripartite motif protein 59 inhibits proliferation, migration and
invasion in breast cancer cells. Biomed Pharmacother. 89:462–467.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan P, He L and Zhou Y: TRIM59 deficiency
curtails breast cancer metastasis through SQSTM1-selective
autophagic degradation of PDCD10. Autophagy. 15:747–749. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu W, Chen J, Wu J, Lin J, Yang S and Yu
H: Knockdown of tripartite motif-59 inhibits the malignant
processes in human colorectal cancer cells. Oncol Rep.
38:2480–2488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shen H, Zhang J, Zhang Y, Feng Q, Wang H,
Li G, Jiang W and Li X: Knockdown of tripartite motif 59 (TRIM59)
inhibits proliferation in cholangiocarcinoma via the PI3K/AKT/mTOR
signalling pathway. Gene. 698:50–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aierken G, Seyiti A, Alifu M and Kuerban
G: Knockdown of tripartite-59 (trim59) inhibits cellular
proliferation and migration in human cervical cancer cells. Oncol
Res. 25:381–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen G, Chen W, Ye M, Tan W and Jia B:
TRIM59 knockdown inhibits cell proliferation by down-regulating the
Wnt/β-catenin signaling pathway in neuroblastoma. Biosci Rep.
39:BSR201812772019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu Y, Zhang Z and Xu G: TRIM proteins in
neuroblastoma. Biosci Rep. 39:BSR201920502019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao R, Lv G, Zhang C, Wang X and Chen L:
TRIM59 induces epithelial-to-mesenchymal transition and promotes
migration and invasion by PI3K/AKT signaling pathway in
medulloblastoma. Oncol Lett. 15:8253–8260. 2018.PubMed/NCBI
|
|
29
|
Wang Y, Zhou Z, Wang X, Zhang X, Chen Y,
Bai J and Di W: Trim59 is a novel marker of poor prognosis and
promotes malignant progression of ovarian cancer by inducing
annexin a2 expression. Int J Biol Sci. 14:2073–2082. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang P, Zhang H, Wang Y, Zhang P and Qi
Y: Tripartite motif-containing protein 59 (TRIM59) promotes
epithelial ovarian cancer progression via the focal adhesion
kinase(FAK)/AKT/Matrix metalloproteinase (MMP) pathway. Med Sci
Monit. 25:3366–3373. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Khatamianfar V, Valiyeva F, Rennie PS, Lu
W, Yang BB, Bauman G, Moussa M and Xuan JW: TRIM59, a novel
multiple cancer biomarker for immunohistochemical detection of
tumorigenesis. BMJ Open. 2:e0014102012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang XP, Li X, Situ MY, Huang LY, Wang J,
He T, Yan QH, Xie XY, Zhang YJ, Gao YH, et al: Entinostat reverses
cisplatin resistance in esophageal squamous cell carcinoma via
down-regulation of multidrug resistance gene 1. Cancer Lett.
414:294–300. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ilson DH: Esophageal cancer chemotherapy:
Recent advances. Gastrointest Cancer Res. 2:85–92. 2008.PubMed/NCBI
|