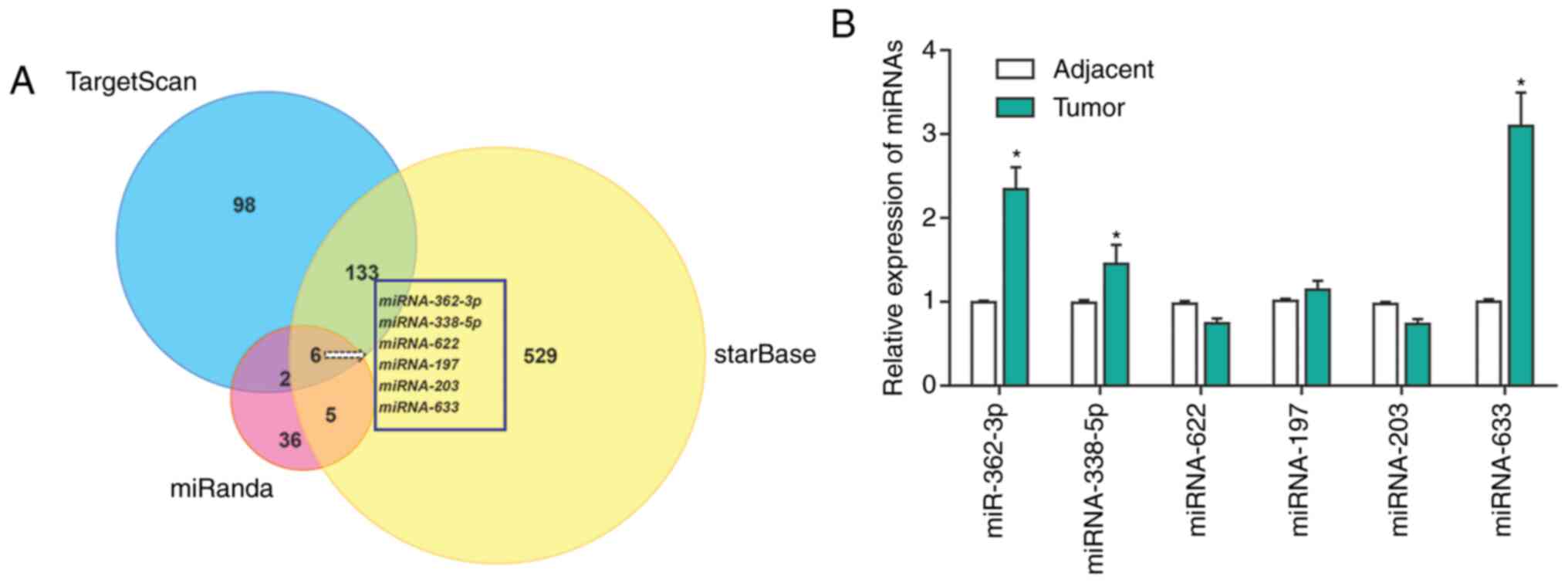
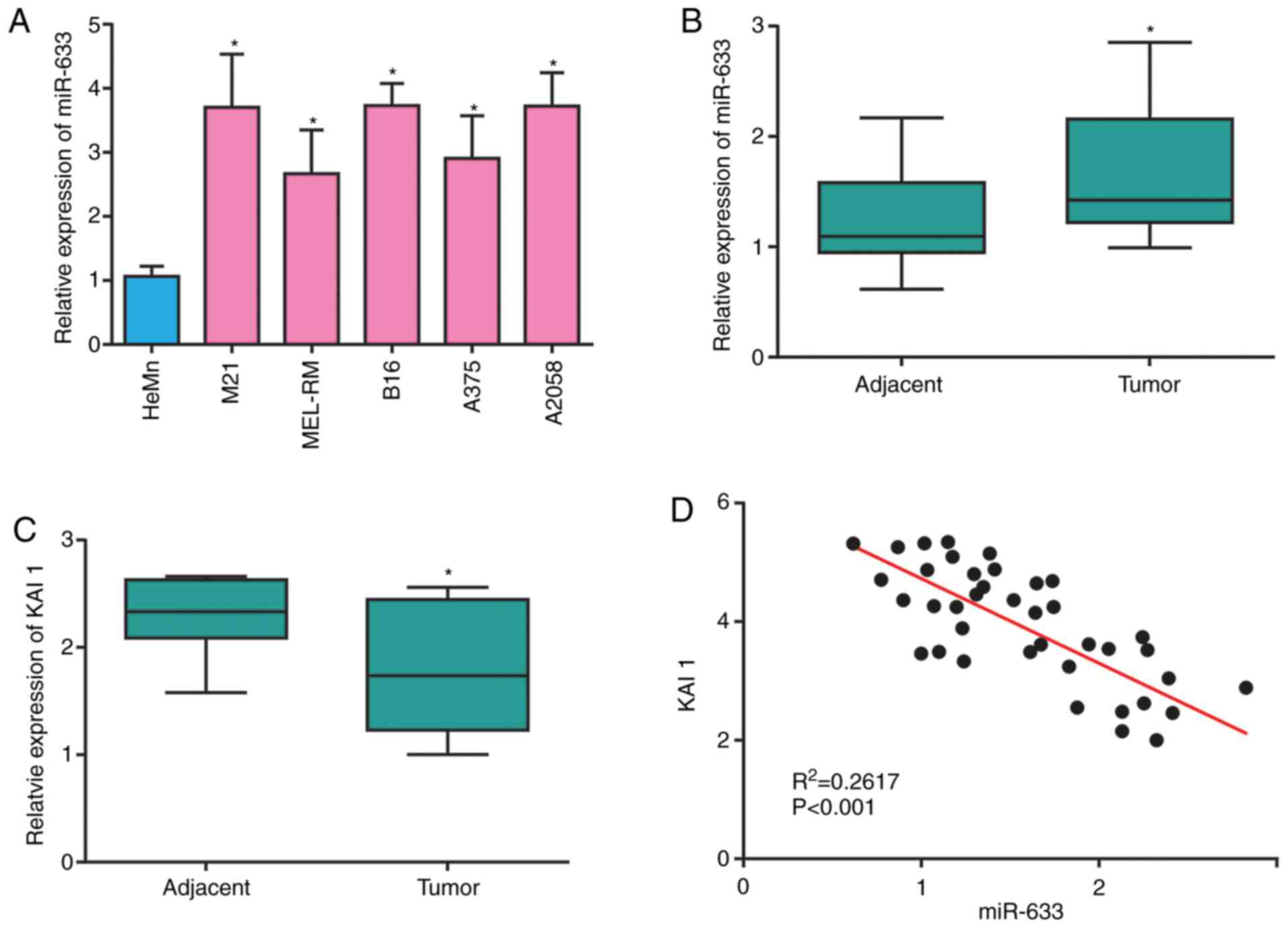
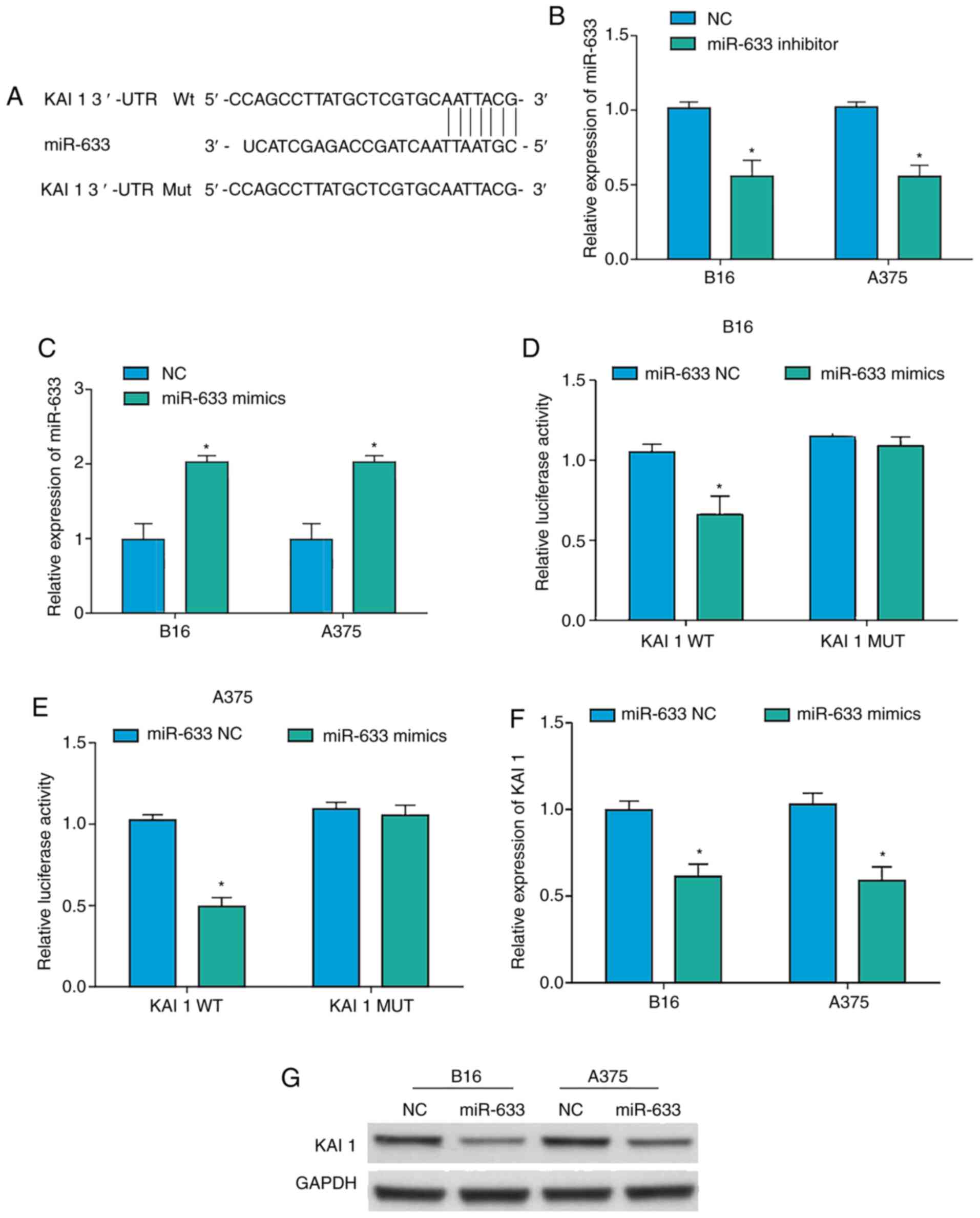
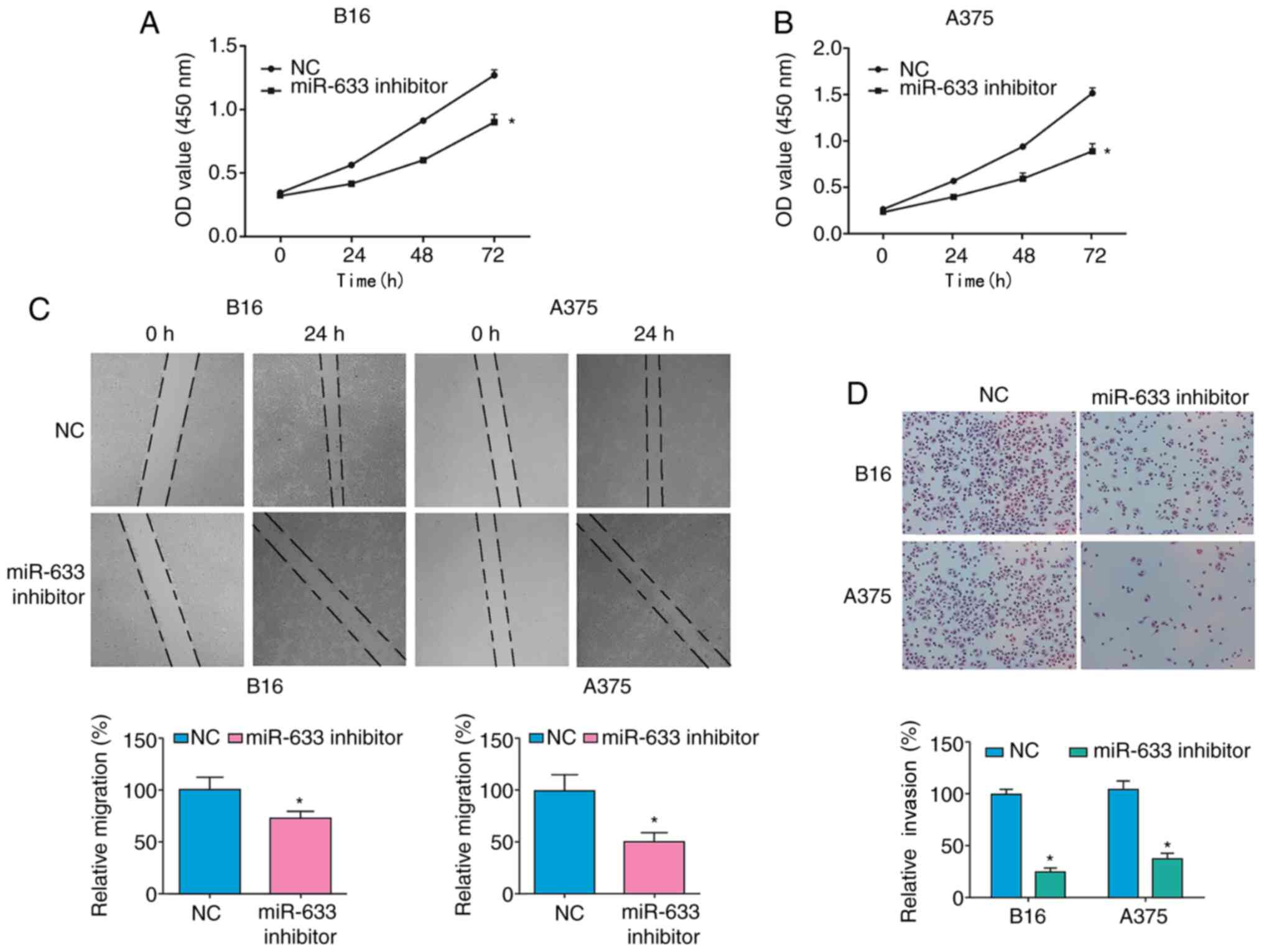
|
1
|
Stark MS, Gray ES, Isaacs T, Chen FK,
Millward M, McEvoy A, Zaenker P, Ziman M, Soyer HP, Glasson WJ, et
al: A panel of circulating MicroRNAs detects uveal melanoma with
high precision. Transl Vis Sci Technol. 8:122019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia Z, Yang C, Yang X, Wu S, Feng Z, Qu L,
Chen X, Liu L and Ma Y: MiR-652 promotes proliferation and
migration of uveal melanoma cells by targeting HOXA9. Med Sci
Monit. 25:8722–8732. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fumagalli MR, Lionetti MC, Zapperi S and
La Porta C: Cross-Talk between circRNAs and mRNAs modulates
MiRNA-mediated circuits and affects melanoma plasticity. Cancer
Microenviron. 12:95–104. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang C, Xia Z, Zhu L, Li Y, Zheng Z, Liang
J and Wu L: MicroRNA-139-5p modulates the growth and metastasis of
malignant melanoma cells via the PI3K/AKT signaling pathway by
binding to IGF1R. Cell Cycle. 18:3513–3524. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sánchez-Sendra B, García-Giménez JL,
González-Muñoz JF, Navarro L, Murgui A, Terrádez L, Pinazo I,
Martin JM and Monteagudo C: Circulating miRNA expression analysis
reveals new potential biomarkers for human cutaneous melanoma
staging. J Eur Acad Dermatol Venereol. 34:e126–e129. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun HW, Yang GL, Wang SN, Zhang YJ, Ding
JX and Zhang XN: MicroRNA-92a regulates the development of
cutaneous malignant melanoma by mediating FOXP1. Eur Rev Med
Pharmacol Sci. 23:8991–8999. 2019.PubMed/NCBI
|
|
7
|
Nakamura K and Okuyama R: Immunotherapy
for advanced melanoma: Current knowledge and future directions. J
Dermatol Sci. 83:87–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reale E, Taverna D, Cantini L, Martignetti
L, Osella M, De Pittà C, Virga F, Orso F and Caselle M:
Investigating the epi-miRNome: Identification of epi-miRNAs using
transfection experiments. Epigenomics. 11:1581–1599. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Afrang N and Honardoost M: Cell cycle
regulatory markers in melanoma: New strategies in diagnosis and
treatment. Med J Islam Repub Iran. 33:962019.PubMed/NCBI
|
|
10
|
Hämäläinen M, Teppo HR, Skarp S,
Haapasaari KM, Porvari K, Vuopala K, Kietzmann T and Karihtala P:
NRF1 and NRF2 mRNA and protein expression decrease early during
melanoma carcinogenesis: An insight into survival and MicroRNAs.
Oxid Med Cell Longev. 2019:26470682019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rossi E, Schinzari G, Maiorano BA,
Pagliara MM, Di Stefani A, Bria E, Peris K, Blasi MA and Tortora G:
Conjunctival melanoma: Genetic and epigenetic insights of a
distinct type of melanoma. Int J Mol Sci. 20:54472019. View Article : Google Scholar
|
|
12
|
Chen L, Karisma VW, Liu H and Zhong L:
MicroRNA-300: A transcellular mediator in exosome regulates
melanoma progression. Front Oncol. 9:10052019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang W, Hou L, Wei J, Du Y, Zhao Y, Deng
X and Lin X: Hsa-MiR-217 inhibits the proliferation, migration, and
invasion in non-small cell lung cancer cells via targeting SIRT1
and P53/KAI1 signaling. Balkan Med J. 37:208–214. 2020.PubMed/NCBI
|
|
14
|
Zhang Q, Huang F, Yao Y, Wang J, Wei J, Wu
Q, Xiang S and Xu L: Interaction of transforming growth
factor-β-smads/microRNA-362-3p/CD82 mediated by M2 macrophages
promotes the process of epithelial-mesenchymal transition in
hepatocellular carcinoma cells. Cancer Sci. 110:2507–2519. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Habibzadeh P, Honarvar B, Silawi M,
Bahramjahan S, Kazemi A, Faghihi MA and Lankarani K: Association
between rs2303861 polymorphism in CD82 gene and non-alcoholic fatty
liver disease: A preliminary case-control study. Croat Med J.
60:361–368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Asada H, Tomiyasu H, Uchikai T, Ishihara
G, Goto-Koshino Y, Ohno K and Tsujimoto H: Comprehensive analysis
of miRNA and protein profiles within exosomes derived from canine
lymphoid tumour cell lines. PLoS One. 14:e02085672019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu CJ, Yang JH, Huang FZ, Yang JH, Liu
CP, Mao XH, Yi WM, Shen XB, Peng C, Chen MF, et al: The role of
miR-99b in mediating hepatocellular carcinoma invasion and
migration. Eur Rev Med Pharmacol Sci. 24:79092020.PubMed/NCBI
|
|
18
|
Long J, Luo J and Yin X: MiR-338-5p
promotes the growth and metastasis of malignant melanoma cells via
targeting CD82. Biomed Pharmacother. 102:1195–1202. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee MS, Lee J, Kim YM and Lee H: The
metastasis suppressor CD82/KAI1 represses the TGF-β 1 and wnt
signalings inducing epithelial-to-mesenchymal transition linked to
invasiveness of prostate cancer cells. Prostate. 79:1400–1411.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li W, Hu M, Wang C, Lu H, Chen F, Xu J,
Shang Y, Wang F, Qin J, Yan Q, et al: A viral microRNA
downregulates metastasis suppressor CD82 and induces cell invasion
and angiogenesis by activating the c-met signaling. Oncogene.
36:5407–5420. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishioka C, Ikezoe T, Pan B, Xu K and
Yokoyama A: MicroRNA-9 plays a role in interleukin-10-mediated
expression of E-cadherin in acute myelogenous leukemia cells.
Cancer Sci. 108:685–695. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu L, Hou Y, Tu G, Chen Y, Du YE, Zhang H,
Wen S, Tang X, Yin J, Lang L, et al: Nuclear drosha enhances cell
invasion via an EGFR-ERK1/2-MMP7 signaling pathway induced by
dysregulated miRNA-622/197 and their targets LAMC2 and CD82 in
gastric cancer. Cell Death Dis. 8:e26422017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jee BK, Park KM, Surendran S, Lee WK, Han
CW, Kim YS and Lim Y: KAI1/CD82 suppresses tumor invasion by MMP9
inactivation via TIMP1 up-regulation in the H1299 human lung
carcinoma cell line. Biochem Biophys Res Commun. 342:655–661. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mine M, Yamaguchi K, Sugiura T, Chigita S,
Yoshihama N, Yoshihama R, Hiyake N, Kobayashi Y and Mori Y: MiR-203
inhibits frizzled-2 expression via CD82/KAI1 expression in human
lung carcinoma cells. PLoS One. 10:e01313502015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang QH, Yao YL, Wu XY, Wu JH, Gu T, Chen
L, Gu JH, Liu Y and Xu L: Anti-MiR-362-3p inhibits migration and
invasion of human gastric cancer cells by its target CD82. Dig Dis
Sci. 60:1967–1976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dai W, Wang C, Wang F, Wang Y, Shen M,
Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al: Anti-MiR-197
inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem
Biophys Res Commun. 446:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Q, Zhang LY, Wu S, Huang C, Liu J, Wang
P and Cao Y: Bioinformatics analysis identifies MicroRNAs and
target genes associated with prognosis in patients with melanoma.
Med Sci Monit. 25:7784–7794. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cooling L: An update on the I blood group
system. Immunohematology. 35:85–90. 2019.PubMed/NCBI
|
|
30
|
Santoni G, Morelli MB, Santoni M, Nabissi
M, Marinelli O and Amantini C: Targeting transient receptor
potential channels by microRNAs drives tumor development and
progression. Adv Exp Med Biol. 1131:605–623. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ylösmäki L, Polini B, Carpi S, Martins B,
Smertina E, Feola S, Fusciello M, Peltonen K, Nieri P, Ylösmäki E
and Cerullo V: Harnessing therapeutic viruses as a delivery vehicle
for RNA-based therapy. PLoS One. 14:e02240722019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sharma A, Biswas A, Liu H, Sen S,
Paruchuri A, Katsonis P, Lichtarge O, Dakal TC, Maulik U, Gromiha
MM, et al: Mutational landscape of the BAP1 locus reveals an
intrinsic control to regulate the miRNA network and the binding of
protein complexes in uveal melanoma. Cancers (Basel). 11:16002019.
View Article : Google Scholar
|
|
33
|
Chekhun VF, Borikun TV, Bazas VМ, Andriiv
AV, Klyusov OM, Yalovenko TM and Lukianova NY: Association of
circulating miR-21, −205, and −182 with response of luminal breast
cancers to neoadjuvant FAC and AC treatment. Exp Oncol. 42:162–166.
2020.PubMed/NCBI
|
|
34
|
Bonazzi VF, Stark MS and Hayward NK:
MicroRNA regulation of melanoma progression. Melanoma Res.
22:101–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu L, Liang Yn, Luo Xq, Liu Xd and Guo Hx:
Association of miRNAs expression profiles with prognosis and
relapse in childhood acute lymphoblastic leukemia. Zhonghua Xue Ye
Xue Za Zhi. 32:178–181. 2011.(In Chinese). PubMed/NCBI
|
|
36
|
Wang Y, Chen H, Fu Y, Ai A, Xue S, Lyu O
and Kuang Y: MiR-195 inhibits proliferation and growth and induces
apoptosis of endometrial stromal cells by targeting FKN. Int J Clin
Exp Pathol. 6:2824–2834. 2013.PubMed/NCBI
|
|
37
|
Arribas AJ, Campos-Martín Y, Gómez-Abad C,
Algara P, Sánchez-Beato M, Rodriguez-Pinilla MS, Montes-Moreno S,
Martinez N, Alves-Ferreira J, Piris MA and Mollejo M: Nodal
marginal zone lymphoma: Gene expression and miRNA profiling
identify diagnostic markers and potential therapeutic targets.
Blood. 119:e9–e21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou X, Liu X, Zhang G, Zhang Q, Chen H,
Wang Y, Fang F and Sun J: Knockdown THOC2 suppresses the
proliferation and invasion of melanoma. Bioengineered. 10:635–645.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
You J, Chang R, Liu B, Zu L and Zhou Q:
Nm23-H1 was involved in regulation of KAI1 expression in
high-metastatic lung cancer cells L9981. J Thorac Dis. 8:1217–1226.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu X, Guo X, Li H, Chen J and Qi X:
Src/STAT3 signaling pathways are involved in KAI1-induced
downregulation of VEGF-C expression in pancreatic cancer. Mol Med
Rep. 13:4774–47778. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bhalla S, Kaur H, Dhall A and Raghava GP:
Prediction and analysis of skin cancer progression using genomics
profiles of patients. Sci Rep. 9:157902019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hou Q, Han S, Yang L, Chen S, Chen J, Ma
N, Wang C, Tang J, Chen X, Chen F, et al: The interplay of
microRNA-34a, LGR4, EMT-associated factors, and MMP2 in regulating
uveal melanoma cells. Invest Ophthalmol Vis Sci. 60:4503–4510.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu T, Chen S, Qu L, Wang Y, Chen HD and He
C: Identification of a five-miRNA signature predicting survival in
cutaneous melanoma cancer patients. Peer J. 7:e78312019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Khan NS, Lukason DP, Feliu M, Ward RA,
Lord AK, Reedy JL, Ramirez-Ortiz ZG, Tam JM, Kasperkovitz PV,
Negoro PE, et al: CD82 controls CpG-dependent TLR9 signaling. FASEB
J. 33:12500–12514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang G, Cheng Y, Chen G, Tang Y, Ardekani
G, Rotte A, Martinka M, McElwee K, Xu X, Wang Q and Zhou Y: Loss of
tumor suppressors KAI1 and p27 identifies a unique subgroup of
primary melanoma patients with poor prognosis. Oncotarget.
6:23026–23035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu N, Liu Z, Liu X and Chen H:
Comprehensive analysis of a competing endogenous RNA network
identifies seven-lncRNA signature as a prognostic biomarker for
melanoma. Front Oncol. 9:9352019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Prabhu VV and Devaraj SN: KAI1/CD82,
metastasis suppressor gene as a therapeutic target for
non-small-cell lung carcinoma. J Environ PatholToxicol Oncol.
36:269–275. 2017. View Article : Google Scholar
|