Introduction
Melanoma is one of the most commonly occurring forms
of cancer, and malignant melanoma is the third most common skin
malignancy (1–6). For most patients with melanoma,
immunotherapy, chemotherapy or small-molecule inhibitor
administration are not effective therapies (7). Furthermore, the outcomes of patients
with advanced-stage disease remain poor (8,9).
MicroRNAs (miRNAs/miRs) are short RNAs (~22
nucleotides in length) that do not encode protein, and yet are
important regulators of oncogenesis (10,11). By
binding to complementary sequences in the 3′-untranslated region
(3′-UTR) of target mRNAs, miRNAs can alter the stability and
translation of these transcripts, thereby influencing phenotypic
outcomes within cells. A single miRNA can target multiple different
mRNAs, giving rise to complex regulatory networks that control
diverse pathological and physiological biological activities
(12,13). The dysregulation of miRNAs is a
common hallmark of oncogenesis and tumor progression (14,15). By
evaluating patterns of miRNA expression, it may be possible to
better diagnose or monitor specific types of cancer. For example,
miR-144 and miR-92a have been shown to serve as valuable and
specific diagnostic biomarkers that can guide the detection of
specific subtypes of colorectal cancer (16). Additionally, miR-221 can suppress the
expression of PHD finger protein 2, thereby influencing liver
cancer invasion, leading researchers to highlight this signaling
axis as a potential target for therapeutic intervention (17). Efforts have also been made to
successfully identify patterns of miRNA dysregulation associated
with melanoma (18). Given their
essential roles as regulators of cancer onset and progression,
further analyses of miRNAs in these oncogenic contexts are
warranted.
Tetraspanin cluster of differentiation 82(CD82),
also known as Kang-Ai 1 (KAI1), is an important tumor suppressor
gene that was first detected based upon analyses of human
metastatic prostate cancer samples (19). There is robust evidence linking KAI1
downregulation with the invasive and metastatic activities of
various tumors based upon histopathological and molecular analyses
(20). KAI1 mutations and associated
loss-of-function are evident in a range of tumor types, reaffirming
the role of this gene as a tumor suppressor (21–23).
KAI1 expression in melanoma has been shown to be associated with
tumor grade and patient prognosis, and it has been validated as a
risk factor for disease progression (24,25).
However, the association between miR-633 and malignant melanoma has
not been elucidated to date. The present study investigated the
role of miR-633 in the proliferation and migration of melanoma
cells. Furthermore, the potential role of miR-633 in regulating
KAI1 expression in melanoma cells was explored.
Materials and methods
Prediction of the candidate miRNA
associated with KAI1
The online target gene prediction databases
TargetScan (http://www.targetscan.org/vert_72), StarBase
(http://starbase.sysu.edu.cn) and miRanda
(http://www.microrna.orgmicrorna) were
used to identify miRNAs that could be associated with KAI1, and to
predict the binding region of miRNA-633 to the 3′-UTR of KAI1.
Sample collection
All cancer tissues (n=11) and paracancerous tissues
(n=10) used in the present study were collected via surgical
resection from patients with melanoma at Cangzhou Central Hospital
(Table I). No patients received any
treatment, including chemotherapy or radiation therapy, prior to
surgery. According to the melanoma treatment guidelines, the
resection range was determined based on the different stage, and
the paracancerous tissues were taken as close to the outer edge as
possible to ensure that it was the adjacent normal tissue. Resected
samples were subjected to pathological confirmation, after which
they were snap-frozen with liquid nitrogen and stored at −80°C.
However, tumor cells were present at the outer edge in one of the
patients, so11 cancer tissues and 10 paracancerous tissues were
used in the present study. Patients and their families were
informed of all study protocols and were asked to sign informed
consent forms. The Ethics Committee of Cangzhou Central Hospital
approved the present study.
 | Table I.Clinicopathological characteristics
of patients with melanoma (n=10). |
Table I.
Clinicopathological characteristics
of patients with melanoma (n=10).
| Characteristic | Number of patients,
n |
|---|
| Total | 10 |
| Age, years |
|
|
<45 | 2 |
|
≥45 | 8 |
| Sex |
|
|
Male | 7 |
|
Female | 3 |
| TNM
classification |
|
|
I+II | 7 |
|
III+IV | 3 |
| Distant
metastasis |
|
| No | 9 |
|
Yes | 1 |
Cell culture and transfection
Normal human primary epidermal melanocytes (HeMn);
(The Cell Bank of Type Culture Collection of Chinese Academy of
Sciences) were maintained in F-12K medium(Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). A375, A2058, B16, MEL-RM and M21 melanoma cell
lines(The Cell Bank of Type Culture Collection of Chinese Academy
of Sciences) were maintained in RPMI-1640 media supplemented with
10% fetal bovine serum (both purchased from Gibco; Thermo Fisher
Scientific, Inc.). All cells were grown in humidified 5%
CO2 incubators at 37°C. A375 and B16 cells were
co-transfected with KAI1 wild-type (WT) or KAI1 mutant (MUT),
and/or with miR-633 inhibitors or inhibitor-negative control (NC)
and miR-633 mimics or mimic-NC. The transfection of cells was
performed with the plasmids (Promega Corporation) using
Lipofectamine®3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol,
with downstream analyses conducted at 24–48 h post-transfection.
The sequences of oligo-ribonucleotides were as follows: miR-633
mimics, 5′-CUAAUAGUAUCUACCACAAUAAA-3′; mimic-NC
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-633 inhibitor,
5′-UUUAUUGUGGUAGAUACUAUUAG-3′; and inhibitor-NC,
5′-CAGUACUUUUGUGUAGUACAA-3′.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from the
cells and tissues, after which reverse transcription was performed
with a PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.).
After the cDNA concentrations were quantified, qPCR was conducted
with SYBR® Premix Ex Taq (Takara Biotechnology Co.,
Ltd.) following the manufacturer's instructions. GAPDH and U6 were
utilized as normalization controls for mRNA and miRNA levels,
respectively. Relative fold changes in expression levels were
calculated using the 2−ΔΔCq method (26). Primers used were as follows: KAI1,
forward 5′-ATCCGATATCCGATCGACATGAGAGGAGTTCGAT-3′ and reverse
5′-CTAGGCGAGATAGACTACCATG-3′; GAPDH, forward
5′-ATCCGATTACCGATACCTAGACC-3′ and reverse
5′-ATGGACTATATCCGACGACGA-3′; miR-633, forward
5′-CCGATACGATGAGAGAAACCCTGA-3′ and reverse
5′-GGACAGAGTTGACTTAAGGCTAGA-3′; and U6, forward
5′-TGCGTTCCCTTTGTCATCCT-3′ and reverse,
5′-AACGCTTCAC-GAATTTGCGT-3′. The thermocycling conditions were as
follows: 95°C for 30 sec, followed by 40 cycles of 92°C for 5 sec
and 60°C for 30 sec, dissociation at 60°C for 1 min and 95°C for 1
sec.
Dual-luciferase reporter assay
Cells were transfected with KAI1WT or KAI1MUT
luciferase reporter plasmids (Promega Corporation), along with
miR-633 mimics or control constructs for 24 h at 37°C using
Lipofectamine®3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol.
Then, a dual-luciferase reporter assay system (Promega Corporation)
was utilized to quantify luciferase activity. Renilla
luciferase activity was used as the internal control, and data are
presented as the ratio of firefly to Renilla luciferase
activities.
Cell Counting Kit-8 (CCK-8) assay
Cells were added to 96-well plates (2×103
cells/well). At the indicated time points, 10 µl CCK-8 solution
(Dojindo Molecular Technologies, Inc.) was added per well, and
absorbance at 450 nm was assessed using a microplate reader
(Bio-Rad Laboratories, Inc.).
Wound healing assay
Transfected cells were seeded into 6-well plates at
a density of 5×105 cells/well and cultured to confluency
in RPMI-1640 medium with 15% FBS at 37°C with 5% CO2. A
linear wound was made using a10 µl sterile pipette tip across the
confluent cell layer, and the plates were washed twice to remove
detached cells and debris. The cells were cultured in RPMI-1640
medium with 2.5% FBS, 24 h after the wound scraping. Then, the size
of the wounds was observed and measured at 0 and 24 h under a light
microscope (magnification, ×20).
Invasion assay
To assess cell invasion, 3×104 cells were
resuspended in serum-free RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) and added to the upper portion of a Transwell
chamber (Corning Inc.) that had been coated with Matrigel™. At
total of 600 µl media containing 20% FBS was added to the lower
chamber, and plates were subsequently incubated for 24 h. Cells
that had invaded through the matrix to the lower chamber were
subsequently fixed for 20 min using 4% paraformaldehyde, after
which they were stained for 20 min with crystal violet. The cells
were then observed using optical light microscopy (Olympus
Corporation). The numbers of invaded cells in five random fields of
view were subsequently quantified for each sample.
Western blotting
RIPA buffer (Beyotime Institute of Biotechnology)
was used to lyse cells, after which a BCA kit (Beyotime Institute
of Biotechnology) was used to quantify protein levels in the
lysates. Protein (40 ug/lane) was subsequently subjected to
electrophoretic separation (12% SDS-PAGE), transferred to PVDF
membranes (EMD Millipore), and blocked with 5% non-fat milk at room
temperature for 2 h. The membranes were then incubated overnight
with primary antibodies (cat. no. 10205-2-A; 1:1,000; ProteinTech
Group, Inc.) at 4°C. Blots were subsequently incubated at room
temperature for 2 h with HRP-conjugated secondary antibodies (cat.
no. KC-4G3; 1:20,000; Kang Chen Biotech, Inc.), after which ECL
(Pierce; Thermo Fisher Scientific, Inc.) was used to detect the
protein bands. Image J software (1.48u version; National Institutes
of Health) was used for densitometric analyses.
Statistical analysis
All statistical analyses were performed using SPSS
21.0 (IBM Corp.) and GraphPad Prism 7.0 (GraphPad Software, Inc.).
Data are expressed as the mean ± SD. Multiple group comparisons
were analyzed using one-way analysis of variance and Tukey's post
hoc test. Independent-samples t-tests were used to analyze the
significance of mRNA levels in tumor tissues and adjacent normal
tissues. Associations between the expression of pairs of genes were
evaluated using Spearman's rank correlation analyses.
Independent-samples t-tests were used to analyze the significance
of cell proliferation, wound healing/migration and Transwell
invasion assay results. All comparisons were two-tailed. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of miRNAs that
potentially target KAI1 in melanoma via bioinformatics prediction
analyses
The potential upstream miRNAs that may target KAI1
were selected using the TargetScan, StarBase and miRanda online
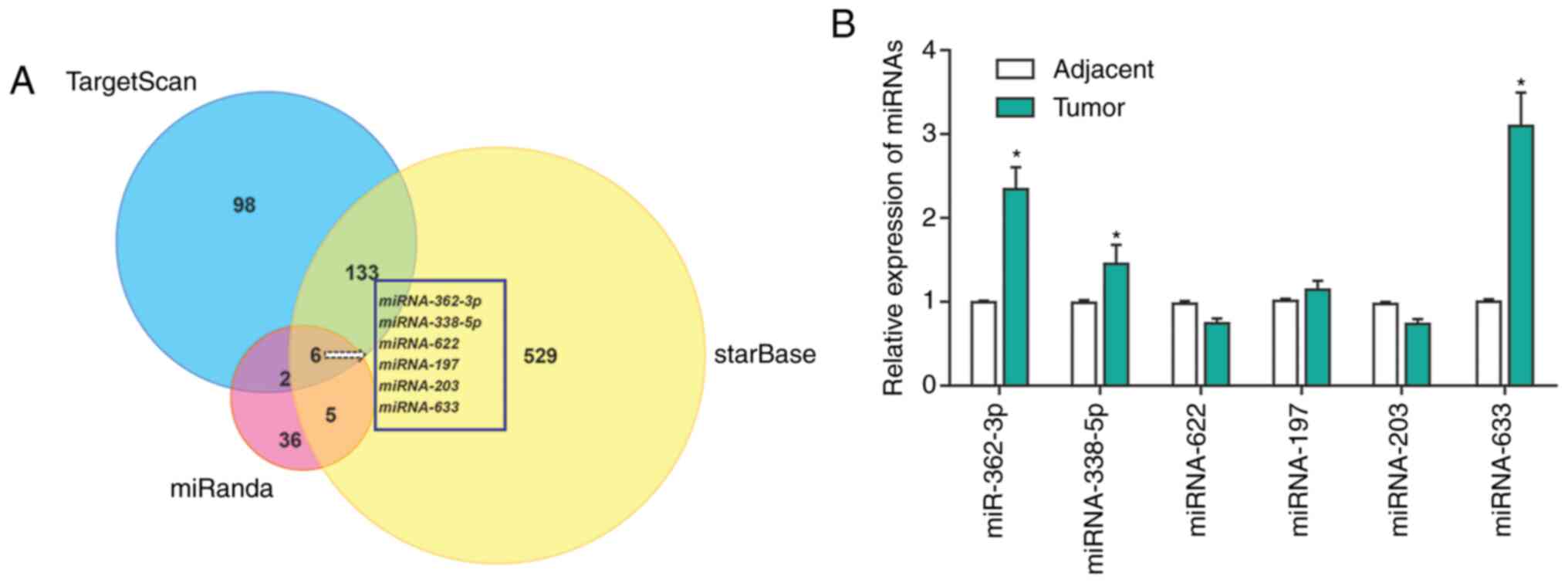
tools (27). In total, 6 putative
regulators of KAI1 (miR-633, miR-362, miR-338, miR-622, miR-203 and
miR-197) were identified using this approach (Fig. 1A). The expression levels of these
miRNAs were subsequently measured in tumors from patients with
melanoma and their matched adjacent normal tissue samples (Fig. 1B). Among the six miRNA candidates,
miR-633 was found to be upregulated the most in melanoma tissues
compared with adjacent normal tissues.
miR-633 is upregulated in melanoma and
is negatively correlated with KAI1 expression
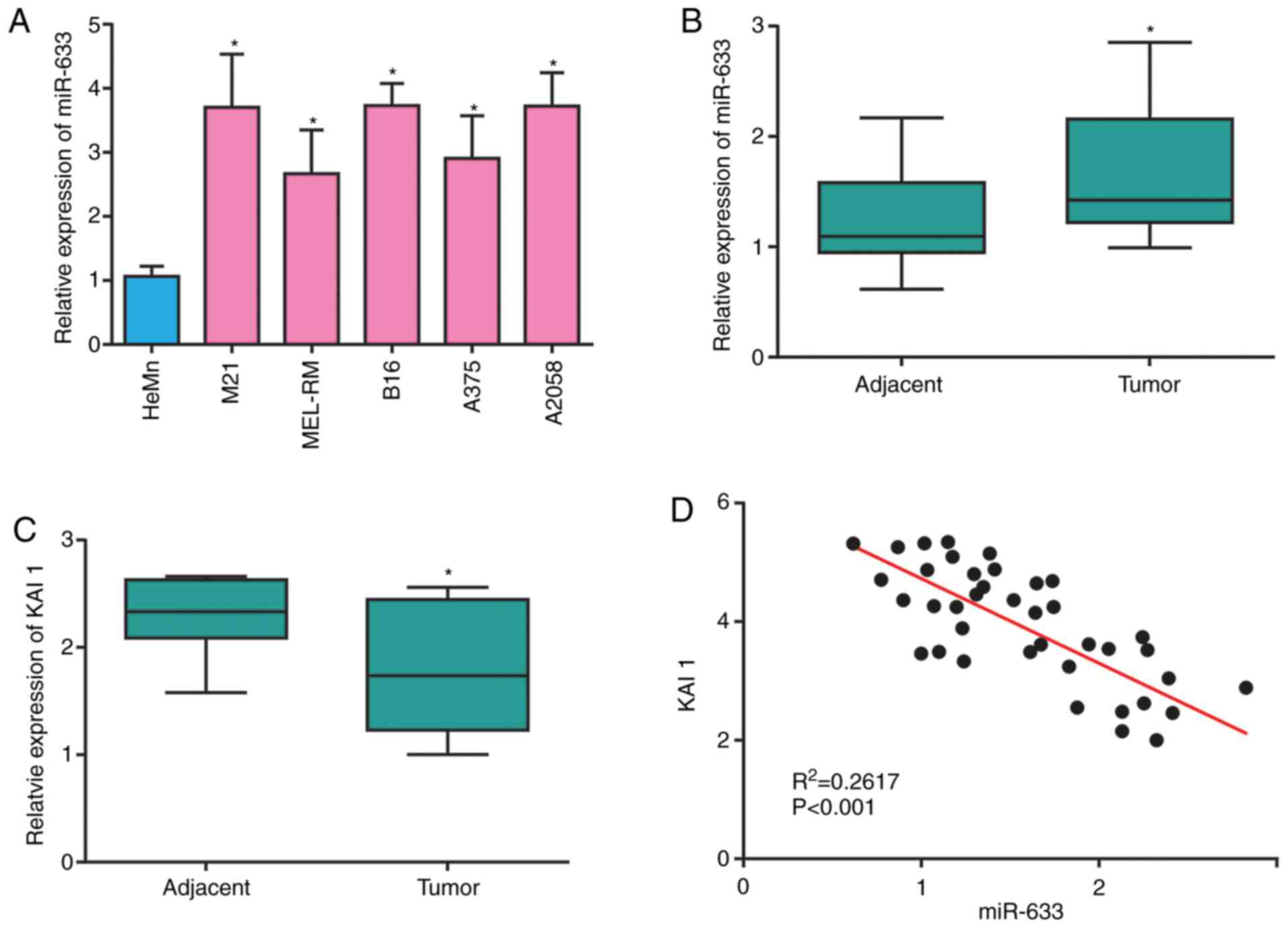
miR-633 was found to be expressed at higher levels
in melanoma cells compared with HeMn control cells (Fig. 2A). Similarly, it was found that
miR-633 was upregulated in melanoma tumor tissues compared with
adjacent normal tissues, the results were re-plotted in Fig. 2B for ease of reference (Fig. 2B). When the mRNA expression levels
for the KAI1 gene were examined in the same samples, the results
exhibited an opposite pattern, with KAI1 mRNA expression levels
significantly decreased in melanoma tissues compared with normal
tissues (Fig. 2C). Pearson
correlation analyses confirmed that miR-633 and KAI1 expression was
negatively correlated in melanoma tissue samples (Fig. 2D).
miR-633 suppresses KAI1 expression in
melanoma
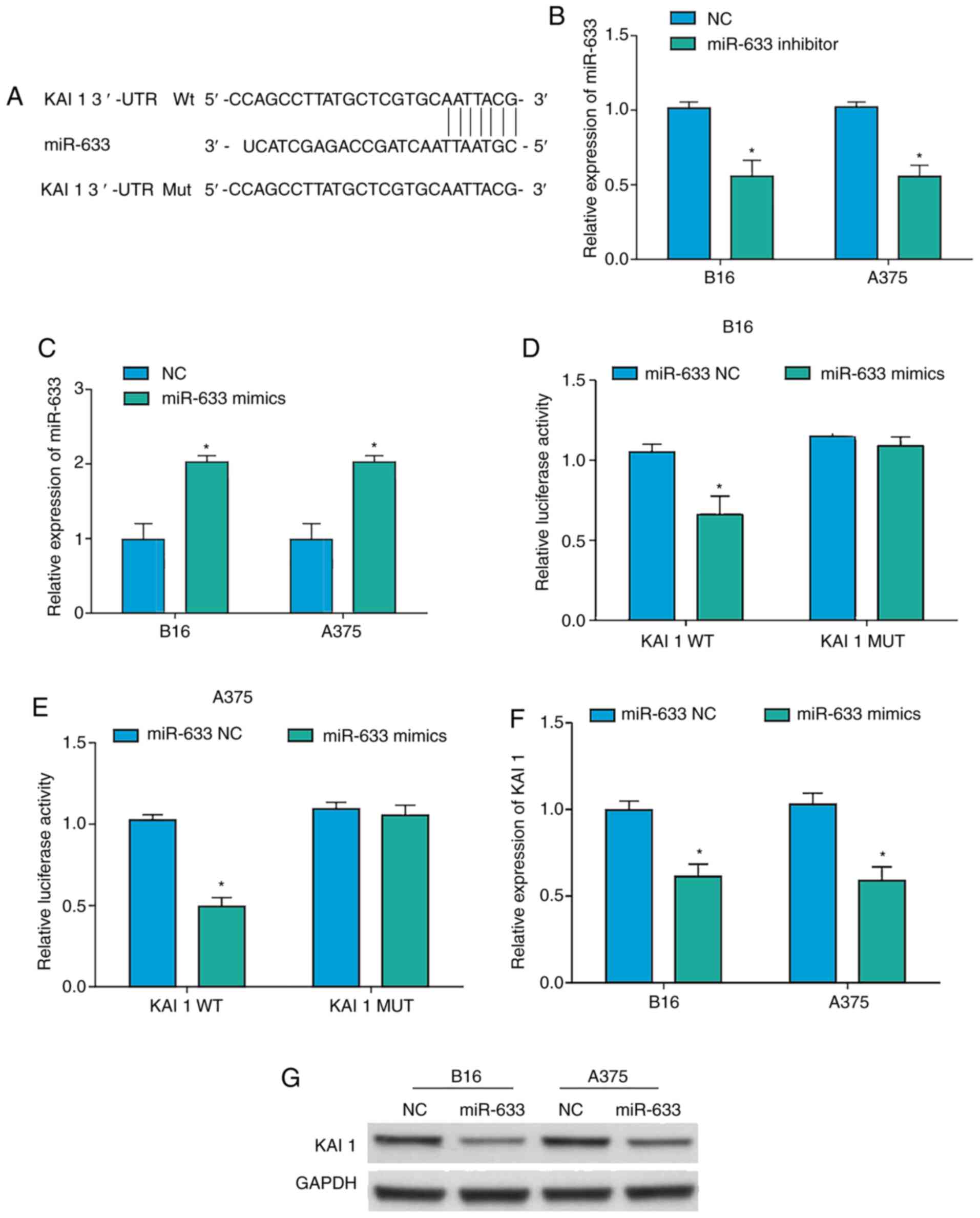
After having identified the predicted miR-633
binding site within the KAI1 3′-UTR, WT and MUT versions of a KAI1
luciferase reporter were constructed (Fig. 3A). Additionally, an miR-633 inhibitor
and mimics were prepared, and their transfection efficiency was
validated in A375 and B16 cells (Fig. 3B
and C). Using luciferase reporter assays, the results confirmed
that miR-633 mimics were able to bind to the WT, but not the MUT,
version of the KAI1 reporter, consistent with a specific binding
interaction between miR-633 and KAI1 (Fig. 3D and E). Subsequently, it was
determined that KAI1 expression was decreased at both the mRNA and
the protein level in A375 and B16 cells overexpressing miR-633
(Fig. 3F and G). Taken together,
these findings confirmed thatmiR-633 may serve as a negative
regulator of KAI1 transcription and translation in melanoma
cells.
miR-633 enhances melanoma cell
proliferation and migration
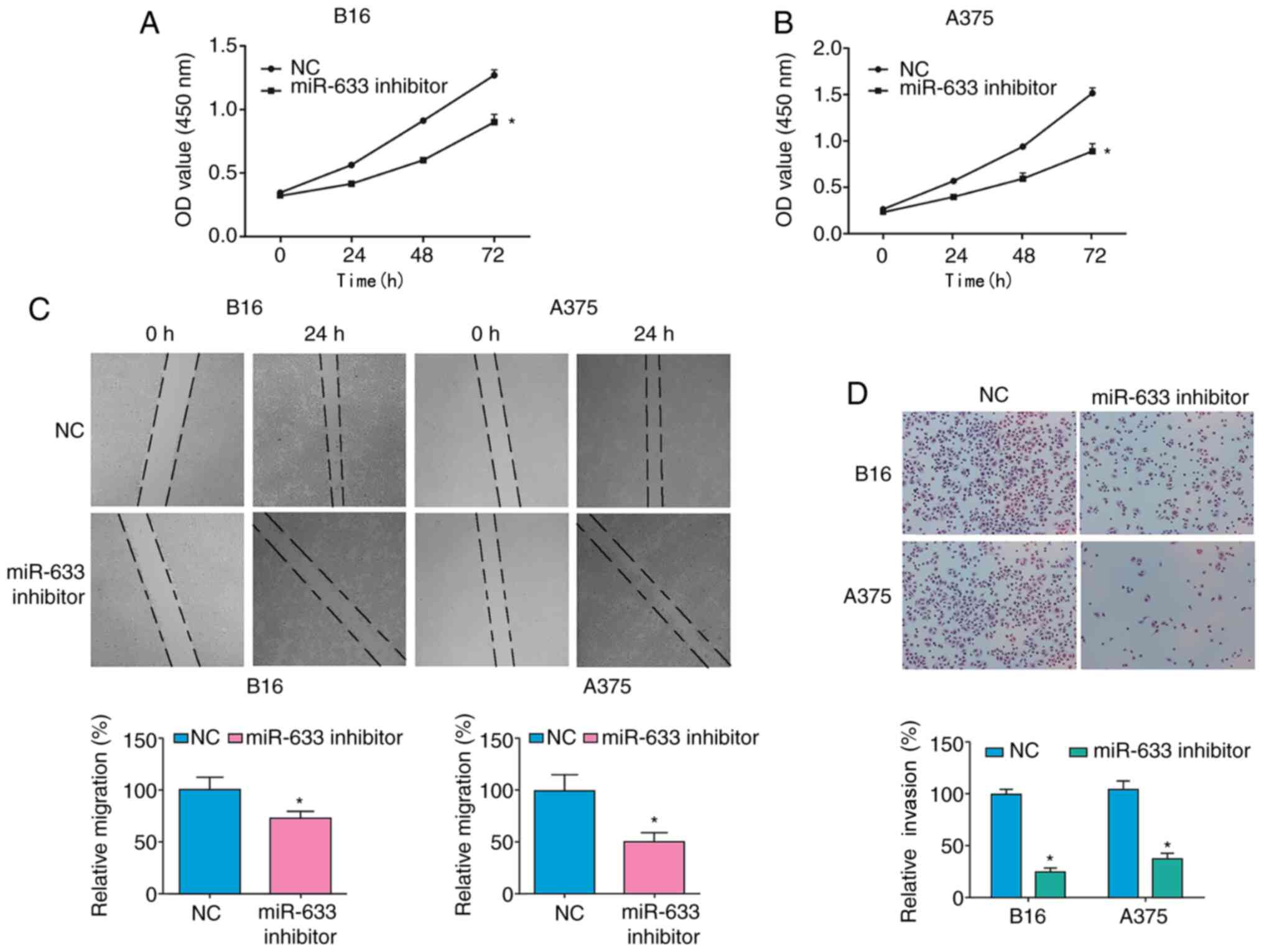
Finally, the biological importance of miR-633 was
assessed in melanoma. Using a CCK-8 assay, it was found that
transfection with the miR-633 inhibitor significantly suppressed
the numbers of melanoma cells over time compared with cells
transfected with NC (Fig. 4A and B),
suggesting a role for this miRNA in the proliferation of melanoma
cells. In addition, miR-633 inhibitor transfection into both the
A375 and B16 melanoma cell lines decreased migration rates in wound
healing assays (Fig. 4C) and cell
invasion in Transwell assays (Fig.
4D). Collectively, these findings demonstrated that miR-633 was
able to promote the migration and proliferation of melanoma
cells.
Discussion
In the present study, it was demonstrated that
miR-633 was significantly upregulated in melanoma tissue compared
with adjacent normal tissue from11patients with melanoma. In
addition, a series of melanoma cell lines exhibited a significantly
higher level of miR-633 expression compared with HeMn cells. In
previous studies, miR-633 was shown to be a functionally important
tumor-associated miRNA in lung and brain cancer (28,29). Its
functional importance in melanoma, however, had yet to be properly
elucidated. The present study used the TargetScan, StarBase and
miRanda algorithms to predict 6 candidate miRNAs that specifically
targeted KAI1.KAI1 was shown to have one potential complementary
miR-633-binding site within its 3′-UTR. Experimental results
demonstrated that over expression of miR-633 led to a significant
reduction in the levels of KAI1 protein, and deregulated expression
of miR-633 significantly altered the proliferation, migration and
invasion of A375 and B16 melanoma cells. Taken together, the
findings of the present study have provided, to the best of the
authors' knowledge, the first evidence thatmiR-633 may function as
a tumor promoter in human melanoma, mediated partly through
targeting KAI1 expression.
A total of 3,657 mature human miRNAs have been
identified to date (miRBase, 2019, http://www.mirbase.org). In prior studies, miRNAs
identified from the integumentary system were successfully used as
diagnostic and prognostic biomarkers in patients with melanoma
(30,31). For example, miR-213 is a suppressor
of malignant melanoma progression, thereby indicating that it may
be a potentially viable therapeutic target or diagnostic biomarker
in patients with this form of cancer (32). Previous studies indicated that
deregulation of miR-21 was associated with pro-apoptotic effects in
pancreatic and breast cancer cell lines (33). In melanoma, a previous study revealed
that miR-205, miR-200 and members of the let-7 family(−125b, −146a,
−155, −21, −25, −23a and −29b) were deregulated (34). Previous studies also suggested that
abnormal miR-633 expression was associated with the prednisone
response early in childhood acute lymphoblastic leukemia relapse
(35). However, abnormal miR-633
expression has only been definitively identified in few types of
tumors to date. The association between miR-633 and malignant
melanoma has not been previously elucidated. The present study
demonstrated that the expression levels of miR-633 were upregulated
in melanoma tissues and cell lines compared with normal tissues and
cells, respectively. Furthermore, the results obtained revealed
that deregulation of miR-633 significantly altered the
proliferation and migration of A375 and B16 melanoma cells.
Therefore, miR-633 may serve as a novel biomarker for melanoma in
the future.
KAI1, also known as CD82, is an important
transcriptional regulator and tumor suppressor gene, and KAI1
mutations are commonly encountered in a range of tumor types
(36). KAI1 regulates chromatin
accessibility, in part via the E-cadherin pathway (37), and prior bioinformatics analyses have
suggested that KAI1 mutations are associated with lung, bone and
brain cancer prognoses, underscoring the relevance of this gene in
oncogenic contexts (38). KAI1 may
attenuate signaling to shut down metastatic colonization through
attenuation of epidermal growth factor receptor signaling and
inhibition of the Wnt signaling pathway. You et al (39) suggested that the KAI1 promoter may be
regulated by the p53 and NME/NM23 nucleoside diphosphate kinase 1
genes. In pancreatic carcinoma, over expressing KAI1 attenuated the
phosphorylation of SRC and STAT3, thereby inhibiting the expression
of vascular endothelial growth factor C and limiting the activity
of pancreatic carcinoma cells (40).
KAI1 has also been shown to serve as a mediator of metabolic
reprogramming in tumor cells. For example, KAI1 can suppress the
progression of cancers of the digestive system via influencing
invasion-associated protein metabolism (41). It also serves as an important tumor
suppressor gene in the context of integumental tumors, with KAI1
inactivating mutations being a common feature in human melanoma
(42). The ability of KAI1 to
suppress oncogenesis is linked to its ability to inhibit the
expression of AP-1, JUNB, poly (ADP-ribose) polymerase (PARP) and
other oncogenes (43). Khan et
al (44) reported that KAI1 is a
key regulator of Toll-like receptor 9 (TLR9) trafficking and
signaling. KAI1 modulates TLR9-dependent NF-κB nuclear
translocation, which is critical for inflammatory cytokine
production. A recent study demonstrated that KAI1 was significantly
associated with poor survival and could act as an independent
prognostic factor in human melanoma for both 5-year and 10-year
survival rates (45). KAI1
deregulation has been observed in melanoma cell lines and tumor
samples, and the over expression of this protein may significantly
reduce melanoma progression (46).
Furthermore, miR-203 inhibited frizzled-2 expression via KAI1
expression in human lung carcinoma cells, andKAI1expression may be
a useful marker for metastatic, invasive cancer, and prognostic
factor in tumors, such as lung cancer (47). However, the specific mechanisms
governing KAI1 downregulation in the context of melanoma had yet to
be clarified.
The present study aimed to investigate the role of
miR-633 in melanoma through its potential interaction with KAI1,
revealing that miR-633 may be an important regulator of malignant
melanoma. The results showed that miR-633 was upregulated in
melanoma cells and tumor tissue samples compared with their
controls. In addition, miR-633 inhibition resulted in impaired
migration and proliferation of A375 and B16 melanoma cells in
vitro. Notably, the results of the dual-luciferase reporter
assay demonstrated that miR-633 directly regulated the expression
of KAI1. Therefore, the data obtained in the present study have
provided important insights into the regulation of KAI1 by miRNAs
in cancer cells. miR-633 may serve as a potential candidate for the
diagnosis and treatment of human melanoma.
In conclusion, the present study has provided novel
evidence to show that inhibition of miR-633 suppresses the
proliferation and migration of the malignant melanoma cell lines
A375 and B16, partly via regulation of KAI1.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
YL and ZW made substantial contributions to the
conception and design of the study. Both authors collected the
samples and clinical data, and contributed significantly to data
analysis. ZW performed the experiments and drafted the initial
manuscript. YL gave final approval of the version to be published.
Both authors agreed to be accountable for all aspects of the work
in ensuring that questions related to the accuracy and integrity of
any part of the work were appropriately investigated and resolved.
Both authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Cangzhou Central Hospital (Cangzhou, China; approval
no. 201903501). Written informed consent was provided by patients
and their families prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stark MS, Gray ES, Isaacs T, Chen FK,
Millward M, McEvoy A, Zaenker P, Ziman M, Soyer HP, Glasson WJ, et
al: A panel of circulating MicroRNAs detects uveal melanoma with
high precision. Transl Vis Sci Technol. 8:122019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia Z, Yang C, Yang X, Wu S, Feng Z, Qu L,
Chen X, Liu L and Ma Y: MiR-652 promotes proliferation and
migration of uveal melanoma cells by targeting HOXA9. Med Sci
Monit. 25:8722–8732. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fumagalli MR, Lionetti MC, Zapperi S and
La Porta C: Cross-Talk between circRNAs and mRNAs modulates
MiRNA-mediated circuits and affects melanoma plasticity. Cancer
Microenviron. 12:95–104. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang C, Xia Z, Zhu L, Li Y, Zheng Z, Liang
J and Wu L: MicroRNA-139-5p modulates the growth and metastasis of
malignant melanoma cells via the PI3K/AKT signaling pathway by
binding to IGF1R. Cell Cycle. 18:3513–3524. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sánchez-Sendra B, García-Giménez JL,
González-Muñoz JF, Navarro L, Murgui A, Terrádez L, Pinazo I,
Martin JM and Monteagudo C: Circulating miRNA expression analysis
reveals new potential biomarkers for human cutaneous melanoma
staging. J Eur Acad Dermatol Venereol. 34:e126–e129. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun HW, Yang GL, Wang SN, Zhang YJ, Ding
JX and Zhang XN: MicroRNA-92a regulates the development of
cutaneous malignant melanoma by mediating FOXP1. Eur Rev Med
Pharmacol Sci. 23:8991–8999. 2019.PubMed/NCBI
|
|
7
|
Nakamura K and Okuyama R: Immunotherapy
for advanced melanoma: Current knowledge and future directions. J
Dermatol Sci. 83:87–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reale E, Taverna D, Cantini L, Martignetti
L, Osella M, De Pittà C, Virga F, Orso F and Caselle M:
Investigating the epi-miRNome: Identification of epi-miRNAs using
transfection experiments. Epigenomics. 11:1581–1599. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Afrang N and Honardoost M: Cell cycle
regulatory markers in melanoma: New strategies in diagnosis and
treatment. Med J Islam Repub Iran. 33:962019.PubMed/NCBI
|
|
10
|
Hämäläinen M, Teppo HR, Skarp S,
Haapasaari KM, Porvari K, Vuopala K, Kietzmann T and Karihtala P:
NRF1 and NRF2 mRNA and protein expression decrease early during
melanoma carcinogenesis: An insight into survival and MicroRNAs.
Oxid Med Cell Longev. 2019:26470682019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rossi E, Schinzari G, Maiorano BA,
Pagliara MM, Di Stefani A, Bria E, Peris K, Blasi MA and Tortora G:
Conjunctival melanoma: Genetic and epigenetic insights of a
distinct type of melanoma. Int J Mol Sci. 20:54472019. View Article : Google Scholar
|
|
12
|
Chen L, Karisma VW, Liu H and Zhong L:
MicroRNA-300: A transcellular mediator in exosome regulates
melanoma progression. Front Oncol. 9:10052019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang W, Hou L, Wei J, Du Y, Zhao Y, Deng
X and Lin X: Hsa-MiR-217 inhibits the proliferation, migration, and
invasion in non-small cell lung cancer cells via targeting SIRT1
and P53/KAI1 signaling. Balkan Med J. 37:208–214. 2020.PubMed/NCBI
|
|
14
|
Zhang Q, Huang F, Yao Y, Wang J, Wei J, Wu
Q, Xiang S and Xu L: Interaction of transforming growth
factor-β-smads/microRNA-362-3p/CD82 mediated by M2 macrophages
promotes the process of epithelial-mesenchymal transition in
hepatocellular carcinoma cells. Cancer Sci. 110:2507–2519. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Habibzadeh P, Honarvar B, Silawi M,
Bahramjahan S, Kazemi A, Faghihi MA and Lankarani K: Association
between rs2303861 polymorphism in CD82 gene and non-alcoholic fatty
liver disease: A preliminary case-control study. Croat Med J.
60:361–368. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Asada H, Tomiyasu H, Uchikai T, Ishihara
G, Goto-Koshino Y, Ohno K and Tsujimoto H: Comprehensive analysis
of miRNA and protein profiles within exosomes derived from canine
lymphoid tumour cell lines. PLoS One. 14:e02085672019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu CJ, Yang JH, Huang FZ, Yang JH, Liu
CP, Mao XH, Yi WM, Shen XB, Peng C, Chen MF, et al: The role of
miR-99b in mediating hepatocellular carcinoma invasion and
migration. Eur Rev Med Pharmacol Sci. 24:79092020.PubMed/NCBI
|
|
18
|
Long J, Luo J and Yin X: MiR-338-5p
promotes the growth and metastasis of malignant melanoma cells via
targeting CD82. Biomed Pharmacother. 102:1195–1202. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee MS, Lee J, Kim YM and Lee H: The
metastasis suppressor CD82/KAI1 represses the TGF-β 1 and wnt
signalings inducing epithelial-to-mesenchymal transition linked to
invasiveness of prostate cancer cells. Prostate. 79:1400–1411.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li W, Hu M, Wang C, Lu H, Chen F, Xu J,
Shang Y, Wang F, Qin J, Yan Q, et al: A viral microRNA
downregulates metastasis suppressor CD82 and induces cell invasion
and angiogenesis by activating the c-met signaling. Oncogene.
36:5407–5420. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nishioka C, Ikezoe T, Pan B, Xu K and
Yokoyama A: MicroRNA-9 plays a role in interleukin-10-mediated
expression of E-cadherin in acute myelogenous leukemia cells.
Cancer Sci. 108:685–695. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu L, Hou Y, Tu G, Chen Y, Du YE, Zhang H,
Wen S, Tang X, Yin J, Lang L, et al: Nuclear drosha enhances cell
invasion via an EGFR-ERK1/2-MMP7 signaling pathway induced by
dysregulated miRNA-622/197 and their targets LAMC2 and CD82 in
gastric cancer. Cell Death Dis. 8:e26422017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jee BK, Park KM, Surendran S, Lee WK, Han
CW, Kim YS and Lim Y: KAI1/CD82 suppresses tumor invasion by MMP9
inactivation via TIMP1 up-regulation in the H1299 human lung
carcinoma cell line. Biochem Biophys Res Commun. 342:655–661. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mine M, Yamaguchi K, Sugiura T, Chigita S,
Yoshihama N, Yoshihama R, Hiyake N, Kobayashi Y and Mori Y: MiR-203
inhibits frizzled-2 expression via CD82/KAI1 expression in human
lung carcinoma cells. PLoS One. 10:e01313502015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang QH, Yao YL, Wu XY, Wu JH, Gu T, Chen
L, Gu JH, Liu Y and Xu L: Anti-MiR-362-3p inhibits migration and
invasion of human gastric cancer cells by its target CD82. Dig Dis
Sci. 60:1967–1976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dai W, Wang C, Wang F, Wang Y, Shen M,
Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al: Anti-MiR-197
inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem
Biophys Res Commun. 446:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Q, Zhang LY, Wu S, Huang C, Liu J, Wang
P and Cao Y: Bioinformatics analysis identifies MicroRNAs and
target genes associated with prognosis in patients with melanoma.
Med Sci Monit. 25:7784–7794. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cooling L: An update on the I blood group
system. Immunohematology. 35:85–90. 2019.PubMed/NCBI
|
|
30
|
Santoni G, Morelli MB, Santoni M, Nabissi
M, Marinelli O and Amantini C: Targeting transient receptor
potential channels by microRNAs drives tumor development and
progression. Adv Exp Med Biol. 1131:605–623. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ylösmäki L, Polini B, Carpi S, Martins B,
Smertina E, Feola S, Fusciello M, Peltonen K, Nieri P, Ylösmäki E
and Cerullo V: Harnessing therapeutic viruses as a delivery vehicle
for RNA-based therapy. PLoS One. 14:e02240722019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sharma A, Biswas A, Liu H, Sen S,
Paruchuri A, Katsonis P, Lichtarge O, Dakal TC, Maulik U, Gromiha
MM, et al: Mutational landscape of the BAP1 locus reveals an
intrinsic control to regulate the miRNA network and the binding of
protein complexes in uveal melanoma. Cancers (Basel). 11:16002019.
View Article : Google Scholar
|
|
33
|
Chekhun VF, Borikun TV, Bazas VМ, Andriiv
AV, Klyusov OM, Yalovenko TM and Lukianova NY: Association of
circulating miR-21, −205, and −182 with response of luminal breast
cancers to neoadjuvant FAC and AC treatment. Exp Oncol. 42:162–166.
2020.PubMed/NCBI
|
|
34
|
Bonazzi VF, Stark MS and Hayward NK:
MicroRNA regulation of melanoma progression. Melanoma Res.
22:101–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu L, Liang Yn, Luo Xq, Liu Xd and Guo Hx:
Association of miRNAs expression profiles with prognosis and
relapse in childhood acute lymphoblastic leukemia. Zhonghua Xue Ye
Xue Za Zhi. 32:178–181. 2011.(In Chinese). PubMed/NCBI
|
|
36
|
Wang Y, Chen H, Fu Y, Ai A, Xue S, Lyu O
and Kuang Y: MiR-195 inhibits proliferation and growth and induces
apoptosis of endometrial stromal cells by targeting FKN. Int J Clin
Exp Pathol. 6:2824–2834. 2013.PubMed/NCBI
|
|
37
|
Arribas AJ, Campos-Martín Y, Gómez-Abad C,
Algara P, Sánchez-Beato M, Rodriguez-Pinilla MS, Montes-Moreno S,
Martinez N, Alves-Ferreira J, Piris MA and Mollejo M: Nodal
marginal zone lymphoma: Gene expression and miRNA profiling
identify diagnostic markers and potential therapeutic targets.
Blood. 119:e9–e21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou X, Liu X, Zhang G, Zhang Q, Chen H,
Wang Y, Fang F and Sun J: Knockdown THOC2 suppresses the
proliferation and invasion of melanoma. Bioengineered. 10:635–645.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
You J, Chang R, Liu B, Zu L and Zhou Q:
Nm23-H1 was involved in regulation of KAI1 expression in
high-metastatic lung cancer cells L9981. J Thorac Dis. 8:1217–1226.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu X, Guo X, Li H, Chen J and Qi X:
Src/STAT3 signaling pathways are involved in KAI1-induced
downregulation of VEGF-C expression in pancreatic cancer. Mol Med
Rep. 13:4774–47778. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bhalla S, Kaur H, Dhall A and Raghava GP:
Prediction and analysis of skin cancer progression using genomics
profiles of patients. Sci Rep. 9:157902019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hou Q, Han S, Yang L, Chen S, Chen J, Ma
N, Wang C, Tang J, Chen X, Chen F, et al: The interplay of
microRNA-34a, LGR4, EMT-associated factors, and MMP2 in regulating
uveal melanoma cells. Invest Ophthalmol Vis Sci. 60:4503–4510.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu T, Chen S, Qu L, Wang Y, Chen HD and He
C: Identification of a five-miRNA signature predicting survival in
cutaneous melanoma cancer patients. Peer J. 7:e78312019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Khan NS, Lukason DP, Feliu M, Ward RA,
Lord AK, Reedy JL, Ramirez-Ortiz ZG, Tam JM, Kasperkovitz PV,
Negoro PE, et al: CD82 controls CpG-dependent TLR9 signaling. FASEB
J. 33:12500–12514. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang G, Cheng Y, Chen G, Tang Y, Ardekani
G, Rotte A, Martinka M, McElwee K, Xu X, Wang Q and Zhou Y: Loss of
tumor suppressors KAI1 and p27 identifies a unique subgroup of
primary melanoma patients with poor prognosis. Oncotarget.
6:23026–23035. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu N, Liu Z, Liu X and Chen H:
Comprehensive analysis of a competing endogenous RNA network
identifies seven-lncRNA signature as a prognostic biomarker for
melanoma. Front Oncol. 9:9352019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Prabhu VV and Devaraj SN: KAI1/CD82,
metastasis suppressor gene as a therapeutic target for
non-small-cell lung carcinoma. J Environ PatholToxicol Oncol.
36:269–275. 2017. View Article : Google Scholar
|