|
1
|
Zamorano JL, Lancellotti P, Rodriguez
Munoz D, Aboyans V, Asteggiano R, Galderisi M, Habib G, Lenihan DJ,
Lip GYH, Lyon AR, et al: 2016 ESC position paper on cancer
treatments and cardiovascular toxicity developed under the auspices
of the ESC committee for practice guidelines: The task force for
cancer treatments and cardiovascular toxicity of the European
society of cardiology (ESC). Eur Heart J. 37:2768–2801. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gyongyosi M, Lukovic D, Zlabinger K,
Spannbauer A, Gugerell A, Pavo N, Traxler D, Pils D, Maurer G,
Jakab A, et al: Liposomal doxorubicin attenuates cardiotoxicity via
induction of interferon-related DNA damage resistance. Cardiovasc
Res. 116:970–982. 2020.PubMed/NCBI
|
|
3
|
Gupta SK, Garg A, Bar C, Chatterjee S,
Foinquinos A, Milting H, Streckfuß-Bömeke K, Fiedler J and Thum T:
Quaking inhibits doxorubicin-mediated cardiotoxicity through
regulation of cardiac circular RNA expression. Circ Res.
122:246–254. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Force T, Krause DS and Van Etten RA:
Molecular mechanisms of cardiotoxicity of tyrosine kinase
inhibition. Nat Rev Cancer. 7:332–344. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Skubitz KM: Cardiotoxicity monitoring in
patients treated with tyrosine kinase inhibitors. Oncologist.
24:e600–e602. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Santoni M, Guerra F, Conti A, Lucarelli A,
Rinaldi S, Belvederesi L, Capucci A and Berardi R: Incidence and
risk of cardiotoxicity in cancer patients treated with targeted
therapies. Cancer Treat Rev. 59:123–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gill D, Georgakis MK, Koskeridis F, Jiang
L, Feng Q, Wei WQ, Theodoratou E, Elliott P, Denny JC, Malik R, et
al: Use of genetic variants related to antihypertensive drugs to
inform on efficacy and side effects. Circulation. 140:270–279.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Griffith M, Griffith OL, Coffman AC,
Weible JV, McMichael JF, Spies NC, Koval J, Das I, Callaway MB,
Eldred JM, et al: DGIdb: Mining the druggable genome. Nat Methods.
10:1209–1210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wagner AH, Coffman AC, Ainscough BJ, Spies
NC, Skidmore ZL, Campbell KM, Krysiak K, Pan D, McMichael JF,
Eldred JM, et al: DGIdb 2.0: Mining clinically relevant drug-gene
interactions. Nucleic Acids Res. 44:D1036–D1044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cotto KC, Wagner AH, Feng YY, Kiwala S,
Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL and Griffith
M: DGIdb 3.0: A redesign and expansion of the drug-gene interaction
database. Nucleic Acids Res. 46:D1068–D1073. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kamat MA, Blackshaw JA, Young R, Surendran
P, Burgess S, Danesh J, Butterworth AS and Staley JR: PhenoScanner
V2: An expanded tool for searching human genotype-phenotype
associations. Bioinformatics. 35:4851–4853. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liao X, Lan C, Liao D, Tian J and Huang X:
Exploration and detection of potential regulatory variants in
refractive error GWAS. Sci Rep. 6:330902016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boyle AP, Hong EL, Hariharan M, Cheng Y,
Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et
al: Annotation of functional variation in personal genomes using
RegulomeDB. Genome Res. 22:1790–1797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mesbah-Uddin M, Elango R, Banaganapalli B,
Shaik NA and Al-Abbasi FA: In-silico analysis of inflammatory bowel
disease (IBD) GWAS loci to novel connections. PLoS One.
10:e01194202015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang E, Zhao H, Zhao D, Li L and Du L:
Functional prediction of chronic kidney disease susceptibility gene
PRKAG2 by comprehensively bioinformatics analysis. Front Genet.
9:5732018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ward LD and Kellis M: HaploReg v4:
Systematic mining of putative causal variants, cell types,
regulators and target genes for human complex traits and disease.
Nucleic Acids Res. 44:D877–D881. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ward LD and Kellis M: HaploReg: A resource
for exploring chromatin states, conservation, and regulatory motif
alterations within sets of genetically linked variants. Nucleic
Acids Res. 40((Database Issue)): D930–D934. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo L, Du Y, Chang S, Zhang K and Wang J:
rSNPBase: A database for curated regulatory SNPs. Nucleic Acids
Res. 42((Database Issue)): D1033–D1039. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lou J, Gong J, Ke J, Tian J, Zhang Y, Li
J, Yang Y, Zhu Y, Gong Y, Li L, et al: A functional polymorphism
located at transcription factor binding sites, rs6695837 near LAMC1
gene, confers risk of colorectal cancer in Chinese populations.
Carcinogenesis. 38:177–183. 2017.PubMed/NCBI
|
|
20
|
Ganguly K, Saha T, Saha A, Dutta T,
Banerjee S, Sengupta D, Bhattacharya S, Ghosh S and Sengupta M:
Meta-analysis and prioritization of human skin
pigmentation-associated GWAS-SNPs using ENCODE data-based
web-tools. Arch Dermatol Res. 311:163–171. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Casper J, Zweig AS, Villarreal C, Tyner C,
Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et
al: The UCSC genome browser database: 2018 update. Nucleic Acids
Res. 46:D762–D769. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Haeussler M, Zweig AS, Tyner C, Speir ML,
Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN,
et al: The UCSC genome browser database: 2019 update. Nucleic Acids
Res. 47:D853–D858. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee CM, Barber GP, Casper J, Clawson H,
Diekhans M, Gonzalez JN, Hinrichs AS, Lee BT, Nassar LR, Powell CC,
et al: UCSC genome browser enters 20th year. Nucleic Acids Res.
48:D756–D761. 2020.PubMed/NCBI
|
|
24
|
McCall MN, Illei PB and Halushka MK:
Complex sources of variation in tissue expression data: Analysis of
the GTEx lung transcriptome. Am J Hum Genet. 99:624–635. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carithers LJ, Ardlie K, Barcus M, Branton
PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J,
Gelfand ET, et al: A novel approach to high-quality postmortem
tissue procurement: The GTEx project. Biopreserv Biobank.
13:311–319. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Karolchik D, Barber GP, Casper J, Clawson
H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L,
Haeussler M, et al: The UCSC genome browser database: 2014 update.
Nucleic Acids Res. 42:D764–D770. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
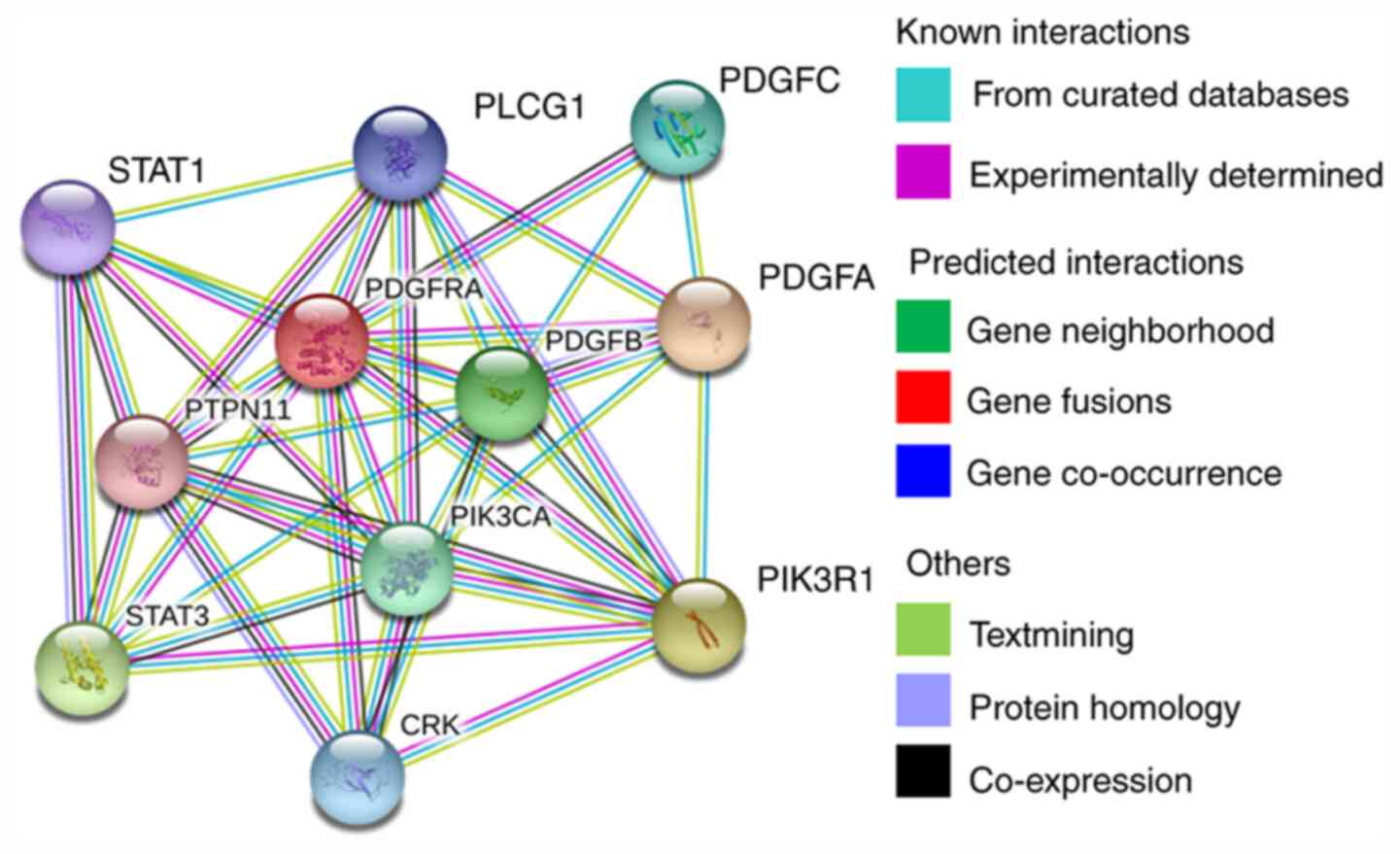
Tallquist MD and Soriano P: Cell
autonomous requirement for PDGFRalpha in populations of cranial and
cardiac neural crest cells. Development. 130:507–518. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hoch RV and Soriano P: Roles of PDGF in
animal development. Development. 130:4769–4784. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hong SP, Song S, Cho SW, Lee S, Koh BI,
Bae H, Kim KH, Park JS, Do HS, Im I, et al: Generation of
PDGFRα+ cardioblasts from pluripotent stem cells. Sci
Rep. 7:418402017. View Article : Google Scholar : PubMed/NCBI
|