Introduction
Liver cancer is the sixth most common type of cancer
and the fourth leading cause of cancer-associated mortality
worldwide, with ~841,000 new cases and 782,000 deaths in 2018
(1). The 5-year survival rate of
liver cancer is estimated to be 18% (2). Liver hepatocellular carcinoma (LIHC)
accounts for 75–85% of primary liver cancers (1) and its overall median survival times is
estimated to be 29.8 months (2.48 years) (3), and this has not improved over time
(3). Current treatment methods do
not ensure effective early diagnosis and treatment of LIHC.
Therefore, improving the prevention and treatment of this disease
is imperative.
MicroRNAs (miRNAs) are small non-coding sequences
with a length of ~20 bp, which regulate gene expression following
transcription. miRNAs control several eukaryotic developmental and
cellular processes (4–6). miRNAs usually bind to the
3′-untranslated region of the target mRNAs to cause their
degradation. A previous study has demonstrated that the majority of
human protein-coding genes contain at least one conserved miRNA
binding site (7), whereas a high
number of miRNAs can regulate multiple mRNAs (4), suggesting that the biological functions
of miRNAs are highly diverse. Therefore, it is not surprising that
disorders of miRNAs are often associated with human diseases,
including cancer (8,9). Increasing evidence has revealed that
miRNAs are closely associated with the abnormal expression of genes
regulating hepatocyte proliferation, cell cycle, metastasis and
apoptosis, which ultimately leads to the development of LIHC
(10,11), and these suggests that miRNAs play an
important role in the occurrence and development of LIHC, and have
potential diagnostic and therapeutic value (12,13).
Among them, let-7c has been identified as an anticancer miRNA in
LIHC (14). Additional research has
indicated that let-7c represses cell proliferation, migration and
invasion, and induces G1 phase arrest and apoptosis of
LIHC cells (15). Furthermore,
let-7c can enhance sorafenib-induced apoptosis of LIHC cells by
targeting Bcl-xL (16).
The PI3K/Akt signaling pathway is highly mutated and
activated in several cancer types, which mediates the
proliferation, survival, migration and angiogenesis of these cancer
cells (17–19) and has become a target for human
cancer treatment. Notably, the PI3K/Akt signaling pathway is
associated with LIHC progression, vascular infiltration and
metastasis, as well as poor prognosis and a low survival rate in
patients with LIHC (20).
Furthermore, the FoxO signaling pathway, which is closely
associated with the PI3K/Akt signaling pathway, serves an important
role in the occurrence and development of hepatocellular carcinoma
(21,22).
Therefore, in the present study, the association
between let-7c and the PI3K/Akt/FoxO signaling pathway, as well as
their roles in the development of LIHC were investigated using The
Cancer Genome Atlas (TCGA) and various public databases. The
effects of let-7c-5p on PI3K/Akt/FoxO signaling pathway-related
target genes were analyzed following overexpression of let-7c-5p in
the MHCC-97H cell line. The results may reveal novel targets and
strategies for the diagnosis and treatment of LIHC.
Materials and methods
Clinical significance of let-7c-5p in
hepatocellular carcinoma as determined by TCGA analysis
The Cancer Genome Mapping (derived from TCGA) data
portal is the largest and most commonly used public resource,
providing datasets, such as somatic mutations, gene expression,
gene methylation and copy number variation, for thousands of tumor
samples (23). In the present study,
the let-7c-5p expression profiles of different types of human
cancer and adjacent normal tissues were obtained from
Tumor-miRNA-Pathway (http://bioinfo.life.hust.edu.cn/miR_path/), an online
TCGA data analysis tool that displays miRNA expression levels in 34
different types of tumor, including LIHC, adrenocortical carcinoma
(ACC), bladder urothelial carcinoma (BLCA), breast invasive
carcinoma (BRCA), cervical squamous cell carcinoma and endocervical
adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon
adenocarcinoma (COAD), lymphoid neoplasm diffuse large b-cell
lymphoma (DLBC), esophageal carcinoma (ESCA), FFPE pilot phase II
FFPE (FPPP), glioblastoma multiforme (GBM), head and neck squamous
cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal
clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma
(KIRP), acute myeloid leukemia (LAML), brain lower grade glioma
(LGG), lung adenocarcinoma (LUAD), lung squamous cell carcinoma
(LUSC), mesothelioma (MESO), ovarian serous cystadenocarcinoma
(OV), pancreatic adenocarcinoma (PAAD), pheochromocytoma and
paraganglioma (PCPG), prostate adenocarcinoma (PRAD), rectum
adenocarcinoma (READ), sarcomav (SARC), skin cutaneous melanoma
(SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors
(TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus
endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS) and
uveal melanoma (UVM) (24). The
patient survival data corresponding to certain cancer types (across
30 cancer types), including LIHC, such as ACC, BLCA, BRCA, CESC,
CHOL, COAD, ESCA, HNSC, KICH, KIRC, KIRP, LUAD, LUSC, MESO, OV,
PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC,
UCS and UVM, that were significantly associated with let-7c-5p
expression were obtained using OncomiR online software (http://www.oncomir.org/) (25). This led to the identification of the
miRNAs associated with tumor formation and patient survival.
Target genes of let-7c-5p and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis
DIANA-TarBase v8 (http://www.microrna.gr/tarbase) is a reference
database devoted to the indexing of experimentally supported miRNA
targets (26). This database was
used to screen the experimentally supported target genes of
let-7c-5p (the filter value is default). Subsequently, the online
biological tool KOBAS 3.0 (http://kobas.cbi.pku.edu.cn), which is a web server
for gene/protein functional annotation (annotate module) and
functional gene set enrichment, was used for KEGG (27) pathway enrichment analysis of the
obtained target genes (28–30). The genes belonging to the PI3K-Akt
and FoxO signaling pathways were selected for further analysis.
ONCOMINE analysis
Oncomine gene expression array datasets (http://www.oncomine.org; an online cancer microarray
database) were used to display the gene summary view to analyze the
mRNA differential expression levels of the let-7c-5p target genes
that belonged to the PI3K-Akt and FoxO signaling pathways in 195
cancer vs. normal analyses (31).
The thresholds were as follows: P<0.05; fold-change >1.5;
gene rank, ALL; data type, mRNA. The cancer specimen and normal
control datasets were compared for each gene. The genes with
different mRNA expression levels in the ONCOMINE analysis were
selected for further analysis.
Kaplan-Meier plotter analysis
Kaplan-Meier plotter (www.kmplot.com) is an online database containing
microarray gene expression data and survival information derived
from Gene Expression Omnibus (32),
TCGA (https://www.cancer.gov/tcga) and the
Cancer Biomedical Informatics Grid (33), which contain mRNA expression data and
survival information of 364 cases of clinical LIHC (34). These databases also contain miRNA
expression data and survival information of 614 cases of clinical
LIHC (35). To analyze overall
survival (OS), all possible cut-off values between the lower and
upper quartiles were computed and the best performing threshold was
used as the cut-off, and the follow up threshold was restricted 60
months (5-year survival). The data were verified using a Kaplan
Meier survival curve. In the present study, the associations
between OS and let-7c-5p and its target genes obtained from
ONCOMINE analysis were evaluated using the Kaplan-Meier plotter.
Genes which were expressed at low levels and were associated with a
high survival rate (HR>1) were selected for further analysis. In
addition, the association between target gene expression data in
different tumor grades and survival data was investigated.
LinkedOmics analysis
The LinkedOmics database (http://www.linkedomics.org/login.php) is a web-based
platform for the analysis of 32 TCGA cancer-related datasets
(36). In the present study, the
correlation between let-7c-5p expression and the expression levels
of its target genes in LIHC was analyzed using LinkedOmics. The
target genes that exhibited a negative correlation with let-7c
expression were selected for further analysis.
UALCAN analysis
UALCAN (http://ualcan.path.uab.edu) is an interactive web
resource based on level 3 RNA sequencing methodology and the
clinical data of 31 cancer types from TCGA. It is used to analyze
the relative expression of genes in different tumor subgroups and
of normal tumor samples with different tumor stages, tumor grades,
ethnicity, weight or other clinicopathological characteristics
(37). UALCAN was used to analyze
the expression levels of the target genes that exhibited a negative
correlation with let-7c-5p in different tumor grades of LIHC.
cBioPortal analysis
The cBioPortal for Cancer Genomics contained
sequencing and pathological data of 30 different types of cancer
(38,39). The hepatocellular carcinoma dataset
(derived from TCGA) containing 366 pathologically reported cases
was selected for further analysis of the target genes obtained from
the LinkedOmics dataset. In the setting of query parameters,
mutations, assumed copy number changes from GISTIC and mRNA
expression Z-scores (RNASeq V2 RSEM) were selected in ‘Select
Genomic Profiles’, and the z-score threshold was ±2.0. The inputted
target gene of let-7c-5p was analyzed using OncoPrint, Genetic
Alteration, Mutual Exclusivity, Co-expression and the OS
plotter.
Culture of MHCC-97H cell line
The LIHC MHCC-97H cell line (iCell Bioscience Inc.,
http://www.icellbioscience.com/cellDetail/568) was
cultured in high-glucose DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin-streptomycin (Solarbio, Ltd.) at 37°C in a
humidified incubator containing 5% CO2.
Cell transfection
Prior to overexpression, MHCC-97H cells were plated
in 6-well culture plates, (1×106/cell). Once the
MHCC-97H cells reached 80% confluence, they were transfected with
hsa-let-7c-5p mimics (cat. no. 4464066, Assay ID: MC10436; Thermo
Fisher Scientific, Inc., the overexpression group) or negative
control miRNA mimic (cat. no. 4464058; Thermo Fisher Scientific,
Inc. the overexpression control group) by using Lipofectamine™
RNAiMAX Transfection Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol (https://assets.thermofisher.com/TFS-Assets/LSG/manuals/Lipofectamine_RNAiMAX_Reag_protocol.pdf).
After transfection, MHCC-97H cells were incubated at 37°C with 5%
CO2 for 48 h. Each experiment was repeated 3 times.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from each group using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. hsa-let-7c-5p was
reverse transcribed and amplified using the TaqMan™ MicroRNA assay
(cat. no. 4427975, Assay ID: 000379; Thermo Fisher Scientific,
Inc.) at the following conditions: 16°C for 30 min, 42°C for 30 min
and 85°C for 5 min (RT PCR), and 95°C for 10 min followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec (qPCR). To determine
the expression levels of cyclin B2 (CCNB2), cyclin E2 (CCNE2),
cyclin dependent kinase 4 (CDK4), casein kinase 1ε (CSNK1E), homer
scaffold protein 1 (HOMER1), heat shock protein 90 α family class A
member 1 (HSP90AA1), neuroblastoma RAS viral oncogene homolog
(NRAS), protein phosphatase 2 catalytic subunit α (PPP2CA), protein
kinase AMP-activated catalytic subunit α2 (PRKAA2) and Rac family
small GTPase 1 (RAC1), cDNAs were synthesized using the
PrimeScript™ RT reagent kit (cat. no. RR037Q; Takara Bio, Inc.),
and the primer sequences used for qPCR are listed in Table SI. qPCR was performed using a
StepOnePlus™ system (Thermo Fisher Scientific, Inc.) and the
TaqMan® Universal Master Mix II (cat. no. 4440043;
Thermo Fisher Scientific, Inc.), with the following thermocycling
conditions: 95°C for 10 min followed by 40 cycles of 95°C for 15
sec and 60°C for 60 sec. U6 (cat. no. 4427975, Assay ID: 001973;
Thermo Fisher Scientific, Inc.) and ACTB served as the internal
controls for normalizing the relative expression levels of
has-let-7c-5p and mRNA, respectively. Relative expression levels
were calculated using the 2−ΔΔCq method (40).
Gene Ontology (GO) function analysis,
KEGG pathway enrichment analysis and construction of
protein-protein and gene-gene interaction networks
The functional analysis tool g:Profiler, gOST (rev
f0f4439; http://biit.cs.ut.ee/gprofiler/gost) was used to
analyze GO functions (41,42) and KEGG pathway enrichment (43,44).
GeneMANIA (http://genemania.org/) was used to
construct gene-gene interaction networks (45,46), and
the Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING) database (version 11.0; http://string-db.org/) was used to construct the
protein-protein interaction networks (47,48).
Statistical analysis
OS was estimated by Kaplan-Meier analysis, and a
log-rank test was performed to evaluate statistical significance.
The co-expression and regression line of let-7c-5p and its target
genes in LIHC tumors was determined by Pearson's correlation
analysis. Co-expression among let-7c-5p target genes was determined
by Spearman's correlation analysis. The association between the
mRNA expression levels of the let-7c-5p target genes and the LIHC
tumor grade was analyzed using Welch's test in a one-way
heteroscedastic ANOVA with Games-Howell post hoc tests. For the
expression difference of target genes between the let-7c-5p
overexpression and control groups, the data are presented as the
mean ± standard deviation and analyzed by unpaired Student's t-test
using SPSS v19 software (IBM Corp.). The data were representative
of three independent experiments performed in triplicate. In all
statistical analyses, P<0.05 was considered to indicate a
statistically significant difference.
Results
let-7c may act as a tumor suppressor
gene in LIHC
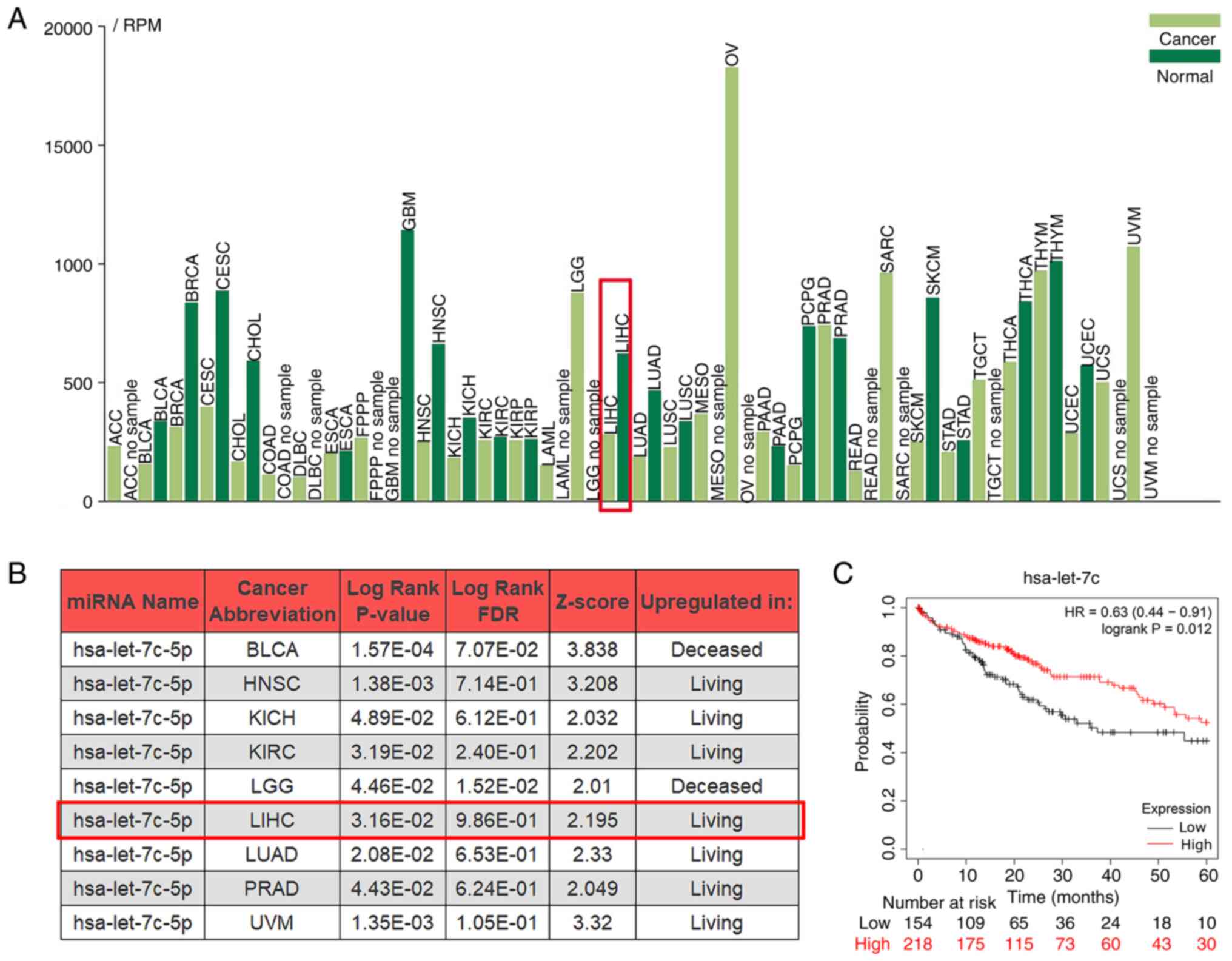
The expression levels of let-7c-5p in different
cancer types and their potential association with the survival rate
were assessed using the online TCGA data analysis tool. The
tumor-miRNA-pathway, OncomiR and the Kaplan-Meier plotter tools
were used. let-7c-5p was differentially expressed in different
cancer types compared with the corresponding expression noted in
their control group, and its expression was decreased in LIHC
(Fig. 1A). The survival rate of
patients was significantly associated with let-7c-5p expression in
9 cancer types, while their performance was different. The
expression of let-7c-5p were upregulated in living cohorts in LIHC
(Fig. 1B). In addition, the
Kaplan-Meier curve revealed that high let-7c-5p expression was
associated with improved OS of patients with LIHC (Fig. 1C). Overall, these results
demonstrated that let-7c-5p may act as a tumor suppressor gene in
LIHC.
Target genes of let-7c are involved in
the PI3K/Akt/FoxO signaling pathway in LIHC
Using TarBase v8.0 (Diana Tools), 2,051 target genes
of let-7c were predicted. These target genes were subsequently used
for KEGG pathway enrichment analysis using KOBAS 3.0. The data
indicated that 58 target genes belonged to the ‘PI3K-Akt signaling
pathway’, whereas 33 target genes belonged to the ‘FoxO signaling
pathway’ (Table I). Among these
genes, 15 target genes were associated with ‘PI3K-Akt signaling
pathway’ and ‘FoxO signaling pathway’. After excluding duplicate
count genes, 76 let-7c-5p target genes associated with the
PI3K/Akt/FoxO signaling pathway were identified.
 | Table I.let-7c-5p target genes in the
PI3K-Akt and FoxO signaling pathways. |
Table I.
let-7c-5p target genes in the
PI3K-Akt and FoxO signaling pathways.
| KEGG pathway | Number of
genes | Corrected
P-value | Genes |
|---|
| PI3K-Akt signaling
pathway | 58 |
5.81×10−13 | ATF2, ATF4, ATF6B,
BCL2, BCL2L11, CCND1, CCND2, CCNE2, CDK2, CDK4, CDK6, CDKN1A,
CDKN1B, CHRM1, CHUK, COL1A1, COL1A2, COL4A1, COL4A2, CREB3L2,
FGFR1, FN1, GNB1, GNB2, GNG5, HSP90AA1, HSP90B1, IGF1R, IL6R, INSR,
ITGAV, JAK1, LAMC1, LPAR1, MAPK1, MDM2, MET, MYC, NR4A1, NRAS,
PKN2, PPP2CA, PPP2CB, PPP2R1B, PPP2R5A, PPP2R5C, PRKAA1, PRKAA2,
PRLR, RAC1, RBL2, RPS6KB2, THBS1, TLR4, TP53, TSC1, YWHAE,
YWHAG |
| FoxO signaling
pathway | 33 |
3.46×10−11 | AGAP2, BCL2L11,
CCNB2, CCND1, CCND2, CDK2, CDKN1A, CDKN1B, CHUK, CSNK1E, FBXO32,
FOXO1, HOMER1, HOMER2, IGF1R, INSR, IRS2, MAPK1, MAPK8, MDM2, NLK,
NRAS, PLK2, PRKAA1, PRKAA2, PRKAB2, PRKAG1, RBL2, SKP2, MST1,
TGFBR1, TGFBR2, TNFSF10 |
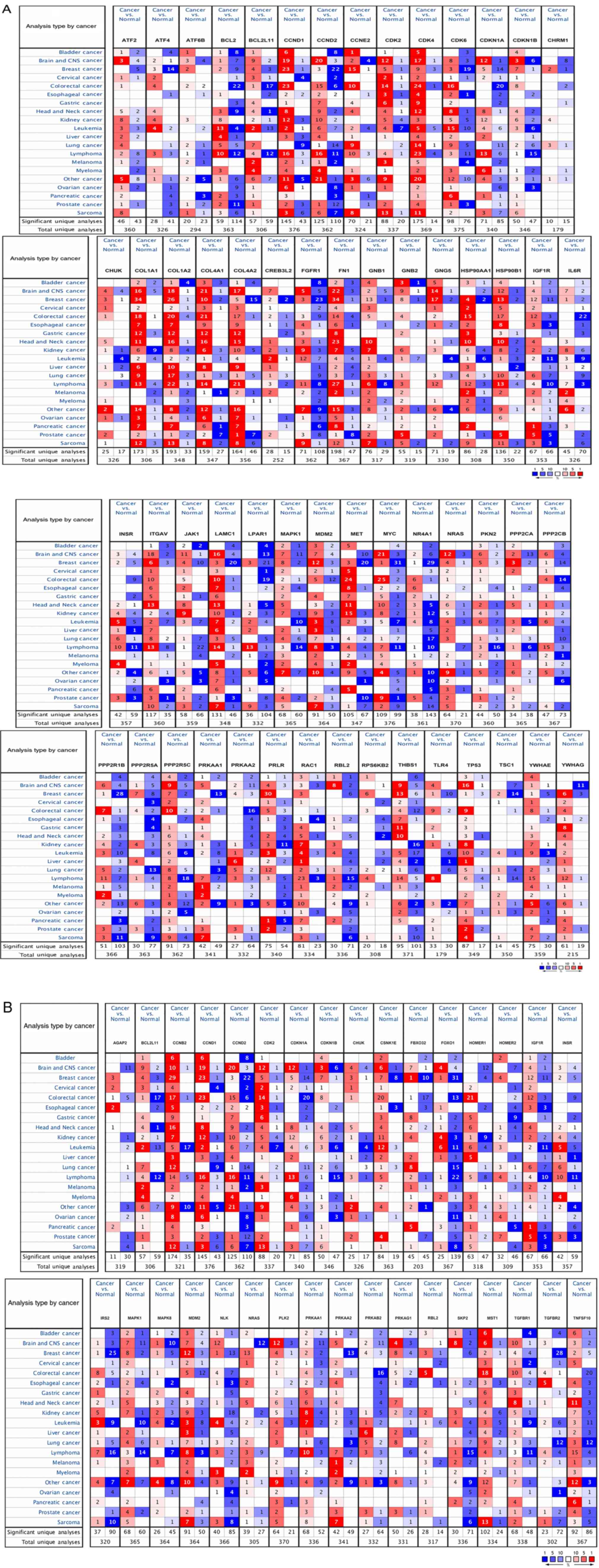
Oncomine analysis was performed to detect the mRNA
levels of these 76 let-7c-5p target genes in different types of
human cancer, including liver cancer. As presented in Fig. 2, the value in the cell presented the
number of datasets with a statistically significant mRNA
differential expression of let-7c-5p target genes. The results
demonstrated that the differential expression of these let-7c-5p
target genes was different between the liver tumor group and the
normal group. For example, there were four upregulated and two
downregulated analyses that met the thresholds for CCND2 and
HSP90B1, respectively. However, for BCL2, there were four
upregulated and one downregulated analysis. The LIHC dataset of
liver cancer was selected to assess the expression of target genes
in detail, and the results demonstrated that 37 of the 76 genes
were upregulated in LIHC, which opposed let-7c-5p expression
(Table SII).
Using the Kaplan-Meier plotter analysis tool, the
prognostic significance of the 37 genes obtained was assessed. It
was identified that increased mRNA expression levels of CCNB2,
CCNE2, CDK2, CDK4, CSNK1E, FBXO32, HOMER1, HSP90AA1, ITGAV, NRAS,
PPP2CA, PRKAA2, RAC1 and SKP2 were associated with poor OS
(Fig. 3), which suggested that the
mRNA expression levels of the target genes of let-7c-5p may be
useful for the prediction of the survival of patients with
LIHC.
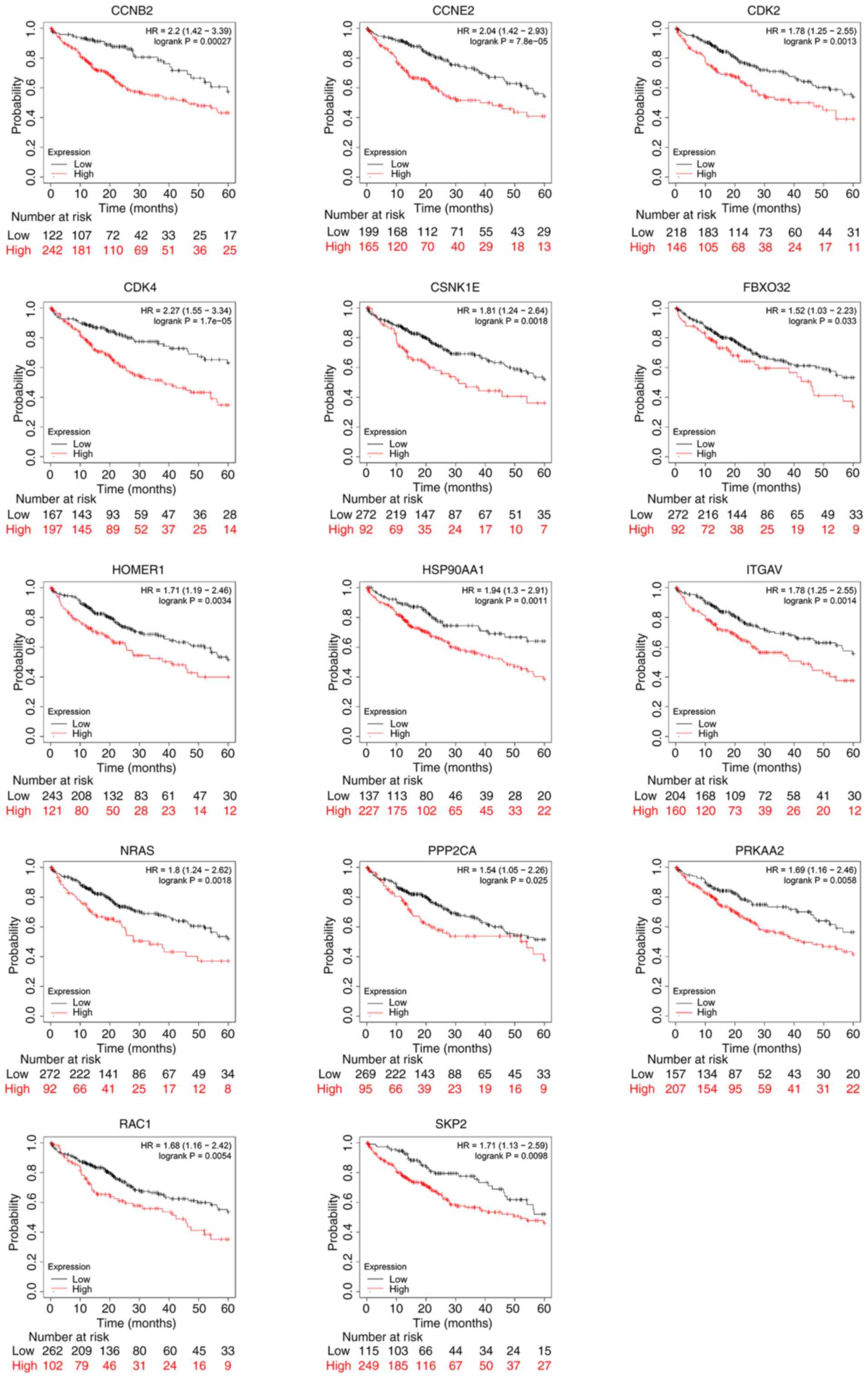 | Figure 3.Prognostic value of let-7c-5p-target
gene mRNA levels in liver hepatocellular carcinoma by using
Kaplan-Meier Plotter. Increased mRNA expression levels of CCNB2,
CCNE2, CDK2, CDK4, CSNK1E, FBXO32, HOMER1, HSP90AA1, ITGAV, NRAS,
PPP2CA, PRKAA2, RAC1 and SKP2 were associated with poor overall
survival (P<0.05). HR, hazard ratio. |
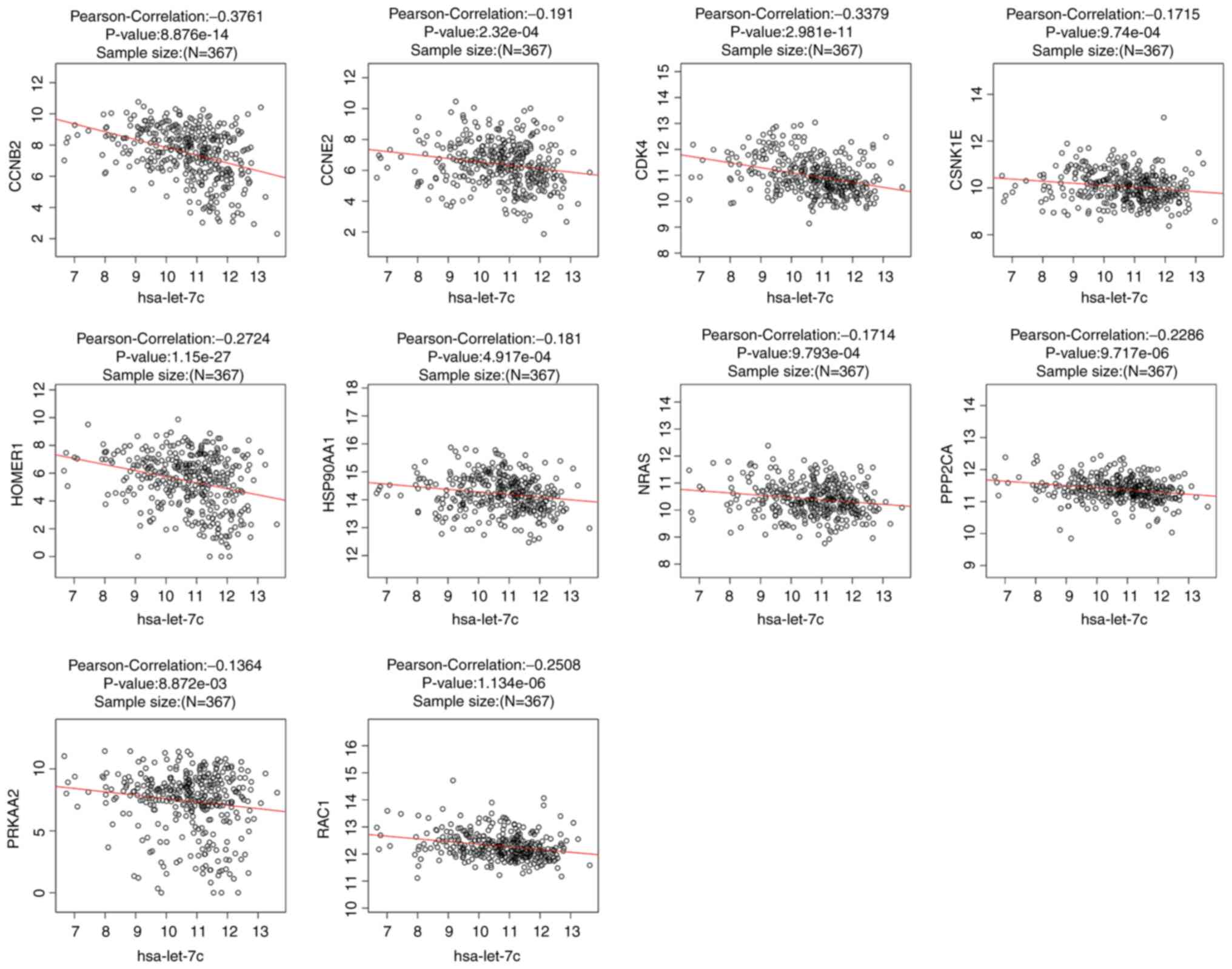
There were 367 overlapping samples with miRNASeq and
RNASeq in the LinkedOmics database, and we used them to investigate
the correlation of expression between let-7c and its target genes.
The data were derived from the Kaplan-Meier plotter analysis of the
LIHC samples. Additionally, Pearson's correlation analysis was used
with P<0.01 as the threshold value. The results indicated that
the expression levels of 10 genes (CCNB2, CCNE2, CDK4, CSNK1E,
HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2 and RAC1) were negatively
correlated with let-7c expression (Fig.
4), which suggested that let-7c-5p may regulate the expression
of these genes in LIHC.
Expression levels of the let-7c-target
genes are closely associated with tumor grade and LIHC
prognosis
The present study aimed to identify potential
candidate biomarkers for OS in patients with LIHC based on the mRNA
expression levels of the target genes of let-7c-5p that were
obtained from the LinkedOmics database. The comparisons between
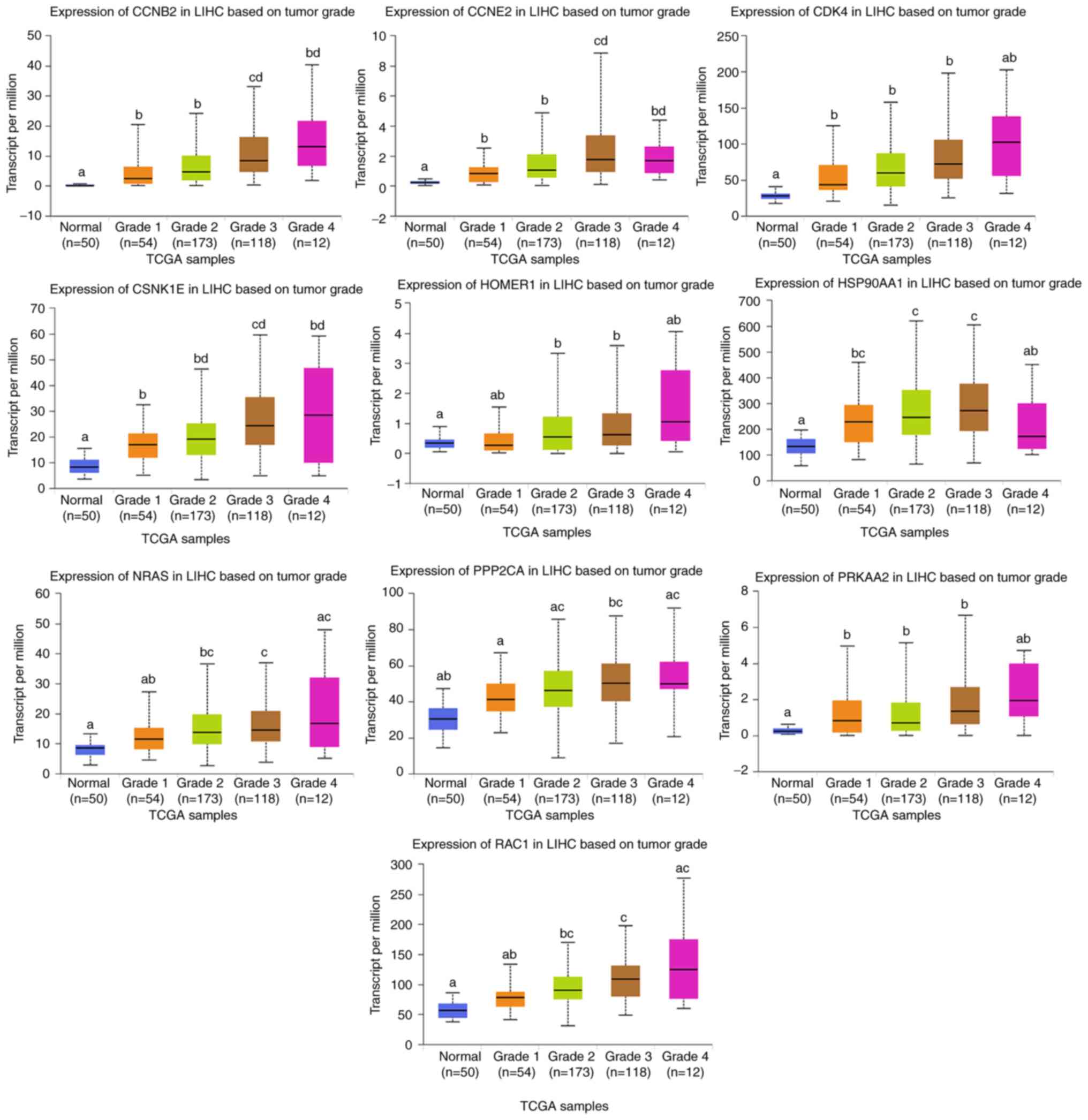
LIHC and liver tissues were performed using UALCAN. Higher mRNA
expression levels of these genes (CCNB2, CCNE2, CDK4, CSNK1E,
HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2 and RAC1) were noted in LIHC
tissues compared with in normal tissues (P<0.05; Fig. 5). Furthermore, the patients with more
advanced stages of LIHC tended to exhibit higher expression levels
of these target genes. Among them, the expression levels of the
CCNB2 and CCNE2 were higher in grade 3 compared with in other
grades (P<0.05). The expression levels of CSNK1E, NRAS, PPP2CA
and RAC1 were higher in grade 3 compared with grade 1. However, the
expression levels of these target genes in tumor grade 4 did not
exhibit this trend like in grade 3, possibly due to the small
sample size of grade 4 tumor samples.
The association between target gene expression data
of different tumor grades and survival data was investigated
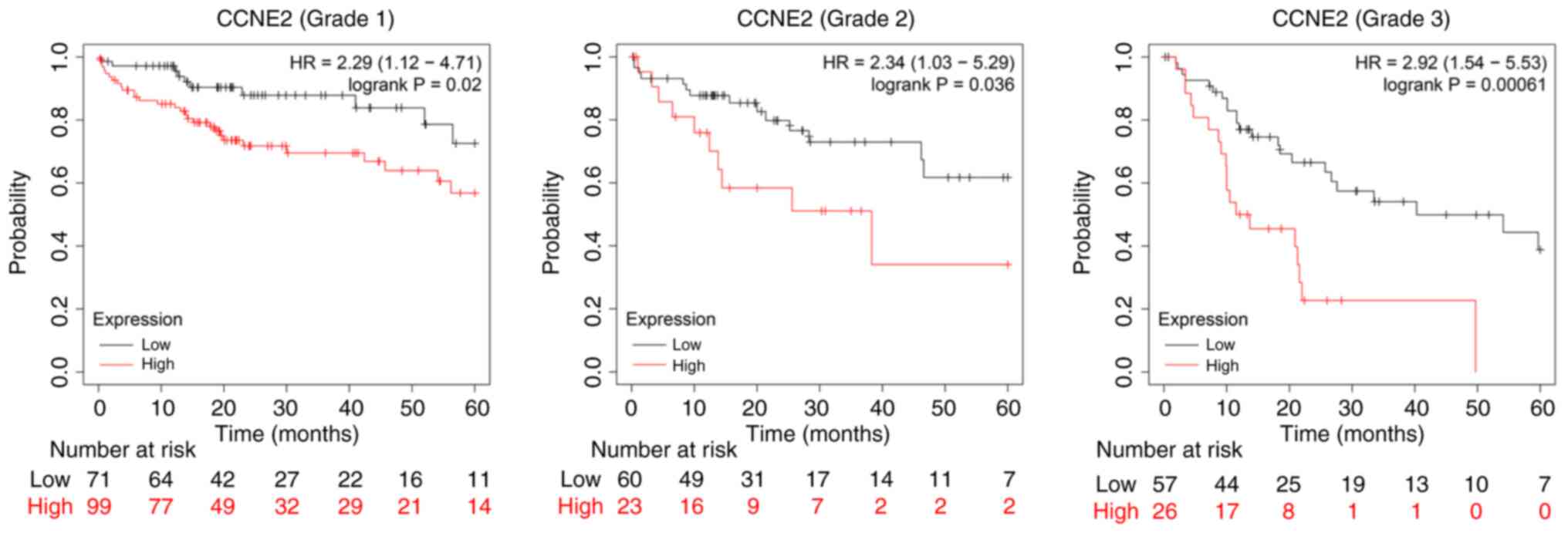
(Fig. 6). High expression levels of
CCNE2 were associated with poor OS in grade 1, 2 and 3 tumors.
However, the expression levels of other genes were only associated
with poor OS in one or two of the grades (data not shown). Overall,
these results indicated that the mRNA expression levels of CCNE2
were associated with different tumor grades in patients with LIHC,
and that their expression levels may be useful for the prediction
of LIHC patient survival.
The performance of these 10 target genes (CCNB2,
CCNE2, CDK4, CSNK1E, HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2 and
RAC1) was assessed in TCGA-LIHC, using cBioPortal. The results
demonstrated that these 10 let-7c-5p target genes were altered in
201 out of 360 patients with LIHC (the total mutation rate was
estimated to be 55.8%; OncoPrint; Fig.
7A). CCNE2, PPP2CA and RAC1 were the three genes with the
highest rate of sequence alterations and their mutation rates were
estimated to be 23, 18 and 12%, respectively. KEGG pathway analysis
demonstrated that 7 of the 10 target genes (CCNE2, CDK4, HSP90AA1,
NRAS, PPP2CA, PRKAA2 and RAC1) belonged to the ‘PI3K-Akt signaling
pathway’ and 5 target genes (CCNB2, CSNK1E, HOMER1, NRAS and
PRKAA2) to the ‘FoxO signaling pathway’. Two of these genes, NRAS
and PRKAA2, were shared by the two pathways (Fig. 7B). Further analysis of the alteration
frequencies revealed that high mRNA levels were the most common
alteration for CCNB2, CDK4, CSNK1E, HOMER1, HSP90AA1, NRAS, PPP2CA,
PRKAA2 and RAC1. For CCNE2, the most common alteration was the
amplification in LIHC tumors (Table
SIII). Subsequently, the correlations of these 10 genes in LIHC
tissues were investigated using Spearman's correlation (Fig. 7C). The results indicated a
significant positive correlation among the majority of these genes,
notably between CCNB2 and CCNE2 or CDK4. Spearman's correlation was
estimated to be 0.63 and 0.62, respectively, and the regression
line is shown in Fig. 7C. The
Spearman's correlation between CCNB2 and CSNK1E or NRAS, CCNE2 and
CDK4 or NRAS, CDK4 and CSNK1E or RAC1 and HOMER1 and NRAS ranged
between 0.3 and 0.6. Additional mutual exclusivity analysis
indicated non-significant mutual exclusivity between two of these
10 genes (CCNB2, CCNE2, CDK4, CSNK1E, HOMER1, HSP90AA1, NRAS,
PPP2CA, PRKAA2 and RAC1), whereas the gene pairs of CCNB2 and CDK4,
CSNK1E or NRAS, CDK4 and CSNK1E, HOMER1 or RAC1, HSP90AA1 and CCNE2
or NRAS, and NRAS and PRKAA2 or RAC1 exhibited significant
(P<0.05) co-occurrence in LIHC (Fig.
7D). The selected samples in the TCGA-LIHC dataset were further
divided into two groups based on whether the 10 target genes were
altered. One group was the altered group: Samples with at least one
alteration in the 10 let-7c-5p target genes in the selected
profiles, and the other group was the unaltered group: Samples
without any alterations in the 10 let-7c-5p target genes in the
selected profiles. Further analysis using the Kaplan-Meier plotter
and a log-rank test indicated that the alterations of these 10
genes were associated with worse OS in patients with LIHC (Fig. 7E). These suggested that alterations
of these 10 genes could have an impact on the prognosis of patients
with LIHC.
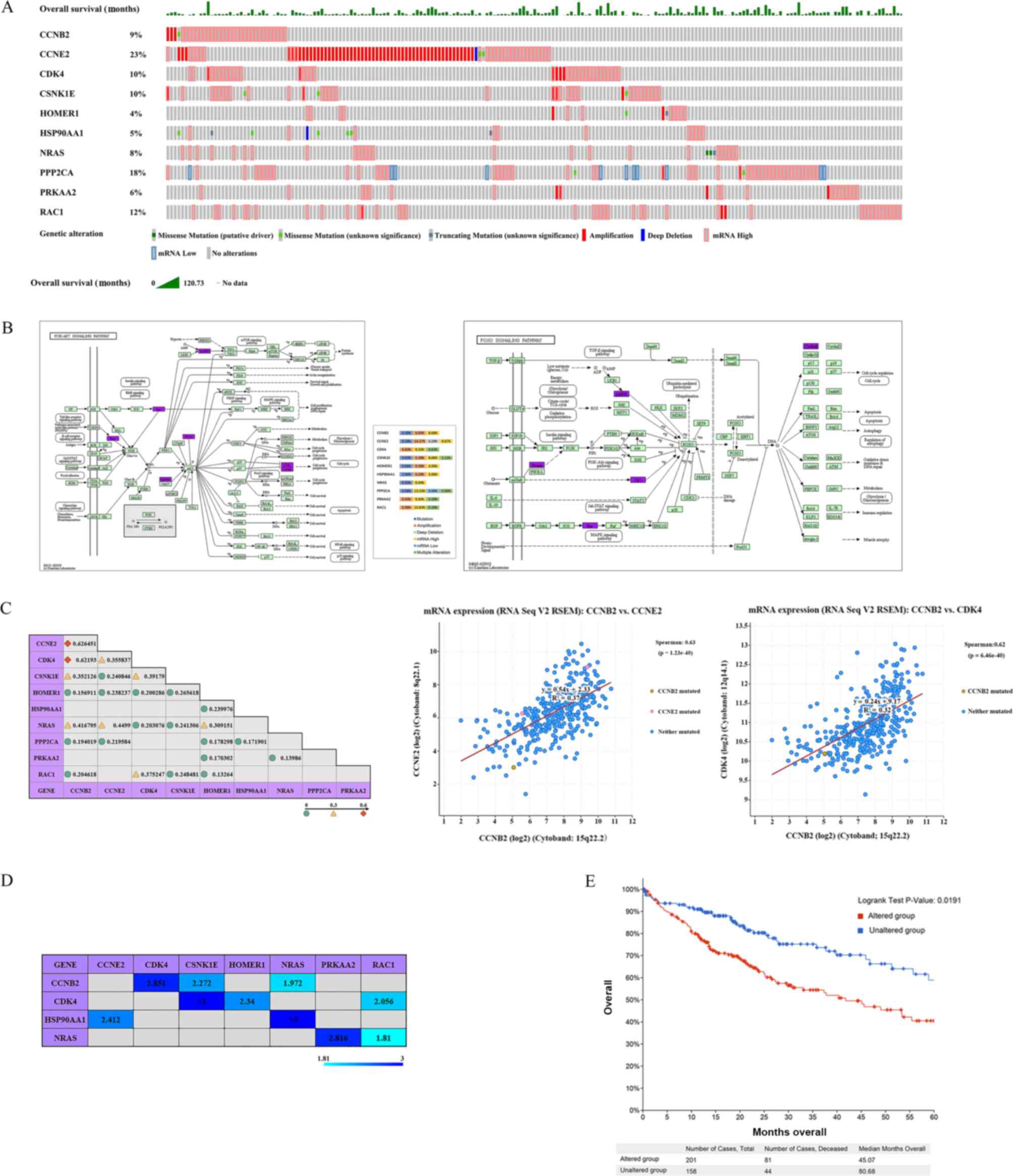 | Figure 7.cBioPortal data visualization and
analysis. (A) Oncoprint of genetic alterations and OS observed for
CCNB2, CCNE2, CDK4, CSNK1E, HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2
and RAC1 in LIHC tumors. (B) Detailed alteration frequencies of
CCNB2, CCNE2, CDK4, CSNK1E, HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2
and RAC1 in LIHC and their position in the PI3K-Akt and FoxO
signaling pathways. KEGG pathway analysis demonstrated that 7 of
the 10 target genes (CCNE2, CDK4, HSP90AA1, NRAS, PPP2CA, PRKAA2
and RAC1) belonged to the ‘PI3K-Akt signaling pathway’ and 5 target
genes (CCNB2, CSNK1E, HOMER1, NRAS and PRKAA2) to the ‘FoxO
signaling pathway’. The genes in purple represent the let-7c-5p
target genes. The alteration frequencies revealed that high mRNA
levels were the most common alteration for CCNB2, CDK4, CSNK1E,
HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2 and RAC1. For CCNE2, the
most common alteration was the amplification in LIHC tumors. (C)
Co-expression of CCNB2, CCNE2, CDK4, CSNK1E, HOMER1, HSP90AA1,
NRAS, PPP2CA, PRKAA2 and RAC1 in LIHC tumors. The regression line
of CCNB2 with CCNE2 and CCNB2 with CDK4 was determined by
Spearman's correlation analysis (Spearman's Correlation >0.6;
P<0.05). (D) Mutual exclusivity of CCNB2, CCNE2, CDK4, CSNK1E,
HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2 and RAC1 in LIHC tumors. The
values in the table represent the log2 Odds Ratio. log2 ratio >0
indicates a tendency towards co-occurrence (P<0.05). (E)
Kaplan-Meier plots comparing OS in cases with/without CCNB2, CCNE2,
CDK4, CSNK1E, HOMER1, HSP90AA1, NRAS, PPP2CA, PRKAA2 and RAC1 gene
alterations in LIHC tumors. The samples were divided into two
groups according to whether the 10 target genes were altered. One
group was the altered group: Samples in which at least one of the
10 target genes was altered (201 in total), and one group was the
unaltered group: Samples with no alteration in all 10 target genes
(158 in total). The analysis using the Kaplan-Meier plotter and a
log-rank test indicated that the alterations of these 10 genes were
associated with worse OS in patients with LIHC. LIHC, liver
hepatocellular carcinoma; OS, overall survival. |
A total of nine genes may be the
target genes of let-7c-5p, and their main function is the
regulation of the cell cycle
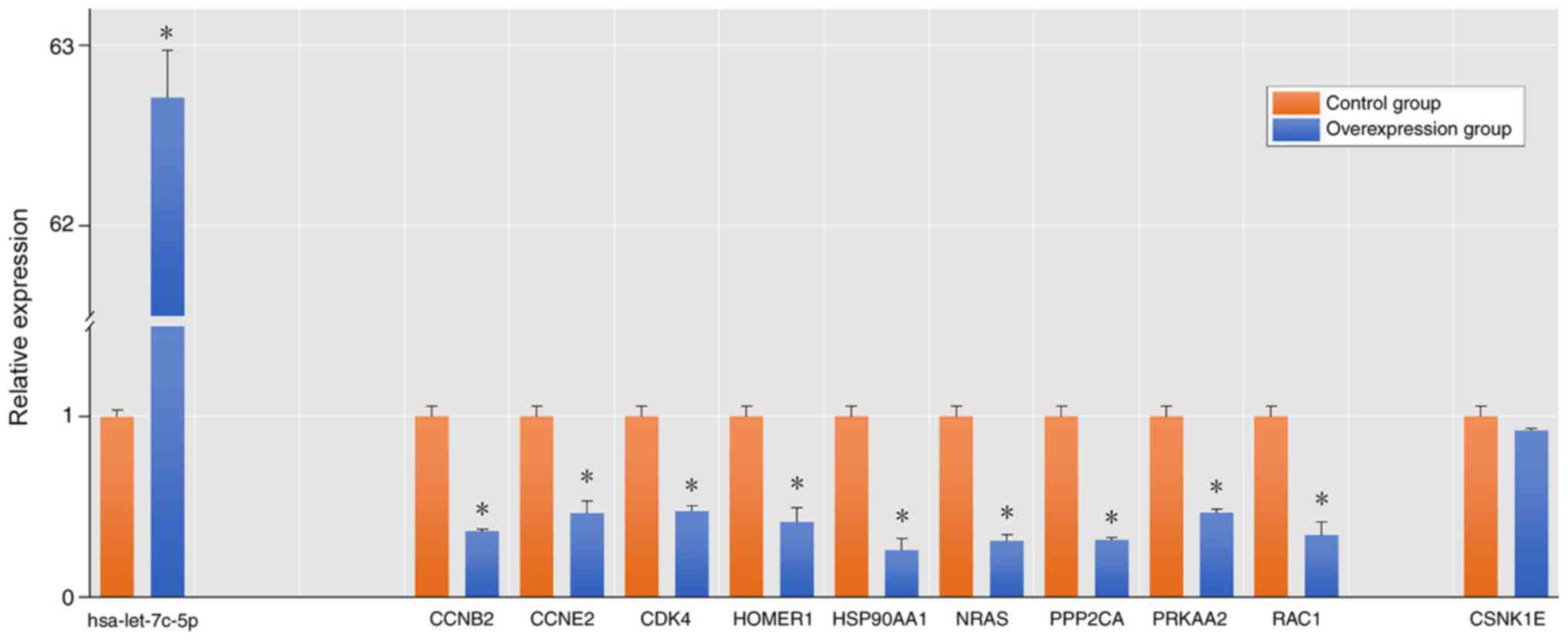
Following let-7c-5p overexpression in MHCC-97H, it
was revealed that there was no significant difference in the
expression levels of CSNK1E compared with the control group.
However, the expression levels of CCNB2, CCNE2, CDK4, HOMER1,
HSP90AA1, NRAS, PPP2CA, PRKAA2 and RAC1 were downregulated to
different degrees, which demonstrated that these genes may be
target genes of let-7c (Fig. 8).
Subsequently, g:Profiler was used to analyze GO function and KEGG
pathway enrichment for these nine genes, while GO analysis was used
to divide gene functions into biological process (BP), molecular
function (MF) and cellular component (CC) categories. According to
the number of genes clustered to the corresponding terms
(Intersection size) and the correlation with LIHC disease, the
results of the present study demonstrated that the genes were
mainly clustered into the following: The MF terms included
‘phosphotransferase activity, alcohol group as acceptor’ and
‘cyclin-dependent protein serine/threonine kinase regulator
activity’. The BP terms included ‘protein phosphorylation’ and
‘cell cycle process’, and the CC terms included ‘cytosol’. Among
the genes, seven belonged to the ‘PI3K-Akt signaling pathway’ and
four belonged to the ‘FoxO signaling pathway’ (Fig. 9).
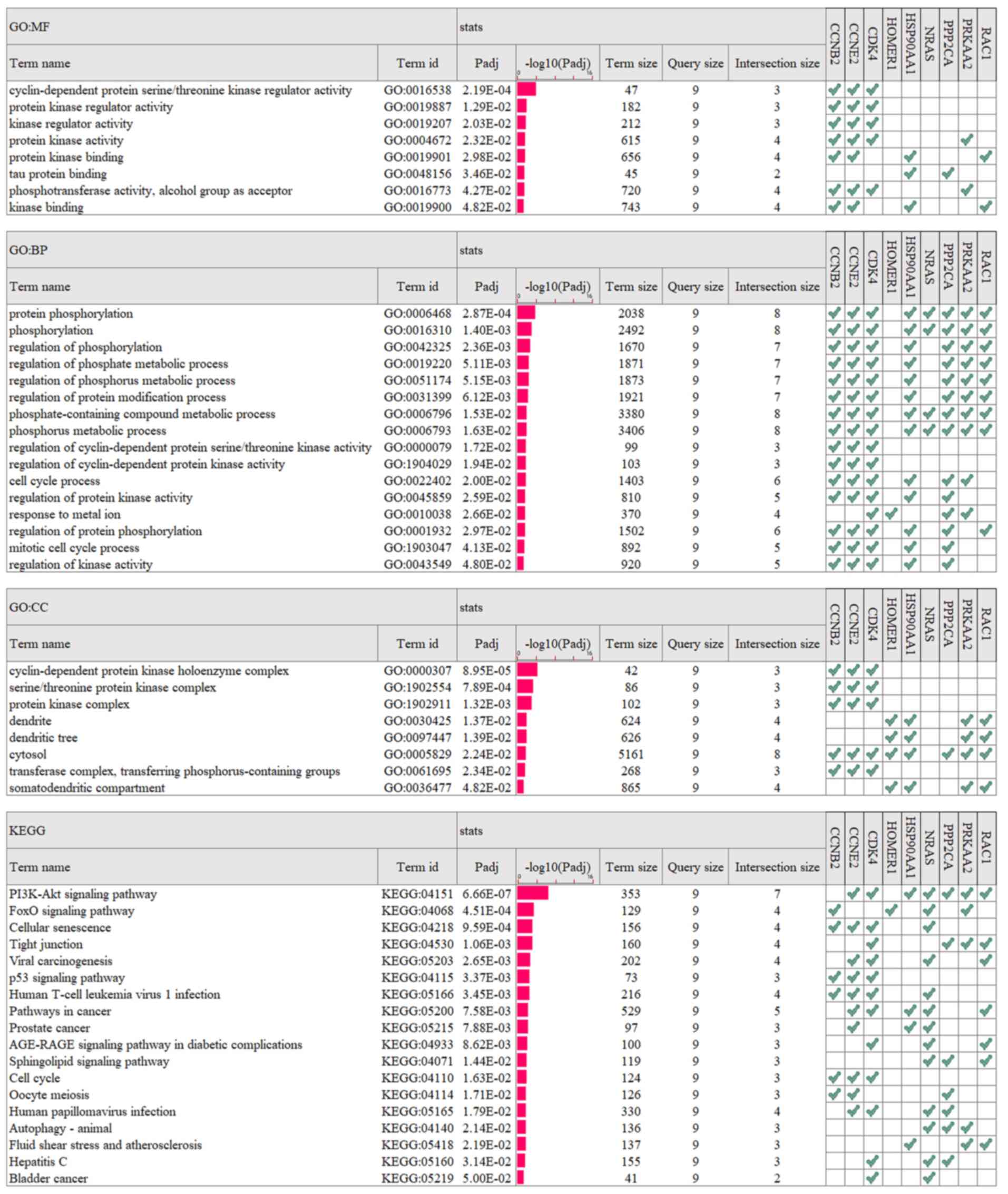 | Figure 9.GO function and KEGG pathway
enrichment for CCNB2, CCNE2, CDK4, HOMER1, HSP90AA1, NRAS, PPP2CA,
PRKAA2 and RAC1 in liver hepatocellular carcinoma tumors. The
results were summarized in the following three main categories: BP,
MF and CC. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and
Genomes; BP, biological process; MF, molecular function; CC,
cellular component. |
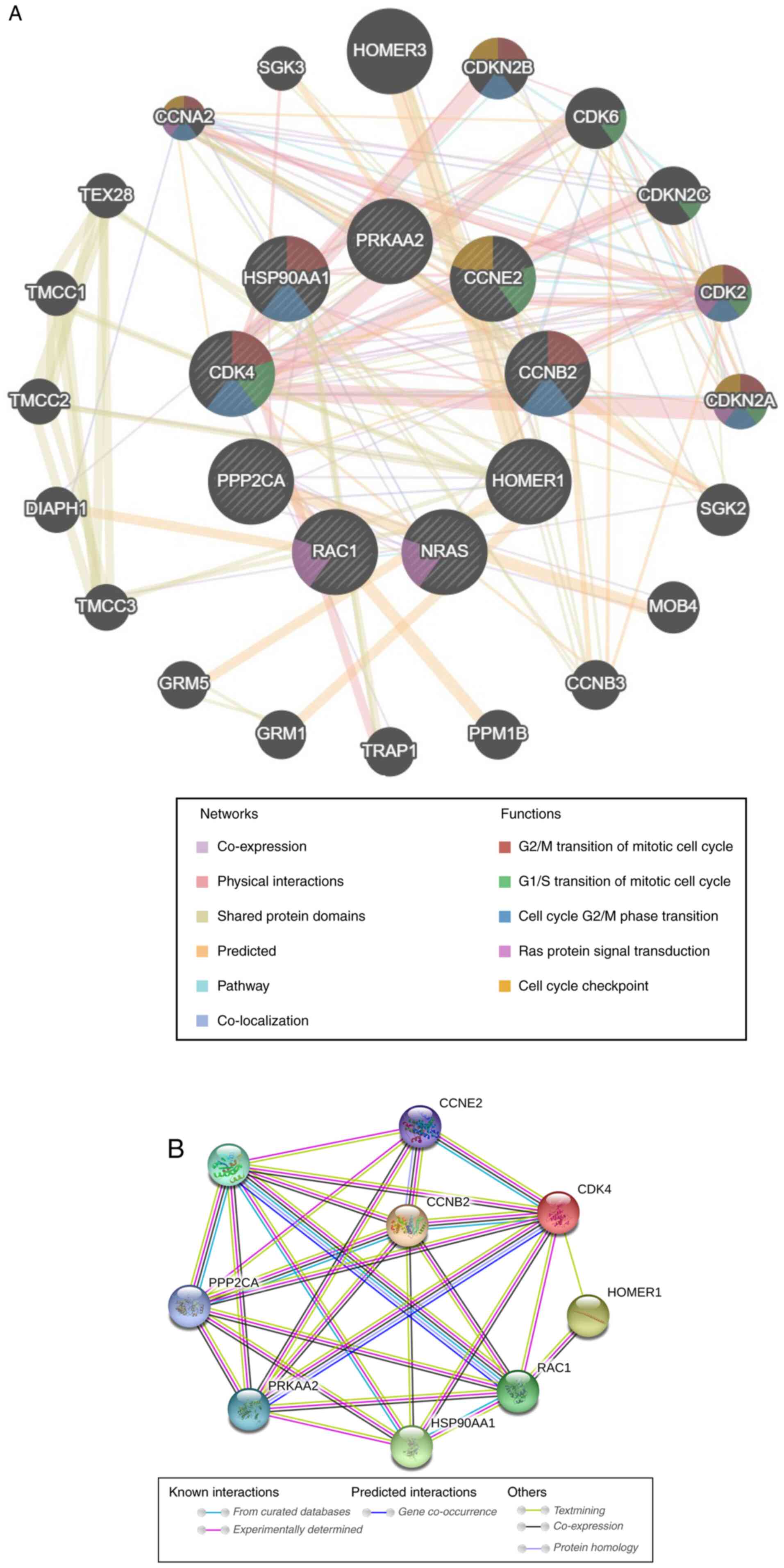
GeneMANIA software was used to analyze associations
in terms of co-expression, physical interactions, shared protein
domains, predicted, pathway and co-localization among these nine
target genes of let-7c-5p (20 additional genes were involved in the
network), and to highlight the functions associated with the
majority of these genes (Fig. 10A).
The results indicated the following processes: ‘G2/M
transition of mitotic cell cycle’, ‘G1/S transition of
mitotic cell cycle’, ‘cell cycle G2/M phase transition’,
‘Ras protein signal transduction’ and ‘cell cycle checkpoint’.
Subsequently, STRING was used to analyze these genes in terms of
their protein expression levels. The results indicated complex
interactions among these genes (Fig.
10B). Overall, these results indicated complex links among the
target genes, whose main function was the regulation of the cell
cycle. Changes in the expression levels of these genes may have an
important effect on the cell cycle.
Discussion
Accumulating evidence has indicated that miRNAs are
abnormally expressed in LIHC tumors and promote or inhibit the
development and progression of LIHC by affecting cell cycle and
proliferation, metastasis and invasion, drug resistance and
inhibition of apoptosis (49–51). It
has been demonstrated that the levels of let-7c are lower in LIHC
tissues than in the corresponding normal adjacent tissues (52). In addition, a previous study
indicated that let-7c inhibits cell proliferation and induces cell
cycle arrest in LIHC tissues (15).
In the present study, let-7c expression was decreased in LIHC
tissues and this was associated with poor OS of LIHC as determined
by the online TCGA data analysis tools Tumor-miRNA-Pathway, OncomiR
and Kaplan-Meier plotter. Jilek et al (53) identified that let-7c bioengineered
preparations could inhibit LIHC development. Overall, these results
indicated that let-7c may be an effective therapeutic target for
LIHC. However, since each miRNA can regulate thousands of target
genes, not all target genes of let-7c expressed in LIHC and are
only regulated by it. Therefore, the exact role of let-7c in the
development and progression of LIHC remains to be elucidated. The
PI3K/Akt signaling pathway has been recognized as one of the most
frequently activated signal transduction pathways in cancer
(17), whereas the FoxO signaling
pathway has been mainly investigated for the potential targeting of
cancer therapeutic molecules and proteins (54). The FoxO signaling pathway is closely
associated with the PI3K/Akt signaling pathway (55). The analysis of the association
between the target genes of let-7c and the PI3K/Akt and FoxO
signaling pathways could further aid the understanding of the role
of let-7c in LIHC and provide a basis for the future application of
let-7c in LIHC therapy. In the present study, the expression levels
and prognostic values of let-7c and its target genes in patients
with LIHC were analyzed using multiple online tools.
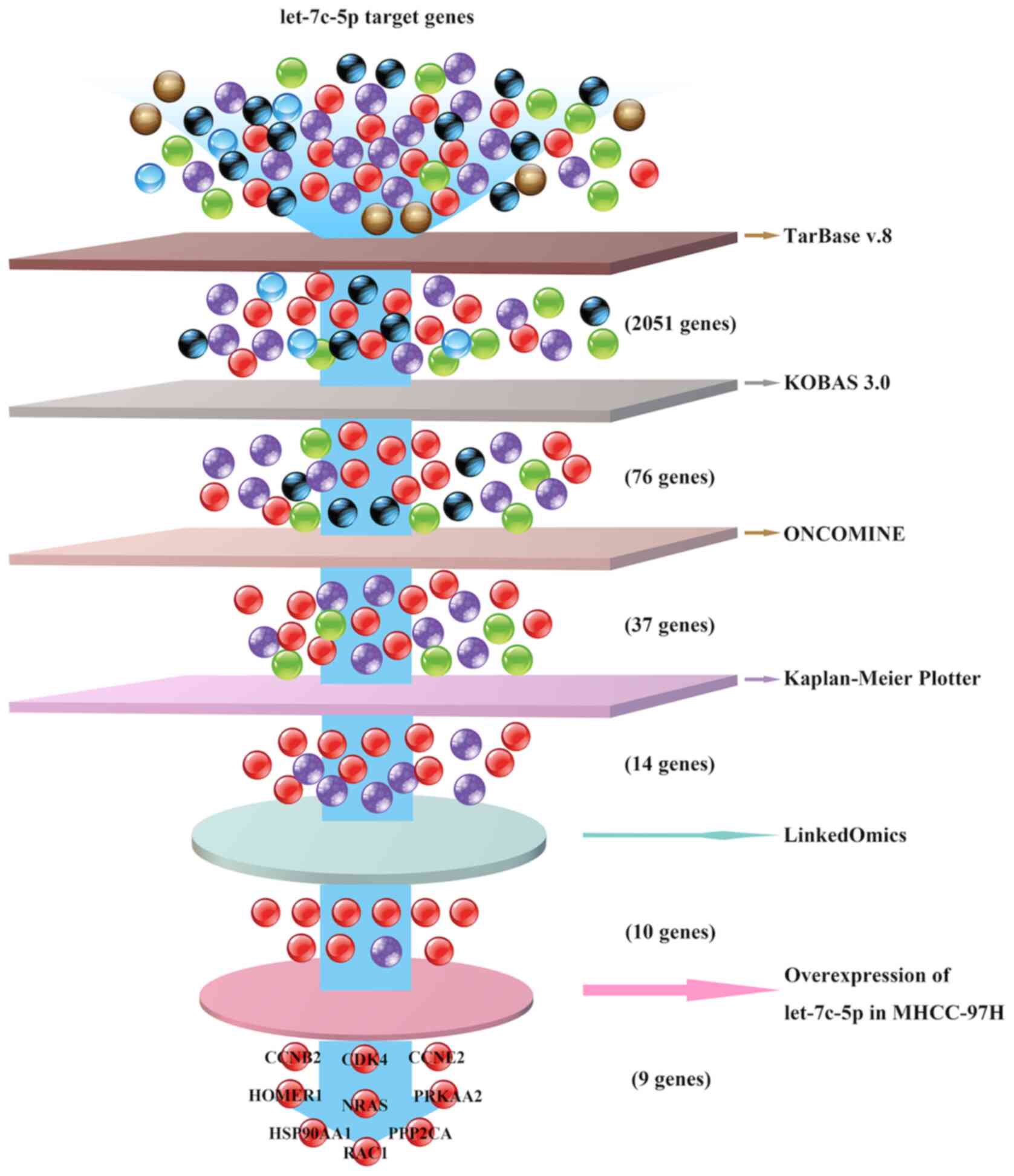
Initially, 2,051 target genes of let-7c were
identified by DIANA-TarBase v8 analysis and 76 target genes were
identified that belonged to the PI3K-Akt and FoxO signaling
pathways using KOBAS 3.0 and KEGG pathway enrichment analysis.
Oncomine was used to analyze mRNA transcription levels, and
Kaplan-Meier plotter was used to evaluate OS. A total of 37 target
genes of let-7c-5p were upregulated in LIHC and 14 genes among them
were associated with poor OS. Finally, 10 genes were negatively
associated with let-7c expression according to LinkedOmics.
Furthermore, in vitro experiments revealed that nine genes
among them were downregulated after let-7c-5p overexpression in
MHCC-97H cells, which suggested that let-7c-5p regulated the
expression of these genes in LIHC (Fig.
11). Additionally, analysis using Kaplan-Meier Plotter
highlighted that the high expression levels of CCNE2 were
associated with poor OS in grade 1, 2 and 3 tumors. The results
suggested that these nine genes, including CCNE2, may be promising
candidate biomarkers for disease and poor prognosis in LIHC. KEGG
analysis demonstrated that let-7c did not directly regulate the
expression of PI3K, Akt and FoxO in LIHC, but mainly that of its
target genes that were upstream molecules of the PI3K-Akt and the
FoxO signaling pathways, which can directly regulate PI3K, Akt and
FoxO, such as NRAS, RAC1, HSP90AA1, PPP2CA, and PRKAA2, or their
downstream molecules, such as CCNB2, CCNE2 and CDK4.
It is well-known that the cell cycle serves an
important role in cancer. GO analysis indicated that six (CCNB2,
CCNE2, CDK4, HSP90AA1, PPP2CA and PRKAA2) of these nine target
genes of let-7c were members of the ‘cell cycle process’ BP term.
Among them, CCNB2 is an important member of the cyclin protein
family and is an important cell cycle regulator associated with
G2/M detection sites (56). High expression levels of CCNB2 are
negatively associated with the OS of patients with LIHC (57). The principal function of CCNE2 is to
be a regulator to facilitate the cell cycle progression from
G0/G1 to S phase (58,59).
Upregulation of CCNE2 is associated with cancer progression and
mortality in several types of cancer (60). CDK4 serves a key role in the
proliferation of mammalian cells and can promote the cell entry to
the S phase of the cell cycle (61).
HSP90AA1 is a chaperone that is highly conserved among eukaryotes
(62). Functional HSP90 is required
for the stability of Akt, which is a key gene of the PI3K-Akt
signaling pathway and serves a crucial regulatory role in
differentiation, cell cycle, transcription, translation, metabolism
and apoptosis (63). PPP2CA serves
critical roles in several cellular processes by dephosphorylating
critical cellular molecules, such as Akt, P53, c-Myc and β-catenin
(64). Notably, downregulation of
PRKAA2 has been reported in breast tumors and other types of cancer
(65,66), which suggests that it possesses tumor
suppressor functions in vivo. However, the data indicated
that PRKAA2 expression was upregulated in LIHC. The role of this
gene in LIHC is not clear and requires further study. Therefore, it
was deduced that the majority of these target genes of let-7c-can
affect the cell cycle of LIHC cells via the PI3K-Akt and FoxO
signaling pathways and that their high expression may aggravate
cell cycle disorders. The data suggest that the high expression
levels of these target genes, notably CCNE2, are associated with
poor prognosis of LIHC. HOMER1 has been reported to interact with
Ca+2 signaling proteins, such as stromal interaction
molecule 1 and ORAI calcium release-activated calcium modulator 1
(67). Alterations in either the
expression or the activity of these proteins have been associated
with the onset and maintenance of tumor phenotypes in LIHC
(68). NRAS is a proto-oncogene that
is associated with several human cancer types, due to its
activating mutation (69)
demonstrated marked upregulation of wild-type NRAS in LIHC cell
lines and patient tissues, whereas NRAS expression is associated
with poor patient survival (70).
RAC1 is a member of the Rac subfamily of GTPases and serves an
important role in several cellular processes, including cell
proliferation, cytoskeletal recombination, antimicrobial
cytotoxicity and epithelial differentiation (71). It has been demonstrated that RAC1
serves an important role in LIHC progression and metastasis
(72,73). Knockdown of RAC1 suppresses the
expansion of LIHC cells (74). These
results suggested that let-7c served an important role, not only in
the regulation of the cell cycle, but also in phenotype progression
and metastasis of LIHC. Correlation analysis between let-7c and its
target genes, as well as the in vitro cell experiments,
indicated that let-7c regulated their expression in LIHC.
Therefore, low expression levels of let-7c in LIHC may be
considered to be an important factor leading to the high expression
of its target genes. The mechanism of action of let-7c may
influence the formation and development of LIHC via the PI3K/Akt
and FoxO signaling pathways. This may be a possible explanation for
the negative association observed between let-7c expression and
poor LIHC prognosis.
In conclusion, the present study demonstrated that
let-7c may be considered to be an anti-oncogene of LIHC, which
mainly affects the cell cycle of LIHC cells via the PI3K-Akt and
FoxO signaling pathways. Among its target genes, CCNE2 may be a
promising candidate biomarker for disease and poor prognosis in
LIHC. The increase in the expression levels of let-7c may be an
effective way to improve the prognosis, survival rate and treatment
of LIHC. The present study provided novel insights into the
mechanisms of LIHC occurrence and development and identified let-7c
as a promising therapeutic target for use in the treatment of
LIHC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data that support the findings of this study are
available from the public databases mentioned in this article but
restrictions apply to the availability of these data, which were
used under license for the current study, and so are not publicly
available. Data are however available from the authors upon
reasonable request and with permission of the public databases
mentioned in this article. The PCR data generated in the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YL and PL designed the present study. PL and NW
performed the experiments. YL, PL and NW analyzed the data and
prepared the figures. YL drafted the initial manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim NG, Nguyen PP, Dang H, Kumari R,
Garcia G, Esquivel CO and Nguyen MH: Temporal trends in disease
presentation and survival of patients with hepatocellular
carcinoma: A real-world experience from 1998 to 2015. Cancer.
124:2588–2598. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gailhouste L and Ochiya T: Cancer-related
microRNAs and their role as tumor suppressors and oncogenes in
hepatocellular carcinoma. Histol Histopathol. 28:437–451.
2013.PubMed/NCBI
|
|
12
|
Li D, Zhang J and Li J: Role of miRNA
sponges in hepatocellular carcinoma. Clin Chim Acta. 500:10–19.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang J, Lu L, Luo Z, Li W, Lu Y, Tang Q
and Pu J: miR-383 inhibits cell growth and promotes cell apoptosis
in hepatocellular carcinoma by targeting IL-17 via STAT3 signaling
pathway. Biomed Pharmacother. 120:1095512019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Au SL, Wong CC, Lee JM, Fan DN, Tsang FH,
Ng IO and Wong CM: Enhancer of zeste homolog 2 epigenetically
silences multiple tumor suppressor microRNAs to promote liver
cancer metastasis. Hepatology. 56:622–631. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu X, Wu L, Yao J, Jiang H, Wang Q, Yang
Z and Wu F: MicroRNA let-7c inhibits cell proliferation and induces
cell cycle arrest by targeting CDC25A in human hepatocellular
carcinoma. PLoS One. 10:e01242662015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shimizu S, Takehara T, Hikita H, Kodama T,
Miyagi T, Hosui A, Tatsumi T, Ishida H, Noda T, Nagano H, et al:
The let-7 family of microRNAs inhibits Bcl-xL expression and
potentiates sorafenib-induced apoptosis in human hepatocellular
carcinoma. J Hepatol. 52:698–704. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo H, German P, Bai S, Barnes S, Guo W,
Qi X, Lou H, Liang J, Jonasch E, Mills GB and Ding Z: The PI3K/AKT
Pathway and renal cell carcinoma. J Genet Genomics. 42:343–353.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Faes S and Dormond O: PI3K and AKT:
Unfaithful partners in cancer. Int J Mol Sci. 16:21138–21152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Q, Lui VW and Yeo W: Targeting the
PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol.
7:1149–1167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hou YQ, Yao Y, Bao YL, Song ZB, Yang C,
Gao XL, Zhang WJ, Sun LG, Yu CL, Huang YX, et al: Juglanthraquinone
C induces intracellular ROS increase and apoptosis by activating
the Akt/Foxo signal pathway in HCC cells. Oxid Med Cell Longev.
2016:49416232016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Q, Yu WN, Chen X, Peng XD, Jeon SM,
Birnbaum MJ, Guzman G and Hay N: Spontaneous hepatocellular
carcinoma after the combined deletion of Akt isoforms. Cancer Cell.
29:523–535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng M, Bragelmann J, Schultze JL and
Perner S: Web-TCGA: An online platform for integrated analysis of
molecular cancer data sets. BMC Bioinformatics. 17:722016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma Z, Liu T, Huang W, Liu H, Zhang HM, Li
Q, Chen Z and Guo AY: MicroRNA regulatory pathway analysis
identifies miR-142-5p as a negative regulator of TGF-β pathway via
targeting SMAD3. Oncotarget. 7:71504–71513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wong NW, Chen Y, Chen S and Wang X:
OncomiR: An online resource for exploring pan-cancer microRNA
dysregulation. Bioinformatics. 34:713–715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46:D239–D245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ai C and Kong L: CGPS: A machine
learning-based approach integrating multiple gene set analysis
tools for better prioritization of biologically relevant pathways.
J Genet Genomics. 45:489–504. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu J, Mao X, Cai T, Luo J and Wei L: KOBAS
server: A web-based platform for automated annotation and pathway
identification. Nucleic Acids Res. 34:W720–W724. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
von Eschenbach AC and Buetow K: Cancer
informatics vision: CaBIG. Cancer Inform. 2:22–24. 2007.PubMed/NCBI
|
|
34
|
Menyhárt O, Nagy Á and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. R Soc Open Sci.
5:1810062018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nagy A, Lanczky A, Menyhart O and Gyorffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
The Gene Ontology C: The Gene Ontology
Resource: 20 years and still GOing strong. Nucleic Acids Res.
47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Reimand J, Kull M, Peterson H, Hansen J
and Vilo J: g:Profiler-a web-based toolset for functional profiling
of gene lists from large-scale experiments. Nucleic Acids Res.
35:W193–W200. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Raudvere U, Kolberg L, Kuzmin I, Arak T,
Adler P, Peterson H and Vilo J: g:Profiler: A web server for
functional enrichment analysis and conversions of gene lists (2019
update). Nucleic Acids Res. 47:W191–W198. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38:W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Max F, Rodriguez H, Lopes C, Zuberi K,
Montojo J, Bader GD and Morris Q: GeneMANIA update 2018. Nuclc
Acids Res. 46:W60–W64. 2018. View Article : Google Scholar
|
|
47
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47:D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Song Y, Wang F, Huang Q, Cao Y, Zhao Y and
Yang C: MicroRNAs contribute to hepatocellular carcinoma. Mini Rev
Med Chem. 15:459–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Han Y, Liu Y, Fu X, Zhang Q, Huang H,
Zhang C, Li W and Zhang J: miR-9 inhibits the metastatic ability of
hepatocellular carcinoma via targeting beta galactoside
alpha-2,6-sialyltransferase 1. J Physiol Biochem. 74:491–501. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Y, Tai Q, Zhang J, Kang J, Gao F,
Zhong F, Cai L, Fang F and Gao Y: MiRNA-206 inhibits hepatocellular
carcinoma cell proliferation and migration but promotes apoptosis
by modulating cMET expression. Acta Biochim Biophys Sin (Shanghai).
51:243–253. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhu XM, Wu LJ, Xu J, Yang R and Wu FS:
Let-7c microRNA expression and clinical significance in
hepatocellular carcinoma. J Int Med Res. 39:2323–2329. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jilek JL, Zhang QY, Tu MJ, Ho PY, Duan Z,
Qiu JX and Yu AM: Bioengineered Let-7c inhibits orthotopic
hepatocellular carcinoma and improves overall survival with minimal
immunogenicity. Mol Ther Nucleic Acids. 14:498–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Luo H, Hao E, Tan D, Wei W, Xie J, Feng X,
Du Z, Huang C, Bai G, Hou Y, et al: Apoptosis effect of
Aegiceras corniculatum on human colorectal cancer via
activation of FoxO signaling pathway. Food Chem Toxicol.
134:1108612019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Farhan M, Wang H, Gaur U, Little PJ, Xu J
and Zheng W: FOXO signaling pathways as therapeutic targets in
cancer. Int J Biol Sci. 13:815–827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Nam HJ and van Deursen JM: Cyclin B2 and
p53 control proper timing of centrosome separation. Nat Cell Biol.
16:538–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang Q, Sun S, Zhu C, Zheng Y, Cai Q,
Liang X, Xie H and Zhou J: Prediction and analysis of weighted
genes in hepatocellular carcinoma using bioinformatics analysis.
Mol Med Rep. 19:2479–2488. 2019.PubMed/NCBI
|
|
58
|
Lauper N, Beck AR, Cariou S, Richman L,
Hofmann K, Reith W, Slingerland JM and Amati B: Cyclin E2: A novel
CDK2 partner in the late G1 and S phases of the mammalian cell
cycle. Oncogene. 17:2637–2643. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hwang HC and Clurman BE: Cyclin E in
normal and neoplastic cell cycles. Oncogene. 24:2776–2786. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Caldon CE and Musgrove EA: Distinct and
redundant functions of cyclin E1 and cyclin E2 in development and
cancer. Cell Div. 5:22010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sherr CJ, Beach D and Shapiro GI:
Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov.
6:353–367. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xiang X, You XM and Li LQ: Expression of
HSP90AA1/HSPA8 in hepatocellular carcinoma patients with
depression. Onco Targets Ther. 11:3013–3023. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Piredda ML, Gaur G, Catalano G, Divona M,
Banella C, Travaglini S, Puzzangara MC, Voso MT, Lo-Coco F and
Noguera NI: PML/RARA inhibits expression of HSP90 and its target
AKT. Br J Haematol. 184:937–948. 2019.PubMed/NCBI
|
|
64
|
Hou C, Li Y, Liu H, Dang M, Qin G, Zhang N
and Chen R: Profiling the interactome of protein kinase C zeta by
proteomics and bioinformatics. Proteome Sci. 16:52018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Fox MM, Phoenix KN, Kopsiaftis SG and
Claffey KP: AMP-activated protein kinase alpha 2 isoform
suppression in primary breast cancer alters AMPK growth control and
apoptotic signaling. Genes Cancer. 4:3–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Vila IK, Yao Y, Kim G, Xia W, Kim H, Kim
SJ, Park MK, Hwang JP, González-Billalabeitia E, Hung MC, et al: A
UBE2O-AMPKα2 axis that promotes tumor initiation and progression
offers opportunities for therapy. Cancer Cell. 31:208–224. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jardin I, Albarran L, Bermejo N, Salido GM
and Rosado JA: Homers regulate calcium entry and aggregation in
human platelets: A role for Homers in the association between STIM1
and Orai1. Biochem J. 445:29–38. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Moccia F, Zuccolo E, Poletto V, Turin I,
Guerra G, Pedrazzoli P, Rosti V, Porta C and Montagna D: Targeting
stim and orai proteins as an alternative approach in anticancer
therapy. Curr Med Chem. 23:3450–3480. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Rajalingam K, Schreck R, Rapp UR and
Albert S: Ras oncogenes and their downstream targets. Biochim
Biophys Acta. 1773:1177–1195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Dietrich P, Gaza A, Wormser L, Fritz V,
Hellerbrand C and Bosserhoff AK: Neuroblastoma RAS viral oncogene
homolog (NRAS) is a novel prognostic marker and contributes to
sorafenib resistance in hepatocellular carcinoma. Neoplasia.
21:257–268. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xiang RF, Stack D, Huston SM, Li SS,
Ogbomo H, Kyei SK and Mody CH: Ras-related C3 botulinum toxin
substrate (Rac) and Src Family Kinases (SFK) Are proximal and
essential for phosphatidylinositol 3-Kinase (PI3K) activation in
natural Killer (NK) Cell-mediated Direct Cytotoxicity against
cryptococcus neoformans. J Biol Chem. 291:6912–6922. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen L, Chan TH, Yuan YF, Hu L, Huang J,
Ma S, Wang J, Dong SS, Tang KH, Xie D, et al: CHD1L promotes
hepatocellular carcinoma progression and metastasis in mice and is
associated with these processes in human patients. J Clin Invest.
120:1178–1191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhao P, Zhang W, Wang SJ, Yu XL, Tang J,
Huang W, Li Y, Cui HY, Guo YS, Tavernier J, et al: HAb18G/CD147
promotes cell motility by regulating annexin II-activated RhoA and
Rac1 signaling pathways in hepatocellular carcinoma cells.
Hepatology. 54:2012–2024. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ran RZ, Chen J, Cui LJ, Lin XL, Fan MM,
Cong ZZ, Zhang H, Tan WF, Zhang GQ and Zhang YJ: miR-194 inhibits
liver cancer stem cell expansion by regulating RAC1 pathway. Exp
Cell Res. 378:66–75. 2019. View Article : Google Scholar : PubMed/NCBI
|