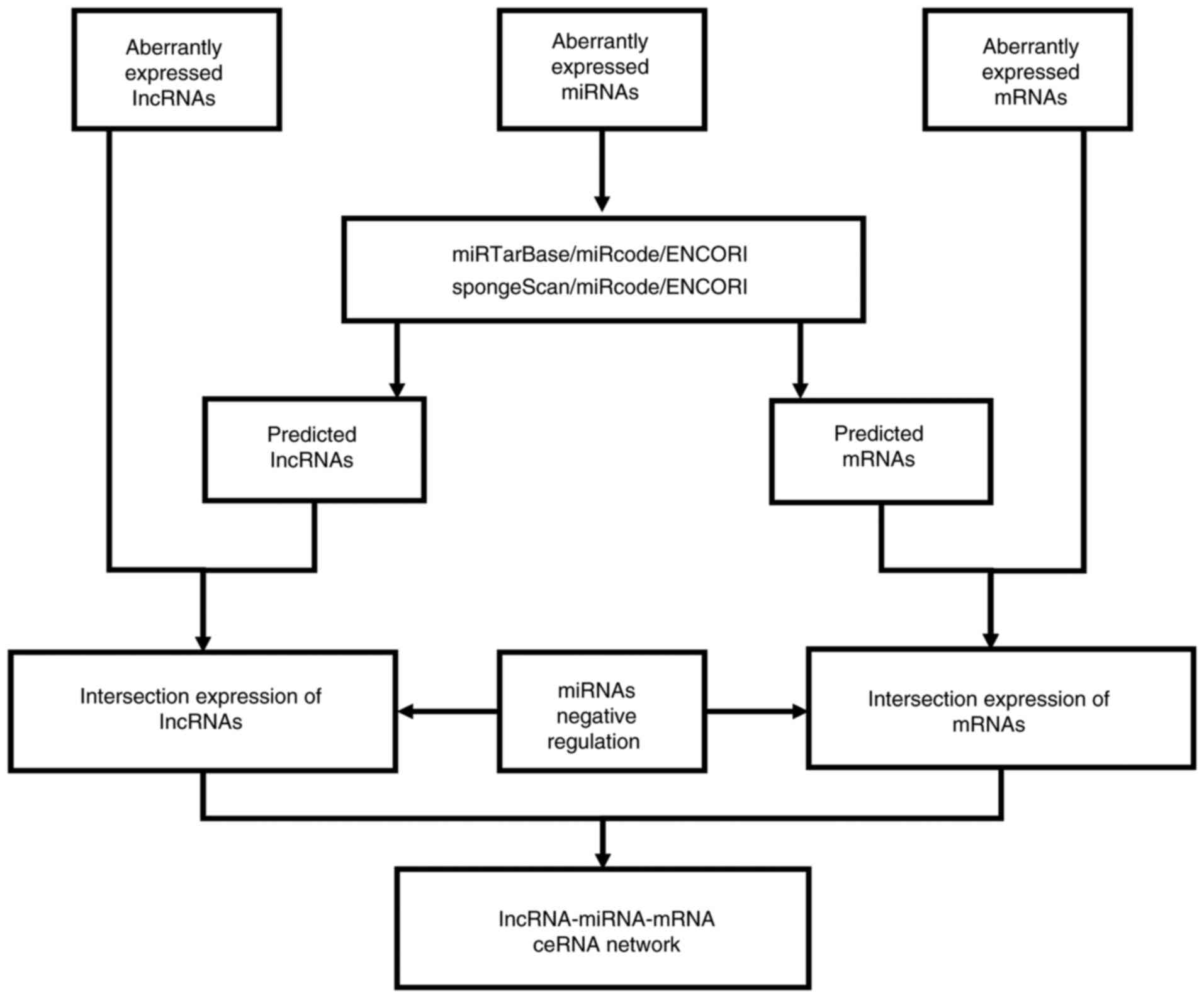
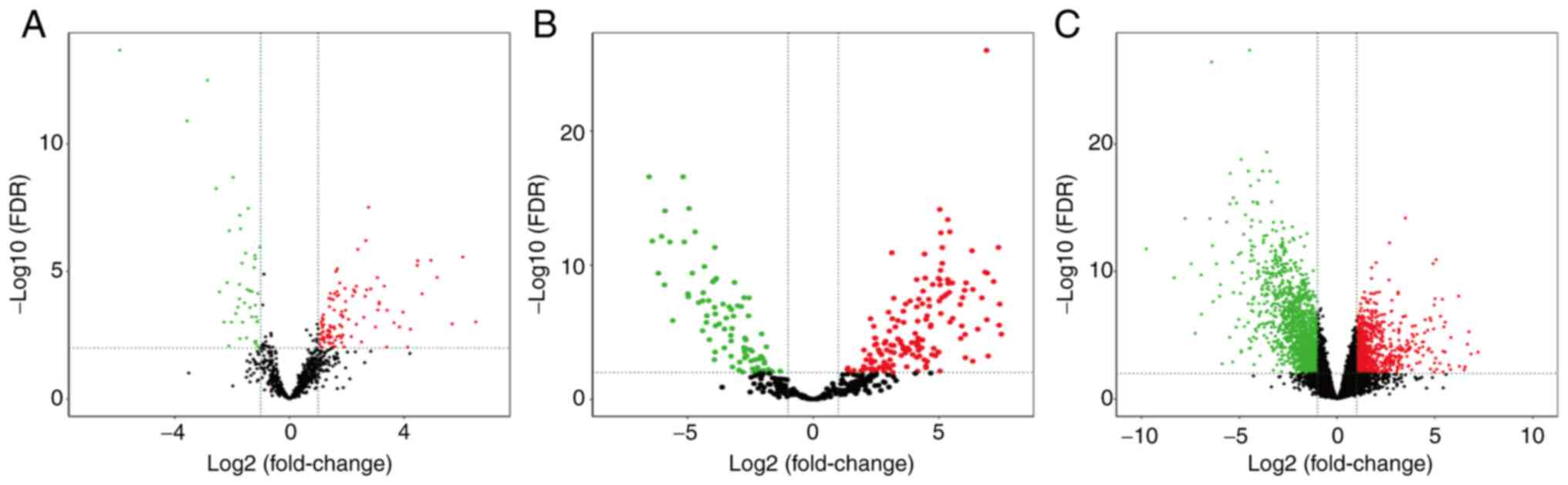
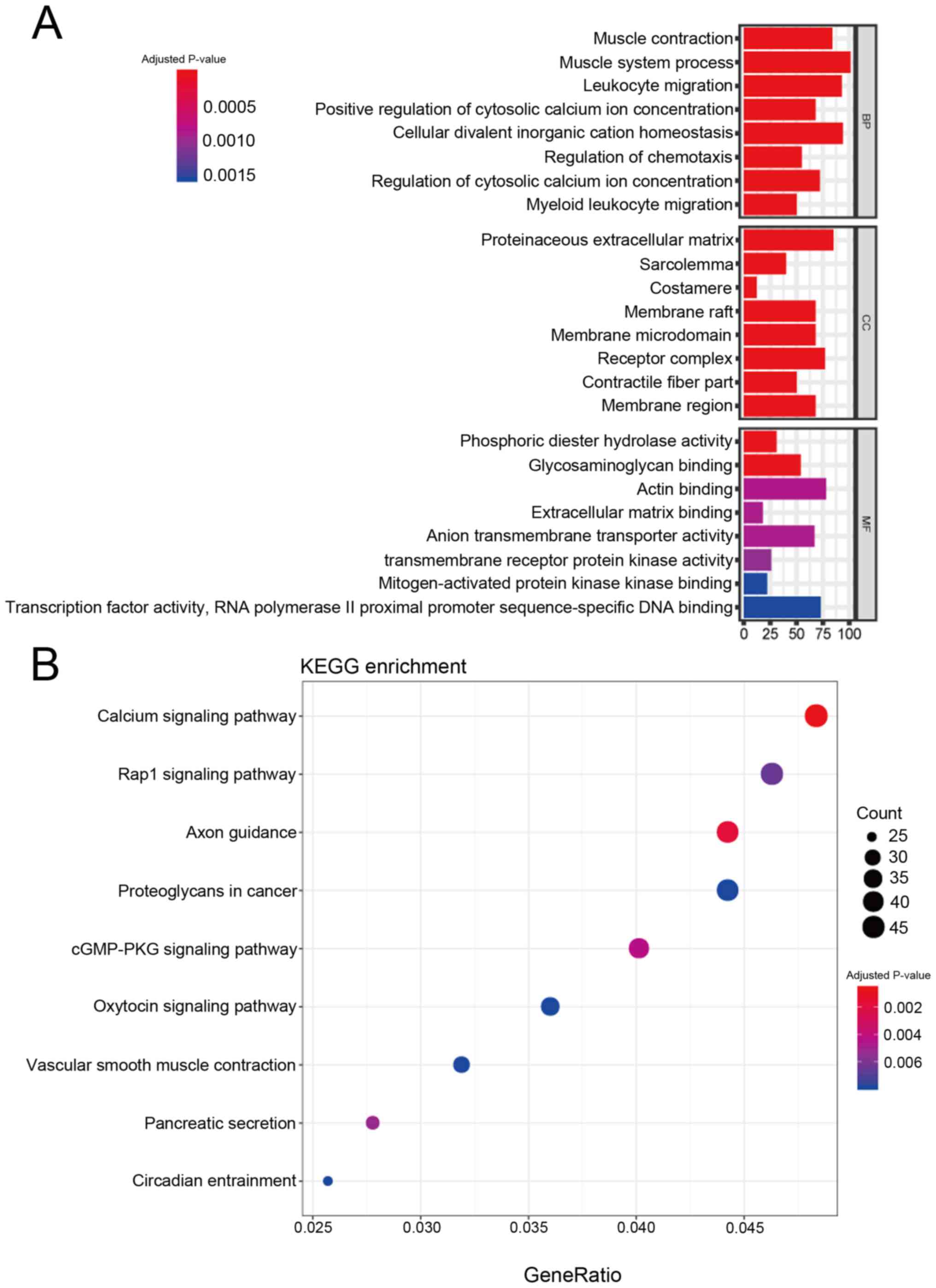
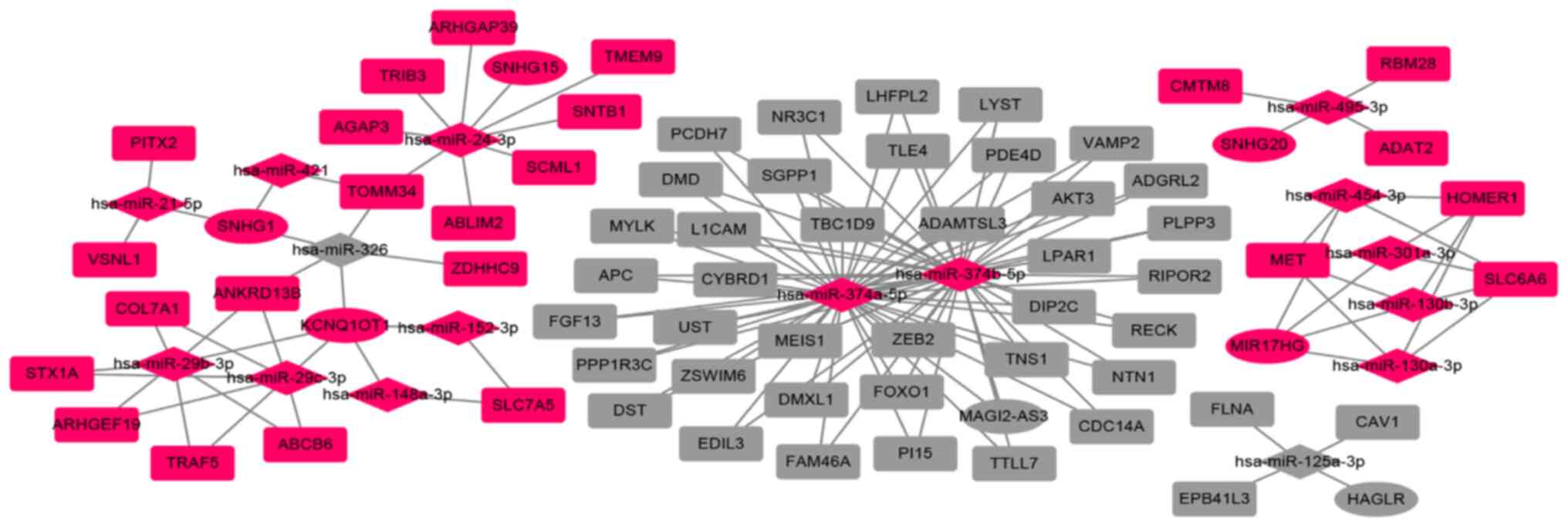
|
1
|
Lee YC, Lee YL, Chuang JP and Lee JC:
Differences in survival between colon and rectal cancer from SEER
data. PLoS One. 8:e787092013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
World Health Organization, . International
classification of diseases for oncology (ICD-O)-3rd edition, 1st
revision. 2013.simplehttps://apps.who.int/iris/handle/10665/96612
|
|
4
|
D'Souza N, de Neree Tot Babberich MPM,
Lord A, Shaw A, Abulafi M, Tekkis P, Wiggers T and Brown G: The
rectosigmoid problem. Surg Oncol. 27:521–525. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Falch C, Mueller S, Braun M, Gani C, Fend
F, Koenigsrainer A and Kirschniak A: Oncological outcome of
carcinomas in the rectosigmoid junction compared to the upper
rectum or sigmoid colon-A retrospective cohort study. Eur J Surg
Oncol. 45:2037–2044. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Käser SA, Froelicher J, Li Q, Müller S,
Metzger U, Castiglione M, Laffer UT and Maurer CA: Adenocarcinomas
of the upper third of the rectum and the rectosigmoid junction seem
to have similar prognosis as colon cancers even without
radiotherapy, SAKK 40/87. Langenbecks Arch Surg. 400:675–682. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ponz de Leon M, Marino M, Benatti P, Rossi
G, Menigatti M, Pedroni M, Di Gregorio C, Losi L, Borghi F,
Scarselli A, et al: Trend of incidence, subsite distribution and
staging of colorectal neoplasms in the 15-year experience of a
specialised cancer registry. Ann Oncol. 15:940–946. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77(15):
3965–3981. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sergeeva OV, Koteliansky VE and Zatsepin
TS: mRNA-based therapeutics-advances and perspectives. Biochemistry
(Mosc). 81:709–722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng Y, Geng L, Wang K, Sun J, Xu W, Gong
S and Zhu Y: Long noncoding RNA expression signatures of colon
cancer based on the ceRNA network and their prognostic value. Dis
Markers. 2019:76367572019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo R, Song J, Zhang W and Ran L:
Identification of MFI2-AS1, a novel pivotal lncRNA for prognosis of
stage III/IV colorectal cancer. Dig Dis Sci. 65:538–3550. 2020.
View Article : Google Scholar
|
|
14
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar
|
|
15
|
Zhang Z, Wang S, Ji D, Qian W, Wang Q, Li
J, Gu J, Peng W, Hu T, Ji B, et al: Construction of a ceRNA network
reveals potential lncRNA biomarkers in rectal adenocarcinoma. Oncol
Rep. 39:2101–2113. 2018.PubMed/NCBI
|
|
16
|
Dong X, Yang Z, Yang H, Li D and Qiu X:
Long Non-coding RNA MIR4435-2HG promotes colorectal cancer
proliferation and metastasis through miR-206/YAP1 Axis. Front
Oncol. 10:1602020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang X, Wen L, Chen S, Zhang J, Ma Y, Hu
J, Yue T, Wang J, Zhu J, Bu D and Wang X: The novel long noncoding
RNA CRART16 confers cetuximab resistance in colorectal cancer cells
by enhancing ERBB3 expression via miR-371a-5p. Cancer Cell Int.
20:682020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, Jessup JM, Brierley JD, Gaspar LE, Schilsky RL, Balch
CM, Winchester DP, Asare EA, Madera M, Gress DM and Meyer LR: AJCC
Cancer Staging Manual. 8th edition. Springer; 2017, View Article : Google Scholar
|
|
19
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lindner P, Paul S, Eckstein M, Hampel C,
Muenzner JK, Erlenbach-Wuensch K, Ahmed HP, Mahadevan V, Brabletz
T, Hartmann A, et al: EMT transcription factor ZEB1 alters the
epigenetic landscape of colorectal cancer cells. Cell Death Dis.
11:1472020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mo S, Zhang L, Dai W, Han L, Wang R, Xiang
W, Wang Z, Li Q, Yu J, Yuan J, et al: Antisense lncRNA LDLRAD4-AS1
promotes metastasis by decreasing the expression of LDLRAD4 and
predicts a poor prognosis in colorectal cancer. Cell Death Dis.
11:1552020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stuckel AJ, Zhang W, Zhang X, Zeng S,
Dougherty U, Mustafi R, Zhang Q, Perreand E, Khare T, Joshi T, et
al: Enhanced CXCR4 expression associates with increased gene body
5-Hydroxymethylcytosine modification but not decreased promoter
methylation in colorectal cancer. Cancers (Basel). 12:5392020.
View Article : Google Scholar
|
|
24
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu J, Li H, Zheng B, Sun L, Yuan Y and
Xing C: Competitive endogenous RNA (ceRNA) regulation network of
lncRNA-miRNA-mRNA in colorectal carcinogenesis. Dig Dis Sci.
64:1868–1877. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei S, Chen J, Huang Y, Sun Q, Wang H,
Liang X, Hu Z and Li X: Identification of hub genes and
construction of transcriptional regulatory network for the
progression of colon adenocarcinoma hub genes and TF regulatory
network of colon adenocarcinoma. J Cell Physiol. 235:2037–2048.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu J, Meng Q, Li X, Yang H, Xu J, Gao N,
Sun H, Wu S, Familiari G, Relucenti M, et al: Long noncoding RNA
MIR17HG promotes colorectal cancer progression via miR-17-5p.
Cancer Res. 79:4882–4895. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sunamura N, Ohira T, Kataoka M, Inaoka D,
Tanabe H, Nakayama Y, Oshimura M and Kugoh H: Regulation of
functional KCNQ1OT1 lncRNA by beta-catenin. Sci Rep. 6:206902016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li F, Li Q and Wu X: Construction and
analysis for differentially expressed long non-coding RNAs and
MicroRNAs mediated competing endogenous RNA network in colon
cancer. PLoS One. 13:e01924942018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao W, Ma X, Liu L, Chen Q, Liu Z, Zhang
Z, Ma S, Wang Z, Li H, Wang Z and Wu J: SNHG20: A vital lncRNA in
multiple human cancers. J Cell Physiol. Jan 15–2019.(Epub ahead of
print). doi: 10.1002/jcp.28143.
|
|
31
|
Li C, Zhou L, He J, Fang XQ, Zhu SW and
Xiong MM: Increased long noncoding RNA SNHG20 predicts poor
prognosis in colorectal cancer. BMC Cancer. 16:6552016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Slattery ML, Herrick JS, Mullany LE,
Valeri N, Stevens J, Caan BJ, Samowitz W and Wolff RK: An
evaluation and replication of miRNAs with disease stage and
colorectal cancer-specific mortality. Int J Cancer. 137:428–438.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang JT, Wang F, Chapin W and Huang RS:
Identification of MicroRNAs as breast cancer prognosis markers
through the cancer genome atlas. PLoS One. 11:e01682842016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Torrejon B, Cristobal I, Rojo F and
Garcia-Foncillas J: Caveolin-1 is markedly downregulated in
patients with early-stage colorectal cancer. World J Surg.
41:2625–2630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang J, Zhu T, Zhao R, Gao D, Cui Y, Wang
K and Guo Y: Caveolin-1 inhibits proliferation, migration, and
invasion of human colorectal cancer cells by suppressing
phosphorylation of epidermal growth factor receptor. Med Sci Monit.
24:332–341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang YX, Zhu HF, Zhang ZY, Ren F and Hu
YH: MiR-384 inhibits the proliferation of colorectal cancer by
targeting AKT3. Cancer Cell Int. 18:1242018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang K, Zhu TN and Zhao RJ: Filamin A
regulates EGFR/ERK/Akt signaling and affects colorectal cancer cell
growth and migration. Mol Med Rep. 20:3671–3678. 2019.PubMed/NCBI
|
|
38
|
Noriega-Guerra H and Freitas VM:
Extracellular matrix influencing HGF/c-MET signaling pathway:
Impact on cancer progression. Int J Mol Sci. 19:33002018.
View Article : Google Scholar
|
|
39
|
Ikegami K, Mukai M, Tsuchida J, Heier RL,
Macgregor GR and Setou M: TTLL7 is a mammalian beta-tubulin
polyglutamylase required for growth of MAP2-positive neurites. J
Biol Chem. 281:30707–30716. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Garnham CP, Roll-Mecak A and Tanner
ME: Phosphinic acid-based inhibitors of tubulin polyglutamylases.
Bioorg Med Chem Lett. 23:4408–4412. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Das V, Kanakkanthara A, Chan A and Miller
JH: Potential role of tubulin tyrosine ligase-like enzymes in
tumorigenesis and cancer cell resistance. Cancer Lett. 350:1–4.
2014. View Article : Google Scholar : PubMed/NCBI
|