Introduction
Glioma is one of the most aggressive and common
malignant intracranial tumors (1–3) and is
characterized by a high invasive ability and a high lethality rate.
In 2014, 56,000 patients died as a result of glioma in China
(4–6). According to the morphological
characteristics of glioma, it can be classified as either
astrocytoma, glioblastoma, oligodendroglioma, mixed
oligoastrocytomas or ependymoma (7).
Despite the development of active therapies (including surgery,
radiotherapy, and chemotherapy), the prognosis of patients with
glioma is still poor (8). A previous
study reported that the median survival time for patients with
high-grade glioma does not exceed 15 months (9). Therefore, elucidating the underlying
mechanism of glioma pathogenesis is beneficial for diagnosis and
treatment.
Long non-coding (lnc)RNAs are defined as a group of
RNAs, with a total length of >200 nucleotides and a lack of
protein-coding ability (10).
lncRNAs, as tumor suppressor genes or oncogenes, play essential
roles in vital cell functions, such as cell growth, proliferation,
and invasion (11,12). Currently, increasing evidence
suggests that lncRNAs function as competitive endogenous RNAs in
cancer progression by sponging and inhibiting the expression of
microRNAs (miRNA/miR) (13). For
example, the lncRNA MIR503HG inhibited cell migration and invasion
by sponging miR-103 in triple-negative breast cancer (14), while the lncRNA MEG3 repressed cell
growth in glioma by modulating the expression level of miR-96-5p
(15). For colorectal cancer (CRC),
the lncRNA MFI2-AS1 facilitated CRC cell proliferation, migration
and invasion by sponging miR-574-5p (16).
The lncRNA MIR22HG is located in the 17p13.3 region;
it primarily exists in the cytoplasm and is frequently
hypermethylated and degraded (17).
It has been hypothesized that the lncRNA MIR22HG could be involved
in the development and progression of different types of tumor,
including hepatocellular carcinoma (18). Previous research has shown that the
lncRNA MIR22HG contributed to the inhibition of gastric cancer by
attenuating NOTCH2 signaling (19).
In endometrial carcinoma, the apoptosis of endometrial carcinoma
cells was strengthened by MIR22HG, which negatively regulated
miR-141-3p and enhanced death-associated protein kinase 1 protein
expression (20). In addition, the
lncRNA MIR22HG plays a tumor suppressor role in lung cancer by
downregulating the oncogenes, Y-box binding protein 1, MET
proto-oncogene receptor tyrosine kinase, and p21 (21).
The cytoplasmic polyadenylation element-binding
protein (CPEB) family includes RNA-binding proteins that mediate
translation through cytoplasmic polyadenylation (22,23). The
CPEB family typically consists of four members, CPEB-1, −2, −3 and
−4 in vertebrates (24). CPEB3
primarily exists in the cytoplasm and plays a vital role in
synaptic plasticity and memory (25,26). In
recent years, it has been reported that CPEB3 participates in
regulating various types of cancer (27,28). In
CRC, the lncRNA SUMO1P3 induced cell proliferation and the cell
cycle, and restricted apoptosis by epigenetically silencing CPEB3
(29).
Recently, the upregulation of CPEB3 was found to
inhibit cell proliferation and growth in a glioma cell line
(30). Furthermore, miR-9 was found
to be a positive regulator in glioma tumorigenesis and angiogenesis
(31). However, the association
between miR-9 and CPEB3 is unclear. In addition, previous findings
indicated that the lncRNA MIR22HG participated in the development
of a variety of different cancers including prostate cancer,
colorectal cancer and glioma (32–34).
However, the exact role and detailed mechanism of the lncRNA
MIR22HG in the development of glioma is unclear.
In the present study, we aimed to explore the
expression of lncRNA MIR22HG, miR-9 and CPEB3. We tried to
investigate the effect of MIR22HG in the progression of glioma and
elucidate the underlying molecular. Our study intended to uncover
the molecular pathological mechanism and provided potential
therapeutic targets for glioma.
Materials and methods
Clinical specimens
All tissues, including 25 human glioma tissues and
15 normal brain tissues (tissues from patients with traumatic brain
injury) were obtained from patients who underwent surgical excision
at the Neurosurgery Department of Tangdu Hospital from January 2017
to January 2018. The mean ages of the patients with glioma and
control patients were 48.5 and 46.8 years, respectively. Patients
with glioma composed of 14 males and 11 females. The control
patients composed of eight males and 7 females. All tissue samples
were pathologically evaluated by senior pathologist according to
the fourth edition of the World Health Organization classification
of tumors of the central nervous system (2016) (35) and stored in liquid nitrogen at −80°C.
The study protocol was approved by the Ethical Committee of Tangdu
Hospital of the Fourth Military Medical University, and written
informed consent was provided from all the patients.
Cell culture
The human glioma cell lines U251 (TCHu 58), U87
(TCHu138; glioblastoma of unknown origin) and the HA1800 normal
human astrocyte cell line (JNO171-101) were purchased from the Cell
Resource Center of the Shanghai Institute of Biological Sciences.
To confirm the identity of the cells over long culture periods, the
U87 cell line was genotyped using short tandem repeat analysis and
the cells were confirmed to be the U87MG cell line from American
Type Culture Collection. All the cell lines were cultured in DMEM,
supplemented with 10% FBS and 100 U/ml penicillin/streptomycin at
37°C in a humidified incubator with 5% CO2.
Starbase and TargetScan analysis
The relationship between MIR22HG and miR-9 was
analyzed using online software Starbase (http://starbase.sysu.edu.cn/). The miRNA-LncRNA tool
was utilized to analyze the relationship. The target miRNA was
miR-9 and the predicted lncRNAs were listed in the software.
Similarly, the relationship between miR-9 and CPEB3 was also
analyzed using TargetScan (http://www.targetscan.org/mamm_31/). The method we
used was miRNA- tool and the target miRNA was also miR-9.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from glioma tissues or cell lines was
extracted using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.). The isolated RNA was reverse transcribed into
complementary dNA using a PrimeScript™ RT master mix (Takara
Biotechnology, Inc.) and TaqMan advanced miRNA assay kit (Thermo
Fisher Scientific, Inc.) at 42°C for 15 min. A 20-µl reaction
system was used: 0.5 µl Sense and antisense primers, 50 ng RT
product and 10 µl 2X SYBR TransStar Green PCR Super mix (cat. no.
B21202; http://www.bimake.com/product/sybr-green-qpcr-master-mix.html)
in an IQ™ 5 Real-Time PCR Detection system (Bio-Rad Laboratories,
Inc.). The thermocycling conditions were: 95°C For 4 min for
denaturation, followed by 40 cycles for 23 sec, and annealing for
28 sec at 60°C and extension at 72°C for 35 sec. miR-103 and GAPDH
served as normalizing controls and the 2−ΔΔCq method was
used for quantification (36,37). The
sequences of the primers were as follows: MIR22HG forward,
5′-CGGACGCAGTGATTTGCT-3′ and reverse, 5′-GCTTTAGCTGGGTCAGGACA-3′;
miR-9 forward, 5′-GAGGCCCGTTTCTCTCTTTG-3′ and reverse,
5′-AGCTTTATGACGGCTCTGTG-3′; CPEB3 forward,
5′-TCAACACAACGACATTGACAAA-3′ and reverse,
5′-CCCTGACACTCGTCACACAT-3′; GAPDH forward,
5′-AAATCCCATCACCATCTTCCAG-3′ and reverse,
5′-TGATGACCCTTTTGGCTCCC-3′; miR-103 forward,
5′-GCCGAGCTGCCTTGTGGAATC-3′ and reverse,
5′-CTGCCTTGTGGAATCACAT-3′.
Cell transfection
The pcDNA3 empty vector and pcDNA3-MIR22HG
overexpression vector, and the miR-9 mimic and negative control
(NC) mimic were constructed by Shanghai GenePharma, Co., Ltd.. The
cells were transfected with 3 µg control or overexpression vector
in one well of 6-well plate. The working concentration of miR-9
mimic was 50 nM. Then, according to the manufacturer's
instructions, the U87 and U251 cell lines were transfected with the
pcDNA3 empty vector, pcDNA3-MIR22HG, miR-9 mimic, or NC mimic using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After transfection for 48 h, the cells were used
for subsequent experiments.
Luciferase reporter assay
The luciferase reporter assay was used to determine
the association between miR-9 and MIR22HG, and CPEB3. Wild-type
(WT) and mutant (MUT) MIR22HG or CPEB3 3′-untranslated region
sequences were inserted into the pmirGLO luciferase reporter
vectors (Promega Corporation) to construct a luciferase reporter
vector. In total, 50 ng WT and MUT luciferase vectors or 50 nM
miR-9 mimic were transfected into U87 or U251 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 20 min at room temperature. After 48 h, the
luciferase activities were detected using a dual-luciferase
reporter assay kit (Promega Corporation). The firefly luciferase
was normalized using Renilla luciferase as the internal
control.
Western blot assay
The collected cells was lysed in the RIPA lysis
buffer (Beyotime Institute of Biotechnology) on the ice for 30 min.
Total protein was extracted from the glioma cell lines and the
protein concentration was determined using a BCA protein
concentration kit (Beyotime Institute of Biotechnology). Then, 20
µg protein was separated per lane using 10% SDS-PAGE and
transferred to PVDF membranes (EMD Millipore). Following which, the
PVDF membranes were blocked with 5% skimmed milk for 2 h, at room
temperature, then incubated with anti-CPEB3 (cat. no. ab10883,
1:1,000 dilution) and β-actin antibodies (cat. no. ab8226, 1:2,000
dilution both from Abcam,) overnight at 4°C. The membranes were
washed with TBST for 5 min three times at room temperature. After
washing with 1X TBST, the membranes were then incubated with
HRP-conjugated goat anti-rabbit IgG H&L (cat. no. as014) or
goat anti-mouse IgG H&L secondary antibodies (cat. no. as003;
1:5,000) (both ABclonal Biotech Co., Ltd.) for 2 h at room
temperature. The signals were visualized using an enhanced
chemiluminescence kit (EMD Millipore).
Cell Counting Kit-8 (CCK-8) assay
The kit was used according the instruction of the
manufacturer. U87 and U251 cells transfected with MIR22HG or/and
miR-9 were seeded in a 96-well plate, at a density of 5,000
cells/well. In total, 10 µl CCK-8 solution (Beyotime Institute of
Biotechnology) was added to each well according to the
manufacturer's instructions. The absorbance was examined at 450 nm
in Multiskan™ FC microplate reader with Skan It software 2.4.2
(both Thermo Fisher Scientific, Inc.).
Matrigel assay
The diluted Matrigel was used to cover the upper
chamber of the Transwell inserts for 5 h at 37°C. Then, 200 µl
serum-free medium containing 3×104 U87 or U251 cells was
added into the upper chamber of the Transwell inserts, while 500 µl
10% FBS-containing complete medium was added to the lower chamber.
The cells were incubated for 24 h (at 37°C in a humidified
incubator with 5% CO2,). After 24 h, the cells were
fixed with 4% paraformaldehyde for 0.5 h at room temperature, then
stained with crystal violet staining solution for 0.5 h at room
temperature. The number of stained cells in five random fields of
view was counted using a light phase contrast microscope with ×10
magnification.
Wound healing assay
The U87 and U251 cell lines were plated in a 6-well
plate until they had reached 90% confluence. The glioma cells were
scraped using a 20-µl pipette tip to generate a wound. Then, the
cells were washed three times with PBS and cultured with serum-free
culture medium for 24 h. Cell migration was observed using a light
phase contrast microscope at ×10 magnification. Images were
captured and analyzed with FL Color Cell Imaging systems version
1.4. (Thermo Fisher Scientific, Inc.).
Statistical analysis
An unpaired Student's t-test was used to analyze
differences between two groups, while one-way ANOVA followed by
Tukey's post hoc test to determine the differences between three or
more groups. SPSS v19.0 (SPSS, Inc.) was used for the analysis. The
data are presented as the mean ± SEM of at least three different
experiments. P<0.05 was considered to indicate a statistically
significant difference.
Results
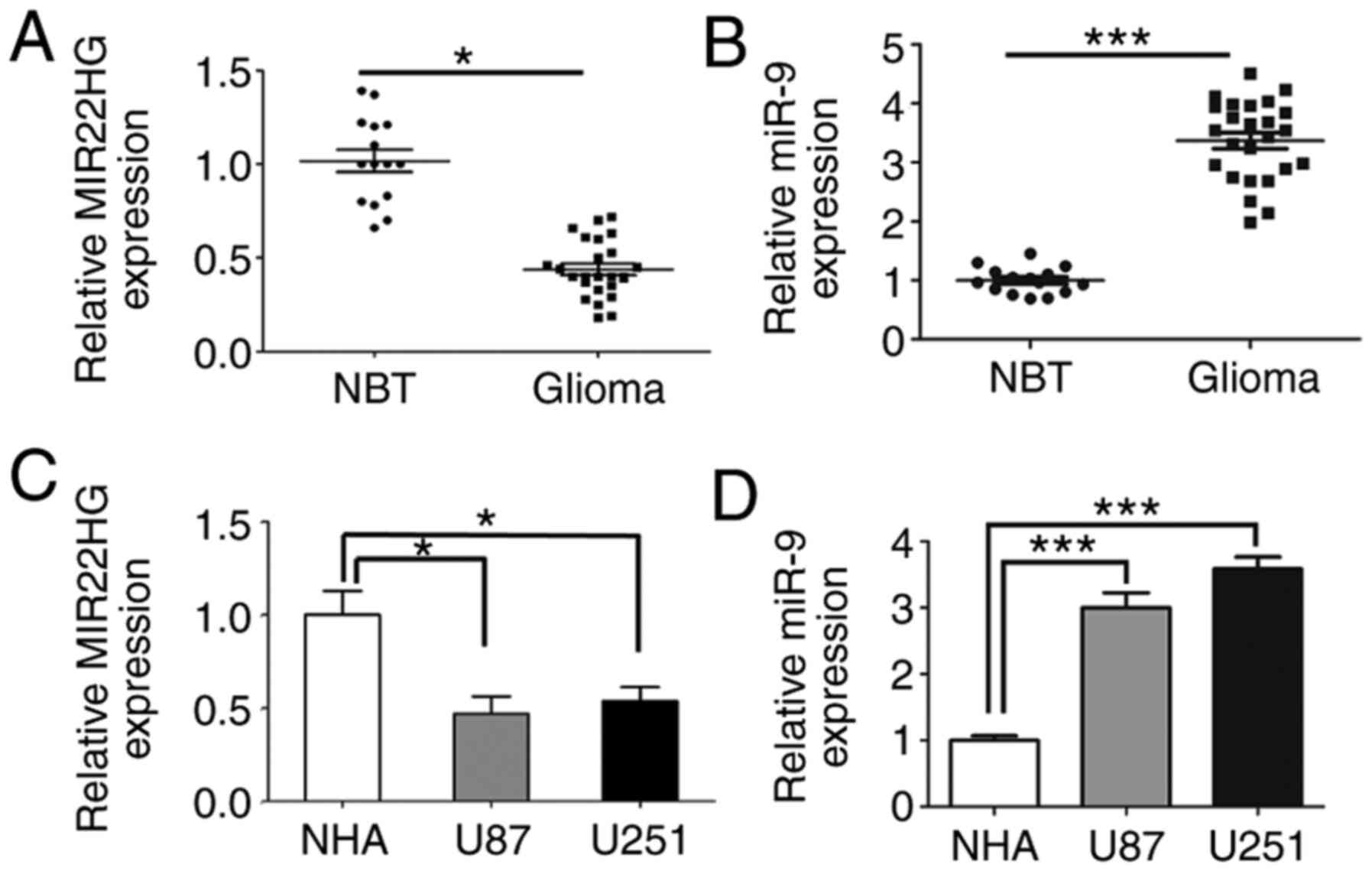
MIR22HG is downregulated and miR-9 is
increased in human glioma tissues and glioma cell lines
To determine the role of MIR22HG in human glioma
tissues, the expression level of MIR22HG was measured using RT-qPCR
and the results showed that the expression level of MIR22HG
significantly decreased in human glioma tissues compared with that
in adjacent normal tissues (Fig.
1A). However, the expression level of miR-9 was increased in
human glioma tissues, which suggested that MIR22HG may be
associated with miR-9 (Fig. 1B).
Therefore there may be a regulatory relationship between them.
Furthermore, the expression level of MIR22HG and miR-9 in the
glioma cell lines was also detected using RT-qPCR and the results
showed that the expression level of MIR22HG was decreased in the
glioma cell lines compared with that in the normal human astrocytes
(Fig. 1C). Conversely, miR-9
expression level was also increased in the glioma cell lines
compared with that in the normal cell lines (Fig. 1D).
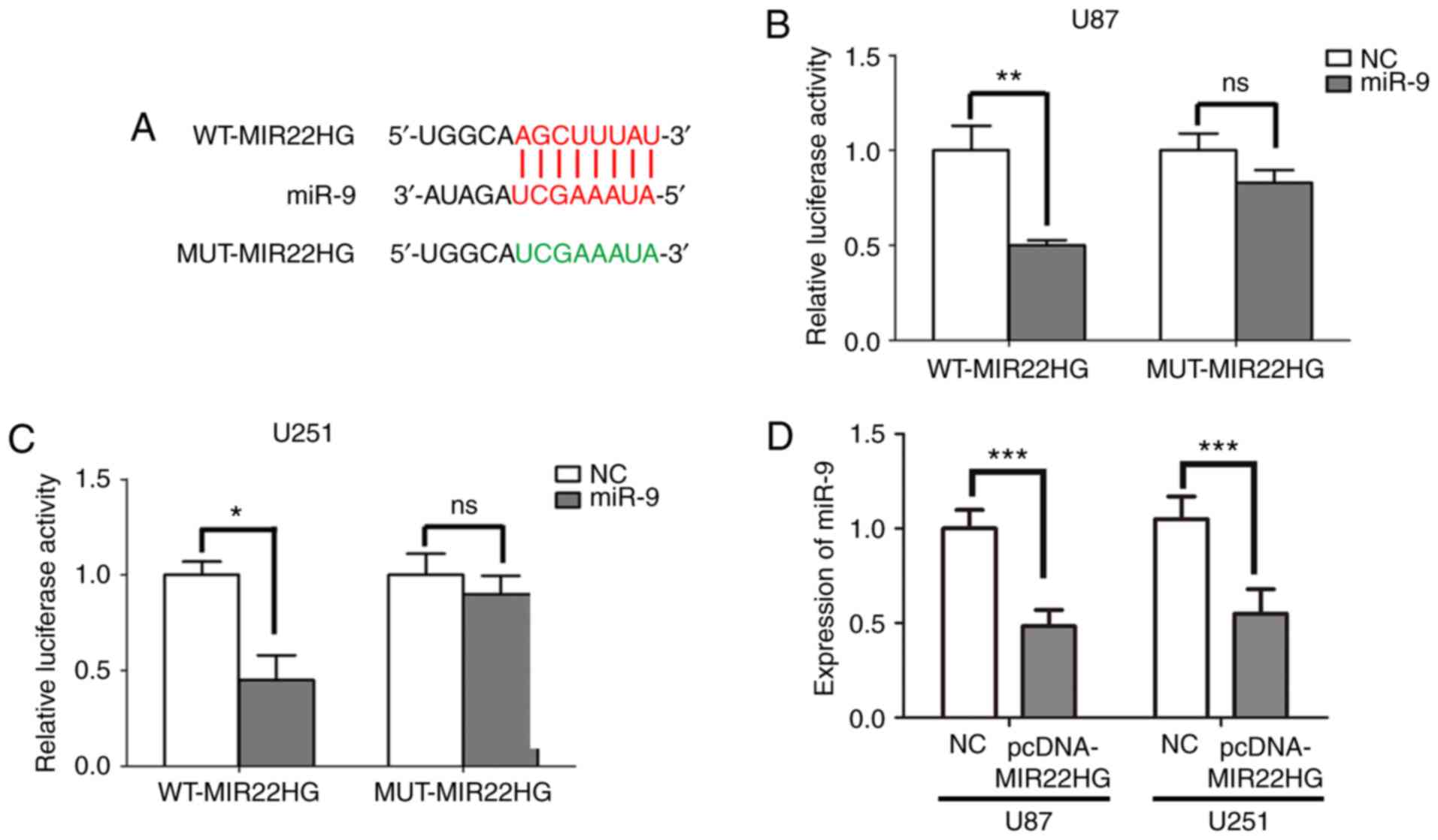
MIR22HG sponges miR-9 in the glioma
cell lines
Previous studies have confirmed that lncRNAs could
act as sponges to regulate miRNAs (38–40). It
was found that MIR22HG contained a potential binding site for miR-9
using the bioinformatics database StarBase v2.0 (Fig. 2A). To further investigate whether
MIR22HG could directly bind to miR-9 in the glioma cell lines, the
U251 or U87 cells were co-transfected with miR-9 mimic and WT or
MUT MIR22HG. The effect of miR-9 mimic in the U87 and U251 cells
was confirmed (Fig. S1A and B).
Then, the luciferase activity of the cells was detected. The
results indicated that the luciferase activity was significantly
reduced when U87 or U251 cells were transfected with the miR-9
mimic and MIR22HG-WT. In contrast, the luciferase activity did not
significantly change when U87 or U251 cells were transfected with
the miR-9 mimic and MIR22HG-MUT (Fig. 2B
and C). These results verified that miR-9 was a direct target
of MIR22HG. Furthermore, MIR22HG was overexpressed in the U87 and
U251 cell lines and the expression level of miR-9 was detected
using RT-qPCR. The results confirmed that MIR22HG was increased
when transfected with pcDNA-MIR22HG (Fig. S1C and D). Furthermore, upregulating
MIR22HG expression suppressed the expression level of miR-9 in the
glioma cell lines (Fig. 2D).
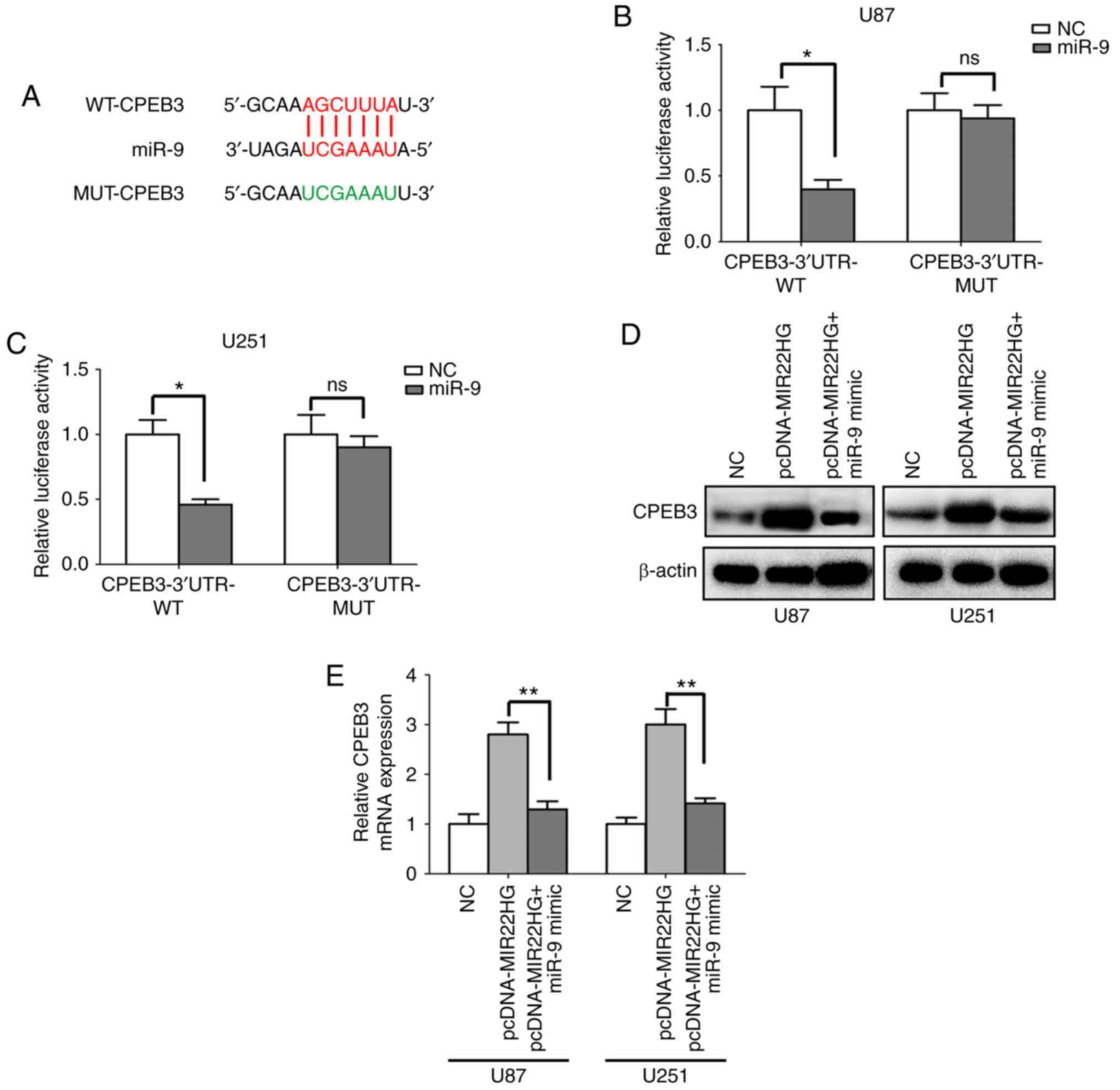
CPEB3 is the target molecule of miR-9
in the glioma cell lines
The present study revealed that miR-9 is the direct
target of MIR22HG in the glioma cell lines; however, the potential
regulatory mechanisms remain unclear. The target gene of miR-9 was
predicted using bioinformatics tools (TargetScan) (Fig. 3A). CPEB3 was identified as the
downstream target of miR-9. The luciferase reporter assay showed
that the luciferase activity was significantly decreased in
CPEB3-WT, while there was no significant difference for CPEB3-MUT
(Fig. 3B and C). Then, the mRNA and
protein expression levels of CPEB3 were determined using RT-qPCR
and western blot analysis, respectively. Transfection with MIR22HG
overexpression vector increased the mRNA and protein expression
level of CPEB3, while the mRNA and protein expression levels of
CPEB3 was reduced when the U87 and U251 cell lines were
co-transfected with miR-9 mimic and MIR22HG overexpression vector
(Fig. 3D and E). In summary, CPEB3
was a direct target of miR-9, and MIR22HG regulated the expression
of CPEB3 via miR-9.
MIR22HG regulates the proliferation,
invasion and migration of the glioma cells through miR-9
It has been reported that MIR22HG is a tumor
suppressor in a variety of different cancer types (33,41,42).
However, the role of MIR22HG in glioma is unknown. To investigate
the function of MIR22HG in the glioma cell lines, MIR22HG
overexpressing vector was transfected into the cell lines with or
without miR-9 mimic. The CCK-8 assay showed that upregulated
MIR22HG expression restricted glioma cell proliferation, but the
miR-9 mimic reversed the inhibitory effects of MIR22HG
overexpression (Fig. 4A and B). In
addition, the overexpression of MIR22HG repressed glioma cell
migration and invasion and the effects were abrogated by
transfection with the miR-9 mimic (Fig.
4C-I). In summary, the aforementioned data suggested that
MIR22HG regulated the proliferation, invasion and migration of
glioma cells through miR-9.
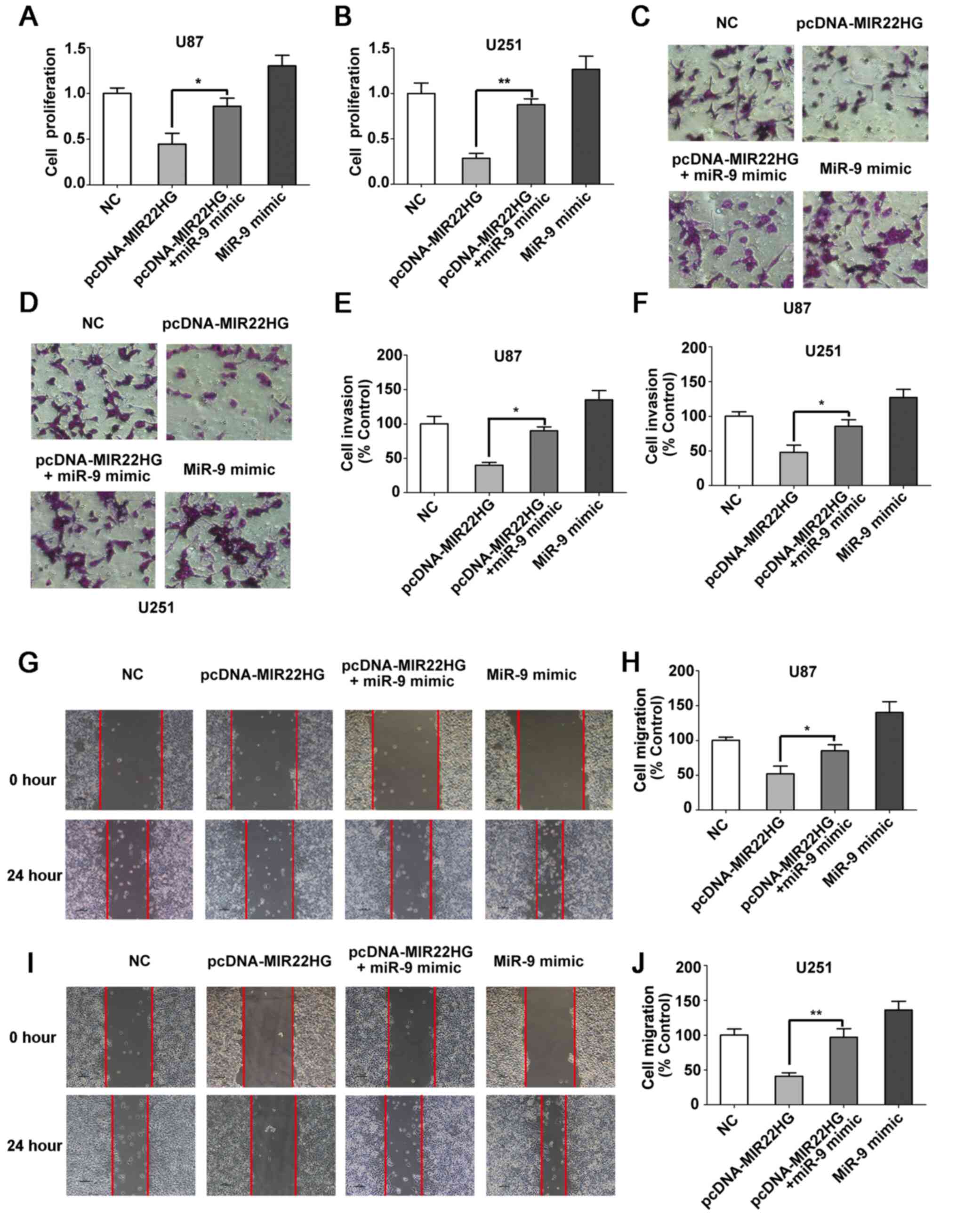 | Figure 4.MIR22HG regulates the proliferation,
invasion and migration of the glioma cells through miR-9. Cell
proliferation was detected in (A) U87 and (B) U251 cell lines
transfected with NC, pcDNA-MIR22HG, pcDNA-MIR22HG + miR-9 mimic, or
with miR-9 mimic using Cell Counting Kit-8 assay. The cell
viability of NC group was set as the control. The data was compared
using one-way ANOVA, followed by Tukey's post hoc test. n=3.
*P<0.05. **P<0.01. Cell invasion was examined in the (C) U87
and (D) U251 cell lines transfected with NC, pcDNA-MIR22HG,
pcDNA-MIR22HG+miR-9 mimic, or miR-9 mimic using Matrigel assay, and
the data was subsequently quantified in (E) U87 and (F) U251 cell
lines. The data was compared using one-way ANOVA, followed by
Tukey's post hoc test. n=3. *P<0.05. Cell migration was
determined in (G, H, I and J) U87 and U251 cell lines transfected
with NC, pcDNA-MIR22HG, pcDNA-MIR22HG + miR-9 mimic, or miR-9
mimic. The data was compared using one-way ANOVA, followed by
Tukey's post hoc test. n=3. *P<0.05. **P<0.01. miR, microRNA;
WT, wild-type; MUT, mutant; NC, negative control. |
Discussion
Glioma is a malignant brain tumor with high
incidence rates, that can immensely impair public health (43). Despite improvements in glioma
therapy, the 5-year survival rate of patients is <6% in 2016
(44). Therefore, clarifying the
detailed molecular mechanisms of glioma would assist with
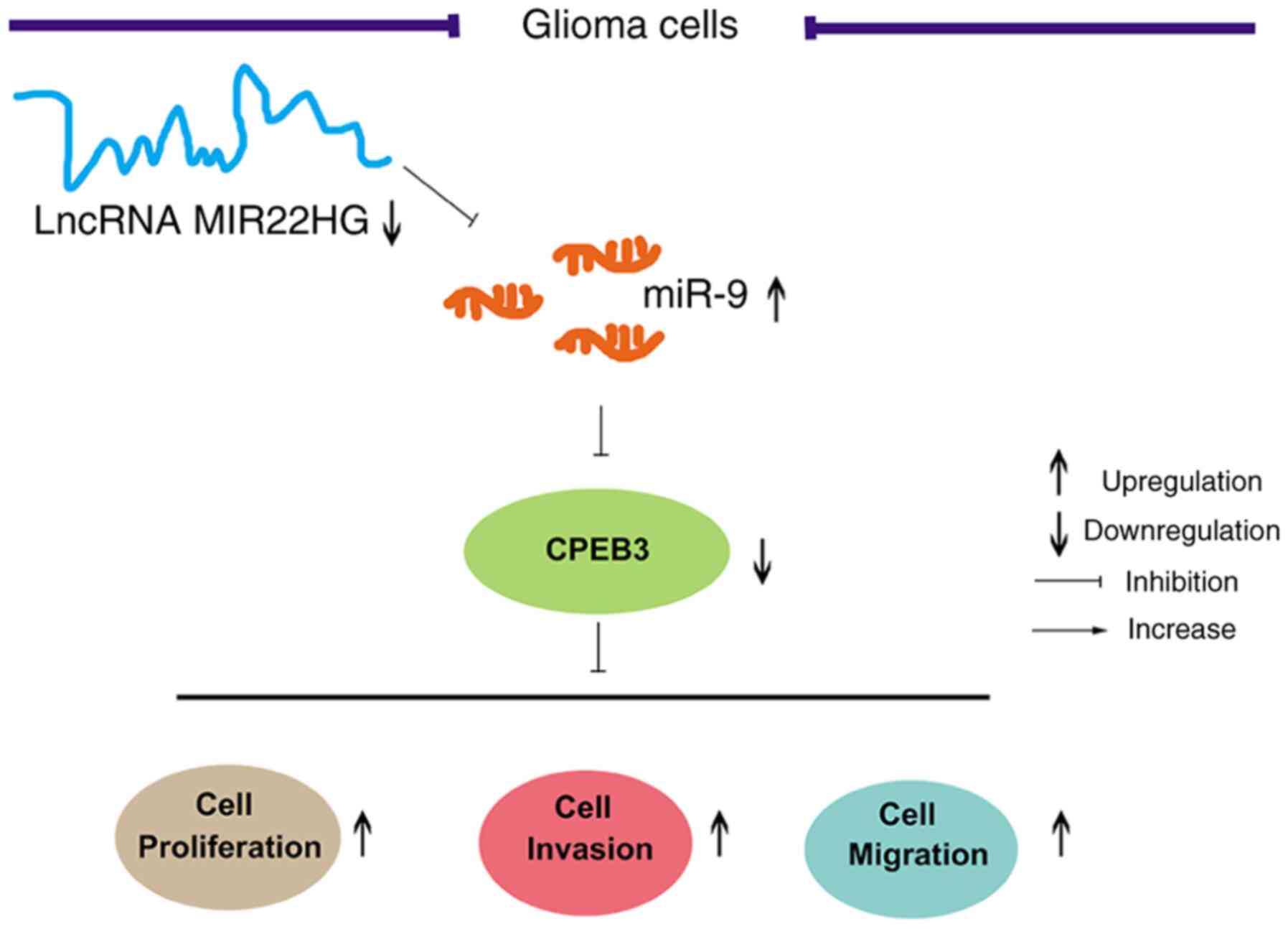
discovering novel therapeutic targets. As showed in Fig. 5, it was found that MIR22HG expression
level was decreased while miR-9 expression level was increased in
glioma tissues and cell lines. Then, MIR22HG was proven to directly
bind to and negatively regulate miR-9 in the glioma cell lines.
Furthermore, it was found that CPEB3 was the target molecule of
miR-9. Functionally, MIR22HG negatively regulated glioma cell
proliferation, invasion and migration by modulating the miR-9/CPEB3
axis.
In the present study, we identified that MIR22HG
expression level was significantly decreased in human glioma
tissues compared with that in adjacent normal tissues. In contrast,
the expression level of miR-9 was increased. It has been reported
that the lncRNA MIR22HG functions as an antioncogene (45) and the expression level was decreased
in multiple types of cancer. For example, the expression level of
the lncRNA MIR22HG was decreased in lung cancer tissues, and was
associated with poor patient survival (21). In thyroid cancer, decreased MIR22HG
expression level was associated with the degree of malignancy of
tumors, which indicated that MIR22HG could be a new diagnostic
marker for thyroid cancer (46).
miR-9 has been found to be an oncogene or a tumor suppressor in
several types of cancer. In prostate cancer cell lines, miR-9
inhibitor transfection could suppress cell proliferation, invasion
and migration (47). Furthermore, a
previous research showed that miR-9 expression level was increased
in glioma tissues and cell lines (48).
Then, the association between MIR22HG and miR-9 was
investigated. The bioinformatics database StarBase v2.0 showed that
MIR22HG contained a potential binding site for miR-9. A luciferase
reporter assay was used to confirm that MIR22HG directly regulated
miR-9 in the glioma cell lines. Next, we investigated the target
gene of miR-9 using TargetScan. CPEB3 was found to be the
downstream target of miR-9 and the luciferase activity confirmed
that CPEB3 was the target molecule of miR-9 in the glioma cell
lines. Moreover, MIR22HG was proved to regulate the and protein
expression level of CPEB3 via miR-9. CPEB3, a member of the CPEB
family, was discovered to monitor cell translation by adjusting
cytoplasmic polyadenylation. It has also been reported that CPEB3
was aberrantly expressed in several types of cancer (23,30,49). In
a study of ovarian cancer cells, miR-301b-3p increased cell
invasion and migration by targeting CPEB3 (50). In addition, CPEB3 plays a crucial
role in glioma pathogenesis. The protein level of CPEB3 was
increased by the lncRNA HCG11, which inhibited glioma cell
proliferation and arrested the cell cycle (30). Thus, MIR22HG and miR-9 may regulate
the development of glioma via targeting CPEB3.
However, the effect of MIR22HG in glioma remains
unclear. To investigate the effect of MIR22HG in the glioma cell
lines, a CCK-8 assay was used to measure cell proliferation. The
results showed that upregulated MIR22HG expression restricted
glioma cell proliferation, but miR-9 mimic reversed the inhibitory
effects of MIR22HG. In addition, the overexpression of MIR22HG
reduced cell migration and invasion. The MIR22HG-induced effects
were abrogated by transfection with miR-9 mimic. It has been
demonstrated that MIR22HG interacts with multiple signaling
pathways and affects the progression of multiple tumors, including
thyroid cancer, gastric cancer, and hepatocellular carcinoma
(19,46,51). The
results from the present study indicated that MIR22HG inhibited the
progression of glioma via negatively modulating the downstream
target miR-9.
MIR22HG was decreased in glioma tissue and cell
lines in the present study, and that the miR-9/CPEB3 axis was the
downstream pathway. However, little is known how MIR22HG expression
level is reduced in glioma. Circular (circ)RNAs have been reported
to regulate the expression of lncRNAs and function as a diagnostic
or prognostic biomarker in laryngeal squamous cell carcinoma and
bladder cancer (52,53). In glioma, it is unknown whether
MIR22HG is modulated by circRNAs, and it also remains unknown which
circRNA directly regulates MIR22HG. This will be investigated in
future studies.
In summary, it was found that MIR22HG expression
level and CPEB3 were reduced while miR-9 expression level was
increased in glioma tissues and cell lines. Then, MIR22HG was found
to negatively modulate miR-9 in the glioma cell lines. In addition,
CPEB3 was proven to be the target molecule of miR-9 in the glioma
cell lines. In addition, upregulated MIR22HG expression level
inhibited the proliferation, invasion and migration of glioma cells
by inhibiting miR-9 expression level. The present study indicated
that MIR22HG regulated the proliferation, invasion and migration of
glioma cells through miR-9 and that CPEB3 is involved in this
mechanism.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by National Natural Science
Foundation Project (grant no. 81571656).
Availability of data and materials
All the data generated and/or analyzed during the
present study are included in this published article.
Author's contributions
XZ and YH conceived and designed the experiments.
YH, HN, LY, TM, MM, BT and SG performed the experiments. YH, HN and
LY collected and analyzed the data. XZ and YH interpreted the
results and wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
This study was performed in accordance with standard
guide-lines and was approved by the Ethics Committee of The Second
Affiliated Hospital of the Fourth Military Medical University. All
patients provided written informed consent prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lv D, Li Y, Zhang W, Alvarez AA, Song L,
Tang J, Gao WQ, Hu B, Cheng SY and Feng H: TRIM24 is an oncogenic
transcriptional co-activator of STAT3 in glioblastoma. Nat Commun.
8:14542017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang
Z, Li Z, Zhang L and Liu Y: Inhibition of TDP43-mediated
SNHG12-miR-195-SOX5 feedback loop impeded malignant biological
behaviors of glioma cells. Mol Ther Nucleic Acids. 10:142–158.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou S, Ding F and Gu X: Non-coding RNAs
as emerging regulators of neural injury responses and regeneration.
Neurosci Bull. 32:253–264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Meir EG, Hadjipanayis CG, Norden AD,
Shu HK, Wen PY and Olson JJ: Exciting new advances in
neuro-oncology: The avenue to a cure for malignant glioma. CA
Cancer J Clin. 60:166–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan X, Curtin J, Xiong Y, Liu G,
Waschsmann-Hogiu S, Farkas DL, Black KL and Yu JS: Isolation of
cancer stem cells from adult glioblastoma multiforme. Oncogene.
23:9392–9400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lino MM, Merlo A and Boulay JL: Notch
signaling in glioblastoma: A developmental drug target? BMC Med.
8:722010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schwartzbaum JA, Fisher JL, Aldape KD and
Wrensch M: Epidemiology and molecular pathology of glioma. Nat Clin
Pract Neurol. 2:494–503, 1–516. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ostrom QT, Gittleman H, Stetson L, Virk SM
and Barnholtz-Sloan JS: Epidemiology of gliomas. Cancer Treat Res.
163:1–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y and Lee CG: MicroRNA and
cancer-focus on apoptosis. J Cell Mol Med. 13:12–23. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang X, Yu H, Sun W, Kong J, Zhang L, Tang
J, Wang J, Xu E, Lai M and Zhang H: The long non-coding RNA CYTOR
drives colorectal cancer progression by interacting with NCL and
Sam68. Mol Cancer. 17:1102018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu J, Dong G, Shi H, Zhang J, Ning Z, Bao
X, Liu C, Hu J, Liu M and Xiong B: LncRNA MIR503HG inhibits cell
migration and invasion via miR-103/OLFM4 axis in triple negative
breast cancer. J Cell Mol Med. 23:4738–4745. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang S and Guo W: Long noncoding RNA MEG3
suppresses the growth of glioma cells by regulating the
miR965p/MTSS1 signaling pathway. Mol Med Rep. 20:4215–4225.
2019.PubMed/NCBI
|
|
16
|
Li C, Tan F, Pei Q, Zhou Z, Zhou Y, Zhang
L, Wang D and Pei H: Non-coding RNA MFI2-AS1 promotes colorectal
cancer cell proliferation, migration and invasion through
miR-574-5p/MYCBP axis. Cell Prolif. 52:e126322019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao X, He M, Wan D, Ye Y, He Y, Han L,
Guo M, Huang Y, Qin W, Wang MW, et al: The minimum LOH region
defined on chromosome 17p13.3 in human hepatocellular carcinoma
with gene content analysis. Cancer Lett. 190:221–232. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang DY, Zou XJ, Cao CH, Zhang T, Lei L,
Qi XL, Liu L and Wu DH: Identification and functional
characterization of long non-coding RNA MIR22HG as a tumor
suppressor for hepatocellular carcinoma. Theranostics. 8:3751–3765.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H and Wang Y: Long noncoding RNA
(lncRNA) MIR22HG suppresses gastric cancer progression through
attenuating NOTCH2 signaling. Med Sci Monit. 25:656–665. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui Z, An X, Li J, Liu Q and Liu W: LncRNA
MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression
and inhibits endometrial carcinoma cells proliferation. Biomed
Pharmacother. 104:223–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su W, Feng S, Chen X, Yang X, Mao R, Guo
C, Wang Z, Thomas DG, Lin J, Reddy RM, et al: Silencing of long
noncoding RNA MIR22HG triggers cell survival/death signaling via
oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res.
78:3207–3219. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang YS, Kan MC, Lin CL and Richter JD:
CPEB3 and CPEB4 in neurons: Analysis of RNA-binding specificity and
translational control of AMPA receptor GluR2 mRNA. Embo J.
25:4865–4876. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Wang Y, Chen B, Shen X and Li W:
Effects of lidocaine-mediated CPEB3 upregulation in human
hepatocellular carcinoma cell proliferation in vitro. Biomed Res
Int. 2018:84031572018.PubMed/NCBI
|
|
24
|
Theis M, Si K and Kandel ER: Two
previously undescribed members of the mouse CPEB family of genes
and their inducible expression in the principal cell layers of the
hippocampus. Proc Natl Acad Sci USA. 100:9602–9607. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chao HW, Tsai LY, Lu YL, Lin PY, Huang WH,
Chou HJ, Lu WH, Lin HC, Lee PT and Huang YS: Deletion of CPEB3
enhances hippocampus-dependent memory via increasing expressions of
PSD95 and NMDA receptors. J Neurosci. 33:17008–17022. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Berger-Sweeney J, Zearfoss NR and Richter
JD: Reduced extinction of hippocampal-dependent memories in CPEB
knockout mice. Learn Mem. 13:4–7. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Yu R and Li L: LINC00641 hinders
the progression of cervical cancer by targeting miR-378a-3p/CPEB3.
J Gene Med. 22:e32122020. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Waku T, Katayama H, Hiraoka M, Hatanaka A,
Nakamura N, Tanaka Y, Tamura N, Watanabe A and Kobayashi A: NFE2L1
and NFE2L3 complementarily maintain basal proteasome activity in
cancer cells through CPEB3-mediated translational repression. Mol
Cell Biol. 40:e00010–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin H, Guo Q, Lu S, Chen J, Li X, Gong M,
Tang L and Wen J: LncRNA SUMO1P3 promotes proliferation and
inhibits apoptosis in colorectal cancer by epigenetically silencing
CPEB3. Biochem Biophys Res Commun. 511:239–245. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Y, Bao C, Zhang X, Lin X, Huang H and
Wang Z: Long non-coding RNA HCG11 modulates glioma progression
through cooperating with miR-496/CPEB3 axis. Cell Prolif.
52:e126152019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du P, Liao Y, Zhao H, Zhang J, MuyitiKerem
u and Mu K: ANXA2P2/miR-9/LDHA axis regulates Warburg effect and
affects glioblastoma proliferation and apoptosis. Cell Signal.
74:1097182020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shen H, Weng XD, Yang D, Wang L and Liu
XH: Long noncoding RNA MIR22HG is down-regulated in prostate
cancer. Math Biosci Eng. 17:1776–1786. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu J, Shao T, Song M, Xie Y, Zhou J, Yin
J, Ding N, Zou H, Li Y and Zhang J: MIR22HG acts as a tumor
suppressor via TGFbeta/SMAD signaling and facilitates immunotherapy
in colorectal cancer. Mol Cancer. 19:512020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Han M, Wang S, Fritah S, Wang X, Zhou W,
Yang N, Ni S, Huang B, Chen A, Li G, et al: Interfering with long
non-coding RNA MIR22HG processing inhibits glioblastoma progression
through suppression of Wnt/beta-catenin signalling. Brain.
143:512–530. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wen PY and Huse JT: 2016 World Health
Organization Classification of central nervous system tumors.
Continuum (Minneap Minn). 23:1531–1547. 2017.PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Inada K, Okoshi Y, Cho-Isoda Y, Ishiguro
S, Suzuki H, Oki A, Tamaki Y, Shimazui T, Saito H, Hori M, et al:
Endogenous reference RNAs for microRNA quantitation in
formalin-fixed, paraffin-embedded lymph node tissue. Sci Rep.
8:59182018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu YL, Liu Y, Cai RP, He SR, Dai RX, Yang
XH, Kong BH, Qin ZB and Su Q: Long non-coding RNA CASC7 is
associated with the pathogenesis of heart failure via modulating
the expression of miR-30c. J Cell Mol Med. 24:11500–11511. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu B, Liu Y, Zhou M, Yao S, Bian Z, Liu
D, Fei B, Yin Y and Huang Z: Comprehensive ceRNA network analysis
and experimental studies identify an IGF2-AS/miR-150/IGF2
regulatory axis in colorectal cancer. Pathol Res Pract.
216:1531042020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu T, Wang S, Wang L, Zhang W, Chen W, Lv
X, Li Y, Hussain Z and Sun W: Long noncoding RNA (lncRNA) CTTN-IT1
elevates skeletal muscle satellite cell proliferation and
differentiation by acting as ceRNA for YAP1 through absorbing
miR-29a in Hu Sheep. Front Genet. 11:8432020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shu J and Wang D: Functional
characterization of the long noncoding RNA MIR22HG as a tumour
suppressor in cervical cancer by targeting IGF2BP2. Eur Rev Med
Pharmacol Sci. 24:7953–7962. 2020.PubMed/NCBI
|
|
42
|
Tang Q, Jiang X, Ma S, Wang L, Li R and Ma
J: MIR22HG regulates miR-486/PTEN axis in bladder cancer to promote
cell proliferation. Biosci Rep. 40:BSR201939912020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lapointe S, Perry A and Butowski NA:
Primary brain tumours in adults. Lancet. 392:432–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shergalis A, Bankhead AR III, Luesakul U,
Muangsin N and Neamati N: Current challenges and opportunities in
treating glioblastoma. Pharmacol Rev. 70:412–445. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen H, Ali M, Ruben A, Stelmakh D and Pak
M: E2F6-mediated downregulation of MIR22HG facilitates the
progression of laryngocarcinoma by targeting the miR-5000-3p/FBXW7
Axis. Mol Cell Biol. 40:e00496–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qin L, Luo JZ, Tang XL and Han CG:
Identification of long noncoding RNA MIR22HG as a Novel biomarker
in thyroid cancer. Pathol Oncol Res. 25:703–710. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen L, Hu W, Li G, Guo Y, Wan Z and Yu J:
Inhibition of miR-9-5p suppresses prostate cancer progress by
targeting StarD13. Cell Mol Biol Lett. 24:202019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen X, Yang F, Zhang T, Wang W, Xi W, Li
Y, Zhang D, Huo Y, Zhang J, Yang A and Wang T: MiR-9 promotes
tumorigenesis and angiogenesis and is activated by MYC and OCT4 in
human glioma. J Exp Clin Cancer Res. 38:992019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhong Q, Fang Y, Lai Q, Wang S, He C, Li
A, Liu S and Yan Q: CPEB3 inhibits epithelial-mesenchymal
transition by disrupting the crosstalk between colorectal cancer
cells and tumor-associated macrophages via IL-6R/STAT3 signaling. J
Exp Clin Cancer Res. 39:1322020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu F, Zhang G, Lv S, Wen X and Liu P:
miRNA-301b-3p accelerates migration and invasion of high-grade
ovarian serous tumor via targeting CPEB3/EGFR axis. J Cell Biochem.
120:12618–12627. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu Y, Zhou Y, Huan L, Xu L, Shen M, Huang
S and Liang L: LncRNA MIR22HG inhibits growth, migration and
invasion through regulating the miR-10a-5p/NCOR2 axis in
hepatocellular carcinoma cells. Cancer Sci. 110:973–984. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lin G, Sheng H, Xie H, Zheng Q, Shen Y,
Shi G and Ye D: circLPAR1 is a novel biomarker of prognosis for
muscle-invasive bladder cancer with invasion and metastasis by
miR-762. Oncol Lett. 17:3537–3547. 2019.PubMed/NCBI
|
|
53
|
Zhao R, Li FQ, Tian LL, Shang DS, Guo Y,
Zhang JR and Liu M: Comprehensive analysis of the whole coding and
non-coding RNA transcriptome expression profiles and construction
of the circRNA-lncRNA co-regulated ceRNA network in laryngeal
squamous cell carcinoma. Funct Integr Genomics. 19:109–121. 2019.
View Article : Google Scholar : PubMed/NCBI
|