Introduction
Oral squamous cell carcinoma (OSCC) is highly
invasive locally and often leads to cervical lymph node metastasis,
with distant metastasis being less frequent (1,2). Factors
related to these properties of OSCC (3,4) include
oncogene products (5–9), mutated tumor suppressor gene products
(10–12), adhesion molecules (13–15),
matrix metalloproteinases (16–18) and
cell cycle proteins (19–21). However, a molecule clearly associated
with the aggressiveness of OSCC has not yet been identified.
Similarly, histopathological features can be linked to superficial
properties of OSCC, but cannot specifically explain its invasive
and metastatic potentials (22).
Given this background, we hypothesized that the identification of
the origin of the tumor or tumor stem cells may be important in
predicting the aggressiveness of OSCC.
Following advances in regenerative medicine, several
somatic stem cells have been identified and the importance in
regenerating certain organs has been recognized. It has been
reported that hepatic and endothelial stem cells are derived from
the bone marrow (BM) and are present in circulating blood (23,24).
Furthermore, BM stem cells may be involved in the regeneration of
the gastrointestinal epithelium (25), and gastric cancer can develop from
these cells (26). Tamai et
al found that BM stem cells (mesenchymal stem cells) are
involved in epithelial repair of skin lesions in patients with
hereditary epidermolysis bullosa (27). It is well known that BM stem cells
include hematopoietic and mesenchymal stem cells.
Multilineage-differentiating stress-enduring (Muse) cells, a small
population of BM mesenchymal stem cells in circulating blood, can
also differentiate into several types of cells, including
epithelial cells (28).
The present study attempted to establish a novel
diagnostic criterion and a method that can determine the
fundamental malignancy of the tumor cells without being confused by
masquerade images or minor genetic abnormalities. The present study
was based on the hypothesis that there are three origins of tumor
cells in OSCC: Epithelial stem cells, oral tissue stem cells from
the salivary gland and BM stem cells. We also hypothesized that
carcinoma derived from less-differentiated stem cells has a higher
malignancy. It was shown that OSCC derived from stem cells in the
salivary gland had a significantly higher metastatic potential and
poorer prognosis among oral carcinomas histopathologically
diagnosed as SCC (29). In addition,
a case of a male patient who underwent hematopoietic stem cell
transplantation (HSCT) from his sister for treating leukemia, and
developed an oral lichenoid lesion due to chronic graft versus host
disease following HSCT was reported. It was demonstrated that oral
lichenoid lesion epithelial cells from the patient originated from
donor BM stem cells, and these cells developed SCC on the
mandibular gingiva (30).
In the present study, sex chromosome analysis was
performed in patients with OSCC developed following HSCT from the
opposite gender, in order to examine whether OSCC originates from
BM stem cells. Gene expression patterns in patients with possibly
BM stem cell-derived OSCC were compared with those in patients with
normally developed OSCC by microarray analysis, in order to examine
whether OSCC with a possible BM origin shows a specific pattern of
the gene expression.
Patients and methods
Patients
Sex chromosome analysis
Six patients (5 males and 1 female) who developed
graft versus host disease (GvHD) in the oral mucosa following HSCT
from a donor of the opposite sex and then developed OSCC were
examined by fluorescence in situ hybridization (FISH) and
G-band staining of sex chromosomes (Tables I and II). The patients (cases 1 to 6) all
presented with a lichenoid lesion, a symptom of chronic GvHD, in
the oral mucosa and subsequently developed OSCC 5 to 18 years after
HSCT. All patients underwent surgery for OSCC and 4 patients (cases
1, 2, 3 and 6) died from primary disease or multiple metastases.
Case 7 was a male patient with OSCC who did not undergo HSCT. Three
patients (cases 8, 9, 10; 2 males, 1 female) presented with a
lichenoid lesion in the oral mucosa following HSCT from an
opposite-sex donor, and were also examined by sex chromosome FISH
(Tables I and II). Two male patients with normal labial
mucosa, who did not undergo HSCT, were also subjected to this
analysis (Table I). All of the above
patients were treated between 2006 and 2018 at the Dokkyo
University (Mibu, Japan) or Ehime University Hospitals (Toon,
Japan).
 | Table I.Clinicopathological characteristics of
patients. |
Table I.
Clinicopathological characteristics of
patients.
| Case no. | Sex | Age | Smoking/Alcohol | Location | Histology | TNM
classification | Tumor cell
differentiation | Anneroth grade | Y-K mode of
invasion | p16 (HPV status) | Prognosis |
|---|
| 1-1 | M | 58 | Never/No | Lower gingiva | SCC | T4aN0M0 | 1 | II | 3 | Negative | Dead |
| 1-2 | M | 65 | Never/No | Esophageal | SCC | T1N0M0 | 2 | II | 2 | Negative | Dead |
| 2 | F | 16 | Never/No | Tongue | SCC | T1N0M0 | 1 | III | 4D | Positive | Dead |
| 3-1 | M | 61 | Ex/No | Tongue | SCC | T2N0M0 | 2 | III | 3 | Negative | Dead |
| 3-2 | M | 62 | Ex/No | Lower gingiva | SCC | T4aN0M0 | 1 | III | 4D | Negative | Dead |
| 4 | M | 50 | Current/Daily | Buccal mucosa | SCC | T3N0M0 | 2 | I | 3 | NE | Alive |
| 5 | M | 30 |
Never/Occasionally | Tongue | SCC | T2N0M0 | 2 | II | 3 | Negative | Alive |
| 6 | M | 41 |
Never/Occasionally | Tongue | SCC | rT0N2cM0 | 1 | II | 4D | Negative | Dead |
| 7 | M | 90 | Current/Daily | Upper gingiva | SCC | T3N0M0 | 3 | III | 3 | Negative | Unknown |
| 8 | F | 38 | Never/No | Tongue | OPMD | NA | NA | NA | NA | NA | NA |
| 9 | M | 17 | Never/No | Buccal | OPMD | NA | NA | NA | NA | NA | NA |
| 10-1 | M | 72 | Never/Daily | Upper gingiva | OPMD | NA | NA | NA | NA | NA | NA |
| 10-2 | M | 72 | Never/Daily | Upper gingiva | OPMD | NA | NA | NA | NA | NA | NA |
 | Table II.Sex chromosomal analysis via XY FISH
in patients with oral SCC who were receiving HSCT. |
Table II.
Sex chromosomal analysis via XY FISH
in patients with oral SCC who were receiving HSCT.
|
|
|
|
|
|
| Oral SCC or
OPMD | Adjacent
normal-appeared oral squamous epithelium |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Case no. | Sex of
donor/recipient | Age at diagnosis
for oral SCC/Age at transplantation | Type of
hematological malignancy | Type of HSCT | Severity of chronic
GvHD | X only (%) | Y containing
(%) | X only (%) | Y containing
(%) |
|---|
| 1-1 | F/M | 58/53 | MLL | Allo-PBSCT | Mild | 96.4 | 3 | 19 | 79 |
| 1-2 | F/M | 65/53 | MLL | Allo-PBSCT | Mild | 86 | 14 | NE | NE |
| 2 | M/F | 16/4, 9 | ALL | UR-BMT | Severe | 100 | 0 | NE | NE |
| 3-1 | F/M | 61/44 | MDS | UR-BMT | Moderate | 66.5 | 33.5 | 3.7 | 96.3 |
| 3-2 | F/M | 62/44 | MDS | UR-BMT | Moderate | 5.7b | 94.3 | NE | NE |
| 4 | F/M | 50/43 | CTCL | Allo-BMT | Moderate | 63.3 | 36.7 | NE | NE |
| 5 | F/M | 30/14 | MDS | Allo-BMT | Mild | 11.7 | 88.3 | 19.8 | 80.2 |
| 6a | F/M | 41/32 | ALL | CBT | Moderate | 5 | 95 | NE | NE |
| 7 | NA | 90/NA | NA | NA | NA | 13 | 87 | NE | NE |
| 8 | M/F | 38/10, 13 | AML | Allo-BMT | Moderate | 89.3 | 10.7 | 90.3 | 9.7 |
| 9 | F/M | 17/16 | SAA | Allo-BMT | Moderate | 7.4 | 92.6 | 9 | 91 |
| 10-1 | F/M | 72/59 | MDS | Allo-BMT | Moderate | 5.2 | 94.8 | NE | NE |
| 10-2 | F/M | 72/59 | MDS | Allo-BMT | Moderate | 6.7 | 93.3 | NE | NE |
| 11 | NA | 10/NA | NA | NA | NA | NA | NA | 0 | 100 |
| 12 | NA | 45/NA | NA | NA | NA | NA | NA | 0.9 | 99.1 |
Assessment by microarray analysis
A total of 16 samples from 14 patients with oral SCC
(11 men and 3 women) were subjected to microarray analysis. One
patient (case 3) had two independent tumors in the oral cavity. Two
samples from different part of 1 tumor of 1 patient (case 17) was
also analyzed. Each tumor was surgically resected at the Dokkyo
Medical University and Ehime University Hospitals between 2017 and
2018. None of the patients had received previous radiotherapy or
chemotherapy. The clinical characteristics of these patients are
shown in Table III.
 | Table III.Clinico-pathological characteristics
of the patients with oral SCC for microarray and clustering
analysis. |
Table III.
Clinico-pathological characteristics
of the patients with oral SCC for microarray and clustering
analysis.
| Case no. | Sex | Age at
diagnosis |
Smoking/Alcohol | Location | Histology | cTNM | pN | Tumor cell
differentiation | Anneroth grade | Y-K mode of
invasion | p16 (HPV
status) | Prognosis |
|---|
| 3-2 | M | 62 | Ex/No | Gingiva | SCC | T4aN0M0 | NDNP | 2 | III | 4D | Negative | Dead |
| 3-3 | M | 63 | Ex/No | Gingiva | SCC | T4aN0M0 | NDNP | 2 | III | 4D | Negative | Dead |
| 4 | M | 50 | Current/Daily | Buccal Mucosa | SCC | T3N0M0 | NDNP | 2 | I | 3 | NE | Alive |
| 13 | F | 55 | Ex/No | Tongue | SCC | T4bN3bM0 | NDNP
(N3b)a | 1 | I | 2 | Negative | Alive |
| 14 | M | 60 | Ex/No | Tongue | SCC | T2N1M0 | N1 | 1 | I | 3 | Negative | Alive |
| 15 | F | 83 | Never/Daily | Buccal Mucosa | SCC | T4aN2bM0 | N0 | 2 | I | 2 | Positive | Alive |
| 16 | M | 87 |
Ex/Occasionally | Tongue | SCC | T2N1M1 | NDNP | 1 | II | 3 | Negative | Unknown |
| 17-1 | M | 65 | Current/Daily | Tongue | SCC | T3N3bM0 | N3b | 1 | II | 4C | Positive | Alive |
| 17-2 | M | 65 | Current/Daily | Tongue | SCC | T3N3bM0 | N3b | 1 | II | 4C | Positive | Alive |
| 18 | M | 71 | Current/Daily | Tongue | SCC | T4aN2bM0 | N0 | 1 | II | 4C | Negative | Alive |
| 19 | F | 86 | Never/No | Gingiva | SCC | T4aN1M0 | N0 | 1 | II | 4C | Negative | Alive |
| 20 | M | 74 | Ex/Daily | Gingiva | SCC | T4bN2bM0 | N0 | 2 | III | 3 | Negative | Alive |
| 21 | M | 69 | Current/Daily | Tongue | SCC | T2N1M0 | N3b | 2 | III | 4D | Negative | Dead |
| 22 | M | 61 | Ex/No | Tongue | SCC | T4aN1M0 | N0 | 1 | III | 4C | Negative | Alive |
| 23 | M | 60 |
Never/Occasionally | Buccal Mucosa | SCC | T3N1M0 | N0 | 1 | II | 3 | Negative | Alive |
| 24 | M | 59 | Ex/Daily | Tongue | SCC | T3N1M0 | N1 | 1 | II | 4C | Negative | Alive |
Histopathological examination
Tissue samples were fixed in formalin and embedded
in paraffin. Sections of 4-µm were stained with hematoxylin and
eosin, and reviewed by experienced pathologists for
histopathological diagnosis. Tumor cell differentiation (WHO
classification) (31), Anneroth
grade (32), Y-K mode of invasion
(22) and pathological lymph node
metastasis were assessed. The histopathologic features of all
patients in the study are shown in Tables I and III.
FISH analysis
Sections (5-µm) were deparaffinized and treated with
0.5% pepsin solution and 0.1 N HCl at 37°C for 12–19 min. Following
neutralization with PBS, the solution was washed with distilled
water and dried. Dig-labeled human X chromosome and FITC-labeled Y
chromosome (both from Chromosome Science Labo, Inc., Hokkaido,
Japan) probe solutions were added, and the sections and probes were
simultaneously denatured on a hot plate at 90°C for 10 min and then
hybridized at 37°C overnight. The sections were stringently washed
with 50% formaldehyde and 2X standard saline citrate (SSC) and 1X
SSC, and X-chromosome signals were detected using anti-Dig-Cy3
(Chromosome Science Labo, Inc.). The sections were counterstained
with DAPI (Merck KGaA) and mounted with an antifade mounting medium
(MountMed; Chromosome Science Labo, Inc.). A CW-4000 cytogenetic
workstation (Leica Microsystems, Inc.) was used to detect the FISH
signals and analyze the data. An objective lens (magnification,
×40) was used for imaging. More than 100 cells were counted for
both X and Y signals in one slide.
Microdissection PCR
Approximately 20 10-µm frozen sections were prepared
from a frozen specimen, and laser capture microdissection was
performed (Fig. 1A) to collect
cancer cells with as little mixed normal epithelia, hemocytes or
stromal tissues as possible. DNA was extracted using a QIAamp Fast
DNA Tissue kit (Qiagen AB). Amelogenin genes in both X and Y
chromosomes (AMELX and AMELY) were simultaneously examined by PCR
using the following primers: 5′-CCCTGGGCTCTGTAAAGAATAGTG-3′ forward
and 5′-ATCAGAGCTTAAACTGGGAAGCTG-3′ reverse (29). PCR products were separated in a
polyacrylamide gel, and sex chromosome patterns were analyzed based
on the different sizes of amplified bands. Sequences of 106 and 112
bp were amplified from X and Y chromosomes, respectively.
Microarray analysis
Microarray analysis was performed as follows. Total
RNA was extracted by lysing the tissues with Isogen (Nippon Gene)
following homogenization with a TissueLyser (Qiagen AB). Total RNA
(500 ng) was used to generate biotin-labeled cRNA using a
GeneChip® 3′ IVT PLUS Reagent kit (Thermo Fisher
Scientific, Inc.). The biotin-labeled RNA was hybridized to Human
Genome U-219 Array Strips (Thermo Fisher Scientific, Inc.). After
washing and staining the strips, the signal was developed and
scanned using GeneAtlas (Thermo Fisher Scientific, Inc.). Data
analysis was conducted using GeneSpring GX 14.9.1 (Agilent
Technologies GmbH). The robust multichip average method was used
with background correction and normalization. Fold change analysis
was performed to identify genes with >3-fold differences, using
a moderated t-test (P<0.05) with Benjamini-Hochberg multiple
correction. Gene expressions of KRT8, ABCC3, GCLC, RYBP, TMEM97,
SLC1A3, IRS2, KYNU, CSAG2, CDA and CCL21 in head and neck SCCs (528
cases) from the cancer genome atlas were analyzed by OncoPrint from
cBioPortal. Microarray data have been deposited in the Gene
Expression Omnibus (GEO; experiment. no. GSE153918) database using
minimum information about microarray experiment guidelines. The
algorithm of the clustering method for the microarray data is shown
in Fig. S1.
Results
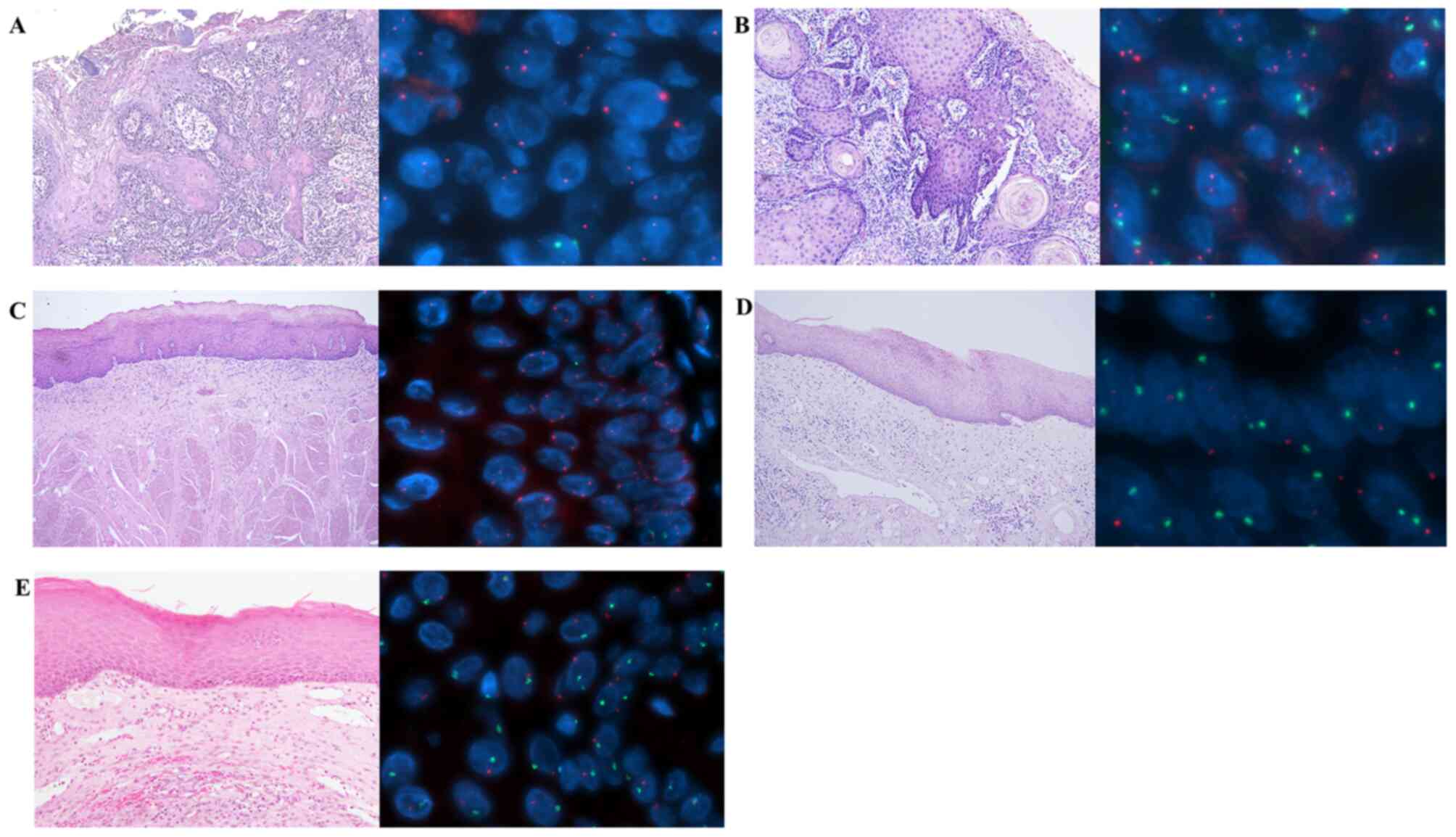
Sex chromosome analysis of OSCC
following HSCT
In three male transplanted patients (cases 1, 3 and
4), more X-only cells were identified than Y-containing cells in
the tissue (Table II), suggesting
the presence of donor-derived cells in OSCC (case 3-1; Fig. 1A). In a second primary cancer that
developed in the gingiva of case 3 (case 3-2), a small number of
cancer cells were found in the FISH specimen, but most were normal
epithelial cells. However, only ~6% were X-only cells; almost all
cancer cells within the observable FISH range were X-only, again
indicating donor-derived cells in OSCC. Distant metastasis was
observed in 2/3 patients, resulting in a poor prognosis.
Oral cancer tissues in 1 female (case 2) and 2 males
(cases 5 and 6) transplanted patients contained cells with sex
chromosomes matching the sex of the recipient (Table II), suggesting that OSCC derived
from recipient cells (case 5; Fig.
1B). In oral cancer tissues from 1 male patient (case 7), who
did not undergo HSCT, there were 13% X-only cells and 87%
Y-containing cells. In oral potentially malignant disorder (OPMD)
epithelial cells from a female patient (case 8) who underwent HSCT,
there were 10.7% Y-containing cells, suggesting that some
epithelial cells were from the donor (Fig. 1C). Adjacent normal-appearing oral
squamous tissues were compared to oral SCC or OPMD tissues in 5
cases. Some normal-appearing oral squamous cells (3.7-19.8%) were
repaired by donor BM stem cells (case 3-1; Fig. 1D). In normal oral mucosa tissues from
two male patients (cases 11 and 12) who did not undergo HSCT, there
were 99.1% (case 11; Fig. 1E) and
100% Y-containing cells, respectively. Most cells in patients with
OSCC had hypertetraploid patterns, including XXXY, XXXYY and
XXYYY.
Sex chromosome analysis in OSCC by
microdissection PCR
Blood from 1 male patient (case 1) showed an X-only
pattern in G-band staining; OSCC cells also showed the same
pattern, suggesting that tumor cells were derived from the donor
(30). The chromosome patterns in
OSCC cells and normal squamous cells were confirmed using
laser-captured microdissection PCR with AMELX and AMELY gene
amplification (Fig. 2A and B). The
results of microdissection PCR showed that OSCC tissues (lane 8) in
a male patient (case 1) and cancer tissues following LCM (lane 9)
had an X-dominant pattern; therefore, these cells were considered
to be derived from the donor (Fig.
2C).
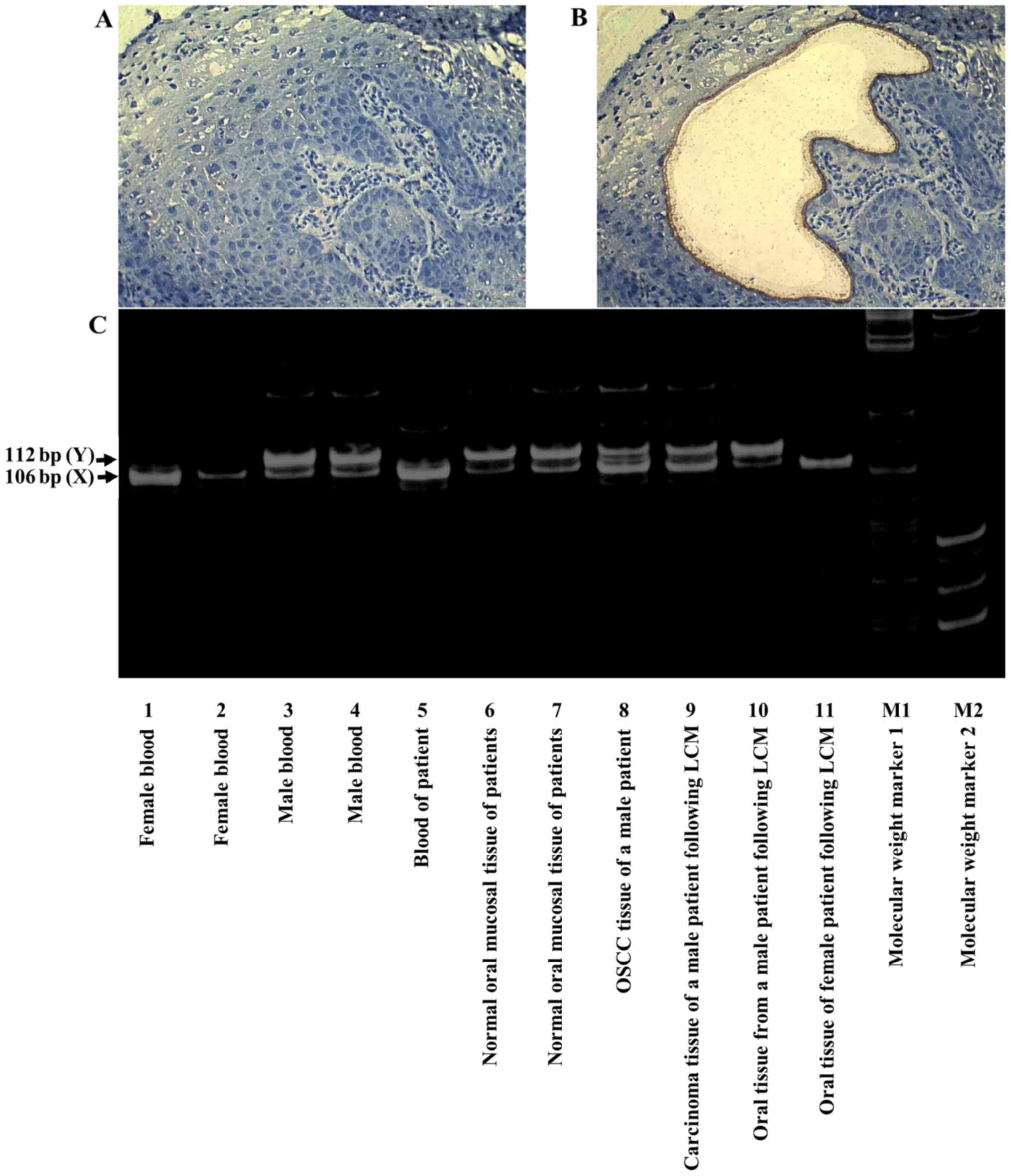 | Figure 2.X- and Y-chromosomes in tumor tissues
identified by microdissection PCR. (A) Before cancer cell
processing by LCM (magnification ×200). (B) After cancer cell
processing by LCM (magnification, ×200). (C) PCR amplification of
Amelogenin genes. Lanes 1 and 2, female blood; lanes 3 and 4, male
blood; 5, blood of patient; 6 and 7, normal oral mucosal tissue of
patients; 8, OSCC tissue of a male patient; 9, carcinoma tissue of
a male patient following LCM; 10, oral tissue from a male patient
following LCM; 11, oral tissue of female patient following LCM; M1,
molecular weight marker 1; M2, molecular weight marker 2. OSCC
(lane 8) and cancer tissues following LCM (lane 9) exhibited an
X-dominant pattern; therefore, these were considered to be
donor-derived cells. LCM, laser captured microdissection; OSCC,
oral squamous cell carcinoma. |
Microarray and cluster analysis
The gene expression patterns of patients with
possibly BM stem cell-derived OSCC (3 samples; cases 3 and 4) were
compared to those of patients with normally developed OSCC (13
samples; cases 13–24). The expression patterns in each group were
evidently different (Fig. 3). In 3
samples of possibly BM stem cell-derived OSCC, high expression
levels (>3-fold on average) were found for KRT8, ABCC3, GCLC,
RYBP, TMEM97, SLC1A3, and IRS2, and low expression levels
(<3-fold) were found for KYNU, CSAG2///CSGA3, CDA, and CCL21.
Cluster analysis based on the signal intensity detected by probes
in 14 OSCC patients (16 samples) identified a cluster in 5 samples
including possibly BM stem cell-derived OSCCs (3 samples, case 3-2,
3–3 and 4; Fig. 3). Two patients
(cases 13 and 20) were included in the cluster in addition to 3
samples from 2 patients (case 3 and 4) used for selecting probes.
These patients (case 13 and 20) had extremely aggressive tumors:
Locally advanced T4b, multiple cervical lymph node metastases and
extranodal extension N3b, or locally advanced T4b. The expressions
of the marker genes identified in this experiment were examined in
head and neck SCC (528 cases) from the cancer genome atlas
(Fig. S2). The alterations of the
individual gene expression were very limited in each case, and no
statistical correlation was identified between the expression
levels of these marker genes and prognosis.
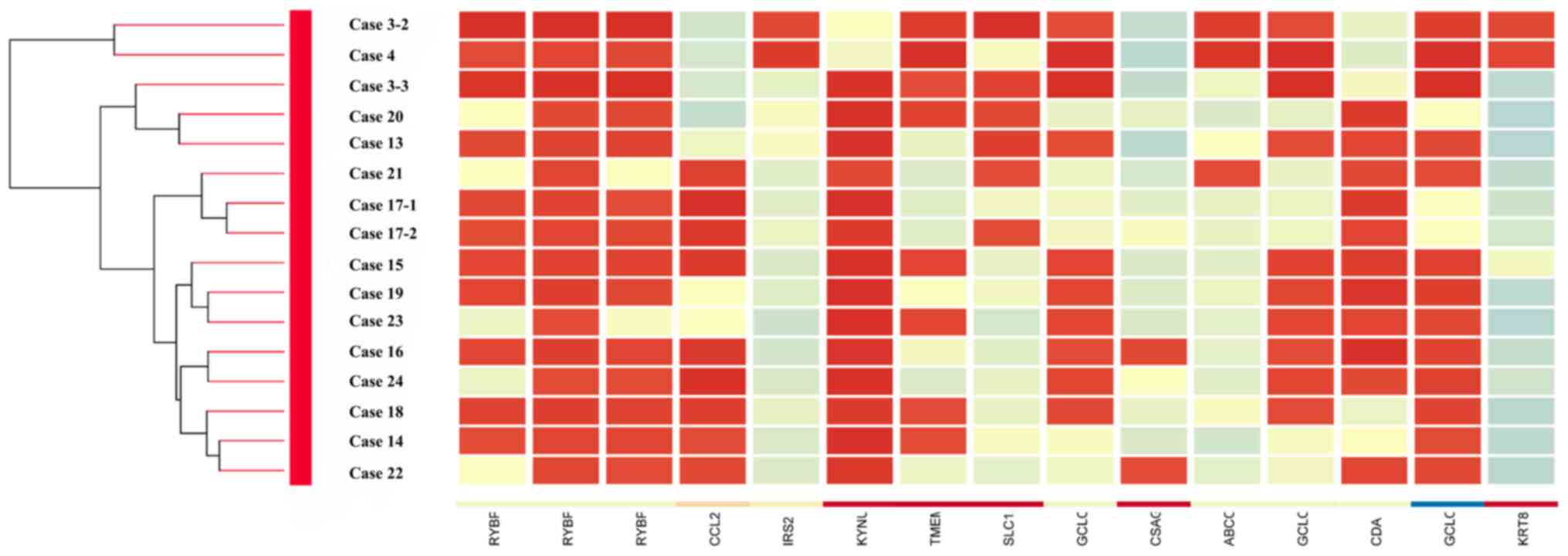 | Figure 3.Clustering analysis from microarray
data. Clustering was performed in 16 OSCC samples (14 patients)
using genes with high (KRT8, ABCC3, GCLC, RYBP, TMEM97, SLC1A3 and
IRS2) and low (KYNU, CSAG2///CSGA3, CDA and CCL21) expression
levels in 3 samples of donor bone marrow-derived stem cells
determined by sex chromosome analysis. Red indicates high
expression and blue indicates low expression. A cluster was formed
in 5 samples (cases 3-2, 3-3, 4, 13 and 20). OSCC, oral squamous
cell carcinoma. |
Discussion
A therapeutic strategy based on the assessment of
tumor grade has previously been used on factors such as
histopathological features, radiographic images, gene mutation and
gene expression. OSCC was routinely assessed by TNM staging,
histopathological grade, HPV status, and analysis of p53 and
relevant molecular abnormalities (11). In a recent study, it was found that
some SCCs derived from salivary gland stem cells were found in
patients with OSCC, significantly affecting prognosis and the true
malignancy of tumor cells (29).
In the present study, it was found that OSCC could
develop from BM-derived stem cells. OSCC occurring from more
undifferentiated stem cells, such as BM-derived stem cells, might
share the characteristics of stem cells (stemness is characterized
by invasion of surrounding tissues, viability of vessels and
ectopic sites, and self-replicating ability); consequently, such
cases had a high malignancy and poor prognosis. A total of 2/3
patients (cases 1 and 3) who developed donor-derived OSCC following
HSCT died from multiple distant metastases, and the other patient
(case 4) had superficial cancer and was being carefully followed
up, although only the 1-year prognosis was good at the time. The
two patients (cases 13 and 20) with OSCC possibly derived from
autologous BM stem cells exhibited significant cervical metastases
that were controlled by extended surgery and intensive
chemotherapy. These patients were also under strict follow up.
In the present study, no statistical differences in
clinicopathological conditions and prognosis between OSCC from
possibly BM-derived stem cells and normally developed OSCC were
observed, due to the small number of cases examined (Kaplan-Meier
analysis; data not shown). Therefore, an attempt was made to access
the expression patterns of genes with >3-fold changes in OSCC
from possibly BM-derived stem cells in 167 OSCC samples from a
public database (GEO; https://www.ncbi.nlm.nih.gov/geo, experiment number,
GSE30784), and cluster analysis was performed. However, no marked
cluster could be found. An attempt was also made to extract cases
from the public database that had similar expression patterns to
those of our 4 patients (5 samples; case 3-2, 3-3, 4, 13 and 20)
with possibly BM stem cell-derived OSCC. However, this was also
unsuccessful, due to the different probe types and analysis
software used. The accumulation of more data from our microarray
analysis is ongoing, in order to develop a method to identify
patients with possibly BM stem cell-derived OSCC among patients
with normally developed OSCC, and to analyze the clinical
characteristics of the possibly BM stem cell-derived OSCC.
Separately, a cluster analysis with a combination of ES
cell-maintaining markers (24 genes) was performed in our OSCC
patients or those from the public database; however, no cluster was
found. The results showed that genes maintaining ES phenotype
(general stemness-maintaining genes) were highly expressed in
almost all OSCCs, indicating the reversion of cancer cells to stem
cells.
As shown by XY-FISH analysis conducted in a very
small population of the cells, a Y-chromosome signal was not
detectable in male patients with OSCC who did not undergo HSCT.
This may be due to technical errors of FISH or Y chromosome dropout
during malignant transformation. However, OSCC from male patients
who underwent HSCT exhibited X-only signal in the majority of the
cells, which might suggest that malignant transformation occurred
in the epithelial cells from female donors. Furthermore, some
epithelial cells in OPMDs of female patients contained Y
chromosomes, which showed that a portion of the epithelial cells in
the lesion might have been derived from BM stem cells of the male
donor. These results suggest that a portion of the epithelial cells
in the oral mucosa were repaired using donor BM stem cells in
several patients following HSCT, possibly resulting in malignant
transformation to SCC. A total of 2/3 (cases 13 and 20) patients
with OSCC derived from donor BM stem cells developed multiple
cancer, so-called ‘field cancerization’, in the upper digestive
tract, where chronic GvHD occurred and epithelial cells were
repaired in BM-derived stem cells. Although the influence of
chronic inflammation and immune-suppressants appeared to be
important for oral carcinogenesis following HSCT, a chronic
persistent inflammation-regeneration (by BM-derived stem
cells)-metaplasia-carcinoma sequence occurs in the oral mucosa. To
the best of our knowledge, this is the first report that focuses on
oral squamous carcinogenesis. Kano et al examined patients
with esophageal SCC that developed from BM-derived stem cells and
found similar results (33). In
order to prove a chronic persistent inflammation-regeneration by BM
derived stem cells-metaplasia-carcinoma sequence in the oral SCC
patients who underwent HSCT, a multi-institutional big study might
be necessary.
BM-derived stem cells consist of hematopoietic and
mesenchymal stem cells, the latter of which can differentiate into
different types of epithelial cells. Among these, Muse cells have
been widely studied and found in the blood, as well as in stromal
tissue from the whole body, leading to differentiation into
tridermic cells. In contrast to ES and iPS cells, Muse cells were
considered as pluripotent stem cells without a potential for
neoplastic transformation (28). The
population of the BM-derived stem cells that lead to OSCC is
unclear; however, it is likely that it is a population of non-Muse
mesenchymal stem cells. We are currently conducting an animal study
to identify the subpopulation of BM-derived stem cells that can
repair the oral epithelium and may lead to the development of OSCC.
To the best of our knowledge, any animal models of oral SCC
carcinogenesis which supported our hypothesis had not been
reported. We recently established the BM stem cells transplantation
model in mice. We will plan to inoculate the sorted sub-population
of the GFP labeled-BM cells to the recipient mice. Then, an oral
carcinogenesis experiment, a chronic persistent
inflammation-regeneration-metaplasia-carcinoma, will be performed
in the recipient mice.
The conventional therapeutic strategy for OSCC was
made according to the clinical and pathological findings,
radiographic features, mutations and expressions of a limited
number of genes, regardless of the origin of the tumor. OSCC might
consist of the carcinomas derived from epithelial stem cells,
salivary gland stem cells, and BM-derived stem cells. If the genes
that can discriminate the origin are identified, OSCC could be
classified into three categories based on cell origin, but not
superficial phenotype. An origin-based diagnosis of cancer cells
can help develop a more specific therapeutic strategy to improve
prognosis. This would be a paradigm shift in the diagnostic and
therapeutic strategies for OSCC.
Supplementary Material
Supporting Data
Acknowledgments
The authors would like to thank Professor Kinuko
Mitani (Department of Hematology and Oncology, Dokkyo Medical
University School of Medicine), Professor Shigemi Yoshihara
(Department of Pediatrics, Dokkyo Medical University School of
Medicine), Professor Keiichi Kubota (Department of
Gastroenterological Surgery, Dokkyo Medical University School of
Medicine) and Professor Kazuyuki Ishida (Department of Molecular
Diagnostic Pathology, Dokkyo Medical University School of Medicine)
for providing the clinical and pathological information of the
patients. The authors would also like to thank Ms. Chiaki
Matsuyama, Ms. Ayako Shimizu (Department of Molecular Diagnostic
Pathology, Dokkyo Medical University School of Medicine) and Mr.
Kazumi Akimoto (Clinical Research Center, Dokkyo Medical University
School of Medicine) for their technical assistance.
Funding
This study was supported by the Young Investigator
Award of Dokkyo Medical University (grant no. 2018-16, 2019-14) and
Japan Society for the Promotion of Science Grant-in-Aid for
Scientific Research [grant no. (C) (17K11887)].
Availability of data and material
The datasets during and/or analyzed during the
current study available from the corresponding author on reasonable
request.
Authors' contributions
THa, KIN, CF, THy, YS, MS, RK, NK, AF, SU and HK
contributed to the study conception and design. THa, THy, YS, MS,
RK and NK collected the data. THa, CF, AF, KIN, DU and HK analyzed
the data. THa drafted the manuscript. THa, KIN, CF, THy, YS, MS,
RK, NK, AF, SU and HK reviewed the final draft of the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The design of the current study was approved by the
Medical Ethical Research Committee of Dokkyo Medical University
Hospital (approval ID, R-25-16J) and Ehime University (approval ID,
2007024). Written informed consent was obtained from all patients
for the use of their samples for research and for publication at
the start of the treatment. The patients could also opt-out, based
on a research plan approved by the Medical Ethical Research
Committee of our institutes. We confirm that no opt-outs were
recorded.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Johnson N, Franceschi S, Ferlay J, Ramadas
K, Schmid S, MacDonald DG, Bouquot JE and Slootweg PJ: Squamous
cell carcinoma. Pathology and Genetics of Head and Neck Tumours.
Barnes L, Eveson JW, Reichart P and Sidransky D: World Health
Organization classification of tumours. 1st edition. IARC Press;
Lyon: pp. 168–175. 2005
|
|
2
|
Shah JP, Zelefsky MJ and O'Malley BB:
Squamous cell carcinoma of the oral cavity. Head and Neck Cancer.
Harrison LB, Sessions RB and Hong WK: Lippincott-Raven; New York,
USA: pp. 411–418. 1999
|
|
3
|
Ginos MA, Page GP, Michalowicz BS, Patel
KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL and Gaffney PM:
Identification of a gene expression signature associated with
recurrent disease in squamous cell carcinoma of the head and neck.
Cancer Res. 64:55–63. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Uchida D, Begum NM, Almofti A, Nakashiro
Ki, Kawamata H, Tateishi Y, Hamakawa H, Yoshida H and Sato M:
Possible role of stromal-cell-derived factor-1/CXCR4 signaling on
lymph node metastasis of oral squamous cell carcinoma. Exp Cell
Res. 290:289–302. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Langer CJ: Exploring biomarkers in head
and neck cancer. Cancer. 118:3882–3892. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sheikh Ali MA, Gunduz M, Nagatsuka H,
Gunduz E, Cengiz B, Fukushima K, Beder LB, Demircan K, Fujii M,
Yamanaka N, et al: Expression and mutation analysis of epidermal
growth factor receptor in head and neck squamous cell carcinoma.
Cancer Sci. 99:1589–1594. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R,
Hammond EH, Fu KK and Milas L: Impact of epidermal growth factor
receptor expression on survival and pattern of relapse in patients
with advanced head and neck carcinoma. Cancer Res. 62:7350–7356.
2002.PubMed/NCBI
|
|
8
|
Sheu JJ, Hua CH, Wan L, Lin YJ, Lai MT,
Tseng HS, Jinawath N, Tsai MH, Chang NW, Lin CF, et al: Functional
genomic analysis identified epidermal growth factor receptor
activation as the most common genetic event in oral squamous cell
carcinoma. Cancer Res. 69:2568–2576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Azuma M, Yoshida H, Kawamata H, Yanagawa
T, Furumoto N and Sato M: Cellular proliferation and ras oncogene
of p21 21,000 expression in relation to the intracellular cyclic
adenosine 3′: 5′-Monophosphate levels of a human salivary gland
adenocarcinoma cell line in culture. Cancer Res. 48:2898–2903.
1988.PubMed/NCBI
|
|
10
|
Shinagawa Y, Kawamata H, Omotehara F,
Nakashiro Ki, Hoque MO, Furihata T, Horiuchi H, Imai Y, Fujimori T
and Fujibayashi T: Evaluation of the chemosensitivity of head and
neck cancer cells based on the diverse function of mutated-p53. Int
J Oncol. 22:383–389. 2003.PubMed/NCBI
|
|
11
|
Tachibana M, Shinagawa Y, Kawamata H,
Omotehara F, Horiuchi H, Ohkura Y, Kubota K, Imai Y, Fujibayashi T
and Fujimori T: RT-PCR amplification of RNA extracted from
formalin-fixed, paraffin-embedded oral cancer sections: Analysis of
p53 pathway. Anticancer Res. 23:2891–2896. 2003.PubMed/NCBI
|
|
12
|
Hoque MO, Kawamata H, Nakashiro KI,
Omotehara F, Hino S, Uchida D, Harada K, Begum NM, Yoshida H, Sato
M and Fujimori T: Dysfunction of the p53 tumor suppressor pathway
in head and neck cancer. Int J Oncol. 21:119–126. 2002.PubMed/NCBI
|
|
13
|
Nagata M, Noman AA, Suzuki K, Kurita H,
Ohnishi M, Ohyama T, Kitamura N, Kobayashi T, Uematsu K, Takahashi
K, et al: ITGA3 and ITGB4 expression biomarkers estimate the risks
of locoregional and hematogenous dissemination of oral squamous
cell carcinoma. BMC Cancer. 13:4102013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kashyap T, Germain E, Roche M, Lyle S and
Rabinovitz I: Role of β4 integrin phosphorylation in human invasive
squamous cell carcinoma: Regulation of hemidesmosome stability
modulates cell migration. Lab Invest. 91:1414–1426. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kurokawa A, Nagata M, Kitamura N, Noman
AA, Ohnishi M, Ohyama T, Kobayashi T, Shingaki S and Takagi R;
Oral, Maxillofacial Pathology, and Surgery Group, : Diagnostic
value of integrin alpha3, beta4, and beta5 gene expression levels
for the clinical outcome of tongue squamous cell carcinoma. Cancer.
112:1272–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mitra RS, Goto M, Lee JS, Maldonado D,
Taylor JM, Pan Q, Carey TE, Bradford CR, Prince ME, Cordell KG, et
al: Rap1GAP promotes invasion via induction of matrix
metalloproteinase 9 secretion, which is associated with poor
survival in low N-stage squamous cell carcinoma. Cancer Res.
68:3959–3969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kawamata H, Uchida D, Hamano H,
Kimura-Yanagawa T, Nakashiro KI, Hino S, Omotehara F, Yoshida H and
Sato M: Active-MMP2 in cancer cell nests of oral cancer patients:
Correlation with lymph node metastasis. Int J Oncol. 13:699–704.
1998.PubMed/NCBI
|
|
18
|
Kawamata H, Nakashiro K, Uchida D, Harada
K, Yoshida H and Sato M: Possible contribution of active MMP2 to
lymphnode metastasis and secreted cathepsin L to bone invasion of
newly established human oral-squamous-cancer cell lines. Int J
Cancer. 70:120–127. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mineta H, Miura K, Suzuki I, Takebayashi
S, Amano H, Araki K, Harada H, Ichimura K, Wennerberg JP and Dictor
MR: Low p27 expression correlates with poor prognosis for patients
with oral tongue squamous cell carcinoma. Cancer. 85:1011–1017.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanazawa T, Iwashita T, Kommareddi P, Nair
T, Misawa K, Misawa Y, Ueda Y, Tono T and Carey TE: Galanin and
galanin receptor type 1 suppress proliferation in squamous
carcinoma cells: Activation of the extracellular signal regulated
kinase pathway and induction of cyclindependent kinase inhibitors.
Oncogene. 26:5762–5771. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Supriatn o, Harada K, Yoshida H and Sato
M: Basic investigation on the development of molecular targeting
therapy against cyclin-dependent kinase inhibitor p27Kip1 in head
and neck cancer cells. Int J Oncol. 27:627–635. 2005.PubMed/NCBI
|
|
22
|
Yamamoto E, Kohama G, Sunakawa H, Iwai M
and Hiratsuka H: Mode of invasion, bleomycin sensitivity and
clinical course in squamous cell carcinoma of the oral cavity.
Cancer. 51:2175–2180. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alison MR, Poulsom R, Jeffery R, Dhillon
AP, Quaglia A, Jacob J, Novelli M, Prentice G, Williamson J and
Wright NA: Hepatocytes from non-hepatic adult stem cells. Nature.
406:2572000. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jackson KA, Majka SM, Wang H, Pocius J,
Hartley CJ, Majesky MW, Entman ML, Michael LH, Hirschi KK and
Goodell MA: Regeneration of ischemic cardiac muscle and vascular
endothelium by adult stem cells. J Clin Invest. 107:1395–1402.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Okamoto R, Yajima T, Yamazaki M, Kanai T,
Mukai M, Okamoto S, Ikeda Y, Hibi T, Inazawa J and Watanabe M:
Damaged epithelia regenerated by bone marrow-derived cells in the
human gastrointestinal tract. Nat Med. 8:1011–1017. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Houghton J, Stoicov C, Nomura S, Rogers
AB, Carlson J, Li H, Cai X, Fox JG, Goldenring JR and Wang TC:
Gastric cancer originating from bone marrow-derived cells. Science.
306:1568–1571. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tamai K, Yamazaki T, Chino T, Ishii M,
Otsuru S, Kikuchi Y, Iinuma S, Saga K, Nimura K, Shimbo T, et al:
PDGFRalpha-Positive cells in bone marrow are mobilized by high
mobility group box 1 (HMGB1) to regenerate injured epithelia. Proc
Natl Acad Sci USA. 108:6609–6614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuroda Y, Kitada M, Wakao S, Nishikawa K,
Tanimura Y, Makinoshima H, Goda M, Akashi H, Inutsuka A, Niwa A, et
al: Unique multipotent cells in adult human mesenchymal cell
populations. Proc Natl Acad Sci USA. 107:8639–8643. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kinouchi M, Izumi S, Nakashiro K, Haruyama
Y, Kobashi G, Uchida D, Hasegawa T and Kawamata H: Determination of
the origin of oral squamous cell carcinoma by microarray analysis:
Squamous epithelium or minor salivary gland? Int J Cancer.
143:2551–2560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Arai Y, Arai H, Aoyagi A, Yamagata T,
Mitani K, Kubota K, Kawamata H and Imai Y: A solid tumor of donor
cell-origin after allogeneic peripheral blood stem cell
transplantation. Am J Transplant. 6:3042–3043. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pindborg JJ, Reichart PA, Smith CJ and van
der Waal I: Histological Typing of Cancer and Precancer of the Oral
Mucosa, 2nd edition. Springer; Berlin: 1997, PubMed/NCBI
|
|
32
|
Anneroth G, Batsakis J and Luna M: Review
of the literature and a recommended system of malignancy grading in
oral squamous cell carcinomas. Scand J Dent Res. 95:229–249.
1987.PubMed/NCBI
|
|
33
|
Kano Y, Ishii H, Konno M, Yamasaki M,
Miyata H, Nishikawa S, Hamabe A, Ogawa H, Takahashi H, Ohta K, et
al: Cells of origin of squamous epithelium, dysplasia and cancer in
the head and neck region after bone marrow transplantation. Int J
Oncol. 44:443–450. 2014. View Article : Google Scholar : PubMed/NCBI
|