Introduction
Oral squamous cell carcinoma (OSCC) is the most
prevalent malignancy of the oral cavity, accounting for >90% of
oral cancer cases, with 354,864 estimated new cases and 177,384
mortalities worldwide in 2018 (1,2). OSCC,
characterized by a high recurrence, a poor prognosis and high
morbidity, severely affects the quality of life of patients.
Therefore, OSCC poses a burden to global health. OSCC is preceded
in 67% of cases by oral premalignant lesions (OPLs), of which 1–18%
undergo malignant transformation into OSCC (3). Patients with early-stage squamous cell
carcinoma (ESCC; TNM I and II) survive longer than those with
advanced-stage squamous cell carcinoma (ASCC; TNM III and IV), with
survival rates of 64.2 and 30.1% for early and late stages,
respectively (4). Despite
improvements in treatment modalities, the 5-year overall survival
rate has improved only marginally, with 33% of cases surviving
between 1973 and 2014, compared with 41% between 2006–2011
(5,6). Delayed diagnosis and the lack of
accurate and timely treatment, derived from the bias of the
standards of clinical decision-making based on the clinical
experience and subjective judgment of doctors, are considered to be
the major reasons for the poor prognosis. A minimally invasive,
reliable and sensitive marker is urgently required to provide an
objective basis for clinical decision-making.
Long non-coding RNAs (lncRNAs) are a class of RNA
molecules with transcripts of >200 nucleotides in lengths, which
were first discovered as ‘transcriptional noise’ in 1989. However,
increasing evidence has suggested that lncRNAs are involved in gene
expression regulations at the epigenetic, transcriptional and
post-transcriptional levels, and are essential in physiological
events (7–9). The aberrant expression of lncRNAs may
directly or indirectly lead to the development of various diseases,
including cancer (10,11). lncRNAs may be promising biomarkers in
cancer diagnosis and prognosis (12,13). For
example, MALAT1 may be used as a marker for the early diagnosis of
prostate cancer (14), the
upregulation of HOTAIR expression is indicative of a poor prognosis
in colon and breast cancer (15),
and the downregulation of GAS5 expression is indicative of a poor
prognosis in gastric cancer (16).
lncRNAs have also been revealed to be differentially expressed in
tissues and salivary samples of the normal oral mucosa, in OPLs and
OSCC (17–20). However, to the best of our knowledge,
the differential expression profiles of lncRNAs in the plasma of
patients with OSCC has not yet been reported.
In the present study, the differential expression
profiles of plasma lncRNAs in OSCC were first investigated using
microarray analysis to screen candidate lncRNAs, followed by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis. Subsequently, the target lncRNAs were further
validated by RT-qPCR to estimate the diagnostic efficacy of plasma
lncRNAs from patients with OPL, ESCC and ASCC. The results of the
present study may provide novel targets for the early diagnosis and
staging of OSCC, which may also provide an objective basis for
clinical decision-making for the early diagnosis, reasonable
implementation of the treatment plan and prognosis evaluation of
OSCC.
Materials and methods
Samples
A total of 67 patients with OSCC (39 men and 28
women; age range, 47–75 years; mean age, 63.5 years), 16 patients
with OPL (3 cases of mild dysplasia, 7 cases of moderate dysplasia
and 6 cases of severe dysplasia) and 19 healthy control individuals
(H group) were recruited between December 2013 and May 2015 from
Capital Medical University Beijing Stomatological Hospital. A total
of three patients with TNM III/IV OSCC and 3 healthy controls were
selected for microarray analysis (Table
SI), and the remaining samples were used for PCR validation,
including 64 patients with OSCC (39 males and 25 females; age
range, 47–75 years; mean age, 63.3 years; TNM staging is presented
in Table SII), 16 patients with
OPLs (9 males and 7 females; age range, 43–72 years; mean age, 58.1
years; all with epithelial dysplasia) and 16 healthy controls (8
males and 8 females; age range, 37–61 years; mean age, 48.6 years).
The recruited subjects had no medical history of other types of
cancer. Blood samples were collected in vacuum tubes with EDTA
anticoagulant and were isolated by centrifugation at 1,000 × g for
10 min at 4°C to obtain the plasma. The collected plasma was stored
at −80°C into a separate RNase-free tube prior to further analysis,
which was divided into 400–500 µl/tubes. Blood samples with
hemolysis were excluded and samples with absence of hemolysis were
included. The present study was approved by the Ethics Committee of
the Capital Medical University Beijing Stomatological Hospital
(Beijing, China; approval no. 201314), and written informed consent
was provide by all participants prior to the study start.
Microarray assay
The differential expression profiles of lncRNAs and
mRNAs in the frozen plasma of patients with OSCC were analyzed by
KangChen Biotechnology Co., Ltd. using Arraystar Human LncRNA
Microarray V4.0 (Agilent Technologies, Inc.). Total RNA was
extracted from 400 µl plasma using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), purified using the
RNasey Mini kit (Qiagen, Inc.), and amplified and labeled using the
Quick Amp Labeling kit One-Color (Agilent Technologies, Inc.),
according to the manufacturer's protocol. The aforementioned steps
were repeated until the cRNA production was >1.65 µg and the
specific activity was >9.0 pmol Cy3/µg cRNA. An equal amount of
labeled cRNAs from each sample was then hybridized using the
Agilent Gene Expression Hybridization kit (Agilent Technologies,
Inc.) at 65°C for 17 h.
Microarray data analysis
The acquired array images were analyzed using
Agilent Feature Extraction software (version 11.0.1.1; Agilent
Technologies, Inc.). Quantile normalization and subsequent data
processing were performed using the GeneSpring GX v12.1 software
package (Agilent Technologies, Inc.). The differential expression
of lncRNAs or mRNAs with statistical significance between the two
groups was screened by P-value/False discovery rate (FDR). The
P-value was calculated using a t-test and modified according to the
Benjamin Hochberg FDR method. The screening criteria were |fold
change|≥2.0 and FDR <0.05. The differentially expressed lncRNAs
or mRNAs between the two samples were screened by fold-change (FC),
and the screening criteria were |log2FC|≥1 and
P<0.05. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analyses were performed for the
differentially expressed lncRNAs or mRNAs, as previously described
(20).
Candidate lncRNA screening and RT-qPCR
validation
From the microarray data, 14 lncRNAs were selected
to perform RT-qPCR verification experiments with the array
homologous plasma samples. The screening strategies were as
follows: i) The top 5 gold level lncRNAs in the general list in the
descending order of FC with the original expression ≥200, namely
LOC101927358, GAS5-AS1, LOC100507156, RP11-539G18.2 and
ARHGEF26-AS1; ii) lncRNAs with |FC|≥5 and original
expression ≥200 in the sub-category analysis list with
tissue-specific set to head-neck, disease-specific set to cancer,
bio-process set to metastasis, namely CTD-2008L17.1 and
LINC01539; iii) lncRNAs with |FC|≥5 and co-existing in the
general list and the differentially expressed lncRNA list in OPL
based on SAGE (21), namely
LINC00665 and NEAT1; iv) lncRNAs with |FC|≥10 and
co-existing in the general list and the differentially expressed
lncRNA list in OSCC based on the microarray (20), namely RP11-250B2.3 and
AP001347.6; and v) the top 3 lncRNAs with the largest FC
coexisting in the general list and the tumor-related lncRNA list
downloaded from the Lnc2Cancer database (http://www.bio-bigdata.com/lnc2cancer/down.jsp),
namely HOTAIR_4, BCAR4 and MNX1-AS1.
The primers were designed using primer premier 5.0
software (Premier Biosoft International) (Table SIII). The reverse transcription of
total RNA was performed in a 20 µl volume containing 500 ng total
RNA, 1 µl 10 µM primers, 1.6 µl of 2.5 mM dNTPs mixture, 4 µl 5X
First-Strand Buffer, 1 µl 0.1 M DTT, 0.3 µl RNase inhibitor, 0.2 µl
SuperScript III RT (Invitrogen; Thermo Fisher Scientific, Inc.) and
14.5 µl water. The program was as follows: 50°C for 60 min, 70°C
for 5 min, and 4°C hold. The ViiA 7 Real-time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used for the
RT-qPCR assay. A total of 2 µl of the cDNA product was used as a
template in 10 µl reaction on a 384-well plate containing 5 µl of
2X PCR master mix (Arraystar), 1 µl of 10 µM specific primer, 2 µl
of RNase-free water. The conditions were as follows: A denaturation
step for 10 min at 95°C, followed by 40 cycles of 10 sec at 95°C
and 60 sec at 60°C. Following amplification, the operation of the
instrument was performed according to the procedure (95°C, 10 sec;
60°C, 60 sec; 95°C, 15 sec) and slowly heated from 60°C to 99°C
(-Ramp Rate was 0.05°C/sec). Each experiment was repeated in
triplicate. The housekeeping gene used was β-actin. The
2−ΔΔCq method was used to measure relative expression
levels (RELs) (22).
Target lncRNA screening and RT-qPCR
validation
A total of 4 lncRNAs were selected to be measured
and validated by RT-qPCR in a large cohort. The screening
strategies were as follows: i) The two lncRNAs with the top FC
among the aforementioned 14 lncRNAs, namely ENST00000412740
and ENST00000588803; ii) the lncRNA with the largest FC in
the general list, namely NR_038323; and iii) the key lncRNA
in OPL, namely NR_131012. The RT-qPCR procedure was the same
as that described earlier, and the primers used are listed in
Table SIII.
Statistical analysis and evaluation of
the 4 lncRNAs as diagnostic markers for the early diagnosis and
staging of OSCC
The relative expression of lncRNAs was calculated
using the 2−ΔΔCq method, where ΔCt=Ct (target gene)-Ct
(β-actin), ΔΔCt=ΔCt (experiment sample)-ΔCt (control sample). R
language (version 3.3.2, http://www.r-project.org/) was used for data
processing. The Shapiro-Wilk test was used to assess the normality
of distribution and the Levene test (two sides) was used to assess
the homogeneity of variance. If the data were normally distributed,
analysis of variance was used to perform variance analysis among
groups and the Tukey's HSD test was used to determine significant
differences between groups. If the data were not normally
distributed, the Kruskal-Wallis test rank sum test was used among
groups, and the Dunn's test was used for post-hoc analysis. The
Benjamin-Hochberg correction was performed to determine the
P-values among groups.
Receiver operating characteristic (ROC) curve
analysis and the area under the ROC curve (AUC) were used to
evaluate the sensitivity and specificity of lncRNAs as novel
diagnostic tools for the early diagnosis and staging of OSCC. A ROC
curve was drawn using the ROC package in R language, and the
comparison of AUC was performed using the DeLong test. Firth's
Bias-Reduced Logistic Regression analysis was performed using the
logistf package of R language, and variables were screened by the
stepwise optimization method to determine the lncRNA combination
with a high diagnostic efficiency. All tests were two-sided and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Differentially expressed profiles of
lncRNAs and mRNAs in the plasma of patients with OSCC
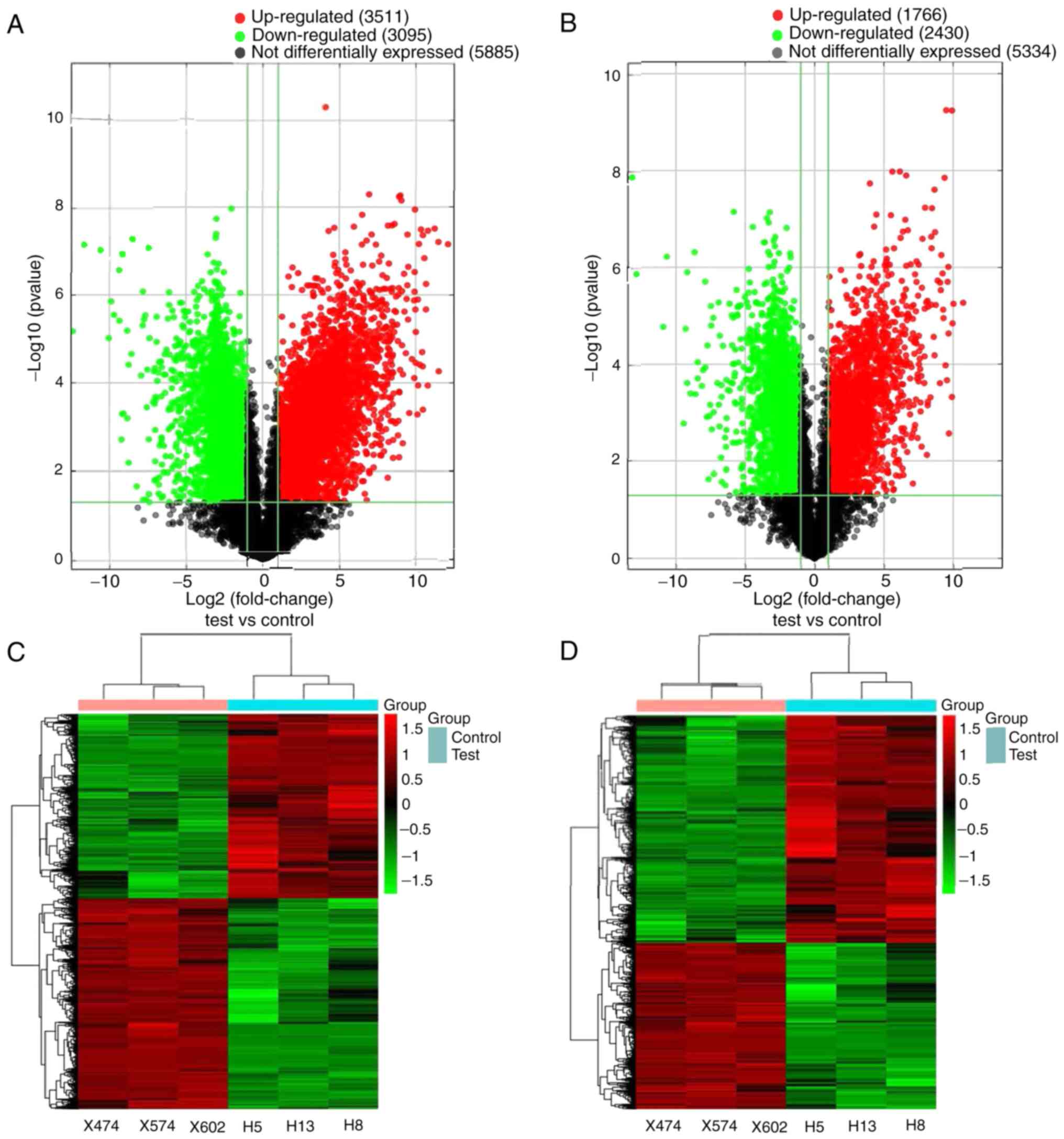
Following image acquisition and data analysis, the
expression matrices of lncRNAs and mRNAs were obtained. The volcano
plot indicated that several lncRNAs and mRNAs were differentially
expressed between the OSCC and normal samples (Fig. 1A and B). According to the screening
standard, a total of 6,606 lncRNAs and 4,196 mRNAs were
differentially expressed in the plasma of patients with OSCC.
Furthermore, 3,511 lncRNAs and 1,766 mRNAs were upregulated, and
3,095 lncRNAs and 2,430 mRNAs were downregulated. The top 20
dysregulated lncRNAs and mRNAs are presented in Tables SIV and SV, respectively. Hierarchical clustering
analysis revealed that the expression profiles exhibited a good
clustering effect on OSCC and normal plasma (Fig. 1C and D). The results of GO and KEGG
analyses are presented in Tables
SVI–SIX. The data have been
deposited in NCBI's Gene Expression Omnibus and are accessible
through GEO series accession no. GSE97251 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97251).
Validation of candidate lncRNAs by
RT-qPCR
Compared with the H group, 14 candidate lncRNAs were
all differentially expressed in OSCC (Table I). Apart from NR_024050, which
was downregulated (no statistical significance), the remaining 13
lncRNAs were all upregulated, and the FC value of 9 lncRNAs
exhibited a statistically significant difference. These results
were consistent with those of the microarray.
 | Table I.REL and FC of 14 candidate lncRNAs
(OSCC/H). |
Table I.
REL and FC of 14 candidate lncRNAs
(OSCC/H).
| lncRNA | OSCC | H | FP | P-value | FM | FDR |
|---|
|
ENST00000412740 | 3.94E-02 | 5.39E-03 | 7.30 | 0.000066 | 1.22 | 0.025648541 |
|
ENST00000427048 | 4.02E-02 | 2.62E-02 | 1.54 | 0.064870 | 1.50 | 0.009596388 |
|
ENST00000428809 | 2.35E-02 | 9.32E-03 | 2.52 | 0.046177 | 2.81 | 0.011765579 |
|
ENST00000533736 | 3.72E-02 | 1.25E-02 | 2.98 | 0.000137 | 3.39 | 0.00325342 |
|
ENST00000588803 | 2.93E-02 | 7.37E-03 | 3.98 | 0.000044 | 2.61 | 0.003797829 |
|
NR_024050 | 4.88E-04 | 6.88E-04 | 0.71 | 0.151403 | 3.90 | 0.022136802 |
|
NR_037605 | 2.57E-03 | 1.50E-03 | 1.72 | 0.093041 | 3.22 | 0.001056383 |
|
NR_037901 | 9.75E-02 | 3.57E-02 | 2.73 | 0.000002 | 3.71 | 0.002373187 |
|
NR_038323 | 4.83E-02 | 1.33E-02 | 3.62 | 0.000002 | 5.30 | 0.000625734 |
|
NR_038835 | 6.38E-02 | 1.79E-02 | 3.56 | 0.000004 | 4.64 | 0.002772666 |
|
NR_040026 | 9.28E-03 | 2.40E-03 | 3.87 | 0.000038 | 1.30 | 0.000843384 |
|
NR_121182 | 8.62E-04 | 6.81E-04 | 1.26 | 0.582889 | 3.64 | 0.004240601 |
|
NR_131012 | 3.06E-02 | 9.75E-03 | 3.14 | 0.012689 | 1.71 | 0.000283274 |
|
uc021qyj.1 | 1.13E-02 | 9.76E-03 | 1.15 | 0.651841 | 1.66 | 0.000092149 |
Validation of target lncRNAs by
RT-qPCR
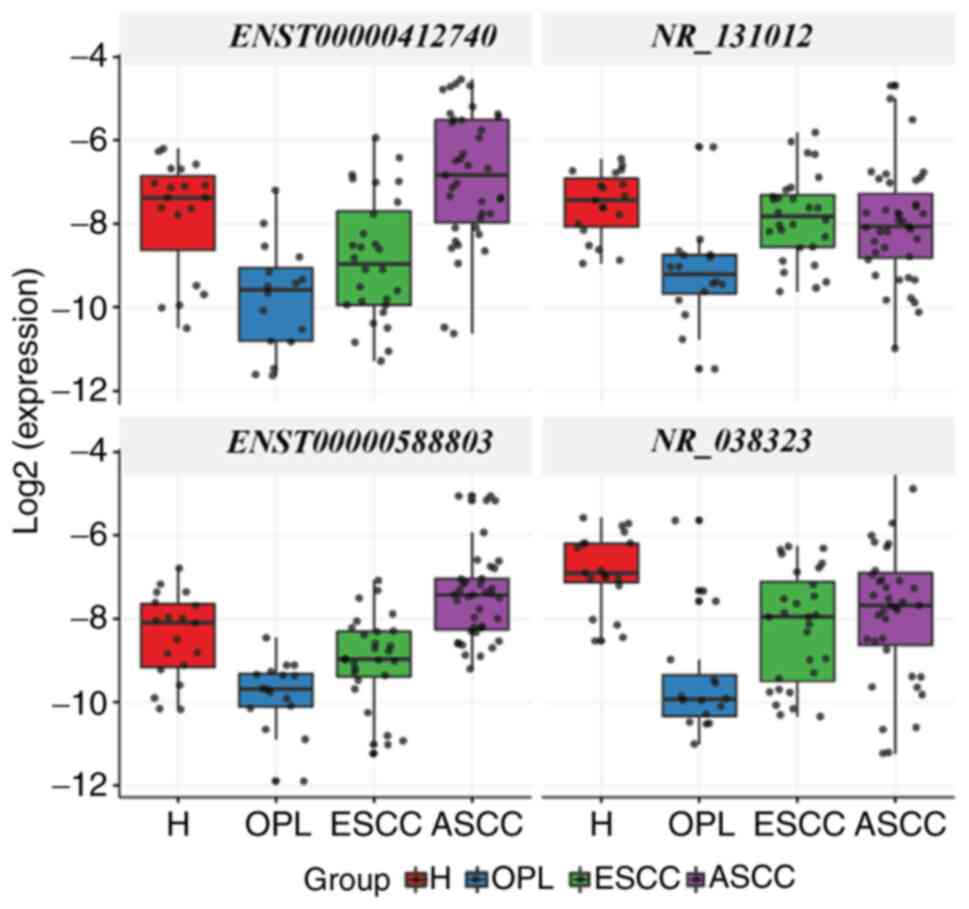
ENST00000412740, NR_131012, ENST00000588803
and NR_038323 were differentially expressed among the H,
OPL, ESCC and ASCC groups (P<0.05). Furthermore, the
differential expression of 4 target lncRNAs was compared between
groups (Fig. 2). Compared with the H
group, ENST00000412740, NR_131012, ENST00000588803 and
NR_038323 were downregulated in the OPL group, and notably,
they were upregulated in the ASCC group compared with the OPL group
(P<0.05). Compared with the H group, NR_038323 was
downregulated in the ESCC and ASCC groups (P<0.05). Compared
with the OPL group, NR_131012 was upregulated in the ESCC
group (P<0.05). Compared with the ESCC group,
ENST00000412740 and ENST00000588803 were upregulated
in the ASCC group (P<0.05).
Screening of diagnostic combination
and evaluation of the diagnostic efficacy of the 4 target
lncRNAs
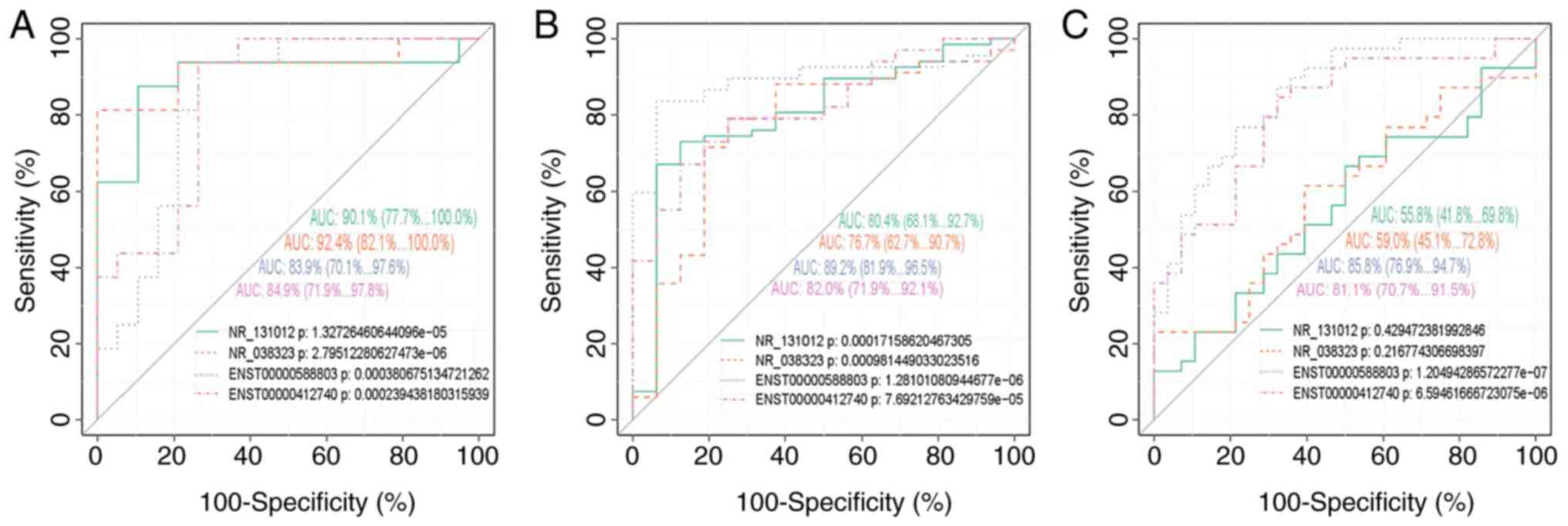
ROC curve analysis revealed that the 4 target
lncRNAs exhibited excellent discriminative ability for OPL vs. H,
OSCC vs. OPL and ASCC vs. ESCC, with an AUC >0.7, apart from
NR_131012 and NR_038323, which were considered as
having moderate discriminative ability only for ASCC vs. ESCC, with
an AUC of 0.558 (95% CI, 0.0418-0.698) and 0.590 (95% CI,
0.451-0.728), respectively (Fig. 3).
However, they exhibited no discriminative ability for OSCC vs. H,
apart from NR_038323 with an AUC >0.7 (Fig. S1). The logistic regression model
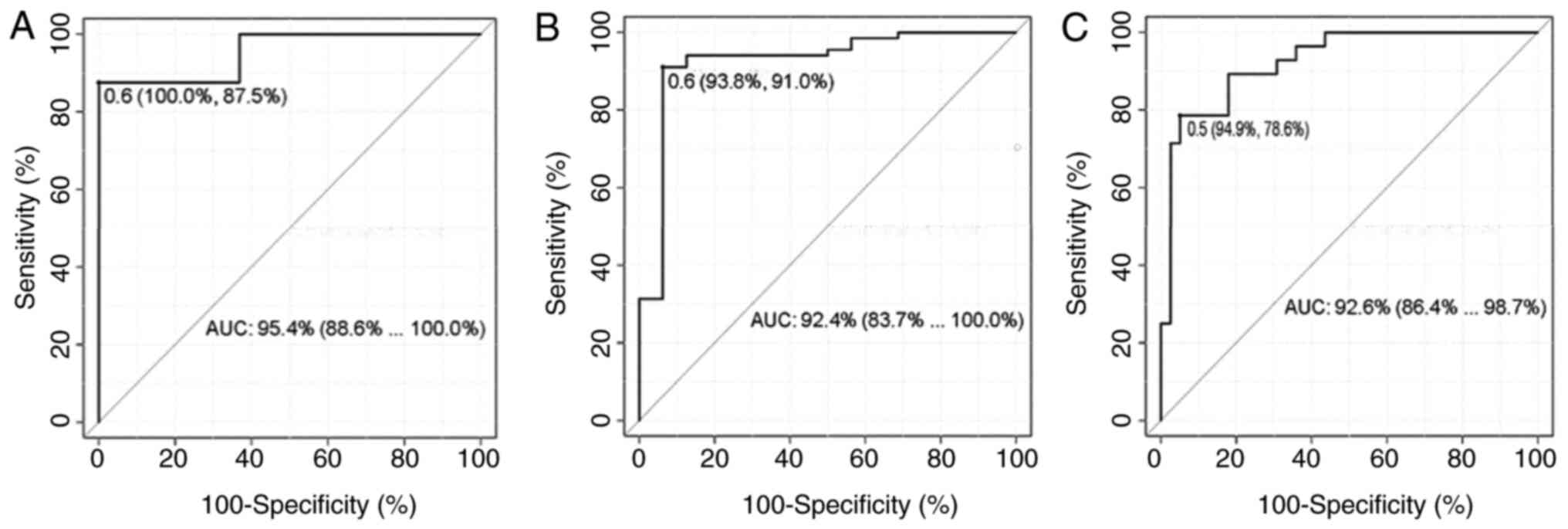
revealed that the combined lncRNAs provided a more prominent
diagnostic efficacy than a single lncRNA, particularly for ASCC vs.
ESCC (Table II). The sensitivity,
specificity and cut-off value of each combination of lncRNAs are
illustrated in Fig. 4.
 | Table II.The results of Delong test of
receiver operating characteristic curve for the combination of
lncRNAs. |
Table II.
The results of Delong test of
receiver operating characteristic curve for the combination of
lncRNAs.
| Comparison | D | Group | P-value |
|---|
| Combind2 vs.
NR_038323 | 2.515717 | OSCC vs. OPL | 0.011879 |
| Combind3 vs.
NR_131012 | 4.764378 | ASCC vs. ESCC | 0.000002 |
| Combind3 vs.
NR_038323 | 4.631511 | ASCC vs. ESCC | 0.000004 |
| Combind3 vs.
ENST00000588803 | 2.027116 | ASCC vs. ESCC | 0.042651 |
| Combind3 vs.
ENST00000412740 | 2.677019 | ASCC vs. ESCC | 0.007428 |
Discussion
To the best of our knowledge, the present study was
the first to identify the differential expression profiles of
plasma lncRNAs in OSCC by microarray analysis. The reliability of
microarray and quality of the array samples were verified to be
credible by RT-qPCR using the array homologous samples. The results
revealed that the profile of lncRNAs in plasma from patients with
OSCC differed significantly from that of the healthy controls. The
majority of the differentially expressed genes have been proven to
be involved in the biological process of OSCC by GO and KEGG
analyses (20,21). However, there are only limited
studies available on the diagnostic role of circulating lncRNAs in
OSCC (23).
In the present study cohort, patients with TNM I/II
stage OSCC accounted for 39.58% of primary OSCC cases, which was
22% of those involving the posterior third of the tongue reported
in the literature (24), indicating
that the early diagnosis of OSCC remains relatively low. However,
patients with TNM I/II stage OSCC accounted for 47.37% of recurrent
OSCC cases, which was slightly higher than that of primary OSCC,
which may be associated with regular follow-up after surgery. A
specialist may improve the early diagnosis of OSCC; however, this
remains insufficient. The early diagnosis and staging of OSCC may
aid doctors in determining effective and appropriate treatment
strategies, including the scope of surgery, radiotherapy,
chemotherapy and other adjuvant therapy, which has an important
impact on the quality of life and prognosis of patients. These
decisions are largely dependent on the clinical experience and
subjective judgment of doctors; however, the lack of objective
indicators leads to a bias in the making of these decisions.
Therefore, an objective, accurate and minimally invasive biomarker
is urgently required.
To date, >1,000 lncRNAs have been proven to be
involved in various biological processes, and an increasing number
of studies have demonstrated that plasma lncRNAs have great
potential for use in tumor diagnosis, prognosis and in the
evaluation of the therapeutic effects (14,25–29).
Circulating lncRNAs are derived from apoptosis, necrotic tissue and
the active secretion of cells and lysis of circulating cells.
Endogenous circulating lncRNAs are bound with proteins, which may
be stable at room temperature and may endure multiple cycles of
freezing and thawing (30,31). According to Schlosser et al
(32), the level of lncRNAs in
plasma has a certain association with its level in tissues, and
lncRNAs may partly be derived from tissues. In the present study,
when target lncRNAs were screened, the profiles of plasma lncRNAs
and tissue lncRNAs in OSCC were compared and it was identified that
only some of the differentially expressed lncRNAs was the same,
which also indicated that the differentially expressed lncRNAs in
the plasma were derived partly from tumor tissues. The expression
of lncRNAs is tissue-specific (32–34).
Therefore, the analysis of plasma lncRNA expression levels may be
used as a minimally invasive diagnostic method for diseases.
In the present study, the four target lncRNAs were
significantly downregulated in the plasma of patients with OPLs and
gradually increased with the malignant transformation process. The
differential expression of these four lncRNAs in different stages
of OSCC indicated that they had the potential to be used as
diagnostic markers for OPL and OSCC staging. The single lncRNAs
ENST00000412740, NR_131012, ENST00000588803 or
NR_038323 may distinguish OPL from the healthy controls,
with an AUC of 0.901, 0.924, 0.839 and 0.849, respectively, but was
not effective for the determination of OSCC stage. To further prove
the efficacy of the four lncRNAs for the diagnosis of OPLs and
OSCC, ROC curve and logistic regression analyses were performed
with optimal combinations. The results revealed that the AUCs of
the combined lncRNAs were generally larger than those of single
lncRNAs in distinguishing OSCC and OPLs, with a high sensitivity
(93.8%) and specificity (91.0%), particularly in distinguishing
ESCC from ASCC more effectively than all single lncRNAs with a high
sensitivity (94.9%) and specificity (78.6%). The sensitivity of all
combinations was far greater than that of the most well-known
available biomarker, SCCA, with a sensitivity of 38.1% (35). Therefore, they may be very promising
biomarkers for the early diagnosis and staging of OSCC. However,
the expression levels of four lncRNAs in the ESCC group were
similar to those of the H group; therefore, the dynamic monitoring
of lncRNAs needs to combined with clinical examinations to
distinguish the difference between the H group and ESCC group.
NEAT1 (NR_131012) is essential for the
assembly and structural integrity of nuclear subunit paraspeckles
(36) and is regulated by
TP53. p53 and pRb pathway disruptions are an
important step in the early stage of oral carcinogenesis (37), which may lead to the immortalization
of oral epithelial cells (38).
Among these, p53 is the ‘guardian’ of genome integrity,
which has been found to upregulate NEAT1 expression. In oral
premalignant lesions, TP53 mutation damages the p53
signaling pathway (39) and the
expression of NEAT1 is downregulated. With the malignant
transformation process of cells, p53 becomes activated under
the effects of replication stress to upregulate NEAT1
expression, which promotes the formation of nuclear paraspeckles
and the growth of highly divided cancer cells. Furthermore,
NEAT1 promotes ATR signaling in response to replication
stress to inhibit replication-related DNA damage and p53
activation, thereby forming a negative feedback loop that
attenuates the activation of p53 in cells with DNA damage
(40). This indicates that
NEAT1 is downregulated in OPL and upregulated in ESCC and is
expressed in ASCC. NEAT1 is highly expressed in various
types of cancer, and its expression is associated with tumor size,
TNM stage and distant metastasis; it is also a risk factor for a
shorter overall survival (41).
However, to the best of our knowledge, there are no studies
available to date on the molecular mechanisms of the other 3
lncRNAs.
However, the exact mechanisms of these lncRNAs in
the occurrence and development of OSCC remain unclear and
cytological experiments are required to verify their functions. In
addition, the sample size of the present study was small. For
cross-sectional analysis, the validation sample needs to be further
expanded, and the prognosis of patients requires follow-up, in
order to make a comprehensive and accurate assessment of the
clinical value of lncRNAs as diagnostic markers.
In conclusion, the present study demonstrated that
the expression profiles of plasma lncRNAs are altered in OSCC
compared with normal controls. ENST00000412740, NR_131012,
ENST00000588803 and NR_038323 were differentially
expressed in different stages of OSCC and their expression became
altered with the malignant progression of OSCC. This suggests that
these four lncRNAs may be promising biomarkers for the early
diagnosis and staging of OSCC. Furthermore, the diagnostic efficacy
of the combined lncRNAs was more prominent than that of a single
lncRNA.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Professor Xiaofei
Tang and Professor Xinyan Zhang (Institute of Stomatology, Capital
Medical University) for their suggestions and comments, Professor
Zhengxue Han, Dr Yao Liu and Dr Meihua Zhang (Beijing
Stomatological Hospital, Capital Medical University) for their help
in sample collection, and Mr Zhigang Wang (Medical Data Processing
Center, Peking Union Medical College Hospital) for data
processing.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81372897).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
HJ and XW acquired the data, performed the
experiments and drafted the initial manuscript. HJ, XW and ZS
designed the experiments, interpreted the data and analyzed the
results. SZ and HJ revised and approved the final version of the
manuscript. All authors have read and approved the final
manuscript, and agreed to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Capital Medical University Beijing Stomatological
Hospital (Beijing, China; approval no. 201314) and written informed
consent was provided by all participants prior to the study
start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gupta B, Johnson NW and Kumar N: Global
epidemiology of head and neck cancers: A continuing challenge.
Oncology. 91:13–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sinevici N and O'sullivan J: Oral cancer:
Deregulated molecular events and their use as biomarkers. Oral
Oncol. 61:12–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ghani WMN, Ramanathan A, Prime SS, Yang
YH, Razak IA, Abdul Rahman ZA, Abraham MT, Mustafa WMW, Tay KK,
Kallarakkal TG, et al: Survival of oral cancer patients in
different ethnicities. Cancer Invest. 37:275–287. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alonso JE, Han AY, Kuan EC, Strohl M, Clai
JM, St John MA, Ryan WR and Heaton CM: The survival impact of
surgical therapy in squamous cell carcinoma of the hard palate.
Laryngoscope. 128:2050–2055. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bloebaum M, Poort L, Bockmann R and
Kessler P: Survival after curative surgical treatment for primary
oral squamous cell carcinoma. J Craniomaxillofac Surg.
42:1572–1576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kornienk AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lalevee S and Fei R: Long noncoding RNAs
in human disease: Emerging mechanisms and therapeutic strategies.
Epigenomics. 7:877–879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Meng X, Zhu XW, Yang DC, Chen R,
Jiang Y and Xu T: Long non-coding RNAs in Oral squamous cell
carcinoma: Biologic function, mechanisms and clinical implications.
Mol Cancer. 18:1022019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J,
Wei M, Xu C, Wu C, Zhang Z, et al: Long non-coding RNA metastasis
associated in 1ung adenocarcinoma transcript l derived miniRNA as a
novel plasma-based biomarker for diagnosing prostate cancer. Eur J
Cancer. 49:2949–2959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gibb EA, Enfield KS, Stewart GL, Lonergan
KM, Chari R, Ng RT, Zhang L, MacAulay CE, Rosin MP and Lam WL: Long
non-coding RNAs are expressed in oral mucosa and altered in oral
premalignant lesions. Oral Oncol. 47:1055–1061. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang H, Wu Z, Zhang J and Su B: Salivary
lncRNA as a potential marker for oral squamous cell carcinoma
diagnosis. Mol Med Rep. 7:761–766. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang Z, Wu L, Wang L, Yang Y, Meng Y and
Yang HL: Increased expression of the long non-coding RNA UCA1 in
tongue squamous cell carcinomas: A possible correlation with cancer
metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 117:89–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jia H, Wang X and Sun Z: Exploring the
long noncoding RNAs-based biomarkers and pathogenesis of malignant
transformation from dysplasia to oral squamous cell carcinoma by
bioinformatics method. Eur J Cancer Prev. 29:174–181. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia H, Wang X and Sun Z: Exploring the
molecular pathogenesis and biomarkers of high risk oral
premalignant lesions on the basis of long noncoding RNA expression
profiling by serial analysis of gene expression. Eur J Cancer Prev.
27:370–378. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shao T, Huang J, Zheng Z, Wu Q, Liu T and
Lv X: SCCA, TSGF, and the long non-coding RNA AC007271.3 are
effective biomarkers for diagnosing oral squamous cell carcinoma.
Cell Physiol Biochem. 47:26–38. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sciubba JJ: Oral cancer: The importance of
early diagnosis and treatment. Am J Clin Dermalol. 2:239–251. 2001.
View Article : Google Scholar
|
|
25
|
Meseure D, Drak Alsibai K, Nicolas A,
Bieche I and Morillon A: Long noncoding RNAs as architects in
cancer epigenetics, prognostic biomarkers, and potential
therapeutic targets. Biomed Res Int. 2015:3202142015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kladi-Skandali A, Michaelidou K, Scorilas
A and Mavridis K: Long noncoding rnas in digestive system
malignancies: A novel class of cancer biomarkers and therapeutic
targets? Gastroenterol Res Pract. 2015:3198612015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3:52015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tang Q, Ni Z, Cheng Z, Xu J, Yu H and Yin
P: Three circulating long non-coding RNAs act as biomarkers for
predicting NSCLC. Cell Physiol Biochem. 37:1002–1009. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji
J and Sun B: HULC and Linc00152 act as novel biomarkers in
predicting diagnosis of hepatocellular carcinoma. Cell Physiol
Biochem. 37:687–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ge Q, Zhou Y, Lu J, Bai Y, Xie X and Lu Z:
miRNA in plasma exosome is stable under different storage
conditions. Molecules. 19:1568–1575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsui NB, Ng EK and Lo YM: Stability of
endogenous and added RNA in blood specimens, serum, and plasma.
Clin Chem. 48:1647–1653. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schlosser K, Hanson K, Villeneuve PK,
Dimitroulakos J, McIntyre L, Pilote L and Stewart DJ: Assessment of
circulating LncRNAs under physiologic and pathologic conditions in
humans reveals potential limitations as biomarkers. Sci Rep.
6:365962016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mercer TR, Gerhardt DJ, Dinger ME,
Crawford J, Trapnell C, Jeddeloh JA, Mattick JS and Rinn JL:
Targeted RNA sequencing reveals the deep complexity of the human
transcriptome. Nat Biotechnol. 30:99–104. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prensner JR, Iyer MK, Sahu A, Asangani IA,
Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al:
The long noncoding RNA SChLAPI promotes aggressive prostate cancer
and antagonizes the SWI/SNF complex. Nat Genet. 45:1392–1298. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kurokawa H, Yamashita Y, Tokudome S and
Kajiyama M: Combination assay for tumor markers in oral squamous
cell carcinoma. J Oral Maxillofac Surg. 55:964–966. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Smeets SJ, van der Plas M, Schaaij-Visser
TB, van Veen EA, van Meerloo J, Braakhuis BJ, Steenbergen RD and
Brakenhoff RH: Immortalization of oral keratinocytes by functional
inactivation of the p53 and pRb pathways. Int J Cancer.
128:1596–1605. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Graveland AP, Bremmer JF, de Maaker M,
Brink A, Cobussen P, Zwart M, Braakhuis BJ, Bloemena E, van der
Waal I, Leemans CR and Brakenhoff RH: Molecular screening of oral
precancer. Oral Oncol. 49:1129–1135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Adriaens C, Standaert L, Barra J, Latil M,
Verfaillie A, Kalev P, Boeckx B, Wijinhoven PW, Radaelli E, Vermi
W, et al: p53 induces formation of NEAT1 lncRNA-containing
paraspeckles that modulate replication stress response and
chemosensitivity. Nat Med. 22:861–868. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fang J, Qiao F, Tu J, Xu J, Ding F, Liu Y,
Akuo BA, Hu J and Shao S: High expression of long non-coding RNA
NEAT1 indicates poor prognosis of human cancer. Oncotarget.
8:45918–45927. 2017. View Article : Google Scholar : PubMed/NCBI
|