Introduction
Choline kinase (ChK) catalyzes the phosphorylation
of choline by using ATP as a phosphoryl donor. It is the first
enzyme in the CDP-choline pathway for the de novo
biosynthesis of phosphatidylcholine, the most abundant phospholipid
in eukaryotic cell membrane (1).
There are three isoforms of ChK in humans that are encoded by two
separate genes known as chka and chkb. chkb
codes for a single protein (ChKB), while chka undergoes
alternative splicing to produce ChKA1 and ChKA2 isozymes (1). ChKA has been implicated in
carcinogenesis, as this isoform is overexpressed in a variety of
cancer types, including lung, breast, colon, ovarian and prostate
cancer (2). By contrast, ChKB is
crucial for muscle development, mitochondrial function and bone
homeostasis (3).
ChKA serves an essential role in cell proliferation
and transformation, as well as in the regulation of apoptosis and
the cell cycle (4). The level of ChK
and its product, phosphocholine are diagnostic indicators of cancer
and markers for monitoring the tumor response to therapies
(5). Due to the higher level of ChK
expression in different cancer cells, the inhibition of ChKA
activity has become a promising anticancer therapy. Consequently,
the synthesis and testing of various ChKA inhibitors have gained
increasing attention over the past decade (6). However, limited attention has been paid
to the regulation of chk gene expression, particularly by
microRNAs (miRNAs/miRs).
miRNAs are a large family of non-coding RNAs of ~21
nucleotides in length that regulate gene expression either by the
degradation of target mRNAs or the inhibition of protein
translation (7). miRNAs serve
important roles in the immune system, differentiation,
tumorigenesis and cell death (8).
The dysregulation of miRNA expression is common in a number of
types of cancer, and the miRNA profiling of clinical samples may be
used for cancer diagnostic and prognostic applications (9). In humans, miR-195-5p functions as an
anti-oncogene by targeting PHF-19, leading to the suppression of
hepatoma cell invasion, migration and proliferation (10). Similarly, miR-7 has been demonstrated
to exert an anti-metastatic effect on gastric cancer by targeting
insulin-like growth factor-1 receptor (11). In certain cases, including the
suppression of the Myc oncogenic pathway, the combined effects of
several miRNAs have been observed (12). Therefore, a combined miRNA
therapeutics approach to target lung cancer has been proposed
(13). In terms of drug discovery,
the links between miRNAs and numerous human diseases and advances
in anti-miR chemistries (chemical modifications for improved
therapeutic properties) have suggested that the regulation of
miRNAs may lead to the next revolution in pharmaceutical research
(14). Until recently, there were
~20 clinical trials using miRNA- and small interfering RNA
(siRNA)-based therapeutics (15).
Strategies proposed for miRNA therapeutics include the artificial
introduction of miRNAs or antisense oligonucleotides to inhibit
miRNAs (16). In the present study,
miR-367-3p was predicted and experimentally validated for the
potential regulation of chka gene expression in breast cancer MCF-7
cells. The transfection of MCF-7 cells with miR-367-3p
significantly downregulated chka expression, induced
apoptosis and suppressed cell migration.
Materials and methods
Prediction of miRNAs targeting the
human chka mRNA transcript
The prediction of miRNAs targeting the human
chka mRNA transcript was performed using the microRNA.org website by submitting CHKA as the query
gene symbol. The predicted miRNAs were ranked according to the
mirSVR score (17). Minimum free
energy (MFE) for miRNA binding to the target was predicted by
RNAfold (18), mFold (19) and KineFold (20).
Cell lines, miRNA mimics, miRNA
inhibitors, and chka-3′-untranslated region (UTR) firefly
luciferase reporter plasmid
The malignant breast cancer MCF-7 (ATCC®
HTB-22™) cell line, the cervical cancer HeLa
(ATCC®CCL2™) cell line and the hepatoblastoma HepG2
(ATCC®HB8065™) cell line were obtained from the American
Type Culture Collection and cultured in Dulbecco's modified Eagle's
medium (DMEM; Gibco; cat. no. 41965039, Thermo Fisher Scientific,
Inc.), containing 10% heat-inactivated fetal bovine serum (FBS;
Gibco; cat. no. 26140087, Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin (cat. no. P0781, Sigma-Aldrich; Merck KGaA) and 100
mg/ml streptomycin (cat. no. 15140122, Sigma-Aldrich; Merck KGaA)
in an incubator at 37°C and 5% CO2. The mimic for
miR-367-3p (5′-AAUUGCACUUUAGCAAUGGUGA-3′) and its inhibitor were
purchased from Applied Biological Materials (cat. no. MCH01999).
miRIDIAN microRNA mimic Housekeeping Positive Control #2 (GAPDH)
(cat. no. CP-001000-02-05) and miRIDIAN microRNA mimic Negative
Control #1 (cat. no. CN-001000-01-05) were purchased from GE
Healthcare Dharmacon, Inc. The miRNA mimic was reconstituted in 1X
siRNA buffer provided by the manufacturer. The firefly luciferase
reporter construct containing the 3′-UTR of the chka gene
(pMirTarget-chka−3′-UTR) was purchased from OriGene
Technologies, Inc. (cat. no. SC212759).
Cell transfection
The miRNA mimics, inhibitors and plasmids were
transiently transfected into adherent MCF-7 cells cultured in
either 96-well (for miRNA target validation) or 24-well plates (for
other experiments) using Lipofectamine® 3000 (cat. no.
L3000015, Invitrogen; Thermo Fisher Scientific, Inc.). To begin
with, 1×104 and 1×105 cells were seeded into
96-well and 24-well plates, respectively, and cultured for 16–18 h
to achieve 70–80% confluency prior to transfection. The plasmid,
miRNA mimic and miRNA inhibitor were diluted accordingly with
Opti-MEM™ (cat. no. 31985062, Thermo Fisher Scientific, Inc.) to
obtain final transfected concentrations of 200 ng
pMirTarget-chka−3′UTR or pMirTarget empty vector, 25 nM
miRNA mimic and 25 nM miRNA inhibitor. In another tube,
Lipofectamine 3000 reagent (cat. no. L3000015, Invitrogen; Thermo
Fisher Scientific, Inc.) was diluted with Opti-MEM™ according to
the manufacturer's protocol. The two mixtures were mixed at a 1:1
ratio for 20 min at room temperature for the formation of
transfection complexes. For target validation in the 96-well plate,
50 µl the transfection complexes were added to each well containing
50 µl fresh complete DMEM. The medium was replaced after 6 h and
the cells were grown for a further 24 h prior to performing the
firefly luciferase activity assay. For other experiments in the
24-well plate, 100 µl transfection complexes were added to each
well containing 400 µl fresh complete DMEM, following by incubation
for a further 48 h.
Firefly luciferase assay
After transfection with the
pMirTarget-chka−3′-UTR plasmid containing the 3′-UTR of the
chka gene together with mimic for miR-367-3p
(5′-AAUUGCACUUUAGCAAUGGUGA-3′) or microRNA Negative Control #1 (GE
Healthcare Dharmacon, cat. no. CN-001000-01-05) and miR-367-3p
inhibitor (Applied Biological Materials, cat. no. MCH01999) as
described above, a total of 25 µl medium per well was removed from
the 96-well plate and 75 µl the Dual-Glo luciferase reagent (cat.
no. E2920, Promega Corporation) was added, followed by incubation
at room temperature for 10 min. The firefly luciferase activity
(relative light unit) was determined using the GloMax 20/20
luminometer (Promega Corporation). All luciferase assays were
performed in triplicate and the experiment was repeated ≥2 times.
The firefly luciferase activity of miRNA/inhibitor was normalized
with that of miR-NC.
RNA isolation and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). Total RNA was extracted from
the MCF-7, HeLa or HepG2 cells transfected with miR-367-3p,
miRIDIAN (negative control) or miR-GAPDH (positive control) using
the Total RNA Isolation kit (cat. no. K0732, Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol and
genomic DNA removal was performed using the RNase-Free DNase set
(cat. no. 79256, Qiagen GmbH). The RNA concentration was assessed
using a Nano Drop ND-1000 spectrophotometer (Thermo Fisher
Scientific, Inc.). Total RNA (1 µg) was reverse transcribed at 42°C
for 1 h using the RevertAid™ H minus First Strand cDNA Synthesis
kit (cat. no. K1631, Thermo Fisher Scientific, Inc.) in a total
volume of 20 µl. qPCR was performed using an ABI Prism 7500
Sequence Detection system (Thermo Fisher Scientific, Inc.) in a
total reaction volume of 13 µl consisting of 6.25 µl Power
SYBR™-Green PCR Master Μix (cat. no. 4367659, Thermo Fisher
Scientific, Inc.), 0.5 µl each of forward and reverse primers, 1 µl
cDNA and 4.75 µl double-distilled water. The PCR was run with an
initial denaturation at 95°C for 10 min, followed by 40 cycles of
95°C (10 sec) and 60°C (1 min); one cycle consisting of 95°C for 15
sec, 60°C for 1 min and 95°C for 15 sec was introduced to obtain
the dissociation curve for the analysis of PCR specificity. The
primers used were as follows: Tyrosine 3-monooxygenase/tryptophan
5-monooxygenase activation protein ζ (YWHAZ) forward,
5′-TTCTTGATCCCCAATGCTTC-3′ and reverse, 5′-ACTGGGTCTGGCCCTTAACT-3′;
ribosomal protein S18 (RPS18) forward, 5′-TGTGGTGTTGAGGAAAGCA-3′
and reverse, 5′-CTTCAGTCGCTCCAGGTCTT-3′; GAPDH forward,
5′-CAAGGTCATCCATGACAACTTTG-3′ and reverse,
5′-GTCCACCACCCTGTTGCTGTAG-3′; and chka forward,
5′-TCAGAGCAAACATCCGGAAGT−3′ and reverse,
5′-GGCGTAGTCCATGTACCCAAAT-3′. Relative gene expression levels
normalized to the geometric mean of YWHAZ and RPS18 Cq values were
determined using the 2−ΔΔCq method (21).
Western blot analysis
Following 72 h of transfection, MCF-7 cell lysates
were prepared using ProteoJET™ mammalian cell lysis reagent (Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
The protein concentration of the cell lysate was determined using
Bradford assay reagent (cat. no. 5000006, Bio-Rad Laboratories,
Inc.). Protein samples (30 µg) were loaded into the well of 12%
SDS-PAGE. Following separation with 12% SDS-PAGE, the proteins were
transferred onto a nitrocellulose membrane. The membrane was
blocked with 5% (w/v) skimmed milk in TBST buffer for 1 h at room
temperature prior to detection with rabbit primary β-actin
(dilution, 1:5,000; cat. no. ab8227; Abcam) and Chka (dilution,
1:1,000; cat. no. ab88053; Abcam) antibodies overnight at 4°C. The
membrane was washed ≥3 times with TBST (0.1% (v/v) Tween-20 in TBS)
buffer prior to incubation with HRP-conjugated secondary goat
anti-rabbit IgG antibody (dilution, 1:5,000; cat. no. 12–348;
Sigma-Aldrich, Merck KGaA) at room temperature for 1 h. The
membrane was then washed three times prior to detection with
SuperSignal™ West Femto maximum sensitivity substrate (cat. no.
34094, Thermo Fisher Scientific, Inc.). The signal was visualized
using the FUSION FX chemiluminescence imaging system (Vilber
Lourmat) and the band intensities were analyzed using ImageJ 1.49b
software (National Institutes of Health).
Apoptotic and dead cell count
Cellular apoptosis was examined using the Muse™
Annexin V & Dead Cell Assay kit (cat. no. MCH100105, Merck
Millipore) and analyzed using the Muse™ Cell Analyzer from EMD
Millipore, according to the manufacturer's protocol. Transfected
cells were detached by trypsinization, washed with PBS and
resuspended in fresh DMEM (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% FBS to at least 1×106 cells/ml. In one
tube, 100 µl cell suspension was mixed with 100 µl Muse™ Annexin V
& Dead Cell reagent, vortexed using a benchtop vortex mixer and
incubated for 30 min at room temperature in the dark. Stained cells
were counted using a Muse™ Cell Analyzer, according to the
manufacturer's protocol.
Scratch wound healing assay
Following treatment with miRNA mimics as described
earlier, a diameter wide scratch wound was gently created using a
P20 pipette tip into near confluent MCF-7 cells grown in a 6-well
plate. The growth medium containing serum with detached cells after
scratching was then discarded and the cells were washed twice
before 2 ml fresh serum-free medium were added to each well. Images
at the specified time points were captured using a DINO EYE
eye-piece camera (AnMo Electronics Corporation) and the distance
between the two edges of the scratch was measured using ImageJ
1.49b software (National Institutes of Health).
Statistical analysis
Data were analyzed using a Student's t-test or
one-way analysis of variance followed by a Tukey's HSD post
hoc test. P<0.05 was considered to indicate a statistically
significant difference, and all analyses were performed using SPSS
software version 22.0 (IBM Corp.). Data are presented as the mean ±
standard error of mean from 3 independent experiments unless
otherwise stated.
Results
miR-367-3p targets the 3′-UTR of chka
mRNA
A total of 25 potential miRNAs targeting the 3′-UTR
of the human chka mRNA transcript was predicted by
microRNA.org. Hsa-miR-367-3p (miRBase accession no.
MIMAT0000719) exhibited the highest mirSVR score of −1.0819. This
miRNA targets the sequence from nucleotides 1804 to 1825 of the
chka mRNA transcript (NM_001277.2). The analysis of MFE by
RNAfold, mFold and KineFold predicted the strong binding of
miR-367-3p to the target with MFE values of −3.1, −3.0 and −5.2
kcal/mol, respectively. As shown in Fig.
1, the base pairing at the seed region of miR-367-3p with the
target sequence fell into the most favorable category known as
8-mer, which contains perfect Watson-Crick base pairing at position
2–8 of the miRNAs and an adenine nucleotide across from position 1
of the miRNAs (22).
Validation of the predicted miR-367-3p was performed
using firefly luciferase and the 3′-UTR of the chka gene
fusion construct (pMirTarget-chka−3′-UTR) in MCF-7 cells. As
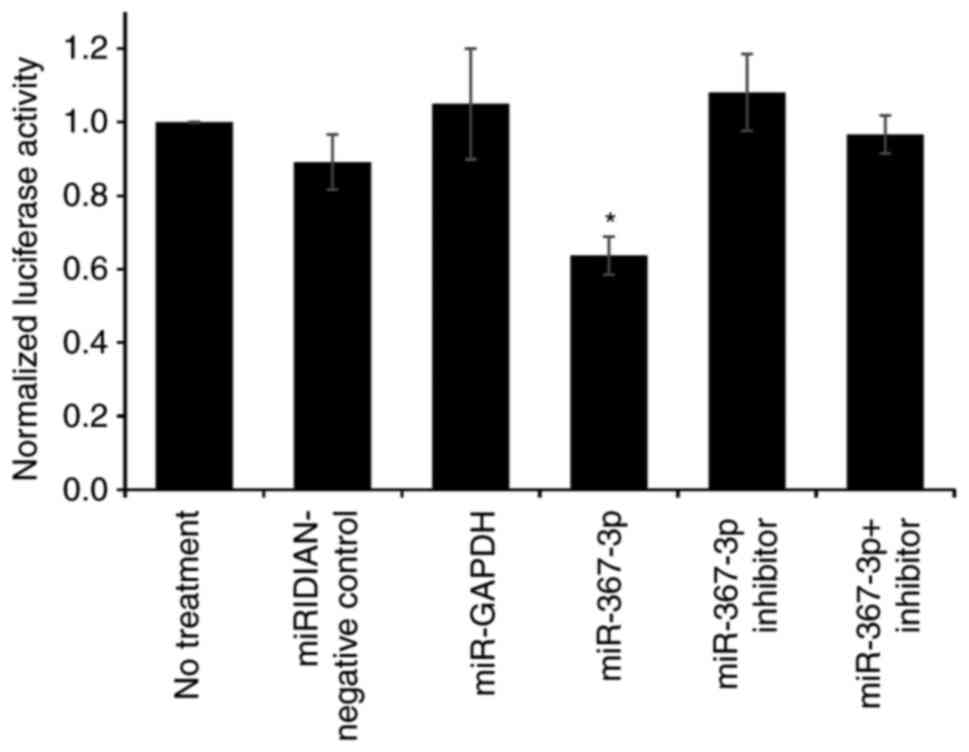
shown in Fig. 2, miR-367-3p
significantly downregulated the relative firefly luciferase
activity by ~40%, compared with the negative control miRNA-treated
cells (P=0.033). Co-treatment with mir-367-3p inhibitor reversed
the effect of its target miRNA, confirming the specific interaction
between this miRNA and this target 3′-UTR. Treatment with the
inhibitor alone slightly increased (not significant, P=0.9715) the
relative firefly luciferase activity compared with the negative
control, possibly due to the inhibition of endogenous
miR-367-3p.
miR-367-3p downregulates the
expression of chka in MCF-7, HeLa and HepG2 cell lines
The mimic of miR-367-3p was subsequently transfected
into MCF-7 cells to assess the potential of this miRNA to
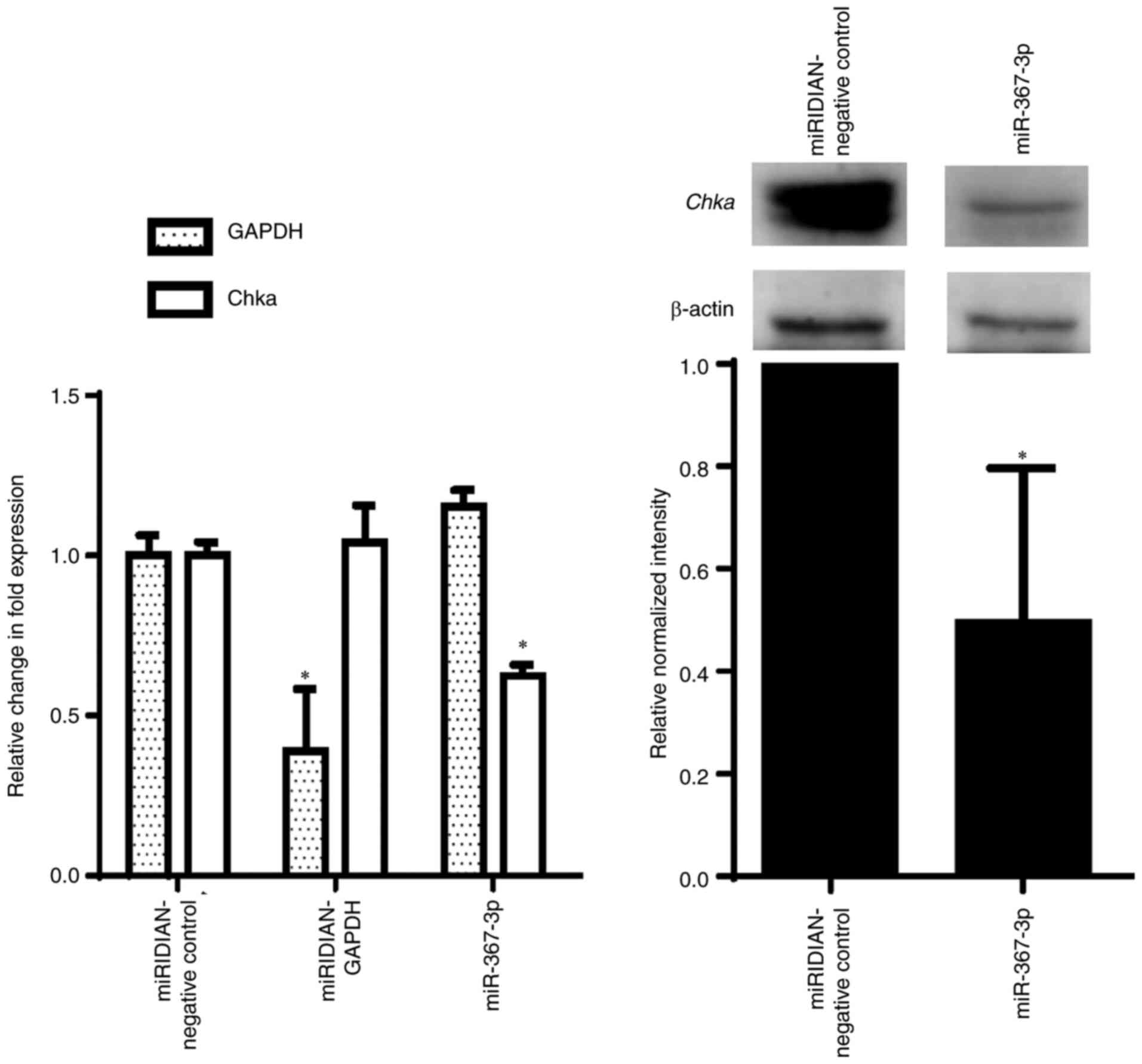
downregulate the expression of the chka gene. As shown in
Fig. 3 (left panel), miR-367-3p
significantly downregulated chka expression to ~60%,
compared with the negative control, which was similar to the level
of downregulation observed in the aforementioned luciferase assay.
Western blot analysis (Fig. 3, right
panel) also revealed a significantly lower Chka protein expression
of ~50%, compared with the negative control. The transfection of
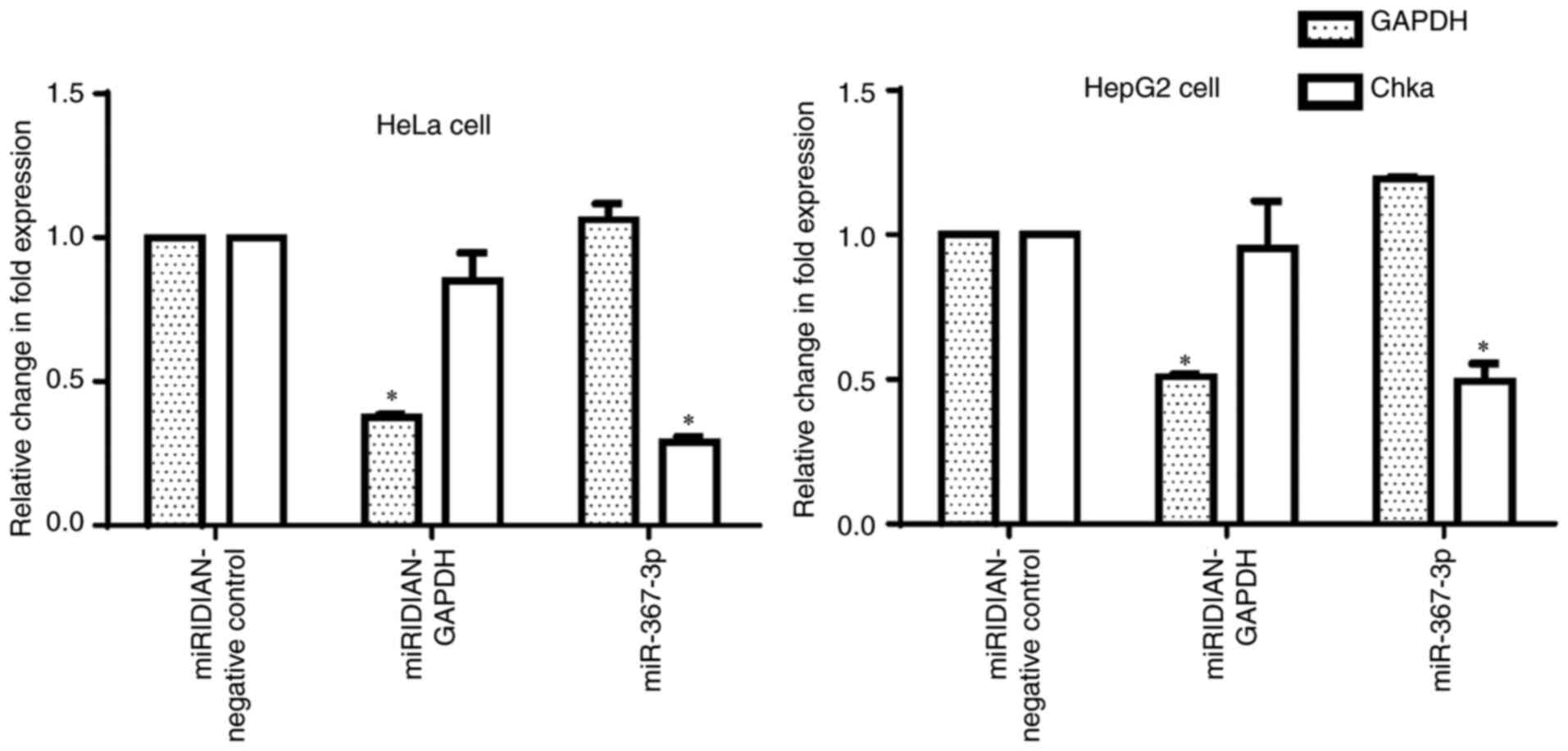
miR-367-3p into HeLa and HepG2 cell lines resulted in the
significant suppression of chka mRNA levels (Fig. 4), as was observed in the MCF-7 cells.
Taken together, these results confirmed the prediction that the
chka mRNA transcript is targeted and downregulated by
miR-367-3p.
miR-367-3p downregulation of chka
expression induces cell death and suppresses cell migration
The inhibition of ChK activity and the knockdown of
chka gene expression by RNA interference (RNAi) have been
reported to induce cancer cell death (4). Therefore, the effect of miR-367-3p
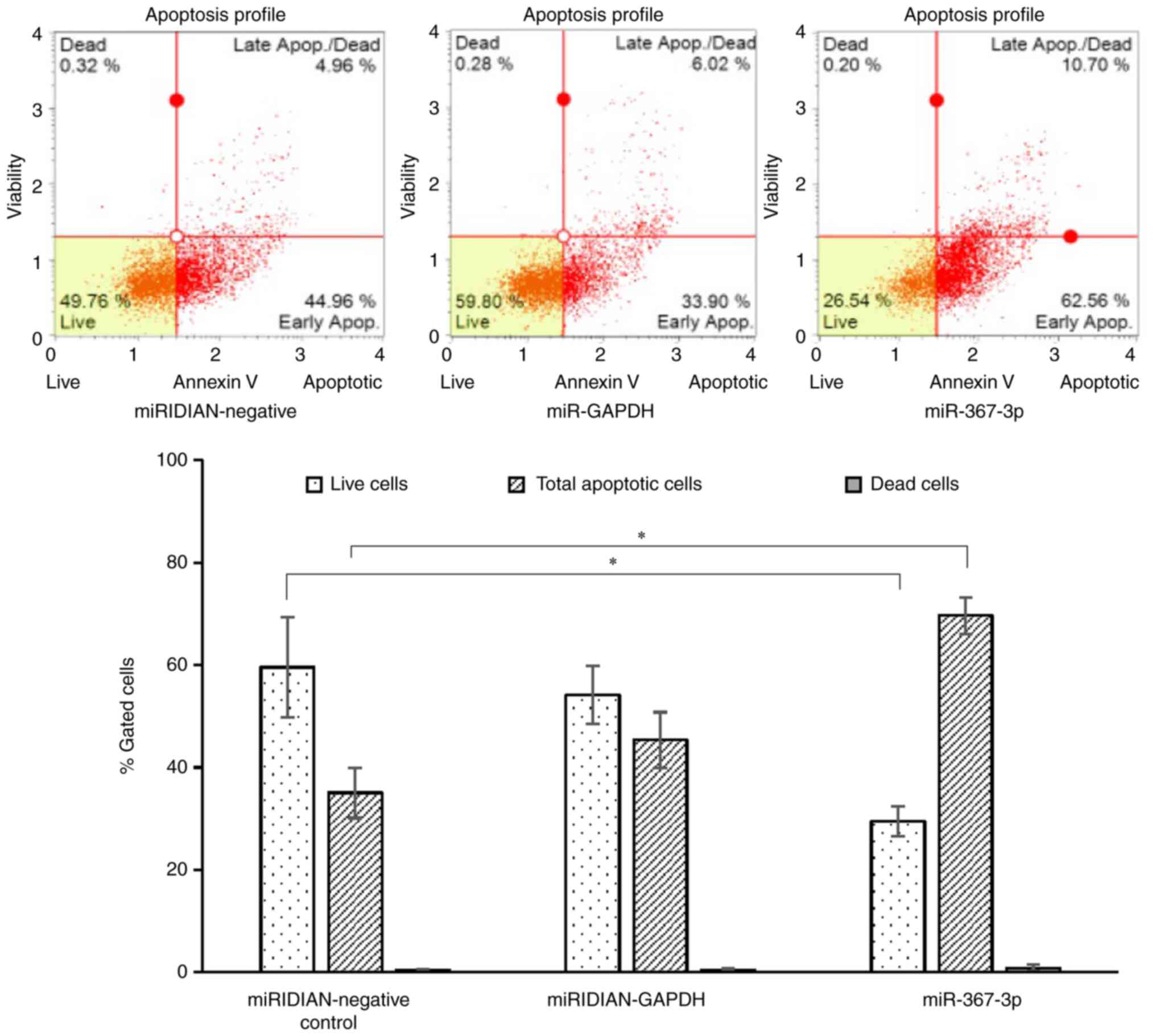
mimic on cellular apoptosis was investigated in the present study.
Based on the results presented in Fig.
5, statistically significant differences were observed in the
percentages of live cells and total apoptotic cells between the
cells transfected with negative control miRNA and the
miR-367-3p-transfected cells. These results suggested that
miR-367-3p induced cell death by targeting chka. The
inhibition of ChKa activity has been demonstrated to decrease the
migration of tumor cells (23,24).
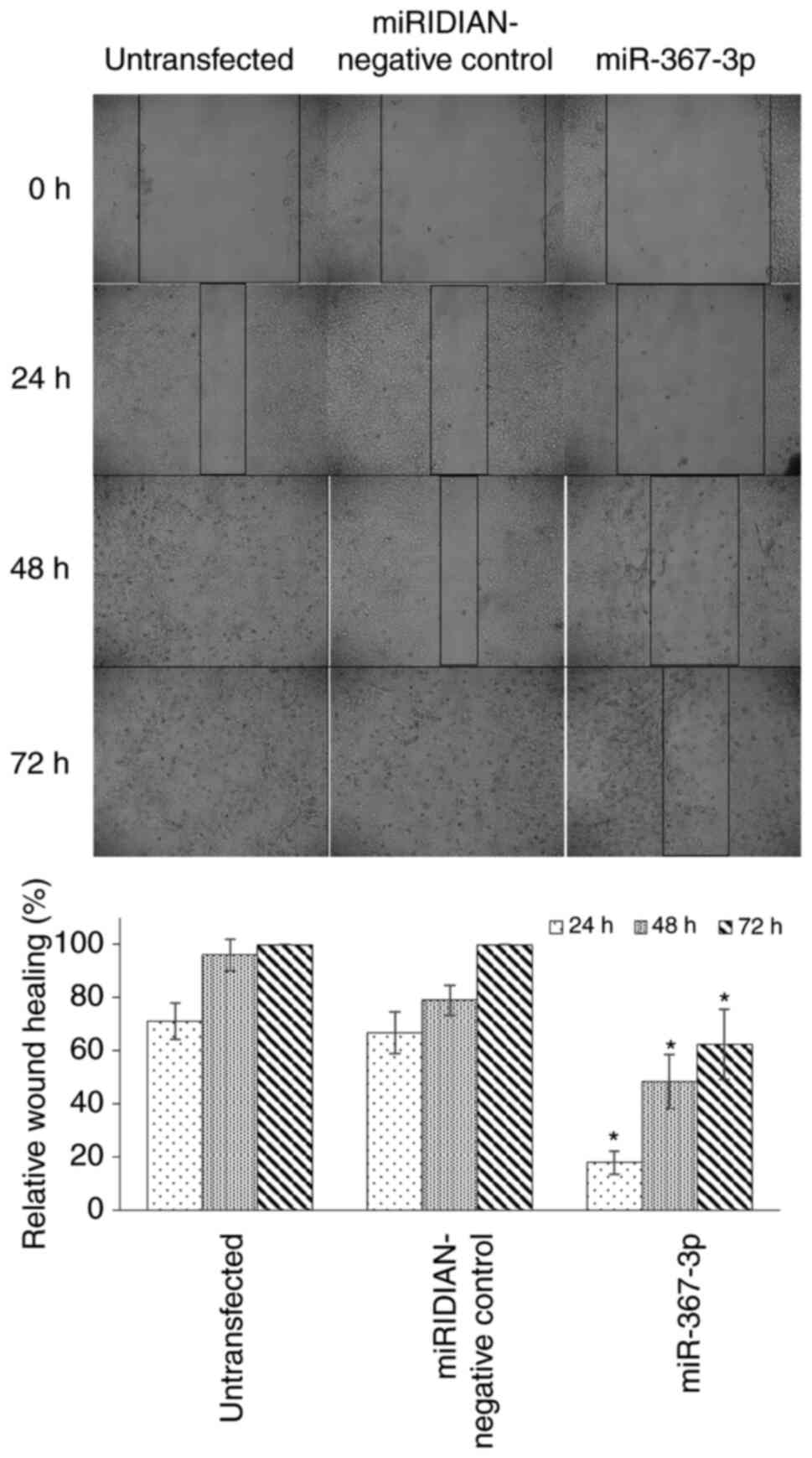
Therefore, the present study investigated whether the
downregulation of chka expression by miR-367-3p was able to
suppress MCF-7 cell migration. As shown in Fig. 6, miR-367-3p treatment significantly
suppressed cell migration, with only ~60% closure of the scratch,
compared with complete closure in the untreated and negative
control cells after 72 h.
Discussion
ChK overexpression has been implicated in various
types of cancer. The inhibition of the activity and the
downregulation of the expression of this enzyme have been applied
as promising anticancer strategies (6). However, the mechanisms that lead to the
higher expression of ChK in cancer cells are not yet well
understood. The present study investigated the possibility of the
regulation of chka gene expression by miRNAs. miR-367-3p was
predicted to target the 3′-UTR of the chka mRNA transcript
with strong affinity, based on low values of mirSVR and MFEs.
Target prediction analysis is crucial as the subsequent
experimental validation of miRNAs is time-consuming and costly
(25). mirSVR offers the optimal
performance among other prediction tools when the ranking of the
output is required (25). MFE of a
miRNA-target interaction is one of the determinants of the
silencing efficiency, as indicated by the improved silencing of the
Arabidopsis MYB81 gene by miR-159 following mutation to
incorporate the sequence for improved binding by miR-159 (26).
miR-367-3p originates from the 3′-end of the hairpin
loop sequence of miR-367, which is expressed as the miR-302/367
cluster. This cluster of miRNAs is mainly expressed in embryonic
stem cells and is able to reprogram somatic cells into pluripotency
(27). miR-367-3p has been reported
to enhance the efficacy of Sorafenib chemotherapy to suppress
hepatocellular carcinoma metastasis by enhancing the pathway
involving the androgen receptor (28). However, the ectopic expression of
miR-367-3p in non-small cell lung cancer cells has been
demonstrated to promote cell proliferation and cell cycle
progression, and inhibit apoptosis (29). According to Yan et al
(30), the same miRNA may serve
different roles in different types of cancer cell by acting as
either an oncogene or tumor suppressor. The contradictory effects
of miR-367 (whether this was miR-367-3p or miR-367-5p was not
specified) on cancer cell growth and the response to drugs have
been reported (31–35). The overexpression of miR-367 in
paclitaxel-sensitive ovarian cancer cells has been demonstrated to
further enhance the sensitivity to this drug (31). The overexpression of miR-367 has also
been demonstrated to inhibit the migration and invasion of gastric
cancer (32). By contrast, a higher
expression of miR-367 has been found to be associated with an
unfavorable prognosis of high-grade glioma (33), resected non-small cell lung cancer
(34) and pancreatic ductal
adenocarcinoma (35). Therefore, the
effects of miR-367-3p on cancer cell survival may be cell
type-dependent. The results of the present study suggested that
this miRNA may be used as a novel therapeutic agent against breast
cancer cells overexpressing ChK. The suppression of Runx2 by
miR-135 and miR-203 has been shown to inhibit breast cancer cell
migration in vitro, as well as tumor growth and metastasis
in vivo (36). The results of
wound healing assay in the present study also demonstrated that the
suppression of chka by miR-367-3p inhibited MCF-7 breast
cancer cell migration in vitro, which may produce the same
anti-metastatic effect in vivo.
Due to the higher level of chka expression in
different cancer cells, the inhibition of Chka activity has become
a promising anticancer therapy. Consequently, the synthesis and
testing of various small-molecule Chka inhibitors have gained
increasing attention over the past decade (6). The inhibition of Chka enzyme activity
or the knockdown of chka gene expression by RNAi
specifically induces the death of cancer cells, but not that of
normal cells (1). The RNAi of
chka has been demonstrated to decrease tumor cell viability
and enhance the efficacy of prodrug enzyme activation treatment in
cancer therapy (37). Recently,
degradable dextran nanopolymer, which is less toxic and more
suitable for cancer therapy, has been synthesized to deliver siRNA
targeting chka in breast cancer cells (38). mir-367-3p, which downregulates the
chka gene, as reported in the present study, may also be
used in a similar manner. Replenishing tumor suppressor miRNAs,
including miR-34 by lipid nanoparticles, miR-200 by liposomal
carriers and miR-26a by the adeno-associated virus-mediated
expression is one the approaches in miRNA therapeutics (39). The downregulation of the chka
gene by RNAi or miRNAs may be more effective than activity
inhibition by small molecules alone in killing cancer cells, based
on the ChKA non-catalytic role in promoting cancer cell survival
(40).
The anti-proliferative effects of miRNAs have
previously been reported in humans. For example, miR-195-5p
functions as an anti-oncogene by targeting PHF-19, leading to the
suppression of hepatoma cell invasion, migration and proliferation
(10). Similarly, miR-7 has been
shown to exert an anti-metastatic effect on gastric cancer by
targeting insulin-like growth factor-1 receptor (11). In the present study, it was confirmed
that the 3′-UTR of the chka transcript was the target of
miR-367-3p and the expression of chka was downregulated by
this miRNA. The decreased level of chka resulted in higher
levels of apoptosis and a lower migration of MCF-7 cells. Although
the expression of miR-367-3p was not determined following the
transfection of its mimic or inhibitor, the downregulation of the
target (chka) gene expression by the miR-367-3p mimic was
consistent throughout all the experiments in the present study. The
results of the present study suggested that miR-367-3p may be a
promising miRNA candidate for further investigation into the roles
of miRNAs in cancer development involving the dysregulation of
chka gene expression. However, further studies are required
to elucidate the effects of chka downregulation by
miR-367-3p on cell proliferation, invasion and anticancer drug
resistance in more cancer cell lines in order to fully understand
the role of chka attenuation by this miRNA in cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Universiti
Sains Malaysia Individual Research University Grant (grant no.
1001/PPSK/8012239) and the Malaysia Ministry of Higher Education
Fundamental Research Grant Scheme (grant no. 203/PPSK/6171222), and
Ms Raikundalia was a PhD candidate supported by the Malaysia
International Scholarship.
Availability of data and materials
The data generated and/or analyzed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
SR and SAFMS performed the experiments and analyzed
the data. LLF designed the experiments, analyzed the data and
drafted the initial manuscript. WCST conceived and designed the
experiments, analyzed the data and was a major contributor in
drafting the manuscript. All authors have read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arlauckas SP, Popov AV and Delikatny EJ:
Choline kinase alpha-putting the choK-hold on tumor metabolism.
Prog Lipid Res. 63:28–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanchez-Lopez E, Zimmerman T, Gomez del
Pulgar T, Moyer MP, Lacal Sanjuan JC and Cebrian A: Choline kinase
inhibition induces exacerbated endoplasmic reticulum stress and
triggers apoptosis via CHOP in cancer cells. Cell Death Dis.
4:e9332013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chang CC, Few LL, Konrad M and See Too WC:
Phosphorylation of human choline kinase beta by protein kinase A:
Its impact on activity and inhibition. PLoS One. 11:e01547022016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gruber J, See Too WC, Wong MT, Lavie A,
McSorley T and Konrad M: Balance of human choline kinase isoforms
is critical for cell cycle regulation: Implications for the
development of choline kinase-targeted cancer therapy. FEBS J.
279:1915–1928. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu G and Vance DE: Choline kinase and its
function. Biochem Cell Biol. 88:559–564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gómez-Pérez V, McSorley T, See Too WC,
Konrad M and Campos JM: Novel 4-amino bis-pyridinium and
bis-quinolinium derivatives as choline kinase inhibitors with
antiproliferative activity against the human breast cancer SKBR-3
cell line. ChemMedChem. 7:663–669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hirschberger S, Hinske LC and Kreth S:
miRNAs: Dynamic regulators of immune cell functions in inflammation
and cancer. Cancer Lett. 431:11–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lan H, Lu H, Wang X and Jin H: MicroRNAs
as potential biomarkers in cancer: Opportunities and challenges.
Biomed Res Int. 2015:1250942015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu H, Hu YW, Zhao JY, Hu XM, Li SF, Wang
YC, Gao JJ, Sha YH, Kang CM, Lin L, et al: MicroRNA-195-5p acts as
an anti-oncogene by targeting PHF19 in hepatocellular carcinoma.
Oncol Rep. 34:175–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao X, Dou W, He L, Liang S, Tie J, Liu
C, Li T, Lu Y, Mo P, Shi Y, et al: MicroRNA-7 functions as an
anti-metastatic microRNA in gastric cancer by targeting
insulin-like growth factor-1 receptor. Oncogene. 32:1363–1372.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bueno MJ, Gómez de Cedrón M, Gómez-López
G, Pérez de Castro I, Di Lisio L, Montes-Moreno S, Martínez N,
Guerrero M, Sánchez-Martínez R, Santos J, et al: Combinatorial
effects of microRNAs to suppress the Myc oncogenic pathway. Blood.
117:6255–6266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kasinski AL, Kelnar K, Stahlhut C,
Orellana E, Zhao J, Shimer E, Dysart S, Chen X, Bader AG and Slack
FJ: A combinatorial microRNA therapeutics approach to suppressing
non-small cell lung cancer. Oncogene. 34:3547–3555. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
van Rooij E, Purcell AL and Levin AA:
Developing microRNA therapeutics. Circ Res. 110:496–507. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chakraborty C, Sharma AR, Sharma G, George
C, Doss CGP and Lee SS: Therapeutic miRNA and siRNA: Moving from
bench to clinic as next generation medicine. Mol Ther Nucleic Acid.
8:132–143. 2017. View Article : Google Scholar
|
|
16
|
Broderick JA and Zamore PD: MicroRNA
therapeutics. Gene Ther. 18:1104–1110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lorenz WA and Clote P: Computing the
partition function for kinetically trapped RNA secondary
structures. PLoS One. 6:e161782011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zuker M: Mfold web server for nucleic acid
folding and hybridization prediction. Nucleic Acids Res.
31:3406–3415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xayaphoummine A, Bucher T and Isambert H:
Kinefold web server for RNA/DNA folding path and structure
prediction including pseudoknots and knots. Nucleic Acids Res.
33:(Web Server Issue). W605–W610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Granata A, Nicoletti R, Tinaglia V, De
Cecco L, Pisanu ME, Ricci A, Podo F, Canevari S, Iorio E, Bagnoli M
and Mezzanzanica D: Choline kinase-alpha by regulating cell
aggressiveness and drug sensitivity is a potential druggable target
for ovarian cancer. Br J Cancer. 110:330–340. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mariotto E, Viola G, Ronca R, Persano L,
Aveic S, Bhujwalla ZM, Mori N, Accordi B, Serafin V, López-Cara LC
and Bortolozzi R: Choline kinase alpha inhibition by EB-3D triggers
cellular senescence, reduces tumor growth and metastatic
dissemination in breast cancer. Cancers (Basel). 10:3912018.
View Article : Google Scholar
|
|
25
|
Oliveira AC, Bovolenta LA, Nachtigall PG,
Herkenhoff ME, Lemke N and Pinhal D: Combining results from
distinct MicroRNA target prediction tools enhances the performance
of analyses. Front Genet. 8:592017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng Z, Reichel M, Deveson I, Wong G, Li
J and Millar AA: Target RNA secondary structure is a major
determinant of miR159 efficacy. Plant Physiol. 174:1764–1778. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rosa A and Brivanlou AH: Regulatory
non-coding RNAs in pluripotent stem cells. Int J Mol Sci.
14:14346–14373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu J, Lin H, Li G, Sun Y, Chen J, Shi L,
Cai X and Chang C: The miR-367-3p increases sorafenib chemotherapy
efficacy to suppress hepatocellular carcinoma metastasis through
altering the androgen receptor signals. EBioMedicine. 12:55–67.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiao G, Gao X, Sun X, Yang C, Zhang B, Sun
R, Huang G, Li X, Liu J, Du N, et al: miR-367 promotes tumor growth
by inhibiting FBXW7 in NSCLC. Oncol Rep. 38:1190–1198. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan W, Jiang X, Wang G, Li W, Zhai C, Chen
S, Shang F, Zhao Z and Yu W: Cyto-biological effects of
microRNA-424-5p on human colorectal cancer cells. Oncol Lett.
20:1202020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen N, Chon HS, Xiong Y, Marchion DC,
Judson PL, Hakam A, Gonzalez-Bosquet J, Permuth-Wey J, Wenham RM,
Apte SM, et al: Human cancer cell line microRNAs associated with in
vitro sensitivity to paclitaxel. Oncol Rep. 31:376–383. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bin Z, Dedong H, Xiangjie F, Hongwei X and
Qinghui Y: The microRNA-367 inhibits the invasion and metastasis of
gastric cancer by directly repressing Rab23. Genet Test Mol
Biomarkers. 19:69–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guan Y, Chen L, Bao Y, Qiu B, Pang C, Cui
R and Wang Y: High miR-196a and low miR-367 cooperatively correlate
with unfavorable prognosis of high-grade glioma. Int J Clin Exp
Pathol. 8:6576–6588. 2015.PubMed/NCBI
|
|
34
|
Campayo M, Navarro A, Vinõlas N, Diaz T,
Tejero R, Gimferrer JM, Molins L, Cabanas ML, Ramirez J, Monzo M
and Marrades R: Low miR-145 and high miR-367 are associated with
unfavourable prognosis in resected nonsmall cell lung cancer. Eur
Respir J. 41:1172–1178. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu Z, Xu Y, Zhao J, Liu Q, Feng W, Fan J
and Wang P: miR-367 promotes epithelial-to-mesenchymal transition
and invasion of pancreatic ductal adenocarcinoma cells by targeting
the Smad7-TGF-β signalling pathway. Br J Cancer. 112:1367–1375.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Taipaleenmäki H, Browne G, Akech J, Zustin
J, van Wijnen AJ, Stein JL, Hesse E, Stein GS and Lian JB:
Targeting of Runx2 by miR-135 and miR-203 impairs progression of
breast cancer and metastatic bone disease. Cancer Res.
75:1433–1444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li C, Penet MF, Wildes F, Takagi T, Chen
Z, Winnard PT, Artemov D and Bhujwalla ZM: Nanoplex delivery of
siRNA and prodrug enzyme for multimodality image-guided molecular
pathway targeted cancer therapy. ACS Nano. 4:6707–6716. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Z, Krishnamachary B and Bhujwalla ZM:
Degradable dextran nanopolymer as a carrier for choline kinase
(Chok) siRNA cancer therapy. Nanomaterials (Basel). 6:342016.
View Article : Google Scholar
|
|
39
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Falcon SC, Hudson CS, Huang Y, Mortimore
M, Golec JM, Charlton PA, Weber P and Sundaram H: A non-catalytic
role of choline kinase alpha is important in promoting cancer cell
survival. Oncogenesis. 2:e382013. View Article : Google Scholar : PubMed/NCBI
|