Introduction
Oral squamous cell carcinoma (OSCC) is the most
common malignancy of the head and neck (1), and is associated with high incidence,
invasion and metastasis rates, as well as a poor prognosis. In the
United States, ~50,000 individuals suffered from OSCC and 8,000
individuals died in 2013 (2). OSCC
has gradually become a serious problem worldwide, despite the
increase in basic research and the rapid development of clinical
treatment in the past few decades. Furthermore, the 5-year survival
rate of this disease has not significantly improved (3). With the identification of a causal link
between Helicobacter pylori (H. pylori) infection and
the occurrence of gastric tumors in the 1900s (4), an increased understanding has been
achieved regarding the association between bacteria and tumors.
Nevertheless, how oral microorganisms influence the tumor
microenvironment (TME), and how they promote tumor development
during tumor progression, remains unknown (1). It has been reported that a large number
of pro-inflammatory cytokines secreted by microorganisms can
transform tumor cells into more aggressive phenotypes by regulating
oncogenes (5). Tumor growth and
metastasis are not only affected by neoplastic cells, but also by
the TME (6). Surgery combined with
chemotherapy is an effective treatment for OSCC (1). However, chemotherapy is not a
first-line therapy for OSCC, since most OSCC cases develop
resistance to chemotherapeutic reagents (7). Periodontitis has been suggested to be
associated with the TME of OSCC, which could be involved in the
development of chemotherapy resistance in OSCC (7). Therefore, further investigations
examining the regulatory mechanism of the TME and identifying novel
biomarkers to optimize patient selection for this therapy have
gained clinical significance and theoretical value.
Porphyromonas gingivalis (P.
gingivalis) is a key pathogen in periodontal disease, and is
known as an independent microorganism risk factor for increased
tumor mortality (8). P.
gingivalis is a special oral pathogen and a potentially
independent microbiological factor that increases the risk of
mortality in patients with OSCC (9).
A previous study revealed the presence of the P. gingivalis
antigen in the tissues of gingival squamous cell carcinoma
(10). An abnormal increase in the
number of P. gingivalis is an important factor in causing
the imbalance of the local microecology of the oral cavity
(7). The dominant bacterial pathogen
in periodontitis is P. gingivalis, which affects the
condition of the TME, increasing the likelihood of OSCC development
(11).
Chemokines are a small molecular protein family that
serve a role in chemical signaling during cell activation and
differentiation, as well as in the process of movement.
Furthermore, chemokines and their signaling receptors are important
in the TME of OSCC (12). According
to a previous study, P. gingivalis induces immune cells to
secrete chemokines [including C-X-C motif chemokine ligand (CXCL)1,
CXCL2, CXCL5 and CXCL8] and promotes tumor growth (13). CXCL2 is a type of small molecular
pro-inflammatory chemokine and a member of four subfamilies (C, CC,
CXC and CX3C), serving a role in coding protein secretion, immune
regulation and the promotion of tumor angiogenesis (14). CXCL2 is highly expressed in breast,
colon, prostate and liver tumors, and is closely associated with
tumor growth, invasion and distant metastasis (15). However, the other biological
functions of CXCL2 in the P. gingivalis-infected TME of OSCC
remain unknown.
Polymorphonuclear neutrophils (PMNs) are the primary
defense line of the immune system, and are the most abundant white
blood cells in the peripheral blood circulation (16). Tumor-associated neutrophils (TANs)
are infiltrates in the TME, and there are two polarized
manifestations of TANs: Defined as the N1 phenotype, one has the
ability of typical tumor inhibition, while another, defined as the
N2 phenotype, has typical tumor promotion, the differentiation of
which depends on the factors of the TME (17,18). In
addition, CD66b+ has been assessed as a PMN marker in
gastric tumors using immunohistochemistry (IHC), and
CD66b+ has been revealed to be a reliable marker to
identify the phenotype of TANs that promote tumor growth (19). It has also been revealed that TANs
are primarily activated by CXC chemokines (20).
In order to analyze the immunoexpression and
clinical significance of CXCL2 and TANs in the TME of P.
gingivalis infection, the immunohistochemistry evaluation,
bioinformatics analyses and statistical analysis were
performed.
Materials and methods
Patient selection
The present study included 205 surgical specimens
from patients [age range, 36–90 years; mean age, 63 years; 103 men
(50.2%); 102 women (49.8%); 76 patients <60 years (37.1%); 129
patients ≥60 years (62.9%)] who underwent surgical treatment for
primary OSCC at the Oncology Department of Oral and Maxillofacial
Surgery of The First Affiliated Hospital of Xinjiang Medical
University (Urumqi, China) between March 2007 and March 2019.
Surgical resection was performed in all 205 patients with OSCC. The
TNM and clinicopathological classification and staging of patients
with OSCC were performed according to the American Joint Committee
on Cancer (AJCC) guidelines (21).
The inclusion criteria were as follows: i) patients with OSCC
located in the tongue, buccal cavity, lip, floor of the mouth, or
gingival and retromolar area as confirmed via biopsy; ii) patients
who did not undergo any treatment before; and iii) patients with
completed clinical data and follow-up. Clinical data, including
age, sex, survival status, differentiation, tobacco and alcohol
consumption, TNM stage, clinical stage, recurrence, periodontal
condition, treatment (surgery, radiotherapy and/or chemotherapy,
post-operative adjuvant radiotherapy or chemotherapy) and tumor
size, were collected. In addition, the present study included 20
cases of benign tumor and non-tumor patients [age range, 23–48
years; mean age, 33 years; 10 men (50.0%); 10 women (50.0%)] who
underwent surgical treatment at the Oncology Department of Oral and
Maxillofacial Surgery of The First Affiliated Hospital of Xinjiang
Medical University (Urumqi, China) between May 2017 and May 2019.
The inclusion criteria were as follows: i) Patients with benign
tumors located in the oral cavity; ii) patients with benign tumors
located in the parotid gland, sublingual gland and submandibular
gland; iii) patients with inflammatory lesions requiring surgical
removal. The present study was approved by the ethics review board
of the First Affiliated Hospital of Xinjiang Medical University
(Urumqi, China) at the original time of data collection (approval
no. IACUC20180411-13) and written consent was obtained at the
original time of data collection.
Bioinformatics analyses
Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) is a public
functional genomics data repository of high-accuracy gene
expression data, chips and microarrays. A total of two gene
expression datasets, GSE87539 (22)
and GSE138206 (23), were downloaded
from GEO (Affymetrix GPL570 platform; Affymetrix Human Genome U133
Plus 2.0 Array; Thermo Fisher Scientific, Inc.). The probes were
converted into the corresponding gene symbols according to the
annotation information in the platform. The GSE87539 dataset
contained 3 samples of non-infected oral epithelial cells and 3
samples of P. gingivalis-infected oral epithelial cells. The
GSE138206 dataset contained 6 OSCC samples and 6 non-cancerous
samples.
The differentially expressed genes (DEGs) between
GSE87539 and GSE138206 samples were screened using GEO2R
(http://www.ncbi.nlm.nih.gov/geo/geo2r). GEO2R is an
interactive web tool that allows users to compare ≥2 datasets in a
GEO series to identify DEGs across the experimental conditions.
Hierarchical clustering of hub genes was performed using UCSU
Cancer Genomics Browser (http://genome-cancer.ucsc.edu) (24). The adjusted P-values (adj. P) and
Benjamini and Hochberg false discovery rate were applied to provide
a balance between the discovery of statistically significant genes
and limitations of false-positives. The probe sets without
corresponding gene symbols or genes with >1 probe set were
removed or averaged, respectively. Log fold-change >1 and adj.
P<0.01 were considered to be statistically significant.
IHC evaluation
A total of 205 OSCC samples from patients were
immersed in 10% neutral buffered formalin for fixation for 3 days
at room temperature. A total of 205 paraffin-embedded OSCC samples
from patients and consecutive 4-µm-thick sections cut from paraffin
blocks were used for IHC evaluation. The expression levels of P.
gingivalis were evaluated, and 119 samples exhibited strong
expression. These 119 samples were used in the evaluation of the
expression levels of CXCL2 and TANs. Non-cancerous samples were
obtained from patients who suffered from benign tumors or
inflammatory disease at the Oncological Department of Oral and
Maxillofacial Surgery, The Affiliated Stomatology Hospital of The
First Affiliated Hospital of Xinjiang Medical University (Urumqi,
China) and maintained at the Institute of Stomatology of Science
and Technology Building of the First Affiliated Hospital of
Xinjiang Medical University (Urumqi, China). The IHC procedure was
as follows: Slides were heated at 65°C for 2 h. Sections that
adhered to the slides were deparaffinized in xylene and rehydrated
in a gradient ethanol (50% ethanol for 10 min, 70% ethanol for 10
min, 80% ethanol for 10 min, 95% ethanol for 10 min, 100% ethanol
for 10 min three times), followed by submerging into EDTA antigenic
retrieval buffer, treatment with 3% hydrogen peroxide for 15 min at
room temperature and incubation with 1% bovine serum albumin
(Gibco; Thermo Fisher Scientific, Inc.) for 30 min at 37°C. The
samples were then incubated with anti-P. gingivalis
(dilution, 1:200; Dia-An, lnc; this antibody is not a commercial
product, and it was prepared at the time), anti-CXCL2 (dilution,
1:500; cat. no. bs-1162R; BIOSS) and anti-TANs (CD66b+;
dilution, 1:500; cat. no. ab197678; Abcam) antibodies overnight at
4°C. After being washed with PBS with 1% Tween, the slides were
incubated with secondary antibodies [Goat Anti-Mouse IgG H & L
(HRP); dilution, 1:2,000; cat. no. ab205719; Abcam; and Goat
Anti-Rabbit IgG H & L (HRP); dilution, 1:2,000; cat. no.
ab205718; Abcam] for 1 h at 37°C, followed by incubation with
streptavidin horseradish peroxidase complex (Thermo Fisher
Scientific, Inc.). The sample sections were then immersed in
3,3′-diaminobenzidine for 5 min at room temperature, counterstained
with 10% Mayer's hematoxylin for 1 min at room temperature,
dehydrated and mounted.
Each tumor specimen was observed by light microscopy
and examined under 10 microscopic fields with a digital camera
(AxiocamMRc; Zeiss AG) attached to a microscope (magnification,
×200; Axioskop 2 Plus Zeiss AG) to evaluate the immunoexpression
levels of P. gingivalis, CXCL2 and TANs. Two experienced
pathologists evaluated the three immunoexpression profiles of
cancerous tissues and non-cancerous tissues. The final score was
determined by multiplying the immunostaining intensity by the
percentage of positive immunostaining cells. Subsequently, the
cancerous samples were classified into three groups: 0, absent
immunostaining; 1, weak immunostaining; and 2, strong
immunostaining. The proportion of positive cells was classified as
follows: 0, 0; 1, 1–25; 2, 26–75; 3, 76–100%. The final staining
score was calculated by multiplying the staining intensity score by
extent of staining score. A final staining score of ≥3 was
considered positive, and others were classified as low
expression.
Statistical analysis
Statistical analysis was performed using SPSS
statistical software (version 21.0; IBM Corp.). Data are presented
as value (%) and the association of P. gingivalis, CXCL2 and
TANs immunoexpression levels in neoplastic cells with the
clinicopathological variables was determined using χ2 or
Fisher's exact tests (two-sided). For these analyses, the absence
and weak immunoexpression levels were grouped together, obtaining
final groups of absent/weak tumor immunoexpression and strong
neoplastic immunoexpression. The cumulative survival rate (CSR)
probability in 10 years was estimated using the Kaplan-Meier
method, and survival curves were compared via a log-rank test. The
follow-up period considered for cumulative survival consisted of
the time between the date of surgery and mortality or the date of
the last information collection regarding the patient. Cox
regression analysis of the survival data was performed to test the
statistical significance of regression coefficients. P<0.05 was
considered to indicate a statistically significant difference.
Results
Study population
Between March 2007 and March 2019, a total of 205
samples from patients who were newly diagnosed with OSCC were
retrospectively analyzed. There were slightly more men than women
(50.2%) and patients with an age of ≥60 years (62.9%). Alcohol
(18.0%) and tobacco (28.3%) consumption were considered as partial
risk factors in patients.
With regard to survival status, the majority of
patients in the present study were alive at the time of
retrospective analysis (71.7%). Furthermore, the patients were
clinically classified as stage I–II (54.1%) and III–IV (45.9%), and
the TNM stages were classified as T1-T2 (65.9%) and T3-T4 (34.1%),
and N0 (70.7%) and N(+) (29.3%). There were no patients with
distant metastases at the time of physical examination (data not
shown).
Among the enrolled patients, 67.3 and 23.9% were
classified as well and moderately differentiated, while 8.8% were
classified as poorly differentiated, according to the
histopathological grade of tumor malignancy as described by Bryne
et al (25). Recurrence
occurred in 14.6% of patients, and there were 101 patients (49.3%)
with poor periodontal condition. Only 13 patients (6.3%) underwent
no therapy, 90 patients (43.9%) underwent surgical therapy, 14
patients (6.8%) received radiotherapy and/or chemotherapy and 88
patients (42.9%) received comprehension therapy (post-operative
adjuvant radiotherapy and/or chemotherapy; Table I).
 | Table I.General information of 205 patients
with oral squamous cell carcinoma. |
Table I.
General information of 205 patients
with oral squamous cell carcinoma.
| Variable | No. of patients
(%) |
|---|
| Sex |
|
|
Male | 103 (50.2) |
|
Female | 102 (49.8) |
| Age, years |
|
|
<60 | 76 (37.1) |
|
≥60 | 129 (62.9) |
| Survival
status |
|
|
Alive | 147 (71.7) |
|
Dead | 58 (28.3) |
| Tobacco
consumption |
|
|
Yes | 58 (28.3) |
| No | 147 (71.7) |
| Alcohol
consumption |
|
|
Yes | 37 (18.0) |
| No | 168 (82.0) |
| Clinical stage |
|
|
I–II | 111 (54.1) |
|
III–IV | 94 (45.9) |
| T
stagea |
|
|
T1-2 | 135 (65.9) |
|
T3-4 | 70 (34.1) |
| N
stagea |
|
| N0 | 145 (70.7) |
|
N(+) | 60 (29.3) |
|
Differentiation |
|
|
Well | 138 (67.3) |
|
Moderate | 49 (23.9) |
|
Poor | 18 (8.8) |
| Recurrence |
|
|
Yes | 30 (14.6) |
| No | 175 (85.4) |
| Periodontal
condition |
|
|
Well | 104 (50.7) |
|
Poor | 101 (49.3) |
| Treatment |
|
|
None | 13 (6.3) |
|
Surgery | 90 (43.9) |
|
Chemotherapy +
radiotherapy | 14 (6.8) |
|
Comprehensive | 88 (42.9) |
IHC expression of P. gingivalis in TME
of OSCC
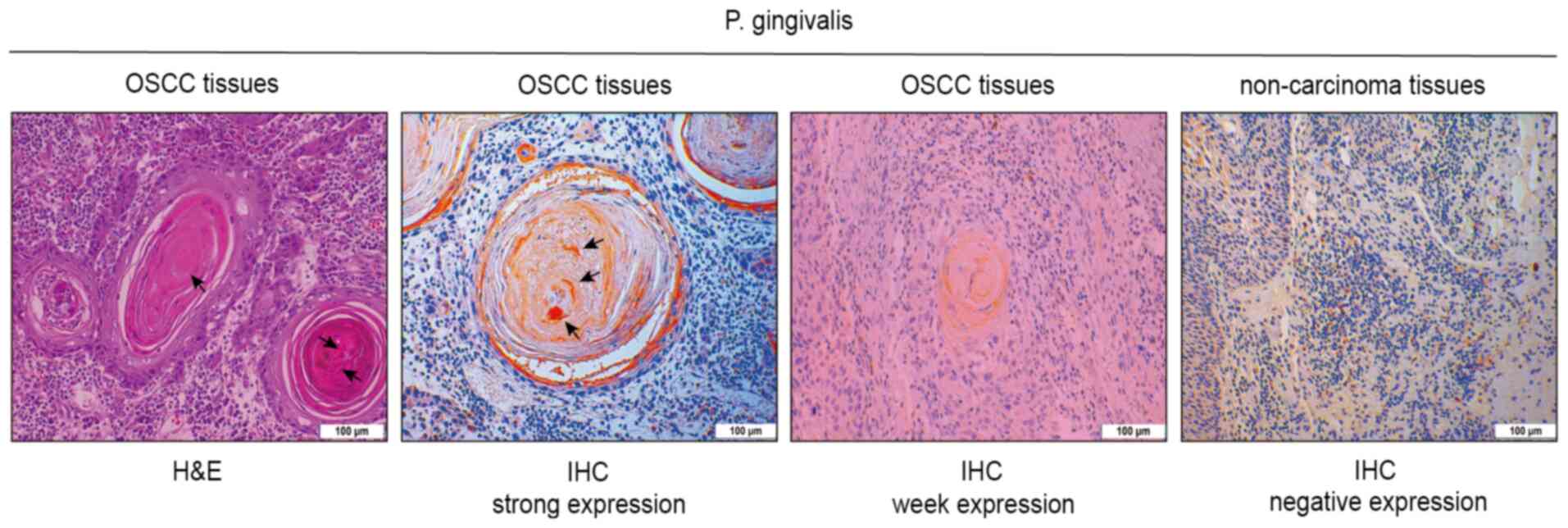
IHC expression of P. gingivalis in OSCC was
weakly positive in 86 samples, while the expression was strongly
positive in 119 samples. P. gingivalis immunoexpression was
predominant in the cytoplasm of neoplastic cells. The strong
immunoexpression of P. gingivalis was detected in carcinoma
nests, while negative immunoexpression was observed in
non-cancerous samples (Fig. 1).
Clinical variables, including the survival status,
differentiation, tobacco consumption, T stage, N stage, clinical
stage, periodontal condition, tumor size and treatment methods. The
result revealed that death, poor differentiation, tobacco
consumption, advanced T stage, N stage and clinical stage, poor
periodontal condition, large size of tumor and no treatment
exhibited significant associations with strong immunoexpression of
P. gingivalis (Table
II).
 | Table II.Immunohistochemical expression of
P. gingivalis in samples from 205 patients with oral
squamous cell carcinoma according to clinical data and
follow-up. |
Table II.
Immunohistochemical expression of
P. gingivalis in samples from 205 patients with oral
squamous cell carcinoma according to clinical data and
follow-up.
|
| P.
gingivalis |
|
|---|
|
|
|
|
|---|
| Variable | Weak, n (%) | Strong, n (%) | P-value |
|---|
| Sex |
|
| 0.233 |
|
Male | 39 (45.3) | 64 (53.8) |
|
|
Female | 47 (54.7) | 55 (46.2) |
|
| Age, years |
|
| 0.361 |
|
<60 | 35 (40.7) | 41 (34.5) |
|
|
≥60 | 51 (59.3) | 78 (65.5) |
|
| Survival
status |
|
|
<0.001a |
|
Alive | 75 (87.2) | 72 (60.5) |
|
|
Dead | 11 (12.8) | 47 (39.5) |
|
|
Differentiation |
|
|
<0.001a |
|
Well | 70 (81.4) | 68 (57.1) |
|
|
Moderate | 16 (18.6) | 33 (27.7) |
|
|
Poor | 0 (0.0) | 18 (15.1) |
|
| Tobacco
consumption |
|
| 0.047a |
|
Yes | 18 (20.9) | 40 (33.6) |
|
| No | 68 (79.1) | 79 (66.4) |
|
| Alcohol
consumption |
|
| 0.346 |
|
Yes | 12 (14.0) | 24 (20.2) |
|
| No | 74 (86.0) | 95(79.8) |
|
| T
stageb |
|
|
<0.001a |
|
T1-2 | 73 (84.9) | 62 (52.1) |
|
|
T3-4 | 13 (15.1) | 57 (47.9) |
|
| N
stageb |
|
| 0.011a |
| N0 | 69 (80.2) | 76 (63.9) |
|
|
N(+) | 17 (19.8) | 43 (36.1) |
|
| Clinical stage |
|
|
<0.001a |
|
I–II | 63 (73.3) | 48 (40.3) |
|
|
III–IV | 23 (26.7) | 71 (59.7) |
|
| Recurrence |
|
| 0.301 |
|
Yes | 10 (11.6) | 20 (16.8) |
|
| No | 76 (88.4) | 99 (83.2) |
|
| Periodontal
condition |
|
| 0.018a |
|
Well | 52 (60.5) | 52 (43.7) |
|
|
Poor | 34 (39.5) | 67 (56.3) |
|
| Tumor size, cm |
|
| 0.007a |
|
<2.9 | 51 (59.3) | 48 (40.3) |
|
|
≥2.9 | 35 (40.7) | 71 (59.7) |
|
| Treatment |
|
| 0.007a |
|
None | 2 (2.3) | 11 (9.2) |
|
|
Surgery | 49 (57.0) | 41 (34.5) |
|
|
Chemotherapy +
radiotherapy | 4 (4.7) | 10 (8.4) |
|
|
Comprehensive | 31 (36.0) | 57 (47.9) |
|
Identification of CXCL2 as a DEG in
the TME of OSCC infected with P. gingivalis by bioinformatics
analyses
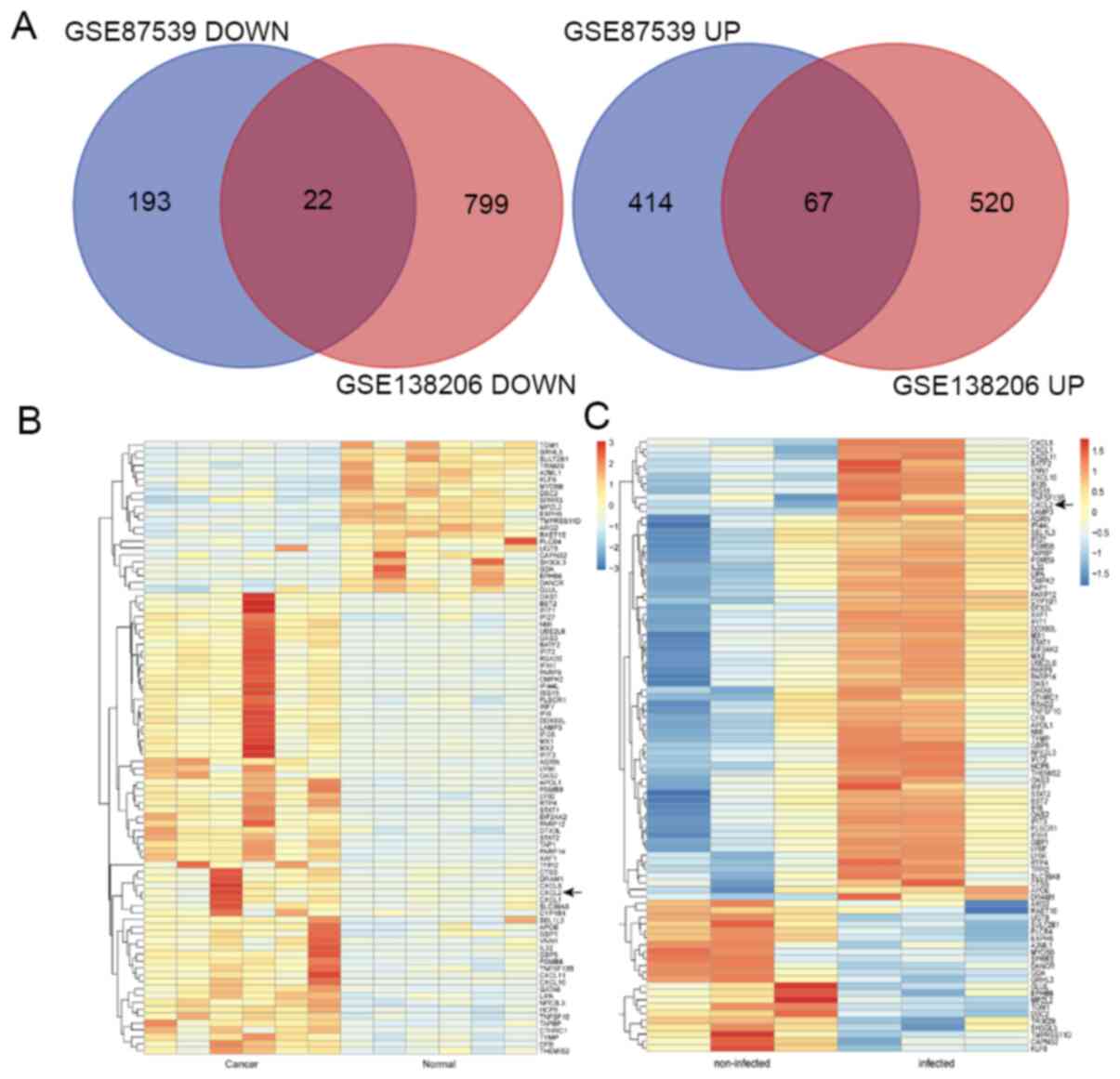
After standardizing the microarray results, the DEGs
(26,469 in GSE87539 and 9,443 in GSE138206) were identified (data
not shown). The overlap between the two datasets containing 89
genes is presented in the Venn diagram (Fig. 2A), and included 67 upregulated genes
and 22 downregulated genes.
Hierarchical clustering demonstrated that CXCL2 was
a DEG between GSE87539 (P. gingivalis-infected oral
epithelial samples compared with non-infected oral epithelial
samples) and GSE138206 (cancerous samples compared with
non-cancerous samples; Fig. 2B and
C).
IHC expression of CXCL2 and TANs in P.
gingivalis-infected TME of OSCC
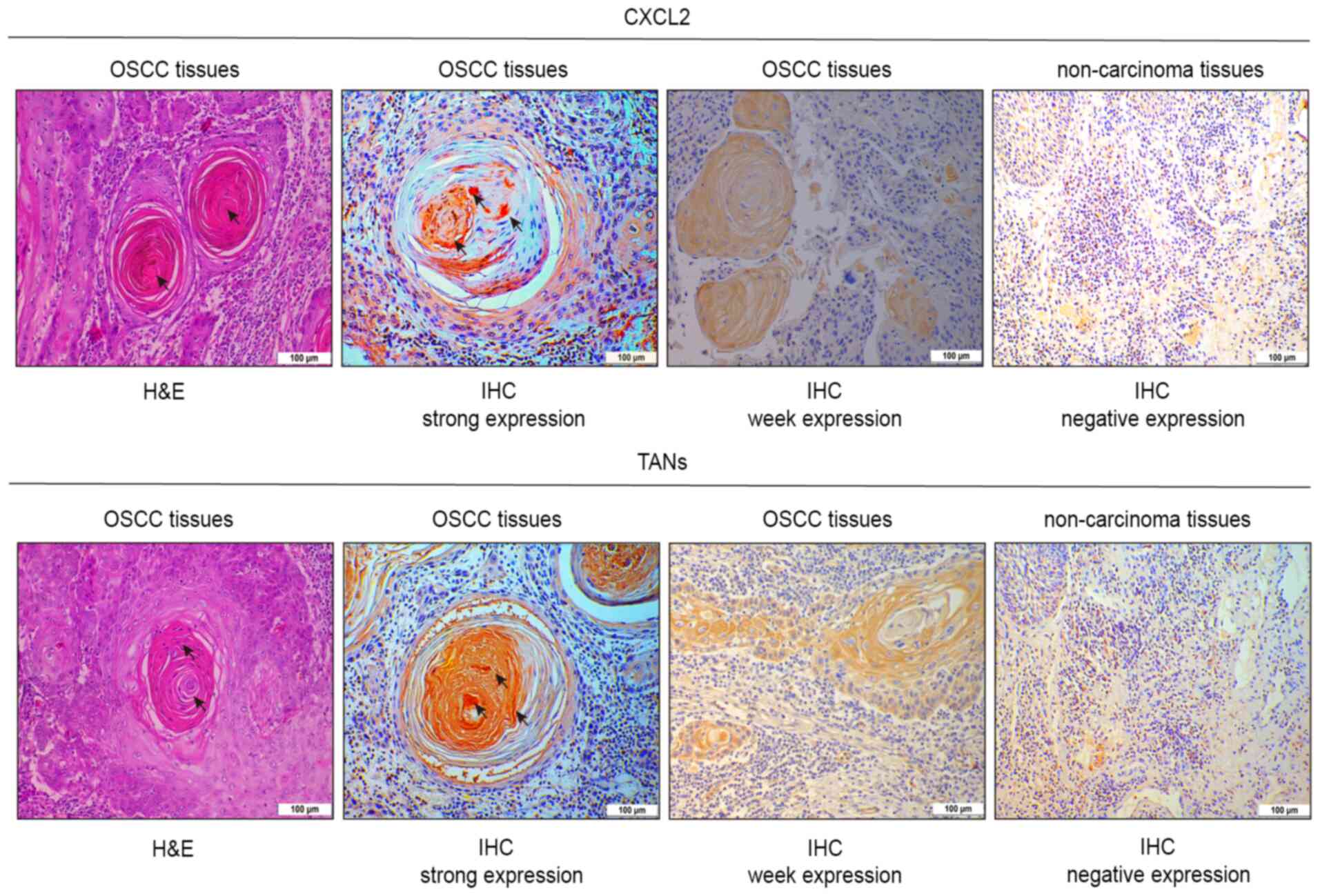
A total of 119 samples with high expression levels
of P. gingivalis were selected for IHC examination of CXCL2
and TANs. In 119 samples with high P. gingivalis expression,
CXCL2 and TANs were weakly positively expressed in 30 and 38
samples, respectively, but were strongly positively expressed in 89
and 81 samples, respectively. CXCL2 was strongly positively
expressed in the OSCC samples, but was negatively expressed in the
non-cancerous samples (Fig. 3).
CD66b+ TANs are associated with a poor
prognosis of patients with cervical cancer (26). In the present study,
CD66b+ TANs exhibited strong positive expression in OSCC
samples and negative expression in non-cancerous samples (Fig. 3).
Clinical variables, including age, survival status,
T stage, tumor size and treatment. The result revealed that high
age, death, advanced T stage, large size of tumor and no treatment
exhibited significant associations with strong CXCL2
expression.
In the present study, clinical variables, including
advanced T stage and clinical stage, were statistically associated
with strong expression of TANs (Table
III). The present results indicated that the expression levels
of P. gingivalis were positively associated with the
expression levels of CXCL2 and TANs (Table IV).
 | Table III.Immunohistochemical expression of
CXCL2 and TANs in 119 samples with strong staining for
Porphyromonas gingivalis from patients with oral squamous
cell carcinoma according to clinical data and follow-up. |
Table III.
Immunohistochemical expression of
CXCL2 and TANs in 119 samples with strong staining for
Porphyromonas gingivalis from patients with oral squamous
cell carcinoma according to clinical data and follow-up.
|
| CXCL2 |
| TANs |
|
|---|
|
|
|
|
|
|
|---|
| Variable | Weak, n (%) | Strong, n (%) | P-value | Weak, n (%) | Strong, n (%) | P-value |
|---|
| Sex |
|
| 0.631 |
|
| 0.824 |
|
Male | 15 (50.0) | 49 (55.1) |
| 21 (55.3) | 43 (53.1) |
|
|
Female | 15 (50.0) | 40 (44.9) |
| 17 (44.7) | 38 (46.9) |
|
| Age, years |
|
| 0.038a |
|
| 0.229 |
|
<60 | 15 (50.0) | 26 (29.2) |
| 16 (42.1) | 25 (30.9) |
|
|
≥60 | 15 (50.0) | 63 (70.8) |
| 22 (57.9) | 56 (69.1) |
|
| Survival
status |
|
| 0.003a |
|
| 0.226 |
|
Alive | 25 (83.3) | 47 (52.8) |
| 26 (68.4) | 46 (56.8) |
|
|
Dead | 5 (16.7) | 42 (47.2) |
| 12 (31.6) | 35 (43.2) |
|
|
Differentiation |
|
| 0.279 |
|
| 0.556 |
|
Well | 20 (66.7) | 48 (53.9) |
| 24 (63.2) | 44 (54.3) |
|
|
Moderate | 8 (26.7) | 25 (28.1) |
| 10 (26.3) | 23 (28.4) |
|
|
Poor | 2 (6.7) | 16 (18.0) |
| 4 (10.5) | 14 (17.3) |
|
| Tobacco
consumption |
|
| 0.628 |
|
| 0.460 |
|
Yes | 9 (30.0) | 31 (34.8) |
| 11 (28.9) | 29 (35.8) |
|
| No | 21 (70.0) | 58 (65.2) |
| 27 (71.1) | 52 (64.2) |
|
| Alcohol
consumption |
|
| 0.189 |
|
| 0.158 |
|
Yes | 7 (23.3) | 17 (19.1) |
| 5 (13.2) | 19 (23.5) |
|
| No | 23 (76.7) | 72 (80.9) |
| 33 (86.8) | 62 (76.5) |
|
| T
stageb |
|
| 0.023a |
|
| 0.015a |
|
T1-2 | 21 (70.0) | 41 (46.1) |
| 26 (68.4) | 36 (44.4) |
|
|
T3-4 | 9 (30.0) | 48 (53.9) |
| 12 (31.6) | 45 (55.6) |
|
| N
stageb |
|
| 0.091 |
|
| 0.053 |
| N0 | 23 (76.7) | 53 (59.6) |
| 29 (76.3) | 47 (58.0) |
|
|
N(+) | 7 (23.3) | 36 (40.4) |
| 9 (23.7) | 34 (42.0) |
|
| Clinical stage |
|
| 0.093 |
|
| 0.002a |
|
I–II | 16 (53.3) | 32 (36.0) |
| 23 (60.5) | 25 (30.9) |
|
|
III–IV | 14 (46.7) | 57 (64.0) |
| 15 (39.5) | 56 (69.1) |
|
| Recurrence |
|
| 0.589 |
|
| 0.075 |
|
Yes | 6 (20.0) | 14 (15.7) |
| 3 (7.9) | 17 (21.0) |
|
| No | 24 (80.0) | 75 (84.3) |
| 35 (92.1) | 64 (79.0) |
|
| Periodontal
condition |
|
| 0.369 |
|
| 0.068 |
|
Well | 11 (36.7) | 41 (46.1) |
| 12 (31.6) | 40 (49.4) |
|
|
Poor | 19 (63.3) | 48 (53.9) |
| 26 (68.4) | 41 (50.6) |
|
| Tumor size, cm |
|
| 0.011a |
|
| 0.141 |
|
<2.9 | 18 (60.0) | 30 (33.7) |
| 19 (50.0) | 29 (35.8) |
|
|
≥2.9 | 12 (40.0) | 59 (66.3) |
| 19 (50.0) | 52 (64.2) |
|
| Treatment |
|
|
<0.001a |
|
| 0.848 |
|
None | 3 (10.0) | 8 (9.0) |
| 4 (10.5) | 7 (8.6) |
|
|
Surgery | 11 (36.7) | 30 (33.7) |
| 13 (34.2) | 28 (34.6) |
|
|
Chemotherapy +
radiotherapy | 0 (0.0) | 10 (11.2) |
| 2 (5.3) | 8 (9.9) |
|
|
Comprehensive | 16 (53.3) | 41 (46.1) |
| 19 (50.0) | 38 (46.9) |
|
 | Table IV.Association between P.
gingivalis immunohistochemical expression and CXCL2 and TANs in
205 patients. |
Table IV.
Association between P.
gingivalis immunohistochemical expression and CXCL2 and TANs in
205 patients.
|
| CXCL2 |
| TANs |
|
|---|
|
|
|
|
|
|
|---|
| P.
gingivalis | Weak, n (%) | Strong, n (%) | P-value | Weak, n (%) | Strong, n (%) | P-value |
|---|
| Weak | 69 (80.2) | 17
(19.8) |
<0.001a | 57 (66.3) | 29
(33.7) |
<0.001a |
| Strong | 30 (25.5) | 89
(74.8) |
| 38 (31.9) | 81
(68.1) |
|
| Total | 99 (48.3) | 106 (51.7) |
| 95 (46.3) | 110 (53.7) |
|
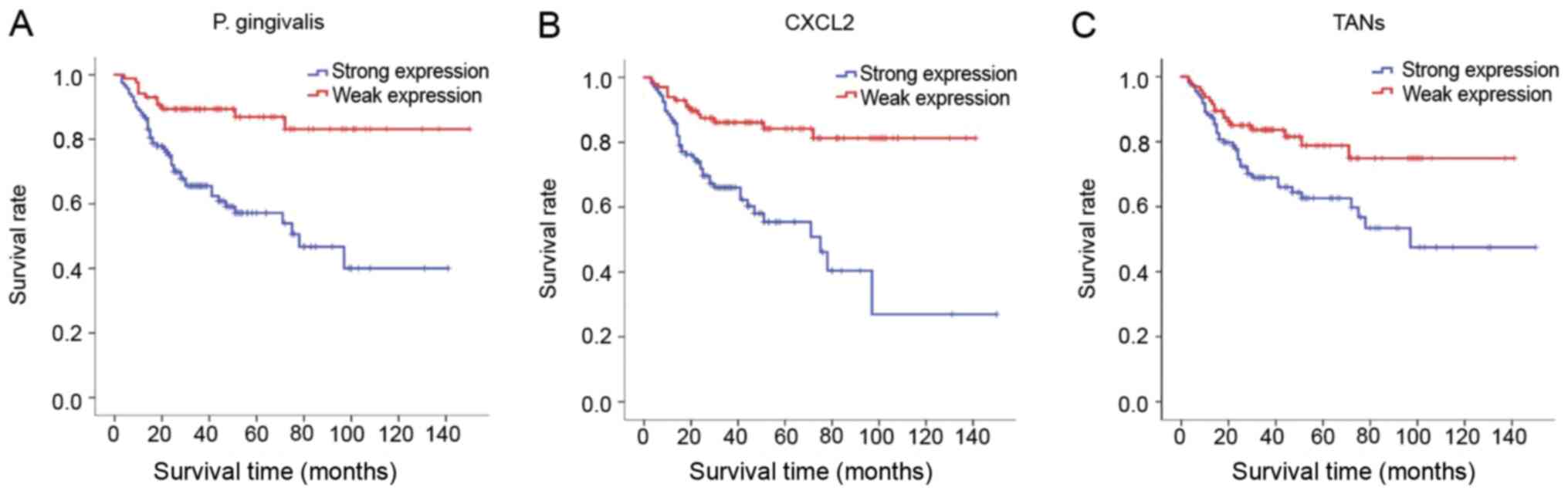
Cumulative survival analysis
In univariate analysis of the 10-year CSR, the
following parameters exhibited an association with the survival
rate: Age, tumor size, body mass index, alcohol consumption,
recurrence and neck dissection. Furthermore, the immunoexpression
levels of P. gingivalis, CXCL2 and TANs were significantly
associated with the 10-year CSR. Patients with a high
immunoexpression of P. gingivalis had an increased risk with
a lower 10-year CSR [hazard ratio (HR), 0.260; 95% CI, 0.135–0.503;
P<0.001], and those with high immunoexpression levels of CXCL2
(HR, 0.283; 95% CI, 0.156–0.514; P<0.001) and TANs (HR, 0.494;
95% CI, 0.283–0.862; P=0.013) also had an enhanced risk with a
lower 10-year CSR compared with patients with low immunoexpression
levels of P. gingivalis, CXCL2 and TANs (Table V).
 | Table V.Univariate analysis of Cox risk ratio
model regression in 205 patients with oral squamous cell
carcinoma. |
Table V.
Univariate analysis of Cox risk ratio
model regression in 205 patients with oral squamous cell
carcinoma.
| Variable | No. of patients
(%) | P-value | HR | HR (95% CI) |
|---|
| Sex |
| 0.684 | 0.898 | 0.535–1.507 |
|
Male | 103 (50.2) |
|
|
|
|
Female | 102 (49.8) |
|
|
|
| Age, years |
| 0.023a | 0.503 | 0.277–0.911 |
|
<60 | 76
(37.1) |
|
|
|
|
≥60 | 129 (62.9) |
|
|
|
| Clinical stage |
| 0.502 | 1.194 | 0.711–2.006 |
|
I–II | 111 (54.1) |
|
|
|
|
III–IV | 94
(45.9) |
|
|
|
| Tumor size, cm |
| 0.027a | 0.820 | 0.069–1.097 |
|
<2.9 | 99
(48.3) |
|
|
|
|
≥2.9 | 106 (51.7) |
|
|
|
| BMI |
| 0.003a | 0.453 | 0.270–0.761 |
|
<22.5 | 72
(35.1) |
|
|
|
|
≥22.5 | 133 (64.9) |
|
|
|
| Tobacco
consumption |
| 0.941 | 1.022 | 0.579–1.803 |
| No | 147 (71.7) |
|
|
|
|
Yes | 58
(28.3) |
|
|
|
| Alcohol
consumption |
| 0.019a | 0.511 | 0.231–1.134 |
| No | 169 (82.4) |
|
|
|
|
Yes | 36
(17.6) |
|
|
|
| Recurrence |
|
<0.001a | 0.801 | 0.587–1.939 |
| No | 175 (85.4) |
|
|
|
|
Yes | 30
(14.6) |
|
|
|
| T
stageb |
| 0.132 | 1.501 | 0.884–2.547 |
|
T1-2 | 135 (65.9) |
|
|
|
|
T3-4 | 70
(34.1) |
|
|
|
| N
stageb |
| 0.847 | 1.059 | 0.594–1.888 |
| N0 | 145 (70.7) |
|
|
|
|
N1-3 | 60
(29.3) |
|
|
|
| Neck
dissection |
| 0.003a | 2.206 | 0.495–0.858 |
|
None | 88
(42.9) |
|
|
|
|
Yes | 117 (57.1) |
|
|
|
| Periodontal
condition |
| 0.636 | 1.133 | 0.676–1.898 |
|
Poor | 101 (49.3) |
|
|
|
|
Well | 104 (50.7) |
|
|
|
| P.
gingivalis |
|
<0.001a | 0.260 | 0.135–0.503 |
|
Weak | 86
(42.0) |
|
|
|
|
Strong | 119 (58.0) |
|
|
|
| CXCL2 |
|
<0.001a | 0.283 | 0.156–0.514 |
|
Weak | 99
(48.3) |
|
|
|
|
Strong | 106 (51.7) |
|
|
|
| TANs |
| 0.013a | 0.494 | 0.283–0.862 |
|
Weak | 95
(46.3) |
|
|
|
|
Strong | 110 (53.7) |
|
|
|
In the multivariate survival analysis of the 10-year
CSR, the results demonstrated that recurrence, BMI, CXCL2
expression, P. gingivalis levels, TANs expression and
alcohol consumption were independent risk factors for the prognosis
of patients with OSCC (Table VI;
Fig. 4).
 | Table VI.Multivariate analysis using Cox risk
ratio regression model in 205 patients with oral squamous cell
carcinoma. |
Table VI.
Multivariate analysis using Cox risk
ratio regression model in 205 patients with oral squamous cell
carcinoma.
|
|
|
|
|
|
| HR (95% CI) |
|---|
|
|
|
|
|
|
|
|
|---|
| Variable | B | SE | Wald | P-value | HR | Upper | Lower |
|---|
| Age (<60 vs. ≥60
years) | 0.510 | 0.361 | 1.994 | 0.158 | 0.600 | 0.296 | 1.219 |
| Recurrence (no vs.
yes) | 1.370 | 0.328 | 17.446 |
<0.001a | 0.254 | 0.134 | 0.483 |
| BMI (<22.5 vs.
≥22.5) | 0.665 | 0.311 | 4.574 | 0.030a | 0.550 | 0.320 | 0.944 |
| Tumor size (<2.9
vs. ≥ 2.9) | 0.303 | 0.300 | 1.015 | 0.314 | 0.739 | 0.410 | 1.331 |
| Neck dissection (no
vs. yes) | 0.093 | 0.174 | 0.287 | 0.592 | 0.911 | 0.648 | 1.281 |
| CXCL2 (weak vs.
strong) | 0.854 | 0.362 | 5.549 | 0.018a | 0.426 | 0.209 | 2.866 |
| P.
gingivalis (weak vs. strong) | 0.688 | 0.399 | 2.975 | 0.035a | 0.503 | 0.230 | 1.098 |
| TANs (weak vs.
strong) | 0.688 | 0.310 | 0.549 | 0.039a | 0.795 | 0.433 | 1.459 |
| Alcohol consumption
(no vs. yes) | 0.134 | 0.063 | 4.494 | 0.034a | 0.543 | 0.610 | 1.094 |
Discussion
As aforementioned, there are a number of factors,
including tobacco and alcohol use, as well as microbiological
agents, that serve important roles in the progression of OSCC
(20). Microorganisms are recognized
as the primary risk factors for OSCC carcinogenesis (27), and the role of microorganisms in the
promotion of OSCC has gradually become a novel area of research. An
epidemiological study has demonstrated that 16–18% of carcinomas
occur due to inflammation (28).
Previous studies have also demonstrated that microbial pathogens,
including Epstein-Barr virus, and hepatitis B and C virus, serve a
role in tumor development (29–31).
Furthermore, bacterial pathogens are associated with carcinogenesis
with H. pylori infection, which is a causative factor in
chronic gastritis, and aids in the development of gastric cancer
(4).
It has been reported that P. gingivalis is
associated with the progression and metastasis of OSCC (32). P. gingivalis invades and
exists in the neoplastic cells, reproduces and survives in the
cytoplasm of infected cells, and spreads to the neighboring cells
(33). In addition, following
invasion, P. gingivalis evades the immune clearance
mechanism of the host, meaning it survives, reproduces and affects
the biological functions of immune cells (34). When P. gingivalis invades OSCC
tissues, it is able to recruit myeloid-derived suppressor cells
(MDSCs) by expressing factors, such as CXCL2 and IL-6 (35). Simultaneously, P. gingivalis
promotes tumor progression by recruiting MDSCs by increasing the
secretion of IL-6 and CXCL2 from infected oral dysplastic
keratinocytes (35). Entry of
microbial metabolites into the TME promotes tumor progression by
eliciting tumor-potentiating immune cell responses (35).
In the present study, staining of P.
gingivalis in OSCC samples was observed, and this was present
to a lesser extent in the non-carcinoma samples. Furthermore, it
was identified that staining was mainly localized in the cytoplasm
of malignant cells. These findings indicated that, at the
histological site, the bacteria have the ability to invade
neoplastic cells in vivo. In addition, stronger P.
gingivalis staining was identified to be associated with a poor
prognosis.
Associations between oral cancer and tooth loss or
periodontal disease have been reported in a previous study
(36). IL-6 and IL-8 have been
identified as vital cytokines involved in periodontitis under P.
gingivalis infection (37). IL-6
has been verified as a biomarker to illustrate the role of P.
gingivalis infection in favor of OSCC initiation and
progression (38). IL-8 is commonly
secreted in the TME, which promotes tumor progression via the
chemotaxis of MDSCs (39). P.
gingivalis expresses multiple types of virulence factors that
serve different roles in subverting the host immune response
(40). Different secreted virulence
factors of P. gingivalis may exert contrasting influences on
the production of IL-8 via various mechanisms (41). CXCL2 has been reported to promote the
generation of monocytic MDSCs (42).
Furthermore, CXCL5 and CCL5 are associated with tumor progression
(43). The present results suggested
that CXCL2 and TANs were markedly increased in OSCC tissues and the
TME that were infected by P. gingivalis, indicating that
P. gingivalis and CXCL2 are involved in the recruitment of
TANs, which contributes to the progression of OSCC. However, using
IHC, the association between the immunoexpression levels of related
proteins in the TME of P. gingivalis infection and clinical
data remains unclear, despite increased research that suggests a
close association between P. gingivalis and OSCC (44).
In the present study, two mRNA microarray datasets
were analyzed to identify DEGs. A total of 89 DEGs were identified
between the two datasets, which included 22 downregulated genes and
67 upregulated genes. Higher mRNA expression levels of CXCL2 were
associated with the TME of OSCC infected by P. gingivalis.
Cancer cells with high CXCL2 expression due to transcriptional
hyperactivation are primed for survival in the metastatic sites
(14). CXCL2 expression appears to
be involved in OSCC-induced bone destruction and promotes tumor
progression (12). Furthermore,
increased CXCL1 expression has been observed in different OSCC cell
lines and tumor specimens, and is associated with leukocyte
infiltration and lymph node metastasis (12). CXCL2 attracts CD66b+ TANs
into the tumor, producing chemokines that enhance cancer cell
survival (45). Furthermore, the
chemokine CXCL2 is the core cytokine that mediates lung metastasis
and chemoresistance in breast cancer. When the CXCL2 receptor is
blocked, the effects of chemotherapy against breast cancer and
metastasis are effectively augmented (46). However, to the best of our knowledge,
clinical research regarding the involvement of CXCL2 in promotion
of cancer metastasis in OSCC has not been reported.
In recent years, the outlook on OSCC has changed and
the tumor is no longer considered as a bulk of neoplastic cells,
but rather as an entirety comprising a complex TME and neoplastic
cells, and it has been suggested that tumor progression occurs via
the TME and neoplastic cell interaction (47). The stromal component of the TME
involves different cell types, such as TANs, cancer-associated
fibroblasts and macrophages (47).
In early tumors, TANs may be able to stimulate T cell responses
(48), but in established tumors,
TANs are immunosuppressive and associated with a more protumor
phenotype with tumor progression (49). These subpopulation of cells interact
with each other, as well as with neoplastic cells, via complex
communication networks through secreted cytokines and chemokines
(50). It has been reported that
H. pylori is causally associated with the malignancies of
gastric epithelia (51). H.
pylori causes inflammation of the gastric mucosa by inducing
gastric epithelial cells to secrete IL-8, resulting in the
recruitment of inflammatory cells at the site of infection
(52). Furthermore, IL-8, other
chemokines and their receptors have been implicated in tumor
development and metastatic progression. It has also been revealed
that IL-8 enhances the generation of CD163+ M2
macrophages, and CD163+ macrophages exhibit a
significant association with poor overall survival in patients with
OSCC (53).
The present results suggested that, in OSCC, the TME
infected with P. gingivalis exhibits increased CXCL2
secretion, which recruits TANs to the site of neoplastic cells and
further promotes tumor development. It was demonstrated that the
immunoexpression levels of CXCL2 were increased in patients with
high expression levels of P. gingivalis. Another study with
similar findings to the present study reported that P.
gingivalis contributed to the accelerated secretion of CXCL8
and CCL2 (54). The number of
CD66b+ TANs in the TME of OSCC infected by P.
gingivalis proportionally increased with the secretion of
CXCL2, and the expression levels of P. gingivalis were
associated with the expression levels of CXCL2 and TANs.
Furthermore, all three of these factors were associated with the
poor prognosis of patients. However, studies regarding the
association between the TME of OSCC and P. gingivalis are
considered to be at early stages, and future investigations are
warranted to determine the underlying molecular mechanism.
In conclusion, to the best of our knowledge, to gain
an increased understanding of the participation of P.
gingivalis in the TME of OSCC, the association between the
immunoexpression of P. gingivalis in OSCC and
clinicopathological features and patient prognosis was analyzed for
the first time in the present study. Using bioinformatics analysis,
it was identified that CXCL2 was a DEG in the TME of OSCC infected
by P. gingivalis. In addition, the associations of the
immunoexpression levels of CXCL2 and TANs in OSCC with
clinicopathological features and patient prognosis were further
analyzed. The present study suggested a mutual interaction between
P. gingivalis and the TME of OSCC, and that the course of
chronic periodontal inflammation may create a tumor-favorable
microenvironment that promotes tumor development and progression.
Furthermore, strong immunoexpression levels of P.
gingivalis, CXCL2 and TANs may be associated with a poor
prognosis in patients with OSCC. Therefore, it is necessary to
develop prevention strategies for patients with OSCC with chronic
oral infection.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZCGo conceived and designed the study. ZCGu
conducted the experiments. SJ, SLJ, XYJ and LLH performed the
statistical analysis. ZCGu wrote the manuscript. ZCGu reviewed and
edited the manuscript. All authors agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved. ZCGo and ZCGu confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Stomatological Hospital of Xinjiang
Medical University (approval no. IACUC20180411-13; Urumqi, China).
Written consent was obtained at the time of the initial data
collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
P. gingivalis
|
Porphyromonas gingivalis
|
|
CXCL2
|
C-X-C motif chemokine ligand 2
|
|
TME
|
tumor microenvironment
|
|
IHC
|
immunohistochemistry
|
|
OSCC
|
oral squamous cell carcinoma
|
|
DEGs
|
differentially expressed genes
|
|
TANs
|
tumor-associated neutrophils
|
|
GEO
|
Gene Expression Omnibus
|
|
CSR
|
cumulative survival rate
|
References
|
1
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma-an update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA: A Cancer J Clin. 63:11–30. 2013.
|
|
3
|
Li P, Cao Q, Shao P, Cai H, Zhou H, Chen
J, Qin C, Zhang Z, Ju X and Yin C: Genetic polymorphisms in HIF1A
are associated with prostate cancer risk in a Chinese population.
Asian J Androl. 14:864–869. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SS, Ruiz VE, Carroll JD and Moss SF:
Helicobacter pylori in the pathogenesis of gastric cancer
and gastric lymphoma. Cancer Lett. 305:228–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Whitmore SE and Lamont RJ: Oral bacteria
and cancer. PLos Pathog. 10:e10039332014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gershkovitz M, Fainsod-Levi T, Zelter T,
Sionov RV and Granot Z: TRPM2 modulates neutrophil attraction to
murine tumor cells by regulating CXCL2 expression. Cancer Immunol
Immunother. 68:33–43. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song JM, Woo BH, Lee JH, Yoon S, Cho Y,
Kim YD and Park HR: Oral Administration of Porphyromonas
gingivalis, a major pathogen of chronic periodontitis, promotes
resistance to paclitaxel in mouse xenografts of oral squamous cell
carcinoma. Int J Mol Sci. 20:24942019. View Article : Google Scholar
|
|
8
|
Gao JL, Kwan AH, Yammine A, Zhou X,
Trewhella J, Hugrass BM, Collins D, Horne J, Ye P, Harty D, et al:
Structural properties of a haemophore facilitate targeted
elimination of the pathogen Porphyromonas gingivalis. Nat
Commun. 9:40972018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Inaba H, Sugita H, Kuboniwa M, Iwai S,
Hamada M, Noda T, Morisaki I, Lamont RJ and Amano A:
Porphyromonas gingivalis promotes invasion of oral squamous
cell carcinoma through induction of proMMP9 and its activation.
Cell Microbiol. 16:131–145. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lanzós I, Herrera D, Santos S, O'Connor A,
Peña C, Lanzós E and Sanz M: Microbiological effects of an
antiseptic mouth rinse in irradiated cancer patients. Med Oral
Patol Oral Cir Bucal. 16:e1036–e1042. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wen L, Mu W, Lu H, Wang X, Fang J, Jia Y,
Li Q, Wang D, Wen S, Guo J, et al: Porphyromonas gingivalis
Promotes oral squamous cell carcinoma progression in an immune
microenvironment. J Dent Res. 99:666–675. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
da Silva JM, Soave DF, Moreira Dos Santos
TP, Batista AC, Russo RC, Teixeira MM and da Silva TA: Significance
of chemokine and chemokine receptors in head and neck squamous cell
carcinoma: A critical review. Oral Oncol. 56:8–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vashishta A, Jimenez-Flores E, Klaes CK,
Tian S, Miralda I, Lamont RJ and Uriarte SM: Putative periodontal
pathogens, Filifactor alocis and Peptoanaerobacter
Stomatis, induce differential cytokine and chemokine production
by human neutrophils. Pathogens. 8:592019. View Article : Google Scholar
|
|
14
|
Hardaway AL, Herroon MK, Rajagurubandara E
and Podgorski I: Marrow adipocyte-derived CXCL1 and CXCL2
contribute to osteolysis in metastatic prostate cancer. Clin Exp
Metastasis. 32:353–368. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang H, Ye YL, Li MX, Ye SB, Huang WR,
Cai TT, He J, Peng JY, Duan TH, Cui J, et al: CXCL2/MIF-CXCR2
signaling promotes the recruitment of myeloid-derived suppressor
cells and is correlated with prognosis in bladder cancer. Oncogene.
36:2095–2104. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fridlender ZG, Sun J, Mishalian I, Singhal
S, Cheng G, Kapoor V, Horng W, Fridlender G, Bayuh R, Worthen GS
and Albelda SM: Transcriptomic analysis comparing tumor-associated
neutrophils with granulocytic myeloid-derived suppressor cells and
normal neutrophils. PLoS One. 7:e315242012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hagerling C and Werb Z: Neutrophils:
Critical components in experimental animal models of cancer. Semin
Immunol. 28:197–204. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Powell DR and Huttenlocher A: Neutrophils
in the tumor microenvironment. Trends Immunol. 37:41–52. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li S, Cong X, Gao H, Lan X, Li Z, Wang W,
Song S, Wang Y, Li C, Zhang H, et al: Tumor-associated neutrophils
induce EMT by IL-17a to promote migration and invasion in gastric
cancer cells. J Exp Clin Cancer Res. 38:62019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sahingur SE and Yeudall WA: Chemokine
function in periodontal disease and oral cavity cancer. Front
Immunol. 6:2142015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carey LA, Metzger R, Dees EC, Collichio F,
Sartor CI, Ollila DW, Klauber-DeMore N, Halle J, Sawyer L, Moore DT
and Graham ML: American Joint Committee on cancer
tumor-node-metastasis stage after neoadjuvant chemotherapy and
breast cancer outcome. J Natl Cancer Inst. 97:1137–1142. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Geng F, Liu J, Guo Y, Li C, Wang H, Wang
H, Zhao H and Pan Y: Persistent exposure to Porphyromonas
gingivalis promotes proliferative and invasion capabilities,
and tumorigenic properties of human immortalized oral epithelial
cells. Front Cell Infect Microbiol. 7:57–61. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qiu X, Lei Z, Wang Z, Xu Y, Liu C, Li P,
Wu H and Gong Z: Knockdown of LncRNARHPN1-AS1 inhibits cell
migration, invasion and proliferation in head and neck squamous
cell carcinoma. J Cancer. 10:4000–4008. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Lei Q, Zhang S, Kong L and Qin B:
Screening and identification of key biomarkers in hepatocellular
carcinoma: Evidence from bioinformatic analysis. Oncol Rep.
38:2607–2618. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bryne M, Koppang HS, Lilleng R, Stene T,
Bang G and Dabelsteen E: New malignancy grading is a better
prognostic indicator than Broders' grading in oral squamous cell
carcinomas. J Oral Pathol Med. 18:432–437. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carus A, Ladekarl M, Hager H, Nedergaard
BS and Donskov F: Tumour-associated CD66b+ neutrophil count is an
independent prognostic factor for recurrence in localised cervical
cancer. Br J Cancer. 108:2116–2122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Akinkugbe AA, Garcia DT, Brickhouse TH and
Mosavel M: Lifestyle risk factor related disparities in oral cancer
examination in the U.S: A population-based cross-sectional study.
BMC Public Health. 20:153–163. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Danaei G: Global burden of
infection-related cancer revisited. Lancet Oncol. 13:564–565. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Iqbal J, McRae S, Banaudha K, Mai T and
Waris G: Mechanism of hepatitis C virus (HCV)-induced osteopontin
and its role in epithelial to mesenchymal transition of
hepatocytes. J Biol Chem. 293:200102018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Horikawa T, Yang J, Kondo S, Yoshizaki T,
Joab I, Furukawa M and Pagano JS: Twist and epithelial-mesenchymal
transition are induced by the EBV oncoprotein latent membrane
protein 1 and are associated with metastatic nasopharyngeal
carcinoma. Cancer Res. 67:1970–1978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chandrakesan P, Roy B, Jakkula LU, Ahmed
I, Ramamoorthy P, Tawfik O, Papineni R, Houchen C, Anant S and Umar
S: Utility of a bacterial infection model to study
epithelial-mesenchymal transition, mesenchymal-epithelial
transition or tumorigenesis. Oncogene. 33:2639–2654. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee J, Roberts JS, Atanasova KR, Chowdhury
N, Han K and Yilmaz Ö: Human primary epithelial cells acquire an
epithelial-mesenchymal-transition phenotype during long-term
infection by the oral opportunistic pathogen, Porphyromonas
gingivalis. Front Cell Infect Microbiol. 7:4932017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Woo BH, Kim DJ, Choi JI, Kim SJ, Park BS,
Song JM, Lee JH and Park HR: Oral cancer cells sustainedly infected
with Porphyromonas gingivalis exhibit resistance to Taxol
and have higher metastatic potential. Oncotarget. 8:46981–46992.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wen L, Mu W, Lu H, Wang X, Fang J, Jia Y,
Li Q, Wang D, Wen S, Guo J, et al: Porphyromonas gingivalis
Promotes oral squamous cell carcinoma progression in an immune
microenvironment. J Dent Res. 99:666–675. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Peres MA, Macpherson LMD, Weyant RJ, Daly
B, Venturelli R, Mathur MR, Listl S, Celeste RK, Guarnizo-Herreño
CC, Kearns C, et al: Oral diseases: A global public health
challenge. Lancet. 394:249–260. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yee M, Kim S, Sethi P, Duzgunes N and
Konopka K: Porphyromonas gingivalis stimulates IL-6 and IL-8
secretion in GMSM-K, HSC-3 and H413 oral epithelial cells.
Anaerobe. 28:62–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Geng F, Wang Q, Li C, Liu J, Zhang D,
Zhang S and Pan Y: Identification of potential candidate genes of
oral cancer in response to chronic infection with Porphyromonas
gingivalis using bioinformatical analyses. Front Oncol.
9:912019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bunt SK, Yang L, Sinha P, Clements VK,
Leips J and Ostrand-Rosenberg S: Reduced inflammation in the tumor
microenvironment delays the accumulation of myeloid-derived
suppressor cells and limits tumor progression. Cancer Res.
67:10019–10026. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zenobia C and Hajishengallis G:
Porphyromonas gingivalis virulence factors involved in
subversion of leukocytes and microbial dysbiosis. Virulence.
6:236–243. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y and Li X:
Lipopolysaccharide-regulated production of bone sialoprotein and
interleukin-8 in human periodontal ligament fibroblasts: The role
of toll-like receptors 2 and 4 and the MAPK pathway. J Periodontal
Res. 50:141–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shi H, Han X, Sun Y, Shang C, Wei M, Ba X
and Zeng X: Chemokine (C-X-C motif) ligand 1 and CXCL2 produced by
tumor promote the generation of monocytic myeloid-derived
suppressor cells. Cancer Sci. 109:3826–3839. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ban Y, Mai J, Li X, Mitchell-Flack M,
Zhang T, Zhang L, Chouchane L, Ferrari M, Shen H and Ma X:
Targeting autocrine CCL5-CCR5 axis reprograms immunosuppressive
myeloid cells and reinvigorates antitumor immunity. Cancer Res.
77:2857–2868. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lafuente Ibáñez de Mendoza I, Maritxalar
Mendia X, García de la Fuente AM, Quindós Andrés G and Aguirre
Urizar JM: Role of Porphyromonas gingivalis in oral squamous
cell carcinoma development: A systematic review. J Periodontal Res.
55:13–22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ohms M, Moller S and Laskay T: An attempt
to polarize human neutrophils toward N1 and N2 phenotypes in vitro.
Front Immunol. 11:5322020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Feliciano P: CXCL1 and CXCL2 link
metastasis and chemoresistance. Nat Genet. 44:8402012. View Article : Google Scholar
|
|
47
|
Eckert AW, Wickenhauser C, Salins PC,
Kappler M, Bukur J and Seliger B: Correction to: Clinical relevance
of the tumor microenvironment and immune escape of oral squamous
cell carcinoma. J Transl Med. 16:402018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Eruslanov EB, Bhojnagarwala PS, Quatromoni
JG, Stephen TL, Ranganathan A, Deshpande C, Akimova T, Vachani A,
Litzky L, Hancock WW, et al: Tumor-associated neutrophils stimulate
T cell responses in early-stage human lung cancer. J Clin Invest.
124:5466–5480. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, Wang
Z, Wang C, Zhang Z, Xia W, et al: γδT17 cells promote the
accumulation and expansion of myeloid-derived suppressor cells in
human colorectal cancer. Immunity. 40:785–800. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Peltanova B, Raudenska M and Masarik M:
Effect of tumor microenvironment on pathogenesis of the head and
neck squamous cell carcinoma: A systematic review. Mol Cancer.
18:632019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yasunaga JI and Matsuoka M: Oncogenic
spiral by infectious pathogens: Cooperation of multiple factors in
cancer development. Cancer Sci. 109:24–32. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wen J, Wang Y, Gao C, Zhang G, You Q,
Zhang W, Zhang Z, Wang S, Peng G and Shen L: Helicobacter pylori
infection promotes Aquaporin 3 expression via the
ROS-HIF-1α-AQP3-ROS loop in stomach mucosa: A potential novel
mechanism for cancer pathogenesis. Oncogene. 37:3549–3561. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nagarsheth N, Wicha MS and Zou W:
Chemokines in the cancer microenvironment and their relevance in
cancer immunotherapy. Nat Rev Immunol. 17:559–572. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Damgaard C, Kantarci A, Holmstrup P,
Hasturk H, Nielsen CH and Van Dyke TE: Porphyromonas
gingivalis-induced production of reactive oxygen species, tumor
necrosis factor-α, interleukin-6, CXCL8 and CCL2 by neutrophils
from localized aggressive periodontitis and healthy donors:
Modulating actions of red blood cells and resolvin E1. J
Periodontal Res. 52:246–254. 2017. View Article : Google Scholar : PubMed/NCBI
|