Introduction
A major challenge with conventional cancer
treatments, including chemotherapy, is the genotypic and phenotypic
heterogeneity of cancer cells, and non-selective cytotoxicity of
these treatment regimens, and this limits their use, increasing the
risk of cancer reoccurrence (1,2).
Therefore, experimental evidences need to be accumulated on
plant-derived chemopreventative strategies, which may be used as
adjuvants for improved cancer management. Chemopreventative agents
(synthetic and natural) consist of nonsteroidal anti-inflammatory
drugs (sulindac, aspirin and celecoxib) (3) and FDA-approved drugs, including
Tamoxifen, Raloxifene and HPV vaccines (Cervarix and Gardasil),
which all show potential anticancer effects. However, these
adjuvants have also demonstrated adverse side effects in patients
(4–6). Considering the constraints of these
agents, studies are focused on identifying alternatives based on
plant-derived compounds, which exhibit decreased toxicity with
higher efficacy for the treatment of cancer (7–9).
Notably, an epidemiological study proposed that 30%
of cancer types can be avoided by a healthy diet (2), and the consumption of certain fruits
and vegetables may decrease the risk of cancer onset (5,10,11).
Several flavonoids extracted from various plants have been shown to
exert anticancer properties (5,7),
exhibiting regulatory effects on several molecular mechanisms,
including their ability to increase the activity of free radical
scavenging enzymes, and increase anti-proliferative and
anti-inflammatory responses (7,12,13).
Furthermore, flavones, a subclass of flavonoids have been
demonstrated to possess anticancer effects through decreasing
cancer cell proliferation by downregulation of various
pro-apoptotic proteins, including nuclear factor-κB along with
modulation of aberrant PI3K/AKT and MAPK/P38 signaling (14–18).
Apoptosis is the desired outcome when treated with these agents,
and if the means by which apoptosis is induced is determined, the
efficacy of novel agents targeting these mechanisms may be
assessed. Apoptosis is primarily mediated by the death receptor
pathway and mitochondrial pathway. The death receptor family, which
is comprised of tumor necrosis factor receptors, including
TRAIL-R2/DR5, TRAIL-R1/DR4, TNFRI/TNFRSF1A, Fas/TNFRSF6/CD95, Fas
and DR6 may initiate the extrinsic pathway, and BCL2 family
members, including Bad, Bak, Bax, Bid, Bik (pro-apoptotic
molecules), and Bcl-XL and Bcl-2 (anti-apoptotic members) are
involved in the mitochondrial pathway (19–21).
Luteolin (3,4,5,7-tetrahydroxy flavone) is a flavone
found in vegetables and fruits, including parsley, carrots,
artichoke, celery and several spices (including thyme and oregano)
(14,16,17,22–25).
Several studies have demonstrated the anti-inflammatory,
anti-microbial, anti-diabetic and anti-carcinogenic properties of
luteolin (23,26–28).
Luteolin has been demonstrated to exhibit anticancer effects
against several types of cancer cell lines, including liver
(HepG2), colon (HT29), lung (LNM35) and breast cancer (MDA-MB-231)
cells, and is a potent HDAC inhibitor (16,19).
Luteolin induces apoptosis and cell cycle arrest by increasing
expression of Bax, Caspase 3 and Caspase 9, as well as the MEK-ERK
pathway, whilst concomitantly decreasing Bcl-2 expression in A549
lung cancer cells (18). Luteolin
also possesses anti-proliferative effects against A549 lung cancer
cells by arresting the cell cycle at the G2 phase and initiating
programmed cell death via the mitochondrial pathway, which is
mediated though activation of JNK and inhibition of NF-κB (22,29).
Luteolin suppresses the MAPK/AKT/PI3K pathway, and NF-kB and STAT3
signalling in several types of cancer cell lines, and also
decreases the expression of matrix metalloproteins and β3 integrin
in B16F10 and A431 melanoma cell lines, thereby inhibiting EMT
(17,20,22,24,30,31).
Notably, flavonoids including luteolin have been
reported to inhibit tumor cell proliferation and apoptosis
(14–19) in various cancer cell lines e.g.,
HPV-18-associated cells and lung cancer cell (17,18).
However, there is lack of scientific evidence to review luteolin as
a probable anticancer agent. Therefore, the present study aimed to
investigate the extensive molecular mechanism through which
luteolin induces anticancer effects on HeLa cell as an apoptosis
inducer by targeting various molecular targets; tumor inhibitors
and promoters: DAPK1, TP53, TP73, PTPRRR and TERT;
cell cycle regulatory genes: CDK2, CCNE2, CCND2 and
CCND; pro and anti-apoptotic genes: BAD, BID, BOK, BAX,
Bcl2, Caspases 3, 7, 8 and 9; receptors and pathways
genes: FASLG, FAS, TNFRS10-B, MTOR, MAPK, PIK3CA, AKT1, ELK1,
FOXO1 and FOXO3, and to further associate it with the
modulation of phosphorylated proteins in luteolin-treated HeLa
cells to enhance the acceptance of luteolin as a potential
anticancer agent.
Material and methods
Cell culture and drug preparation
Information of every important process that ensues
in human cells whether normal or abnormal has depended to a large
extent on using HeLa cells and they have also proved to be the best
model for cervical cancer (32);
therefore, HeLa cells were used for this study. Human cervical
cancer HeLa cells were cultured in Dulbecco's modified Eagle's
medium (DMEM; Sigma-Aldrich; Merck KGaA), supplemented with 10% FBS
(Sigma-Aldrich; Merck KGaA), 100 µg/ml amphotericin (Sigma-Aldrich;
Merck KGaA) and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA),
and cells were cultured at 37°C in a humidified incubator with 5%
CO2.
Luteolin (molecular weight, 286.24 g/mol) was
purchased from Sigma-Aldrich (Merck KGaA) and diluted to a
concentration of 69.87 mM using dimethyl sulfoxide (DMSO; stock
solution), which was then further diluted using DMEM (without FBS)
to a concentration of 1 mM, which was used to make the working
concentrations of 1–40 µM.
MTT assay
MTT assay (Sigma-Aldrich; Merck KGaA) was used to
study the cytotoxic effects of luteolin on HeLa cells. In brief,
1×104 cells/well were plated onto a 96-well plate and
treated as follows: Blank control, DMSO control (0.11%), positive
control (cisplatin 5 µM) and luteolin-treated (1–40 µM); and
treatment was performed for 24 or 48 h, after which images were
captured. Subsequently, plates were decanted, and the cells were
incubated with MTT at final concentration of 5 mg/ml (dissolved in
PBS) at 37°C for 2–4 h, the MTT was discarded, 100 µl DMSO was
added to the wells and plates were incubated at 37°C for 30 min in
the dark. Viable cells convert the tetrazolium salt into insoluble
formazan violet crystals that is quantified using a colorimeter at
570 nm. A graph was plotted by dividing the absorbance of
luteolin-treated cells with that of DMSO controls. Data are
presented as the mean ± standard deviation of three independent
repeats.
Nuclear staining of HeLa cells with
propidium iodide (PI) following luteolin treatment
PI staining of cells treated with luteolin for
different concentrations was performed to assess morphological
variations in the nucleus in cells undergoing apoptosis. In total,
~2.5×105 cells were seeded onto glass coverslips and
treated with luteolin for 24 or 48 h. The cells were then washed
with 1× PBS (pH 7.4) twice and a mixture of ice cold
methanol:acetone (1:1) was used to fix the cells at −20°C for 10
min followed by staining with PI (10 mg/ml in PBS) for 30 sec in
the dark at room temperature, and the cover slips were subsequently
mounted on glass slides. The images were captured at 515 nm using a
fluorescent microscope (Olympus Corporation) at ×40
magnification.
DNA fragmentation assay
Inter-nucleosomal DNA disintegration is one of the
primary features of apoptosis. During apoptosis, nucleases are
activated causing fragmentation of nuclear chromatin into 50–300
kbp fragments (33). The ApoTarget™
Quick Apoptotic DNA Ladder Detection kit (cat. no. #KHO1021;
Invitrogen; Thermo Fisher Scientific, Inc.) was used for rapid
extraction of chromosomal DNA of untreated cells, and cells treated
with 5, 10 and 20 µM luteolin for 48 h, according to the
manufacturer's protocol. The fragmented DNA was extracted from
cells and run on a horizontal electrophoresis gel containing
ethidium bromide and the DNA was resolved on a 1.2% agarose
gel.
Tetramethyl rhodamine, ethyl ester
(TMRE) mitochondrial membrane potential assay
A TMRE Mitochondrial Membrane Potential assay kit
(cat. no. ab113852; Abcam) was used to assess the mitochondrial
membrane potential. In total, ~5×103 HeLa cells were
plated onto clear bottom 96-well plates and treated with 5, 10 and
20 µM luteolin for 48 h at room temperature. TMRE at a final
concentration of 400 nM was used and the cells were incubated in
the dark for 30 min at room temperature. Following washing several
times with the wash buffer provided with the aforementioned kit,
the plate was read on a fluorescence microplate spectrophotometer
(Ex/Em=549/575 nm; Synergy H1 Bioteck Plate Reader) and images were
captured using a fluorescent microscope (Progress Fluorescent
Microscope; Olympus Corporation) at ×40 magnification.
Apoptosis detection using
FITC-conjugated Annexin V/PI
Quantitation of the apoptosis was performed using a
FITC-conjugated Annexin V/PI assay kit (cat. no. ab 14085; Abcam)
and analyzed using a flow cytometer. In brief, 2×105
cells were plated onto six-well plates, and cells were treated with
5, 10 and 20 µM luteolin for 48 h at 37°C. Treated and untreated
cells were collected and washed with PBS and resuspended in 500 µl
binding buffer, followed by staining with PI (50 µg/ml) and
FITC-conjugated Annexin V (10 mg/ml) for 15 min at room temperature
in the dark, and the cells were analyzed using a BD FACS-Aria™ III
Flow cytometer (BD Biosciences). Flow cytometry data were analyzed
using the FlowJo software (FlowJo LLC; Version 10.1).
Cell cycle analysis by flow
cytometry
HeLa cells were treated with 0, 5, 10 and 20 µM
luteolin for 48 h, and all the cells (floating and attached) were
collected. This was followed by washing with 1× PBS and fixed with
absolute ethanol at −20°C overnight. The following day, cells were
washed with 1× PBS and staining with PI (1 mg/ml), containing 0.1%
citrate buffer, Triton X-100 and 0.5% RNase for 30 min at room
temperature. BD FACS-Aria™ III Flow cytometer (BD Biosciences) was
used to analyze the cells for DNA content, and FlowJo software
(FlowJo LLC; version 10.1) was used for data analysis.
Gene expression analysis using TaqMan
arrays
RNA extraction was performed on luteolin-treated
HeLa cells (10 and 20 µM for 48 h at 37°C) and untreated cells
according to the manufacturer's protocol using a GenElute Mammalian
Genomic Total RNA kit (Sigma-Aldrich; Merck KGaA). Extracted total
RNA was quantified using Nanodrop (Thermo Fisher Scientific, Inc.).
RNA was then subjected to first strand synthesis, according to the
manufacturer's protocol, using an Applied Biosystems™ High-Capacity
cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
TaqMan® Gene Expression assays are a
comprehensive collection of pre-designed, pre-formulated primer and
probe sets to perform quantitative gene expression studies
(Apoptosis Array and oncogene array (cat. nos. 4414072 and
4391524). TaqMan® Gene Expression assays that were
manufactured and stocked in advance were used to detect the
expression of genes associated with cell cycle signaling pathways
and apoptosis in the treated and untreated cells. A total of 10 µl
cDNA with a concentration of 100 ng per well and 10 µl master mix
provided in the kit was used. Expression analysis was performed
according to the manufacturer's protocol. The qPCR array was run on
a thermocycler (QuantiStudio3, Applied Biosystems, USA) with the
following reaction conditions: Enzyme activation at 95°C for 10 min
and 40 cycles, denaturation at 95°C for 15 sec, annealing at 60°C
for 1 min. Results were analyzed by the 2−ΔΔCq method
using DataAssist™ software version 3.01 (Thermo Fisher Scientific,
Inc.) as previously described (34).
Caspase 3 activity
Caspases are the final executioners of apoptosis and
Caspase-3 serves a role in the extrinsic and intrinsic pathways of
apoptosis (35). A Caspase-3
Activity kit (cat. no. CASP 3C; Sigma-Aldrich; Merck KGaA) was
used. A total of 1 ×106 cells were plated and treated
(5, 10 and 20 µM luteolin at 37°C for 48 h) with various
concentrations of luteolin. Cell pellets were taken and resuspended
in the lysis buffer (1X) at a concentration of 1X 107
cell/100 µl and incubated for 30 min, prior to being centrifuged at
12,000 × g for 20 min at 4°C. The lysates of the DMSO control and
treated (5, 10 and 20 µM luteolin) HeLa cells and the assay were
set up according to the manufacturers' protocol. The plate was
incubated overnight at room temperature and read at 405 nm, and
fold-changes were calculated by comparing the treated sample
reading with the DMSO control. The experiment was repeated 3 times
and all results are expressed as mean ± standard deviation of three
independent experiments.
Determination of protein expression
using a proteome profiler array
To determine the expression of genes associated with
apoptosis and cell cycle progression at the protein level, a
protein profiler array was purchased from R&D Systems, Inc.
(cat. no. ARY009). The cells were treated with 10 and 20 µM
luteolin for 48 h at 37°C and the lysis was performed with lysis
buffer. The assay was performed according to the manufacturer's
protocol to assess the expression of all the proteins associated
with apoptosis. Protein quantitation of lysates was performed using
a Pierce™ BCA Protein assay kit (Thermo Fisher Scientific, Inc;
cat. no. 23225). A further 400 µg protein of the diluted cell
lysate was used for each membrane, the membranes were incubated
with the lysates on a rocking platform overnight at 2–8°C. The
signal was developed using Streptavidin-HRP and chemiluminescent
detection reagents. Signal intensity was measured using a
chemiluminescent detector Gel Doc system (Bio-Rad Laboratories,
Inc.), and analyzed using Image Lab software (version 6.1; Bio-Rad
Laboratories, Inc.). The experiment was repeated three times and
the results are expressed as the mean ± standard deviation from
three independent experiments (*P<0.005).
Analysis of expression of
phosphorylated AKT pathway proteins
To determine the effect of luteolin treatment on
proteins involved in the AKT pathway, a phosphorylated AKT pathway
array was obtained from Ray-Biotech, Co., Ltd. (cat. no.
AAH-AKT-1-8). The lysates of treated and untreated cells were
quantitated using a Pierce BCA assay (Thermo Fisher Scientific,
Inc.) and ~500 µg lysate was used on each nitrocellulose membrane.
To begin with, the membranes were blocked with blocking buffer from
the aforementioned kit, followed by incubation with the cell lysate
for 24 h, and the assay was performed according to the
manufacturer's protocol. The chemiluminescent gel doc system
(BioRad Laboratories, Inc.) was used to capture the image of the
blot and analysis was performed using Image Lab software (version
6.1).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 21; IBM Corp.). The data was analyzed using
one-way analysis of variance followed by Tukey's HSD post
hoc test. All experiments were performed in triplicate. Results
are expressed as the mean ± standard deviation of three separate
experiments *P<0.05 was considered to indicate a statistically
significant difference.
Results
Luteolin inhibits the growth and
proliferation of HeLa cells
HeLa cells treated with varying concentrations of
luteolin (1–40 µM for 24 or 48 h) exhibited a dose and
time-dependent decrease in the viability of HeLa cells when
compared with the untreated DMSO control. When cells were treated
with 20 µM luteolin for 48 h, ~50% cell death was observed
(Fig. 1A). Luteolin induced
substantial morphological changes in HeLa cells, including rounding
off of the cells and subsequent detachment of the cells from the
surface, and the changes became more significant as the dose of
luteolin was increased (Fig.
1B).
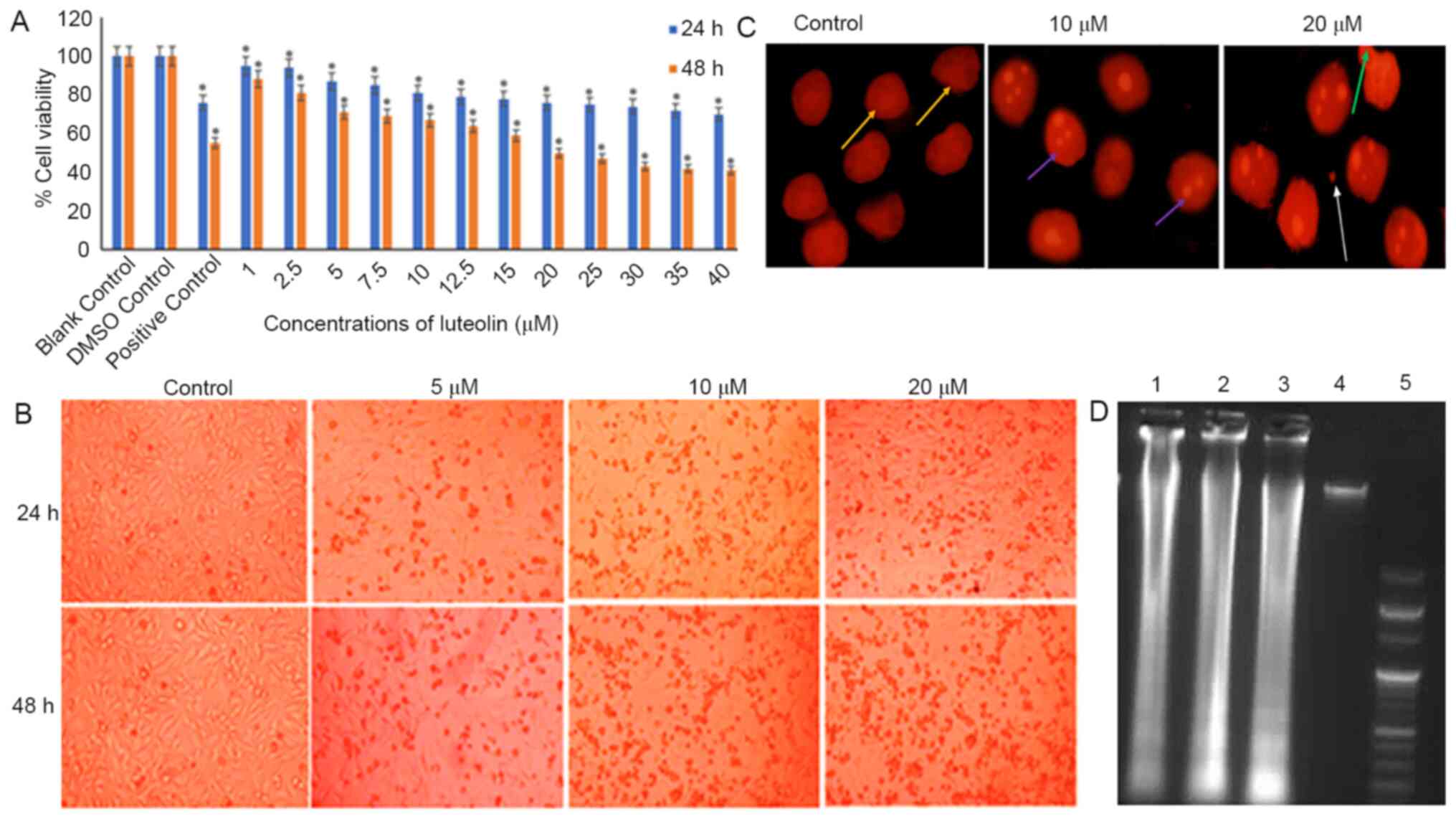 | Figure 1.Anti-proliferative effect of luteolin
on HeLa cells. (A) Induction of cytotoxicity by luteolin at various
concentrations and time points. The graph represents the dose- and
time-dependent decrease in cell viability of HeLa cells treated
with 1–40 µM luteolin for 24 and 48 h. For comparison, 0.11% DMSO
was used as the loading control and 5 µM cisplatin was used as the
positive control. The 24 and 48 h treated samples were compared
with 24 and 48 h DMSO controls, respectively. The data are
expressed as the mean ± standard deviation of three independent
experiments. *P<0.05. The IC50 of luteolin was found
to be 20 µM at 48 h. (B) Microscopic images of HeLa cells treated
with 10, 15 and 20 µM luteolin for 24 and 48 h. Cells exhibited a
characteristic rounding off of the cells, indicating apoptosis.
Magnification, ×10. (C) Changes in nuclear morphology of treated
HeLa cells (10 and 20 µM luteolin) compared with the untreated
controls. These nuclear changes were observed following staining
with propidium iodide under a fluorescence microscope
(magnification, × 40). A dose-dependent increase in the apoptotic
index, characterized by features such as nuclear condensation,
nuclear blebbing, nuclear fragmentation and apoptotic bodies, was
observed. Yellow, large and prominent nuclei; purple, nuclear
fragmentation; green, blebbing; white, apoptotic bodies. (D)
Luteolin induces DNA fragmentation in a dose-dependent manner in
HeLa cells. Lane 5 shows the 100 bp ladders; lanes 1, 2, 3 and 4
show fragmentation of DNA in cells treated with 5, 10 and 20 µm
luteolin and the control, respectively. The experiment was
performed three times. DMSO, dimethyl sulfoxide. |
Luteolin induces nuclear morphological
abnormalities and causes DNA fragmentation in HeLa cells
HeLa cells treated with 10 and 20 µM luteolin
exhibited condensation and disintegration of nuclear material,
blebbing and formation of apoptotic bodies and nuclear debris,
which increased with concentration, while the untreated HeLa cells
possessed a distinct and prominent nucleus (Fig. 1C). Furthermore, the observation that
luteolin induced apoptosis was supported by the results of a DNA
laddering assay in HeLa cells. Treatment of HeLa cells with 5, 10
and 20 µM luteolin for 48 h resulted in fragmentation of DNA. A
typical DNA ladder-like pattern was observed in all the treated
samples when compared with the controls upon gel electrophoresis
(Fig. 1D).
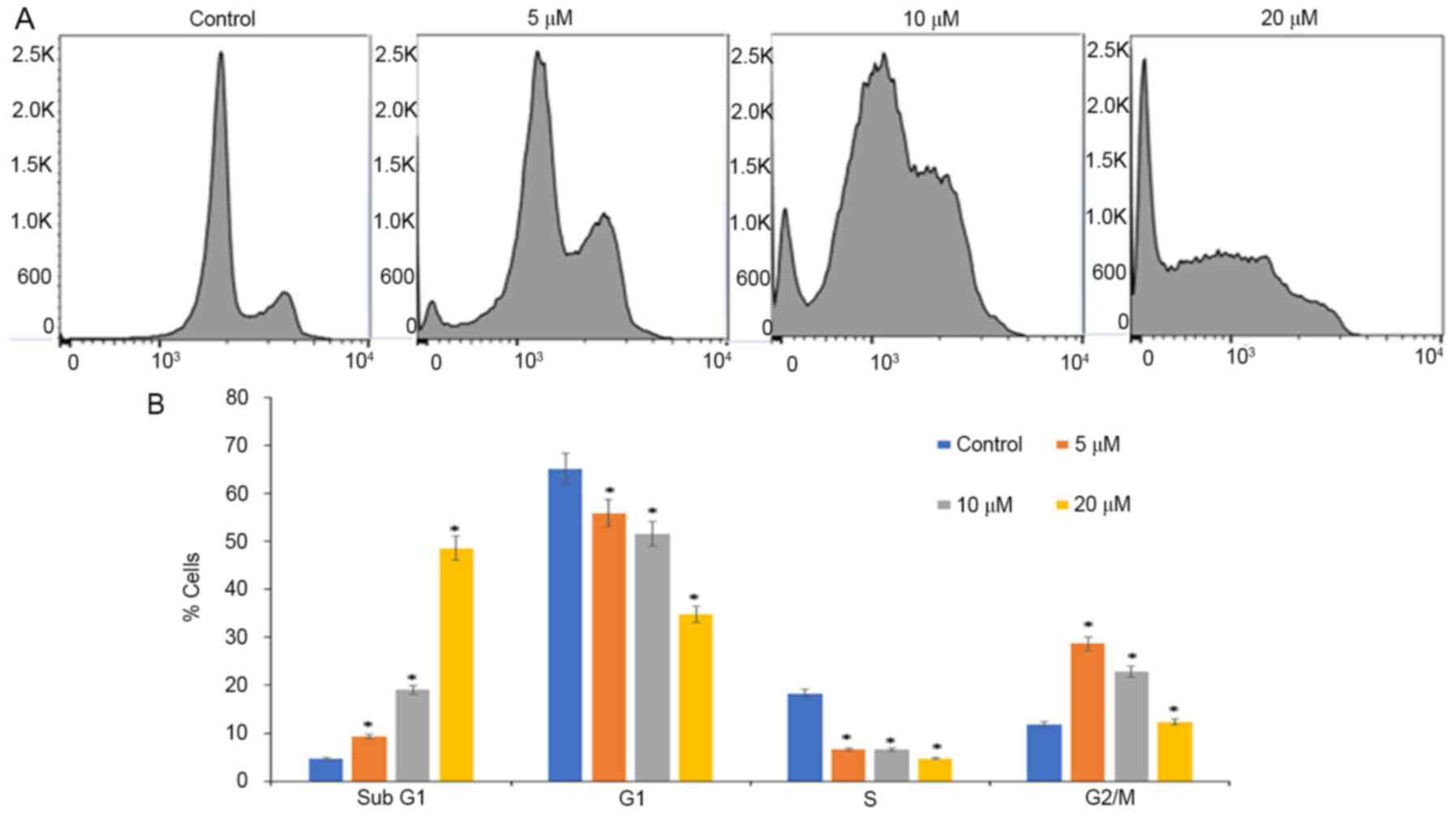
Luteolin treatment arrests cell cycle
progression of HeLa cells at sub-G1 phases
Inhibition of proliferation and growth may be a
result of cell cycle arrest. Analysis of cell cycle progression was
performed on luteolin-treated cells using PI staining and flow
cytometry. Luteolin induced accumulation of cells at the G2/M phase
when treated with 5 µM, and when treated with 10 and 20 µM,
accumulation of cells was observed at sub-G1 phases, and ~50% of
cells were arrested when treated with 20 µM luteolin for 48 h, with
a decrease in the number of cells in the S and G2/M phases when
compared with controls wherein a normal distribution of cells was
observed (Fig. 2). In the present
study, luteolin treatment increased the sub-G1 peak, which is an
indicator of endonuclease activation and apoptosis induction due to
DNA fragmentation, which appears to be the crucial
anti-proliferative mechanism of luteolin treatment (36).
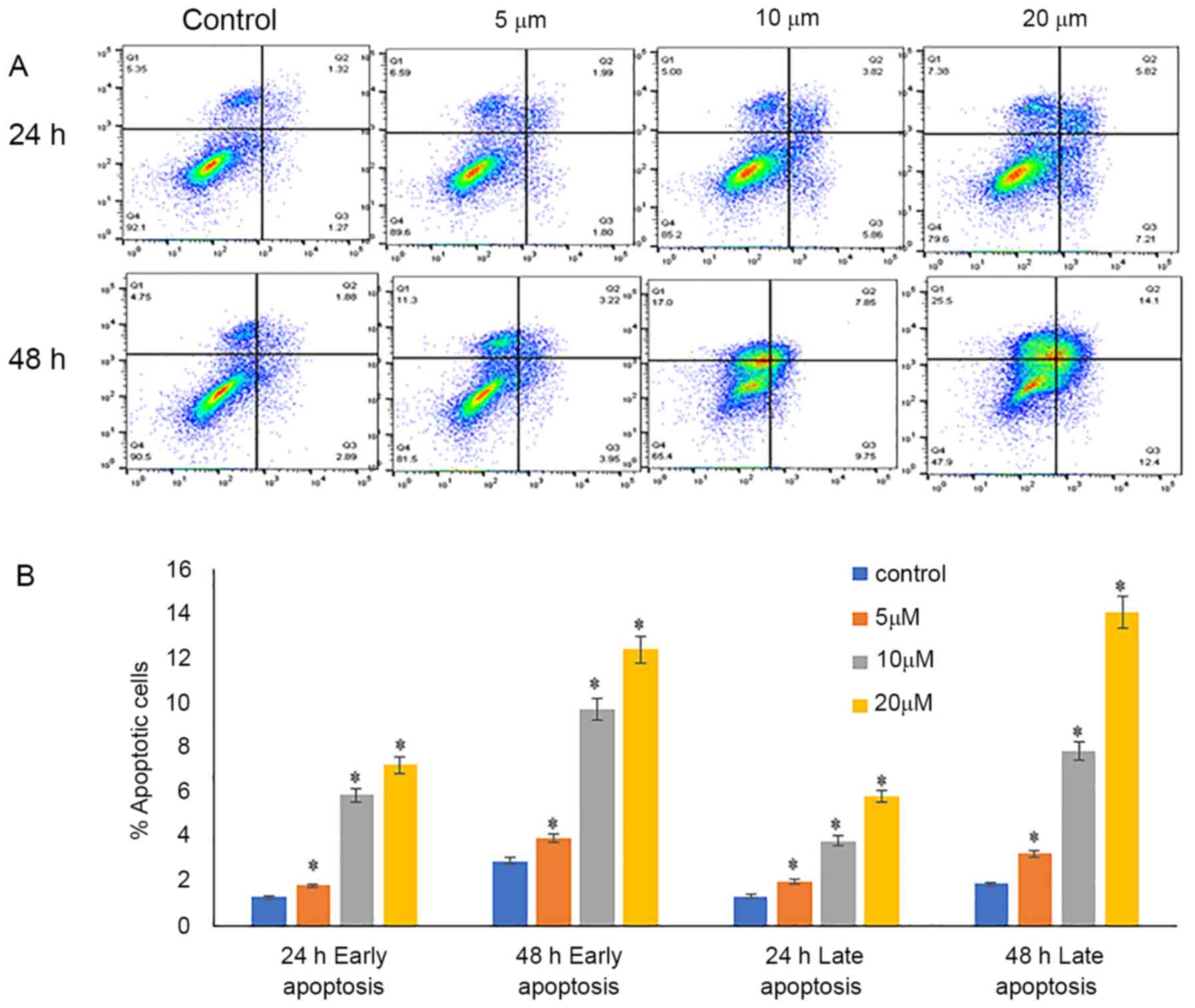
Luteolin induces early and late
apoptosis
Annexin V-FITC and PI were used for the detection of
early and late apoptosis. Annexin V-stained cells were considered
early apoptotic cells, and cells stained with PI and annexin V and
were considered late apoptotic cells. The percentage of viable
cells decreased from 90.5% in the control cells to 65.4 and 47.9%
when treated with 10 and 20 µM luteolin, respectively, for 48 h.
Additionally, there was an increase in early apoptotic cells from
2.89% in control samples to 9.75 and 12.4% when treated with 10 and
20 µM luteolin. A notable increase in the proportion of late
apoptotic cells was detected, increasing from 1.89% in in the
untreated samples to 7.85 and 14% when treated with 10 and 20 µM
luteolin 48 h, respectively (Fig.
3).
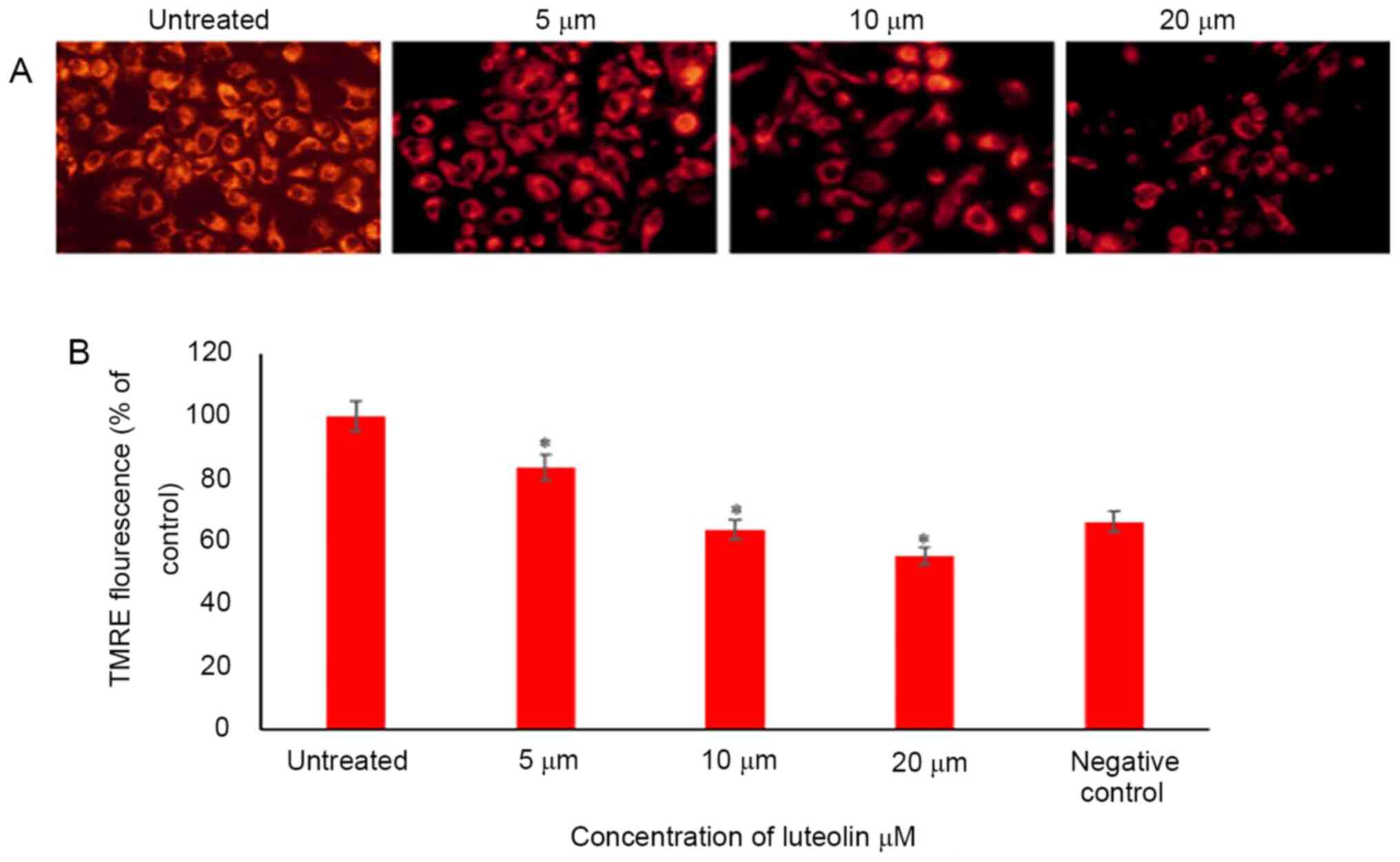
Luteolin decreases the mitochondrial
membrane potential of HeLa cells
Mitochondrial membrane potential is a reliable
measure of cell stress and apoptosis, as it is compromised early
during the stress response and a good indicator of ongoing cell
death. Several dyes may be used to detect mitochondrial membrane
potential, which allows flexibility in wavelengths and use in
combination with other fluorescence markers. In the present study,
TMRE was used. The percentage of TMRE fluorescence was calculated
compared with the control; 84, 64 and 56% of cells fluoresced with
TMRE when HeLa cells were treated with 5, 10 and 20 µM luteolin for
48 h, respectively (Fig. 4).
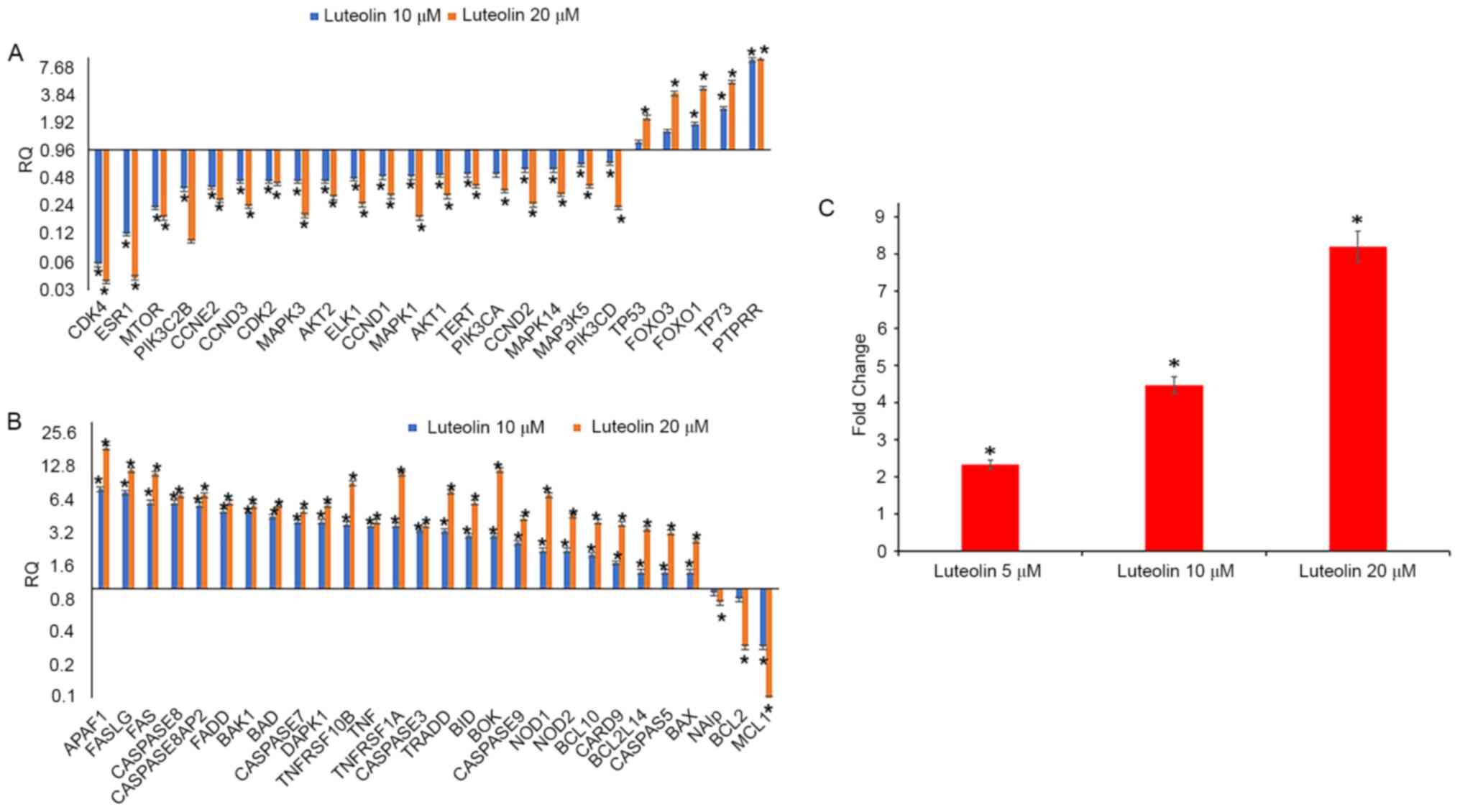
Luteolin inhibits cell cycle
progression by modulation of cell cycle regulatory genes
Following treatment of HeLa cells with 5, 10 and 20
µM luteolin for 48 h, the untreated and treated cells were
subjected to cell cycle progression analysis. The results
demonstrated that 5 µM luteolin induced G2/M phase arrest in HeLa
cells, and when treated with 20 µM luteolin, there was a
substantial accumulation of cells at sub G1 phases. This was
further verified by assessing the expression of cell cycle
regulatory genes (oncogene array, cat. no. 4369514); downregulation
of cell cycle regulatory genes, including CCND1, CCND2, CCND3,
CCNE2, CDKN1A, CDKN2B, CDK4 and CDK2, was observed. All
these genes serve an important role in the G1 phase transition.
Additionally, telomerase reverse transcriptase (TERT) expression
was decreased and PTPRR expression was significantly increased when
treated with 10 and 20 µM luteolin. TERT adds repeat sequences of
DNA (TTAGGG) to the end of chromosomes, thereby increasing the
replicative properties, and its inhibition controls proliferation,
while PTPRR inhibits ERK and MAPK pathways. FOXO1 and FOXO3
expression was also upregulated, and they serve an important role
in apoptosis [Fig. 5A; RQ (relative
quantitation)] values are presented in Table I).
 | Table I.Relative quantitation values of cell
cycle regulatory genes and pathway genes in HeLa cells following 10
and 20 µM luteolin treatment for 48 h. |
Table I.
Relative quantitation values of cell
cycle regulatory genes and pathway genes in HeLa cells following 10
and 20 µM luteolin treatment for 48 h.
| Genes | Gene ensemble
no. | Gene
description | Luteolin 10 µM | Luteolin 20 µM |
|---|
| CDK4 | Hs00175935_m1 | Cyclin dependent
kinase inhibitor 1A | 0.06 | 0.04 |
| ESR1 | Hs01046816_m1 | Estrogen receptor
1 | 0.12 | 0.04 |
| MTOR | Hs00234522_m1 | Mechanistic target
of rapamycin | 0.23 | 0.18 |
| PIK3C2B | Hs00898499_m1 |
Phosphatidylinositol-4-phosphate 3-kinase
catalytic subunit type 2 beta | 0.37 | 0.10 |
| CCNE2 | Hs00372959_m1 | Cyclin E2 | 0.39 | 0.28 |
| CCND3 | Hs00236949_m | Cyclin D3 | 0.45 | 0.24 |
| CDK2 | Hs01548894_m1 | Cyclin-dependent
kinase 2 | 0.45 | 0.42 |
| MAPK3 | Hs00385075_m1 | Mitogen-activated
protein kinase 3 | 0.45 | 0.19 |
| AKT2 | Hs01086102_m1 | AKT
serine/threonine kinase 2 | 0.45 | 0.30 |
| ELK1 | Hs00428286_g1 | ELK1C | 0.48 | 0.25 |
| CCND1 | Hs00765553_m1 | Cyclin D1 | 0.50 | 0.31 |
| MAPK1 | Hs01046830_m1 | Mitogen-activated
protein kinase 1 | 0.50 | 0.18 |
| AKT1 | Hs00178289_m1 | AKT
serine/threonine kinase 1 | 0.52 | 0.31 |
| TERT | Hs00972650_m1 | Telomerase reverse
transcriptase | 0.53 | 0.40 |
| PIK3CA | Hs00907957_m1 |
Phosphatidylinositol-4C | 0.53 | 0.35 |
| CCND2 | Hs00153380_m1 | Cyclin D2 | 0.60 | 0.25 |
| MAPK14 | Hs00176247_m1 | Mitogen-activated
protein kinase 14 | 0.60 | 0.32 |
| MAP3K5 | Hs00178726_m1 | Mitogen-activated
protein kinase kinase kinase 5 | 0.69 | 0.40 |
| PIK3CD | Hs00192399_m1 |
Phosphatidylinositol-4 | 0.71 | 0.23 |
| TP53 | Hs01034249_m1 | Tumor protein
p53 | 1.20 | 2.23 |
| FOXO3 | Hs00818121_m1 | Forkhead box
O3 | 1.60 | 4.10 |
| FOXO1 | Hs00231106_m1 | Forkhead box
O1 | 1.90 | 4.70 |
| TP73 | Hs01056231_m1 | Tumor protein
p73 | 2.80 | 5.40 |
| PTPRR | Hs00373136_m | Protein tyrosine
phosphatase | 9.50 | 10.00 |
Luteolin increases apoptosis in HeLa
cells
HeLa cells treated with 10 and 20 µM luteolin for 48
h along with untreated cells were used to assess gene expression of
apoptotic genes using a TaqMan Array (apoptosis array; cat. no.
4414072). Gene expression levels of APAF1, BID, BAK1, BOK, BAD,
BAX, NOD1, NOD2, BCL10 and BCL2L14, as well as
Caspase-3, Caspase-7 and Caspase-9 increased (10 µM,
RQ≥1.4; 20 µM, RQ≥2), while expression of BCL2 and
MCL-1 expression was decreased in a dose-dependent manner
(20 µM, RQ≤0.5), and NAIP demonstrated a marginal decrease in
expression. Notably, the receptors and ligands responsible for
stimuli-based apoptosis, including FAS, FASL, FADD, TNF,
TNFRSF10B, TNFRSF1A, Caspase-8 and Caspase-8AP2 (RQ≥2)
were found to be increased in a dose-dependent manner following
luteolin treatment as shown in Fig.
5B (RQ values in Table II).
 | Table II.Relative quantitation values of
apoptotic genes in HeLa cells following treatment with 10 and 20 µM
luteolin treatment for 48 h. |
Table II.
Relative quantitation values of
apoptotic genes in HeLa cells following treatment with 10 and 20 µM
luteolin treatment for 48 h.
| Genes | Ensemble gene
no. | Gene
description | Luteolin 10 µM | Luteolin 20 µM |
|---|
| APAF1 | Hs00559441_m1 | Apoptotic peptidase
activating factor 1 | 8.00 | 19.00 |
| FASLG |
-Hs00181225_m1- | Fas ligand
(TNF)superfamily | 7.40 | 12.00 |
| FAS | Hs00163653_m1 | Fas (TNF receptor
superfamily | 6.00 | 11.00 |
| CASPASE 8 | Hs01018151_m1 | Caspase 8 | 6.00 | 7.00 |
| CASPASE 8AP2 | Hs01594281_m1 | CASP8 associated
protein 2 | 5.70 | 7.00 |
| FADD | Hs00538709_m1 | Fas
(TNFRSF6)-associated via death domain | 5.00 | 6.00 |
| BAK1 | Hs00832876_g1 |
BCL2-antagonist/killer 1 | 5.00 | 5.60 |
| BAD | Hs00188930_m1 | BCL2-antagonist of
cell death | 4.50 | 5.70 |
| CASPASE 7 | -Hs00169152_m1 | Caspase 7 | 4.00 | 5.00 |
| DAPK1 | Hs00234480_m1 | Death-associated
protein kinase 1 | 4.00 | 5.70 |
| TNFRSF10B | Hs00366272_m1 | Tumor necrosis
factor receptor superfamily | 3.80 | 9.00 |
| TNF | Hs00174128_m1 | Tumor necrosis
factor (TNF superfamily | 3.70 | 4.00 |
| TNFRSF1A | Hs01042313_m1 | Tumor necrosis
factor receptor superfamily | 3.70 | 11.00 |
| CASPASE 3 | Hs00234387_m1 | Caspase 3 | 3.40 | 3.70 |
| TRADD | Hs00182558_m1 | TNFRSF1A-associated
via death domain | 3.30 | 7.60 |
| BID | Hs00609632_m1 | BH3 interacting
domain death agonist | 3.00 | 6.00 |
| BOK | Hs00261296_m1 | BCL2-related
ovarian killer | 3.00 | 12.00 |
| CASPASE 9 | Hs00154260_m1 | Caspase 9 | 2.60 | 4.30 |
| NOD1 | Hs00196075_m1 | Nucleotide-binding
oligomerization domain 1 | 2.20 | 7.00 |
| NOD2 | Hs00223394_m1 | Nucleotide-binding
oligomerization domain 2 | 2.20 | 4.60 |
| BCL10 | Hs00961847_m1 | B-cell CLL/lymphoma
10 | 2.00 | 4.00 |
| CARD9 | Hs00261581_m1 | caspase recruitment
domain family | 1.70 | 3.80 |
| BCL2L14 | Hs00373302_m1 | BCL2-like 14
(apoptosis facilitator) | 1.40 | 3.50 |
| CASP 5 | Hs00362072_m1 | Caspase 5 | 1.40 | 3.20 |
| BAX | Hs00751844_s1 | BCL2-associated X
protein | 1.40 | 2.70 |
| NAIp | Hs01847653_s1 | NRL family | 0.90 | 0.74 |
| BCL2 | Hs00608023_m1 | B-cell CLL/lymphoma
2 | 0.80 | 0.29 |
| MCL1 | Hs00172036_m1 | Myeloid cell
leukemia sequence 1 (BCL2-related) | 0.29 | 0.10 |
Luteolin increases Caspase-3 activity
in HeLa cells
Caspases are vital proteins that modulate the
apoptotic response. Caspase 3 is the primary effector molecule,
with the crucial role of induction of the extrinsic and intrinsic
pathway of apoptosis (35). Luteolin
treatment upregulated Caspase-3 activity in HeLa cells, and the
activity increased in a dose-dependent manner. Luteolin-treated
HeLa cells exhibited increased expression of Caspase-3 activity by
2-, 4- and 8-fold when treated with 5, 10 and 20 µM luteolin for 48
h, respectively. The increase in Caspase activity following
luteolin treatment, in comparison to the control, is shown in
Fig. 5C.
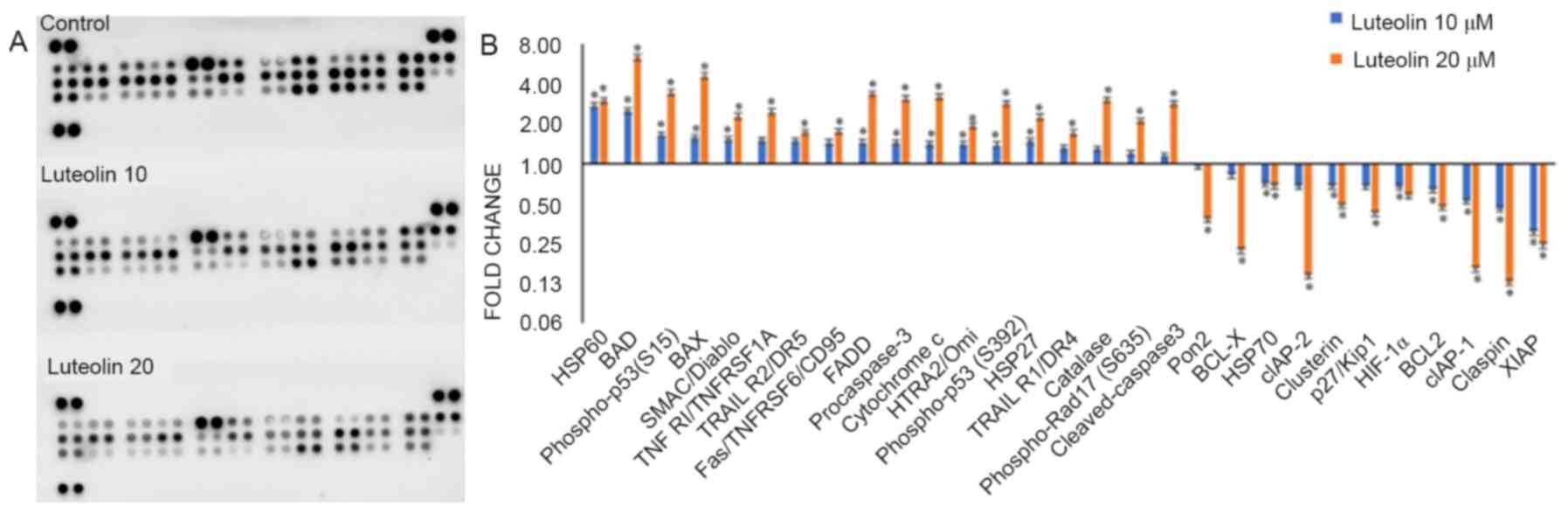
Luteolin alters protein expression of
apoptotic genes
The fold-changes of 35 proteins that are involved in
apoptosis, were investigated using an R&D proteome profiler.
Protein concentration was quantified using a Pierce BCA assay
(Thermo Fisher Scientific, Inc.; cat. no. 23225). Luteolin
significantly upregulated the levels of the pro-apoptotic proteins
involved in mitochondrial apoptosis, including HSP60, BAD, p-p53
(S15), BAX, SMAC/Diablo, pro-Caspase-3, Cytochrome c and HTRA2/Omi,
as well as increasing the expression of receptors and ligands
responsible for receptor based apoptosis, including TNF
RI/TNFRSF1A, TRAIL R2/DR5, Fas/TNFRSF6/CD95, FADD, p-p53 (S392),
p-RAD-17 (S635), HSP27, TRAIL R1/DR4, Catalase, p-Rad17 (S635) and
cleaved-Caspase3 (all exhibiting a fold-change ≥1.5-fold with 20 µM
luteolin treatment), while the levels of the anti-apoptotic
proteins, including Pon2, BCL-X, cIAP-2, Clusterin, p27/Kip1,
HIF-1α, BCL2, cIAP-1, Claspin and XIAP were all downregulated (all
exhibiting a fold-change ≤0.5 with 20 µM luteolin treatment); only
HSP70 expression was marginally decreased (Fig. 6; fold-changes are presented in
Table III).
 | Table III.Fold-change in expression of
apoptotic proteins following luteolin treatment (10 and 20 µM for
48 h). |
Table III.
Fold-change in expression of
apoptotic proteins following luteolin treatment (10 and 20 µM for
48 h).
| Gene | Luteolin 10 µM | Luteolin 20 µM |
|---|
| HSP60 | 2.73 | 2.97 |
| BAD | 2.50 | 6.32 |
| Phospho-p53
(S15) | 1.64 | 3.43 |
| BAX | 1.56 | 4.58 |
| SMAC/Diablo | 1.53 | 2.28 |
| TNF
RI/TNFRSF1A | 1.50 | 2.46 |
| TRAIL R2/DR5 | 1.48 | 1.72 |
|
Fas/TNFRSF6/CD95 | 1.44 | 1.75 |
| FADD | 1.44 | 3.36 |
| Procaspase-3 | 1.43 | 3.08 |
| Cytochrome c | 1.41 | 3.20 |
| HTRA2/Omi | 1.40 | 1.94 |
| Phospho-p53
(S392 | 1.38 | 2.85 |
| HSP27 | 1.47 | 2.24 |
| TRAIL R1/DR4 | 1.31 | 1.71 |
| Catalase | 1.29 | 3.03 |
| Phospho-Rad17
(S635) | 1.20 | 2.10 |
| Cleaved
Caspase3 | 1.15 | 2.85 |
| Pon2 | 0.95 | 0.38 |
| BCL-X | 0.82 | 0.22 |
| HSP70 | 0.70 | 0.67 |
| cIAP-2 | 0.68 | 0.14 |
| Clusterin | 0.67 | 0.48 |
| p27/Kip1 | 0.67 | 0.42 |
|
HIF-1α | 0.66 | 0.57 |
| BCL2 | 0.63 | 0.47 |
| cIAP-1 | 0.52 | 0.16 |
| Claspin | 0.45 | 0.13 |
| XIAP | 0.30 | 0.24 |
Luteolin modulates the Ras-Raf/MAPK
and AKT/MTOR/PIK3 pathways
The MAPK and the PI3K/AKT/MTOR pathways serve a
crucial role in the regulation of cell growth, differentiation and
cell viability. Cancer development is associated with aberrant
activation of genes in these pathways, and this may lead to
increased cell proliferation and survival. Treatment of HeLa cells
with 10 and 20 µM luteolin for 48 h caused decreased expression of
MAPK pathway genes, including MAPK1, MAK14, MAPK3 and
MAP3K5, as well as decreased expression of AKT/MTOR/PIK3
pathway genes, such as AKT1, AKT2, MTOR, PIK3CD, PIK3C2A and
PIK3C2B (all with an RQ≤0.5; Fig.
5A and Table I).
Luteolin alters the phosphorylation of
proteins involved in the AKT signaling pathway
An AKT pathway phosphorylated array (cat. no.
AAH-AKT-1-8) was used to assess the effects of luteolin treatment
on phosphorylation of proteins involved in the AKT pathway. AKT
pathway genes were modulated following treatment with 10 and 20 µM
of luteolin for 48 h. Certain proteins, including P53 (p-ser241)
and P27 (p-Thr198) exhibited a marginal increase in fold-change,
while others, including GSK3a (p-ser21) and GSK3b (p-ser9) were not
affected by luteolin treatment. MTOR (p-ser2448), PRAS 40
(p-Ther246), BAD (p-ser112), PTEN (p-ser380), AKT (p-ser473), ERK2
(p-Y185/Y18, RISK2 (p-ser386), P70S6k (p-Thr421/ser424, PDK1
(p-ser241) and ERK1 (p-T202/Y204) were downregulated. The
fold-changes with respect to the control are presented in Fig. 7 and Table
IV.
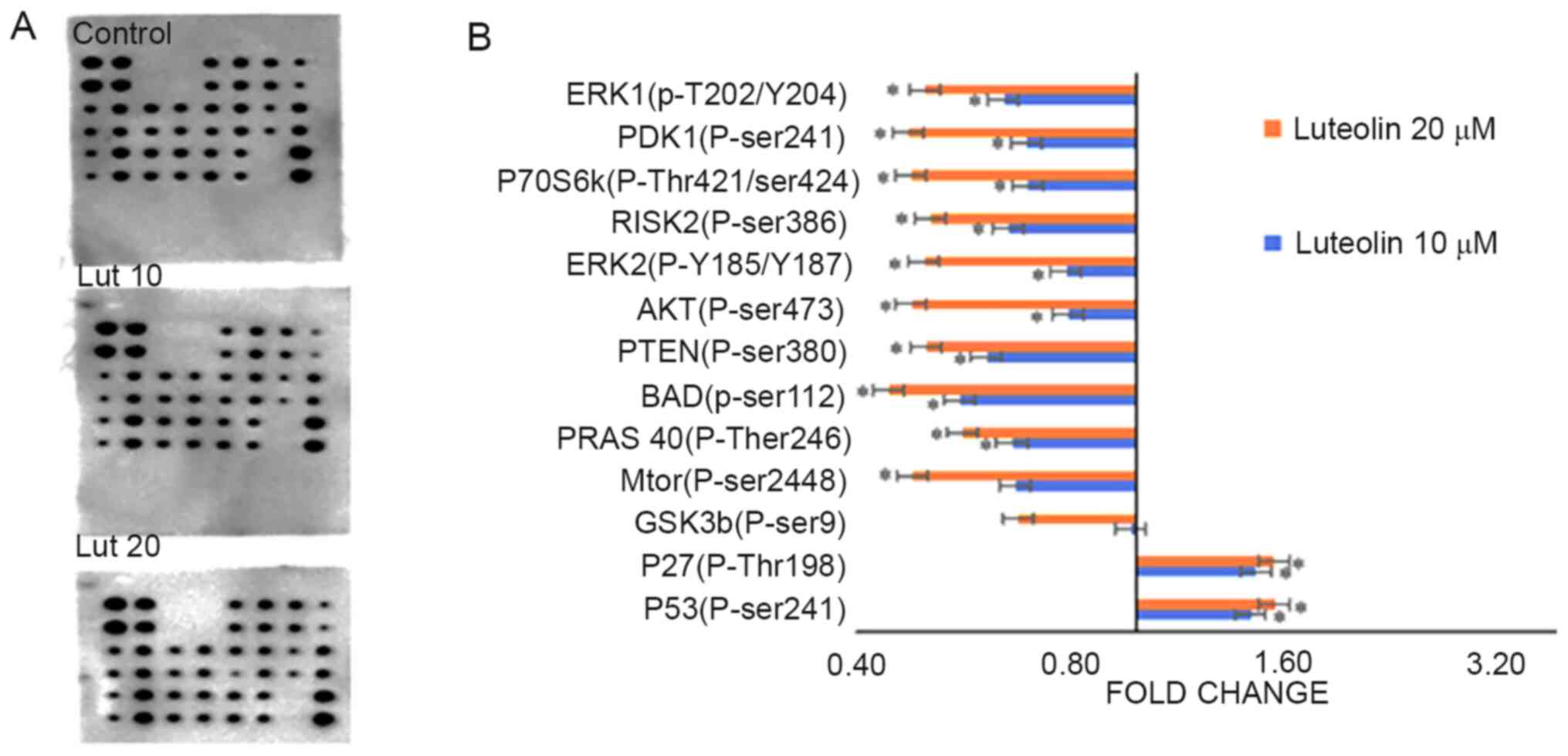 | Figure 7.Differential expression of
phosphorylated proteins in the AKT pathway. (A) Images of the AKT
profiler membranes showing differential expression of the
phosphorylated proteins involved in the AKT signaling pathway at 10
and 20 µM of luteolin treatment for 48 h. (B) Luteolin treatment
decreased expression of phosphorylated proteins involved in the AKT
signaling pathway, including MTOR (p-ser2448), PRAS 40 (p-Ther246),
BAD (p-ser112), PTEN (p-ser380), AKT (p-ser473), ERK2 (p-Y185/187),
RISK2 (p-ser386), P70S6k (p-Thr421/ser424), PDK1 (p-ser241) and
ERK1 (p-T202/Y204), while the expression of P53 (p-ser241) and P27
(p-Thr198) was increased, all in a dose-dependent manner (10 and 20
µM). The fold-changes were calculated by comparing the treated
samples with the DMSO controls. Data are presented as the mean ±
standard deviation of three independent experiments *P<0.05. |
 | Table IV.Fold changes observed following
treatment of HeLa cells with luteolin concentrations of 10 and 20
µM for 48 h. |
Table IV.
Fold changes observed following
treatment of HeLa cells with luteolin concentrations of 10 and 20
µM for 48 h.
| Gene | Luteolin 10 µM | Luteolin 20 µM |
|---|
| P53(P-ser241) | 1.46 | 1.57 |
| P27(P-Thr198) | 1.48 | 1.57 |
| GSK3b(P-ser9) | 0.98 | 0.68 |
|
Mtor(P-ser2448) | 0.67 | 0.48 |
| PRAS
40(P-ther246) | 0.67 | 0.57 |
| BAD(p-ser112) | 0.56 | 0.45 |
| PTEN(P-ser380) | 0.62 | 0.51 |
| AKT(P-ser473) | 0.80 | 0.48 |
|
ERK2(P-Y185/Y187 | 0.80 | 0.50 |
|
RISK2(P-ser386) | 0.66 | 0.51 |
|
P70S6k(P-Thr421/ser424 | 0.70 | 0.48 |
| PDK1(P-ser241) | 0.70 | 0.48 |
|
ERK1(p-T202/Y204) | 0.65 | 0.50 |
Discussion
Despite advances in the early detection of cancer,
and developments in the prevention and treatment of cancer, it
still remains the principal cause of death worldwide (37). Cancer treatment includes a wide range
of chemotherapeutic drugs administered either alone or as an
adjuvant with surgery or radiotherapy, and they are extremely
effective in killing highly proliferative cells, including normal
cells, and this underlies many of the associated side effects
(19,29,37,38).
These drawbacks highlight the requirement for more specific and
less toxic cancer treatment strategies, including plant-derived
chemopreventative agents. Several studies have demonstrated that
plant derived bioactive compounds exert their effects at various
stages of cancer, including initiation, promotion and progression,
and may serve as promising preventative or therapeutic agents.
There is a strong association between a high intake of fruits and
vegetables and a decreased risk of cancer (5,10,11).
Various dietary agents, including curcumin, resveratrol, genistein,
chrysin, sulforaphane and various other flavonoids possess the
ability to reverse or attenuate aberrantly active signaling
pathways associated with carcinogenesis (19,28,38–42).
Flavones exhibit strong anticancer effects in a range of cancer
cell lines via regulation of various molecular mechanisms (14,24,25,43). In
addition to their ability to restore the aberrant signaling of
pathways associated with cancer, flavones may cause cell cycle
arrest, induce apoptosis, and inhibit invasion, metastasis and
inflammation (14,19). Cancer cells may evade apoptosis;
therefore, apoptosis induction by the therapeutic regimen is
required, and this is possible through administration of several
different flavones (14,19,43).
The aim of the present study was to determine the
molecular mechanisms regulated by luteolin in human cervical cancer
cells. Luteolin is the major bioactive compound found in several
different fruits and vegetables, including oregano, parsley and
artichoke, and it possesses anti-neoplastic, anti-inflammatory,
anti-microbial, anti-viral and anti-diabetic properties in
vitro and in vivo (19,24,26,27,44). The
present study demonstrated that luteolin increased cell death in a
dose- and time-dependent manner, and the IC50 was 20 µM
at 48 h. Treatment of HeLa cells with luteolin caused cell death
that was clearly visible using a microscope, with dying cells
exhibiting rounding off and detaching from the culture plates. This
is in agreement with several other studies, which have demonstrated
that luteolin inhibits the growth of cancer cells, with an
IC50 ranging between 10 and 60 µM based on cell line
(24,29,43–47). The
changes to nuclear morphology of HeLa cells treated with 10 and 20
µM for 48 h also justifies the assumption that apoptosis was
induced by luteolin, as nuclear condensation, disintegration,
nuclear blebbing and nuclear debris formation were visualized
following luteolin treatment, while untreated cells did not show
any significant changes in nuclear morphology. Several studies have
reported that flavones, including luteolin, show similar nuclear
morphological changes in several cancer cell lines, including HeLa,
glioma, EC1 and KYSE450 cells (20,24,48).
DNA fragmentation is a characteristic feature of
apoptotic cells (33), and this was
evident in luteolin treated HeLa cells. Exposure of HeLa cells to
5, 10 and 20 µM luteolin for 48 h resulted in disintegration of the
DNA in a ladder-like pattern, while intact DNA was observed in the
untreated control. The degree of fragmentation increased in a
dose-dependent manner when treated for 48 h. Flavonoids, including
chrysin and quercetin, have also been demonstrated to fragment DNA
in different cell lines (20,23,49,50).
Cell cycle analysis revealed that luteolin induced sub-G1 arrest
following treatment with 20 µm for 48 h, in agreement with previous
studies, where arrest at the G2/M or G1 phase was observed when
treated with luteolin in several different cell lines (14,22,24,27,43). The
determination of membrane potential using TMRE and apoptosis
induction by Annexin/PI also supported the notion that apoptosis
was induced by luteolin. Treatment with 5, 10 or 20 µm luteolin for
48 h resulted in mitochondrial depolarization and Annexin/PI
staining showed an increase in the proportion of cells in early and
late apoptosis, and this increased in a dose-dependent manner.
Several studies have reported that flavonoids induce DNA
fragmentation, decrease mitochondrial potential and increase the
proportion of early and late apoptotic cells in various cancer cell
lines (20,24,50,51).
Notably, it has also been demonstrated that luteolin decreased the
expression levels of important cell cycle regulatory genes,
including CCND2, CCND3, CCND1, CCNE2, CDK2, CDK4 and
TERT, and increased the expression of FOXO1 and
FOXO3. The cyclin D proteins, CCNE2 along with
CDK4/CDK6 complex, promotes progression through the G1-S
phase by inhibiting the retinoblastoma protein. hTERT is
responsible for maintenance of the telomeric ends of the
chromosome, and thus, its downregulation assists in the arrest of
cell cycle progression, and similar results have been reported in
different cell lines following treatments with polyphenols
(20,52,53).
FOXO1/3 are considered tumor suppressors that increase
transcription of pro-apoptotic, DNA damage repair and cell cycle
regulatory genes, and their expression is decreased in various
cancer cell lines (20,54). FOXO1 is involved in tumor progression
and is significantly associated with late-stage tumors, and its
expression is important in terms of prognosis of cervical cancer
(20,55).
Pro-apoptotic and anti-apoptotic genes are the
primary proteins involved in regulation of apoptosis, and they form
part of the intrinsic and extrinsic apoptotic pathways. Aberrant
expression of these genes contributes toward the process of
carcinogenesis; therefore, these genes may be targeted for the
prevention and treatment of cancer (20,24,51).
Luteolin treatment of HeLa cells resulted in upregulation of
pro-apoptotic genes involved in the mitochondrial apoptosis
pathway, including APAF1, BAX, BAK1, BAD, BID, BOK, Caspase7,
Caspase9, Caspase 3, NOD1, NOD2, BCL10, CARD9, BCL2L14 and
Caspase 5, as well as extrinsic pathway genes, including
FASLG, FAS, Caspase 8, Caspase 2, FADD, TNFRSF10B, TNF,
TNFRSF1A and TRADD, at the transcriptional level. By
contrast, anti-apoptotic genes, including BCL2, Mcl1 and
NAIP were found to be downregulated in luteolin-treated HeLa
cells. Caspase-3 is the primary effector caspase and serves a
significant role in the induction of apoptosis via regulation of
the extrinsic and intrinsic pathways. Treatment with 5, 10 and 20
µM luteolin for 48 h enhanced the biochemical activity of Caspase-3
by 2, 4 and 8-fold, respectively. The association of caspase 3
activation with decreased cell survival could be hypothesized by
treating the cells with caspase inhibitor (Z-VAD-FMK), but that was
beyond the scope of the present study. However, Islam et al
(35) has reported role of Caspase-3
in EGCG mediated apoptosis of Human Chondrosarcoma Cells (HTB-94).
Numerous studies have demonstrated that flavonoids induce apoptosis
by either the intrinsic or extrinsic pathways (14,19,20,22,48).
Additionally, protein expression of various genes was in agreement
with the changes observed in their gene expression levels.
Treatment of HeLa cells with luteolin resulted in the upregulation
of pro-apoptotic genes at the protein level, including upregulation
of HSP60, BAD, p-p53(S15), BAX, SMAC/Diablo, TNF, RI/TNFRSF1A,
TRAILR2/DR5, Fas/TNFRSF6/CD95, FADD, pro-caspase-3, Cytochrome c,
HTRA2/Omi, p-p53(S392), HSP27, TRAILR1/DR4, Catalase, p-Rad17
(S635), cleaved Caspase3 and p53, which is consistent with earlier
reports on HeLa and leukemia cells treated with the flavonoids
quercetin and luteolin (17,20,23).
Downregulation of antiapoptotic proteins at the mRNA/protein level,
including Pon2, BCL-X, HSP70, cIAP-2, clusterin, p27/Kip1, HIF-1α,
p21/CIP1/CDKN1A and BCL2, as well as upregulation of BAX, p-p53,
release of Cytochrome C, SMAC/DIABLO and cleavage of Caspase 3,
Caspase 9 and PARP1 suggests the involvement of intrinsic
apoptosis. Furthermore, upregulation of receptors and ligands
involved in the extrinsic pathway at the mRNA/protein level,
including TNF, TNFR, RI/TNFRSF1A, TRAIL R1/DR4, TRAIL R2/DR5,
Fas/TNFRSF6/CD95 and Caspase-8/Caspase-8Ap2 highlights the
involvement of the extrinsic pathway. Similar results have been
reported when treated with several different flavonoids and various
cell lines (19,20,56,57).
Based on the aforementioned results, it may be stated that luteolin
activates the extrinsic and the intrinsic pathway of apoptosis, in
agreement with certain previous studies; however, other studies
have reported that luteolin induces either the extrinsic or
intrinsic pathway exclusively (46,47,49). In
cancer cells, alterations of p27 protein-protein interaction alters
p27 from a CDK inhibitor to an oncogene.
Activation of the AKT/RAS-RAF pathway by
phosphorylation averts apoptosis; however, dephosphorylation likely
induces apoptosis (57,58). BAD is phosphorylated at Ser-136 and
Ser-112 by AKT and RAS/RAF, respectively (58). Phosphorylation of BAD leads its
association with 14-3-3 protein, whereas dephosphorylation of BAD
results in its dimerization with BCL2 family proteins, and
accumulation in the mitochondrial membrane, in turn increasing
apoptosis (59). AKT may further
activate other pathways and molecules, including the MTORC pathway
(60), and is initiated by various
growth factors and cytokines through receptor tyrosine kinases,
including HER, and promotes cell survival by inactivating
pro-apoptotic proteins and the forkhead (FoxO1/3a) transcription
factors (61). Luteolin treatment of
HeLa cells decreased the expression levels of important members of
AKT/PI3K signaling; specifically, AKT1, AKT2, MTOR, PIK3CD, PIK3C2A
and PIK3C2B were downregulated. Luteolin also modulated the
phosphorylation of members of the AKT signaling pathway, including
P53 (p-ser241) and P27 (p-Thr198), which were significantly
upregulated, while GSK3b (p-ser9) was slightly downregulated, MTOR
(p-ser2448), PRAS 40 (p-Ther246), BAD (p-ser112), PTEN (p-ser380)
and AKT (p-ser473) were more notably downregulated, and ERK2
(p-Y185/Y187, RISK2 (p-ser386), P70S6k (p-Thr421/ser424), PDK1
(p-ser241) and ERK1 (p-T202/Y204) were significantly downregulated.
Several studies have reported similar results; however, with fewer
phosphorylated proteins assessed. Therefore, the present study is
the first to extensively demonstrate the molecular effects of
luteolin-mediated modification of phosphorylated proteins involved
in the AKT pathway, to the best of our knowledge (14,18,21). The
MAPK pathway serves a potential role in cell proliferation and
differentiation; luteolin treatment of HeLa cells showed
downregulation of various members of the MAPK signaling pathway,
including MAPK1, MAK14, MAPK3 and MAP3K5, a
substantial decrease in gene expression was observed in ELK1 and
ESR1 together with an increase in PTPRR, which is an inhibitor of
the MAPK pathway. Notably, flavonoids have been demonstrated to
inhibit the AKT/MTOR/MAPK signaling pathways through modulation of
various molecules involved in these pathways (20,22,48,49,61).
Plant-derived agents, such as flavonoids, including
luteolin, have proven to be potentially effective options for the
treatment of cancer through modulation of apoptotic pathways.
However, the effect of these agents vary notably when comparing
their effects in vitro and in vivo, and this may be
due to their low bioavailability in vivo. Several studies
have attempted to improve the bioavailability of various agents,
with success in certain cases; quercetin treatment using a nano
delivery system improved its effectiveness in in-vivo
studies (62,63). As the majority of the results cited
regarding luteolin in the present study are based on in
vitro and in vivo studies, and these do not necessarily
always accurately represent the outcomes observed in humans,
additional studies on the different pharmacokinetic parameters are
required, as well as human clinical trials, before it can be
considered a suitable treatment for cancer. Further study on other
cancer cells, including blood tumor cells, will support the use of
luteolin in a broad spectrum.
In conclusion, the various effects of luteolin
highlight its efficacy on the inhibition of cell proliferation,
depolarization of the mitochondrial membrane potential, induction
of cell cycle arrest and impeding the RAS-RAF/MAPK/AKT/PI3K
signaling pathway. Therefore, induction of apoptosis in HeLa cells
may qualify it as a potential chemo-preventive agent. However, it
is first necessary to perform additional in vivo studies and
clinical trials to standardize the dosage and assess the safety of
luteolin, before it can be recommended as a treatment for cervical
cancer treatment.
Acknowledgements
Not applicable..
Funding
The present study was supported by the Zayed
University (RIF grant no. R19056) and a MAHE internal research
grant (grant no. R&DP/MUD/RL-06/2018).
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
RR and SP performed the experiments, collected the
data and wrote the manuscript. NR and SH revised the manuscript and
validated the data. JS and KB analyzed and validated the data. MH
and AH conceived and designed the study, validated the data and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tripathi KD: Essentials of medical
pharmacology. (7th edition). J.P. Medical Ltd. 2013.
|
|
2
|
Masui K, Gini B, Wykosky J, Zanca C,
Mischel PS, Furnari FB and Cavenee WK: A tale of two approaches:
Complementary mechanisms of cytotoxic and targeted therapy
resistance may inform next-generation cancer treatments.
Carcinogenesis. 34:725–738. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Patterson SL, Colbert Maresso K and Hawk
E: Cancer chemoprevention: Successes and failures. Clin Chem.
59:94–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dedeurwaerder S, Defrance M, Calonne E,
Denis H, Sotiriou C and Fuks F: Evaluation of the infinium
methylation 450 K technology. Epigenomics. 3:771–784. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Theodoratou E, Timofeeva M, Li X, Meng X
and Ioannidis JPA: Nature, nurture, and cancer risks: Genetic and
nutritional contributions to cancer. Annu Rev Nutr. 37:293–320.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sauter ER: Breast cancer prevention:
Current approaches and future directions. Eur J Breast Health.
14:64–71. 2018.PubMed/NCBI
|
|
7
|
de Melo FHM, Oliveira JS, Sartorelli VOB
and Montor WR: Cancer chemoprevention: Classic and epigenetic
mechanisms inhibiting tumorigenesis. What have we learned so far?
Front Oncol. 8:6442018.PubMed/NCBI
|
|
8
|
Ko JH, Sethi G, Um JY, Shanmugam MK,
Arfuso F, Kumar AP, Bishayee A and Ahn KS: The role of resveratrol
in cancer therapy. Int J Mol Sci. 18:25892017. View Article : Google Scholar
|
|
9
|
Sur S and Panda CK: Molecular aspects of
cancer chemopreventive and therapeutic efficacies of tea and tea
polyphenols. Nutrition. 43-44:8–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu S, Zhu W, Thompson P and Hannun YA:
Evaluating intrinsic and non-intrinsic cancer risk factors. Nat
Commun. 9:34902018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Willett WC: Diet, nutrition, and avoidable
cancer. Environ Health Perspect. 103 Suppl:8 (Suppl 8):S165–S170.
1995. View Article : Google Scholar
|
|
12
|
Hussain A and Raina R: Flavones combat
cancer via multidimensional molecular approaches. Phytocompounds:
Sources and Bioactivities. Studium Press; 2019
|
|
13
|
Costea T, Hudiță A, Ciolac OA, Gălățeanu
B, Ginghină O, Costache M, Ganea C and Mocanu MM: Chemoprevention
of colorectal cancer by dietary compounds. Int J Mol Sci.
19:37872018. View Article : Google Scholar
|
|
14
|
Koosha S, Alshawsh MA, Looi CY, Seyedan A
and Mohamed Z: An association map on the effect of flavonoids on
the signaling pathways in colorectal cancer. Int J Med Sci.
13:374–385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maru GB, Hudlikar RR, Kumar G, Gandhi K
and Mahimkar MB: Understanding the molecular mechanisms of cancer
prevention by dietary phytochemicals: From experimental models to
clinical trials. World J Biol Chem. 7:88–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Attoub S, Hassan AH, Vanhoecke B, Iratni
R, Takahashi T, Gaben AM, Bracke M, Awad S, John A, Kamalboor HA,
et al: Inhibition of cell survival, invasion, tumor growth and
histone deacetylase activity by the dietary flavonoid luteolin in
human epithelioid cancer cells. Eur J Pharmacol. 651:18–25. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ham S, Kim KH, Kwon TH, Bak Y, Lee DH,
Song YS, Park SH, Park YS, Kim MS, Kang JW, et al: Luteolin induces
intrinsic apoptosis via inhibition of E6/E7 oncogenes and
activation of extrinsic and intrinsic signaling pathways in
HPV-18-associated cells. Oncol Rep. 31:2683–2691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng G, Chai K, Li X, Zhu Y and Huang W:
Luteolin exerts pro-apoptotic effect and anti-migration effects on
A549 lung adenocarcinoma cells through the activation of MEK/ERK
signaling pathway. Chem Biol Interact. 257:26–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Abotaleb M, Samuel SM, Varghese E,
Varghese S, Kubatka P, Liskova A and Büsselberg D: Flavonoids in
cancer and apoptosis. Cancers (Basel). 11:282019. View Article : Google Scholar
|
|
20
|
Kedhari Sundaram M, Raina R, Afroze N,
Bajbouj K, Hamad M, Haque S and Hussain A: Quercetin modulates
signaling pathways and induces apoptosis in cervical cancer cells.
Biosci Rep. 39:BSR201907202019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Badgujar NV, Mistry KN, Rank DN and Joshi
CG: Screening of antiproliferative activity mediated through
apoptosis pathway in human non-small lung cancer A-549 cells by
active compounds present in medicinal plants. Asian Pac J Trop Med.
11:666–675. 2018. View Article : Google Scholar
|
|
22
|
Tuorkey MJ: Molecular targets of luteolin
in cancer. Eur J Cancer Prev. 25:65–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang SW, Chen YR, Chow JM, Chien MH, Yang
SF, Wen YC, Lee WJ and Tseng TH: Stimulation of Fas/FasL-mediated
apoptosis by luteolin through enhancement of histone H3 acetylation
and c-Jun activation in HL-60 leukemia cells. Mol Carcinog.
57:866–877. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen P, Zhang JY, Sha BB, Ma YE, Hu T, Ma
YC, Sun H, Shi JX, Dong ZM and Li P: Luteolin inhibits cell
proliferation and induces cell apoptosis via down-regulation of
mitochondrial membrane potential in esophageal carcinoma cells EC1
and KYSE450. Oncotarget. 8:27471–27480. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park SH, Ham S, Kwon TH, Kim MS, Lee DH,
Kang JW, Oh SR and Yoon DY: Luteolin induces cell cycle arrest and
apoptosis through extrinsic and intrinsic signaling pathways in
MCF-7 breast cancer cells. J Environ Pathol Toxicol Oncol.
33:219–231. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mary V, Haris P, Varghese MK, Aparna P and
Sudarsanakumar C: Experimental probing and molecular dynamics
simulation of the molecular recognition of DNA duplexes by the
flavonoid luteolin. J Chem Inf Model. 57:2237–2249. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Imran M, Rauf A, Abu-Izneid T, Nadeem M,
Shariati MA, Khan IA, Imran A, Orhan IE, Rizwan M, Atif M, et al:
Luteolin, a flavonoid, as an anticancer agent: A review. Biomed
Pharmacother. 112:1086122019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aggarwal R, Jha M, Shrivastava A and Jha
AK: Natural compounds: Role in reversal of epigenetic changes.
Biochemistry (Mosc). 80:972–989. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou Y, Zheng J, Li Y, Xu DP, Li S, Chen
YM and Li HB: Natural polyphenols for prevention and treatment of
cancer. Nutrients. 8:5152016. View Article : Google Scholar
|
|
30
|
Ruan J, Zhang L, Yan L, Liu Y, Yue Z, Chen
L, Wang AY, Chen W, Zheng S, Wang S and Lu Y: Inhibition of
hypoxia-induced epithelial mesenchymal transition by luteolin in
non-small cell lung cancer cells. Mol Med Rep. 6:232–238.
2012.PubMed/NCBI
|
|
31
|
Sundaram MK, Unni S, Somvanshi P, Bhardwaj
T, Mandal RK, Hussain A and Haque S: Genistein modulates signaling
pathways and targets several epigenetic markers in HeLa cells.
Genes (Basel). 10:9552019. View Article : Google Scholar
|
|
32
|
Masters JR: HeLa cells 50 years on: The
good, the bad and the ugly. Nat Rev Cancer. 2:315–319. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shariffa K: Apoptosis-journey of a cell in
life. Univ J Surg Surg Spec. 4(3)2018.
|
|
34
|
Li J, Han J, Hu Y and Yang J: Selection of
reference genes for quantitative real-time PCR during flower
development in tree peony (Paeonia suffruticosa Andr.).
Front Plant Sci. 7:5162016.PubMed/NCBI
|
|
35
|
Islam S, Islam N, Kermode T, Johnstone B,
Mukhtar H, Moskowitz RW, Goldberg VM, Malemud CJ and Haqqi TM:
Involvement of caspase-3 in epigallocatechin-3-gallate-mediated
apoptosis of human chondrosarcoma cells. Biochem Biophys Res
Commun. 270:793–797. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lennarz WJ and Lane MD: Encyclopedia of
biological chemistry. 2nd edition. Academic Press; 2013, PubMed/NCBI
|
|
37
|
Gul-e-Saba, Islamiah M, Ismail N, Mohamad
H, Sung YY and Muhammad TST: Induction of apoptosis by Aaptos sp.,
fractions in human breast cancer cell line, MCF-7. Int J Res Pharm
Sci. 9(2)2018.
|
|
38
|
Lee DH, Park KI, Park HS, Kang SR,
Nagappan A, Kim JA, Kim EH, Lee WS, Hah YS, Chung H, et al:
Flavonoids isolated from Korea Citrus aurantium L. induce G2/M
phase arrest and apoptosis in human gastric cancer AGS cells. Evid
Based Complement Alternat Med. 2012:5159012012.PubMed/NCBI
|
|
39
|
Abdal Dayem A, Choi HY, Yang GM, Kim K,
Saha SK and Cho SG: The anti-cancer effect of polyphenols against
breast cancer and cancer stem cells: Molecular mechanisms.
Nutrients. 8:5812016. View Article : Google Scholar
|
|
40
|
Mocanu MM, Nagy P and Szöllősi J:
Chemoprevention of breast cancer by dietary polyphenols. Molecules.
20:22578–22620. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khan MA, Hussain A, Sundaram MK, Alalami
U, Gunasekera D, Ramesh L, Hamza A and Quraishi U:
(−)-Epigallocatechin-3-gallate reverses the expression of various
tumor-suppressor genes by inhibiting DNA methyltransferases and
histone deacetylases in human cervical cancer cells. Oncol Rep.
33:1976–1984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hussain A, Mohsin J, Prabhu SA, Begum S,
Nusri Qel-A, Harish G, Javed E, Khan MA and Sharma C: Sulforaphane
inhibits growth of human breast cancer cells and augments the
therapeutic index of the chemotherapeutic Drug, gemcitabine. Asian
Pac J Cancer Prev. 14:5855–5860. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xue C, Chen Y, Hu DN, Iacob C, Lu C and
Huang Z: Chrysin induces cell apoptosis in human uveal melanoma
cells via intrinsic apoptosis. Oncol Lett. 12:4813–4820. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cao Z, Zhang H, Cai X, Fang W, Chai D, Wen
Y, Chen H, Chu F and Zhang Y: Luteolin promotes cell apoptosis by
inducing autophagy in hepatocellular carcinoma. Cell Physiol
Biochem. 43:1803–1812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hu C, Cai X, Hu T, Lu W and Cao P:
Mechanism of growth inhibition effect of 3′, 4′, 5,
7-tetrahydroxyflavone on A549 cells. Zhongguo Zhong Yao Za Zhi.
37:1259–1264. 2012.(In Chinese). PubMed/NCBI
|
|
46
|
Aneknan P, Kukongviriyapan V, Prawan A,
Kongpetch S, Sripa B and Senggunprai L: Luteolin arrests cell
cycling, induces apoptosis and inhibits the JAK/STAT3 pathway in
human cholangiocarcinoma cells. Asian Pac J Cancer Prev.
15:5071–5076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
George VC, Naveen Kumar DR, Suresh PK,
Kumar S and Kumar RA: Comparative studies to evaluate relative in
vitro potency of luteolin in inducing cell cycle arrest and
apoptosis in HaCaT and A375 cells. Asian Pac J Cancer Prev.
14:631–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
You Y, Wang R, Shao N, Zhi F and Yang Y:
Luteolin suppresses tumor proliferation through inducing apoptosis
and autophagy via MAPK activation in glioma. Onco Targets Ther.
12:23832019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu X, Li Y, Li X and Aisa HA: Luteolin
induces apoptosis in vitro through suppressing the MAPK and PI3K
signaling pathways in gastric cancer. Oncol Lett. 14:1993–2000.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Samarghandian S, Nezhad MA and Mohammadi
G: Role of caspases, Bax and Bcl-2 in chrysin-induced apoptosis in
the A549 human lung adenocarcinoma epithelial cells. Anticancer
Agents Med Chem. 14:901–909. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tu LY, Bai HH, Cai JY and Deng SP: The
mechanism of kaempferol induced apoptosis and inhibited
proliferation in human cervical cancer SiHa cell: From macro to
nano. Scanning. 38:644–653. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ren X, Zhang Z, Tian J, Wang H, Song G,
Guo Q, Tian J, Han Y, Liao Q, Liu G, et al: The downregulation of
c-Myc and its target gene hTERT is associated with the
antiproliferative effects of baicalin on HL-60 cells. Oncol Lett.
14:6833–6840. 2017.PubMed/NCBI
|
|
53
|
Huang L, Jin K and Lan H: Luteolin
inhibits cell cycle progression and induces apoptosis of breast
cancer cells through downregulation of human telomerase reverse
transcriptase. Oncol Lett. 17:3842–3850. 2019.PubMed/NCBI
|
|
54
|
Song KH, Woo SR, Chung JY, Lee HJ, Oh SJ,
Hong SO, Shim J, Kim YN, Rho SB, Hong SM, et al: REP1 inhibits
FOXO3-mediated apoptosis to promote cancer cell survival. Cell
Death Dis. 8:e25362018. View Article : Google Scholar
|
|
55
|
Zhang B, Gui LS, Zhao XL, Zhu LL and Li
QW: FOXO1 is a tumor suppressor in cervical cancer. Genet Mol Res.
14:6605–6616. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang Y, Chen S, Wei C, Rankin GO, Ye X
and Chen YC: Flavonoids from Chinese bayberry leaves induced
apoptosis and G1 cell cycle arrest via Erk pathway in ovarian
cancer cells. Eur J Med Chem. 147:218–226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Srivastava S, Somasagara RR, Hegde M,
Nishana M, Tadi SK, Srivastava M, Choudhary B and Raghavan SC:
Quercetin, a natural flavonoid interacts with DNA, arrests cell
cycle and causes tumor regression by activating mitochondrial
pathway of apoptosis. Sci Rep. 6:240492016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Fang X, Yu S, Eder A, Mao M, Bast RC Jr,
Boyd D and Mills GB: Regulation of BAD phosphorylation at serine
112 by the Ras-mitogen-activated protein kinase pathway. Oncogene.
18:6635–6640. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Khoo BY, Chua SL and Balaram P: Apoptotic
effects of chrysin in human cancer cell lines. Int J Mol Sci.
11:2188–2199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cardoso MFS, Castelletti CHM, Lima-Filho
JL, Martins DBG and Teixeira JAC: Putative biomarkers for cervical
cancer: SNVs, methylation and expression profiles. Mutat Res.
773:161–173. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Okkenhaug K, Bilancio A, Farjot G, Priddle
H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield
MD, et al: Impaired B and T cell antigen receptor signaling in
p110delta PI 3-kinase mutant mice. Science. 297:1031–1034.
2002.PubMed/NCBI
|
|
62
|
Wang S, Zhang J, Chen M and Wang Y:
Delivering flavonoids into solid tumors using nanotechnologies.
Expert Opin Drug Deliv. 10:1411–1428. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yuan ZP, Chen LJ, Fan LY, Tang MH, Yang
GL, Yang HS, Du XB, Wang GQ, Yao WX, Zhao QM, et al: Liposomal
quercetin efficiently suppresses growth of solid tumors in murine
models. Clin Cancer Res. 12:3193–3199. 2006. View Article : Google Scholar : PubMed/NCBI
|