Introduction
As the third most common malignant disease in the
world, colorectal cancer (CRC) exhibits a 2.27% cumulative risk of
onset between 0–74 years of age (1).
In China, CRC is also one of the 5 most common types of cancer, and
poses a significant disease burden in the aging population
(2). The 5-year relative survival
rate for Chinese patients with CRC has increased from 47.2 to 56.9%
in the past 10 years. However, the possible of mortality rate of
CRC has remained at 50% (3). The
overall prognosis of CRC was improved from the use of adjuvant
chemotherapy and the advance of resection. However, even in the
early stages of CRC, recurrence following surgery remains the
primary, and ultimate, cause of death (4). The development of a potential
therapeutic target, that can aid the detection and treatment of
CRC, is important for the prevention of this disease.
Multiple mitochondrial functions, including
motility, mitochondrial morphology, asymmetric division and
maintenance of ATP levels (5,6) are
necessary for the function of phosphodiesterase-δ. The ADP
ribosylation factor-like GTPase2 (ARL2) assists in the release of
Ras (6). Multiple cell signaling
pathways and various cellular functions are regulated by Ras,
including proliferation, differentiation, vesicle transport,
nuclear assembly, and the regulation of the cytoskeleton are
included (7). Several studies have
shown the contribution of a Ras mutation, such as KRAS-4B, in the
development of human tumors (8–10). It
has been shown that ~21% of all human cancers, and ~30, 45, and 90%
of lung, colon and pancreatic cancers, respectively, originate due
to activating mutations in the Ras family of enzymes (8). It has also been found that the
regulation of ARL2 protein expression could alter the duration and
degree of numerous types of cancer, and the cell migratory and
invasive abilities of cancer cells (11,12).
ARL2 has been shown to regulate p53 localization, resulting in a
chemoresistant phenotype in breast cancer cells (13). As a prognostic marker, ARL2 mRNA
expression was significantly elevated in hepatocellular tumors
(14). In addition, ARL2 acts as an
oncogene in cervical cancer (15).
However, the functional role of ARL2 in CRC is not clear.
AXL is a member of the TAM group of receptors, an
inhibitor of the cytokine receptor-mediated macrophage/monocyte
activation and promoter of apoptotic cell removal. In addition, AXL
is an oncotarget in human CRC (16).
Silencing of AXL gene expression suppresses proliferation,
migration and survival in CRC cells (16). In CRC, AXL is overexpressed and its
elevated protein expression promotes migration, invasion and
epithelial-mesenchymal transition of CRC cell lines (17).
The expression and function of ARL2 in CRC was
investigated in the current study. It was found that ARL2
overexpression or knockdown regulated the proliferation, colony
formation, migration and invasion abilities of CRC cells and it had
a negative association with AXL.
Materials and methods
Patients and samples
The patient samples were collected from The
Affiliated Hospital of Xuzhou Medical University (Jiangsu, China)
from January to June in 2019, including 19 colon samples (6
well-differentiated, 6 moderately differentiated and 7 poorly
differentiated cases). All patients with CRC underwent surgical
resection and histological diagnosis was verified by 2
gastroenterologists (Pathology Department of The Affiliated
Hospital of Xuzhou Medical University) (Xuzhou, China). Patients
with CRC who had undergone preoperative radiotherapy and
chemotherapy were excluded. A total of 19 colon cancer tissues and
3 healthy colorectal tissues (from partial resection in patients
with colorectal trauma. As a regional scientific research and
medical institution, some patients suitable for being assigned to
normal controls were informed and signed informed consent before or
after surgery were used for immunohistochemistry (IHC) analysis.
Surgical resection was performed in all patients with colon cancer,
and the histological diagnosis was confirmed according to 2016
World Health Organization (WHO) guidelines (18). The present study was approved by the
Medical Ethics Committee of the Hospital of The Affiliated Hospital
of Xuzhou Medical University (Jiangsu, China), and written informed
consent was provided from each patient for his/her participation in
the study (approval no. XYFY2019-KL157-01). The clinical
characteristics of the 19 patients with colon cancer are listed in
Table I.
 | Table I.Clinical characteristics of the 19
patients with colorectal cancer. |
Table I.
Clinical characteristics of the 19
patients with colorectal cancer.
|
Characteristics | Number of patients
(%) |
|---|
| Age, years |
|
|
<60 | 5 (26.32) |
|
≥60 | 14 (73.68) |
| Sex |
|
|
Male | 7 (36.84) |
|
Female | 12 (63.16) |
| Cancer
location |
|
|
Proximal | 11 (57.89) |
| Distal
colon | 8 (42.11) |
| Degree of
differentiation |
|
|
Well | 6 (31.58) |
|
Moderately | 6 (31.58) |
|
Poorly | 7 (36.84) |
IHC assay
IHC analysis of ARL2 was performed on 19 colon
cancer and 3 healthy colorectal tissues. The samples were fixed in
10% neutral buffered formalin for ~3-24 h at room temperature,
dehydrated in gradient alcohol solution (30, 50, 70, 80, 90, 95,
100, 100% alcohol each for 30 min) and paraffin-embedded.
Paraffin-embedded tissues were cut into ~3-µm thick continuous
sections. Routine dewaxing, antigen retrieval (microwave thermal
retrieval) and quenching of endogenous peroxidase activity with 3%
H2O2 at room temperature was performed on the
slices. After blocking with 5% goat serum [cat. no. abs933; Absin
(Shanghai) Biotechnology Co., Ltd.] at room temperature for ~1 h,
primary antibody (anti-ARL2; 1:100; cat. no. 188322; Cell Signaling
Technology, Inc.) was added to the sections and incubated overnight
at 4°C. The substrate [primary reactant amplifier; cat. no. abs957;
Absin (Shanghai) Biotechnology Co., Ltd.] for the secondary
antibody was added and incubated at room temperature for 30 min
with the samples. Then the sections were subsequently treated with
a secondary antibody [HRP-labeled goat resistant rabbit IgG;
1:1,000; cat. no. abs957; Absin (Shanghai) Biotechnology Co., Ltd.]
at room temperature for 1 h. The slices were stained with
3′,3′-diaminobenzidine (DAB) at room temperature for about 7 min
[Liquid A and liquid B; 1000:50; cat. no. abs957; Absin (Shanghai)
Biotechnology Co., Ltd.]. Finally, the slides were stained with
0.02% hematoxylin (cat. no. BL702A; Biosharp Life Sciences) and
observed under a light microscope (IX73; Olympus Corporation). The
images were analyzed using Photoshop (Adobe Systems, Inc.).
A total of two pathologists from The Affiliated
Hospital of Xuzhou Medical University (Xuzhou, China) examined the
pathological sections separately under blinded experimental
conditions and all the differences were evaluated by discussion.
High power microscopic fields (two), that were representative of
the samples were selected for each specimen and 200 tumor cells
were counted in each field. The German IHC Score (GIS) (11) was used for scoring. The intensity of
ARL2 immunostaining was scored as follows: 0, negative; 1, weak; 2,
moderate; 3, strong, whereas the percentage of immunoreactive cells
was categorized as 1 (0–25%), 2 (26–50%), 3 (51–75%), and 4
(76–100%). The IHC expression values of ARL2 and the associated
clinical information of each sample are shown in Table II.
 | Table II.ARL2 expression value and
characteristics of the normal healthy and CRC samples used for
IHC. |
Table II.
ARL2 expression value and
characteristics of the normal healthy and CRC samples used for
IHC.
| Sample | Sample number | Age, years | GIS |
|---|
| Normal healthy
tissue | N1 | 35 | 0 |
|
| N2 | 60 | 1 |
|
| N3 | 85 | 1 |
| CRC tissues |
|
|
|
| Well
differentiated | H1 | 66 | 8 |
|
| H2 | 63 | 12 |
|
| H3 | 85 | 12 |
|
| H4 | 40 | 8 |
|
| H5 | 67 | 9 |
|
| H6 | 62 | 12 |
|
Moderately differentiated | M1 | 66 | 4 |
|
| M2 | 80 | 8 |
|
| M3 | 82 | 8 |
|
| M4 | 50 | 4 |
|
| M5 | 78 | 4 |
|
| M6 | 54 | 8 |
| Poorly
differentiated | L1 | 73 | 4 |
|
| L2 | 66 | 4 |
|
| L3 | 37 | 0 |
|
| L4 | 76 | 0 |
|
| L5 | 55 | 6 |
|
| L6 | 70 | 8 |
|
| L7 | 77 | 8 |
Cell lines and culture conditions
The HCT8, HCT116 and 293T cell lines were acquired
from the Cancer Institute, Xuzhou Medical University (Jiangsu,
China), which were originally purchased from the Shanghai Institute
of Biochemistry and Cell Biology, Chinese Academy of Science
(Shanghai, China). The HCT8, HCT116 and 293T cell lines were
authenticated using short tandem repeat DNA profiling, and
subsequently cultured in DMEM containing 10% fetal bovine serum
(cat. no. SH30022.01B; HyClone; Cytiva), 100 µg/ml streptomycin and
100 U/ml penicillin, and incubated at 37°C in a humidified
incubator with 5% CO2.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the HCT8 and HCT116
cells using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.), in line with the manufacturer's protocol. The
total RNA was reverse transcribed into cDNA for PCR amplification.
RT-qPCR was performed using TransStart® Top Green qPCR
SuperMix Assays (cat. no. AQ131; Beijing Transgen Biotech Co.,
Ltd.) and in a thermal cycler (LightCycler® 480II; Roche
Diagnostics). Human mRNA sequences, encoding ARL2 and AXL genes,
were retrieved from the nucleotide database of National Center for
Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/). The primers were
designed using Primer Premier v5.0 (Premier Biosoft) and Basic
Local Alignment Search Tool was used in NCBI to determine the
specificity of the primers. The primers were synthesized by Generay
Biotech Co., Ltd. The following thermocycling conditions were used:
Initial denaturation at 95°C for 30 sec, followed by 40 cycles of a
two-step cycling program (95°C for 5 sec and 60°C for 30 sec). The
mRNA expression was normalized to the expression of β-actin and
calculated using the 2−ΔΔCq method (19). The specific primers for ARL2, AXL and
β-actin were as follows: ARL2 forward, 5′-GGGAGGACATCGACACCA-3′ and
reverse, 5′-AGGACCGCAGGGACTTC-3′; AXL forward,
5′-GTTTGGAGCTGTGATGGAAGGC-3′ and reverse,
5′-CGCTTCACTCAGGAAATCCTCC-3′; and β-actin forward,
5′-TGACGTGGACATCCGCAAAG-3′ and reverse,
5′-CTGGAAGGTGGACAGCGAGG-3′.
Immunofluorescence
ARL2 pcDNA3.1 HCT8 and HCT116 cells, ARL2 shRNA HCT8
and HCT116 cells, and the corresponding control group cells were
fixed with 4% paraformaldehyde at room temperature for 20 min and
blocked in PBS containing 5% goat serum [cat. no. abs933; Absin
(Shanghai) Biotechnology Co., Ltd.]. The cells were then washed
three times, with PBS for 5 min and permeabilized with 0.3% Triton
X-100 for 1 h. An additional washing step with PBS was performed
three times, for 5 min each time, then 5% BSA (cat. no. EPR12774;
Abcam, Inc.) was incubated with the cells for 1 h at room
temperature. The samples were incubated overnight at 4°C with a
primary antibody against ARL2 (cat. no. 188322; 1:100; Cell
Signaling Technology, Inc.). The samples were subsequently washed
with PBS three times for 5 min each time, then they were incubated
with the secondary antibody (goat polyclonal secondary antibody
against rabbit IgG-H&L, Alexa Fluor® 647; cat. no.
ab150083; Abcam). An Olympus IX73 microscope was used to capture
the fluorescence images (Olympus Corporation). For the quantitative
analysis of the intensity of fluorescence expression, the
immunofluorescence pictures were quantitatively analyzed using
Image-Pro Plus 1.8.0 software (Media Cybernetics Inc.) to calculate
the mean gray value of the represented area in each picture.
Western blot analysis
Western blot analysis was performed, as previously
described (20). ARL2 and AXL were
resolved on 12 and 6% SDS-PAGE, respectively, then transferred to
nitrocellulose membranes. Primary antibodies against the following
proteins were used: ARL2 (cat. no. 188322; Cell Signaling
Technology, Inc.), AXL (cat. no. 8661; Cell Signaling Technology,
Inc.), β-actin (cat. no. 66009-1-Ig; ProteinTech Group, Inc.). The
following secondary antibodies were used: HRP-goat anti-mouse
(1:2,000; cat. no. SA00004-1; Proteintech Inc.) and HRP-goat
anti-rabbit (1:2,000; cat. no. SA00004-2; Proteintech Inc.). A
Tanon 5200 automatic chemiluminescence imaging analysis system was
used to detect the protein bands with an enhanced chemiluminescence
reagent (both from Tanon Science and Technology Co., Ltd.). For the
quantitative analysis of the intensity of protein expression, the
pictures were quantitatively analyzed using Image J v.1.8.0
software (National Institutes of Health Inc.) to calculate the
expression levels of protein bands.
Plasmids and viral infection
The vectors ARL2 pcDNA3.1–3×Flag-C (pcDNA3.1) and
ARL2 pLVX-short hairpin (sh) RNA-PURO (shRNA) were purchased from
YouBio Technology Co. Ltd. The ARL2 pcDNA3.1 was transfected into
the HCT8 and HCT116 cells, with the two packing vectors, pMD2G and
psPAX (Shanghai YouBio Technology Co., Ltd.), as previously
described (21). Stable cell lines
overexpressing ARL2 were selected following treatment with 40 µg/ml
G418 (antibiotic) for ~2 weeks. The ARL2 interfering viruses were
generated by co-transfecting 293T cells with the other two packing
vectors (pMD2G and psPAX), and concentrated as previously described
(22). Stable ARL2 knockdown cell
lines were generated by infection of the cells with lentivirus and
clone selection with 2 µg/ml puromycin for ~2 weeks.
Cell proliferation and colony
formation
The Cell Counting Kit (CCK)-8 method was used to
detect cell proliferation according to the manufacturer's protocol
(cat. no. VC5001; VICMED Life Sciences). The in vitro cell
proliferation assay method was performed by diluting
5×103 cells in 100 µl medium. The ARL2 pcDNA3.1 HCT8 and
HCT116 cells, ARL2 shRNA HCT8 and HCT116 cells, and the
corresponding control group cells were transferred into 96-well
plates and incubated at 37°C with 5% CO2 for 6 days. On
days 0, 1, 2, 3 and 4, 10 µl CCK-8 was added to the wells and
incubated for 4 h at 37°C to detect cell proliferation.
Subsequently, the optical density (OD) of each well was measured
using a microplate reader (Tecan Spark 84669JK7573; Tecan Group,
Ltd.) at 450 nm. The growth curve was created according to the OD
of each well. Following ARL2 overexpression or knockdown, the
colony formation assay was performed. The cells were collected,
resuspended as single cells (0.5×105) and seeded into a
6-well plate, which was incubated at 37°C with 5% CO2.
Following 2 weeks of incubation, the cells were washed with PBS
twice and stained with crystal violet solution at room temperature
for ~20 min. The number of colonies (>50 cells) were measured
under a light microscope (IX73; Olympus Corporation).
Wound healing assay
ARL2 pcDNA3.1 HCT8 and HCT116 as well as ARL2 shRNA
HCT8 and HCT116 cells (5×105 per well) were incubated in
6-well plates for 24 h. When the cells were ~100% confluent, the
monolayer was scratched with a 200 µl pipette tip. The suspension
cells were washed with PBS and fresh serum-free medium was added.
The cells were cultured at 37°C with 5% CO2 and the
wound was allowed to heal. The images of the wound were captured at
0 and 24 h. The wound closure rates were measured based on previous
reports (11). The ImageJ software
(v.1.48, National Institutes of Health) was used to evaluate the
wound area, and the wound closure rate was calculated using the
following formula: (original wound area/actual wound area)/area of
the original wound ×100.
Matrigel transwell invasion assay
The assay was performed using an 8-µm filter
membrane (cat. no. 3422; Corning Inc.). A total of 100 µl Matrigel
(cat. no. 356234; Corning Inc.) (diluted with serum-free DMEM by
1:8) was plated in the Transwell chamber and kept in an incubator
at room temperature. Following 4 h of incubation, 200 µl serum-free
medium containing 1×105 cells was added to the upper
chamber of the Transwell insert, while 750 µl DMEM with 10% FBS
(cat. no. 26140079; Gibco; Thermo Fisher Scientific Inc.) was added
to the lower chamber. The cells were incubated at 37°C in 5%
CO2 for 16 h. A cotton swab was used to remove the
apical matrix and the cells. Following fixation with 4%
paraformaldehyde at room temperature for ~10 min, the transmembrane
cells were stained with crystal violet staining solution at room
temperature for ~20 min, and each well was counted at 5 different
high-power microscope fields under a light microscope
(magnification, ×400). The experiment was conducted in
triplicate.
Statistical analysis
All the experiments were performed independently,
three times. The data are presented as the mean ± SD. The number of
repetitions for each experiment is illustrated in the corresponding
figure. Statistical differences between two groups were determined
using a two tailed t-test, and the differences among >2 groups
were analyzed one-way ANOVA followed by the Dunnett's post-test.
SPSS v20.0 (IBM, Corp.) and GraphPad Prism v6.0 (GraphPad Software,
Inc.) were used for statistical analysis. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of ARL2 is increased in
colon cancer tissues
A total of 22 human clinical specimens were
collected, including 19 CRC tissues (6 well-differentiated, 6
moderately-differentiated and 7 poorly differentiation cases) and 3
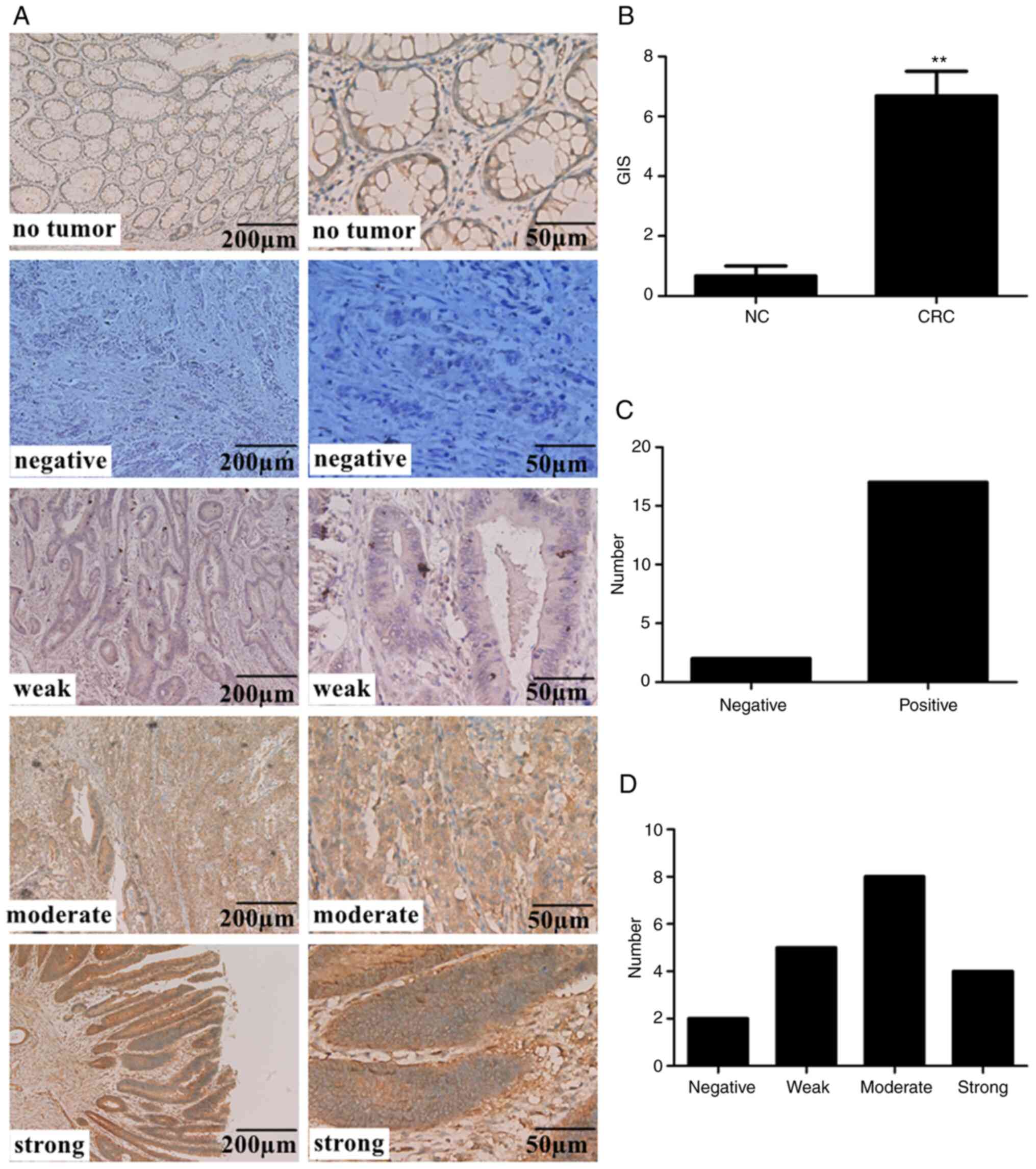
normal healthy colon tissue. In 17 of the 19 specimens (89.47%)
(Fig. 1C), ARL2 was expressed in ≥1%
cancer cells, with different staining intensities: 5 with weak
staining, 8 with moderate staining, and 4 with strong staining
(Fig. 1A and D). There were
significant differences in the expression level of ARL2 between CRC
and normal healthy colon tissue (Fig.
1B).
Taken together, these results demonstrated that the
protein expression levels of ARL2 were significantly increased in
CRC.
Overexpressed ARL2 decreases
proliferation and colony-formation abilities of CRC cells
To investigate the physiological function of ARL2 in
colon cancer, ARL2 overexpressing (pcDNA3.1) and knockdown (shRNA)
cell lines were established using plasmid transfection and viral
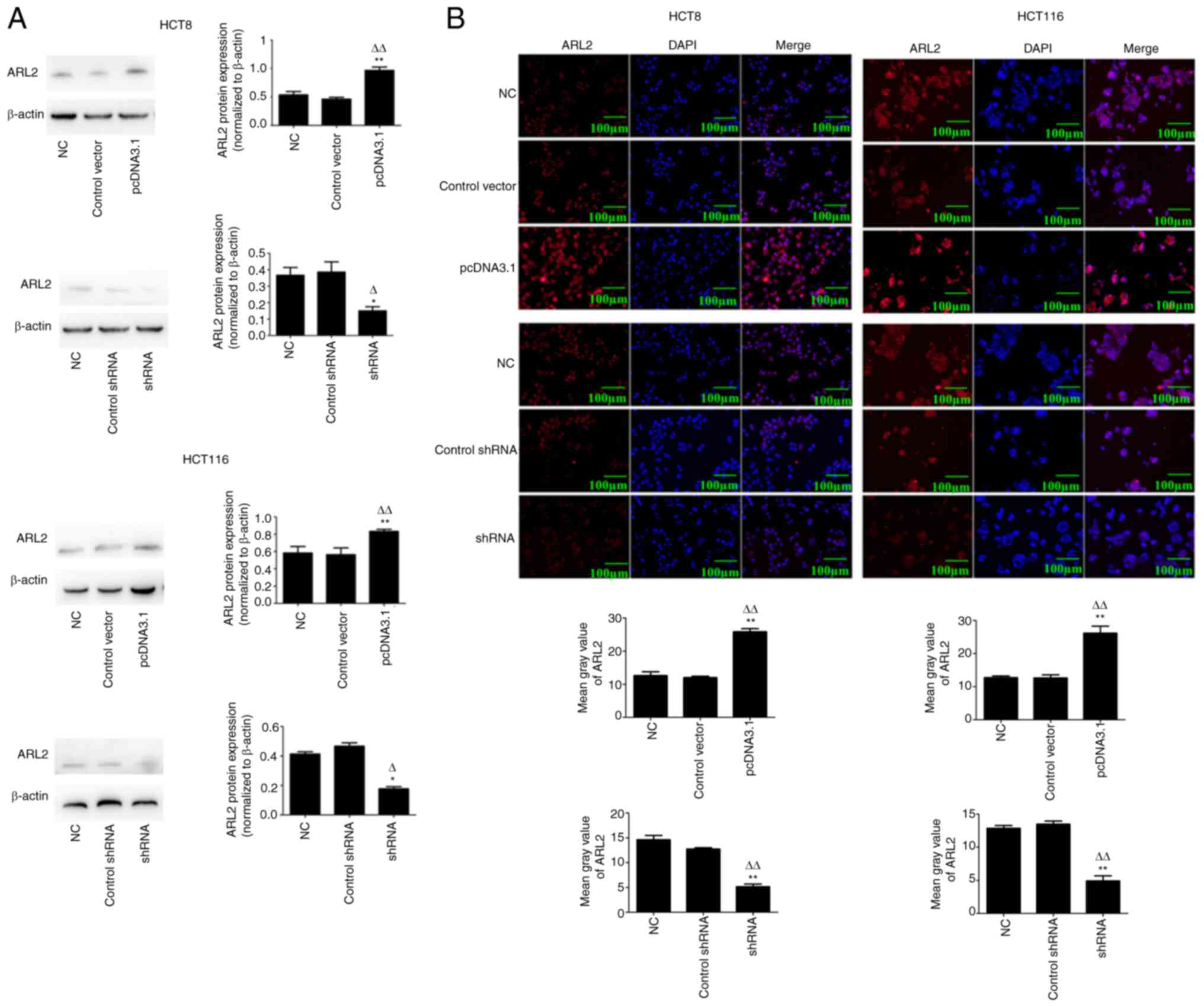
infection in 2 colon cancer cell lines (HCT8 and HCT116). Western
blot analysis indicated that the protein expression levels of ARL2
were significantly increased or decreased, respectively following
transfection with overexpression or knockdown plasmids in both the
cell lines (Fig. 2A; P<0.05).
Furthermore, immunofluorescence confirmed that ARL2 protein
expression level was significantly increased or reduced following
transfection with overexpression or knockdown plasmids,
respectively, in both the cell lines (Fig. 2B).
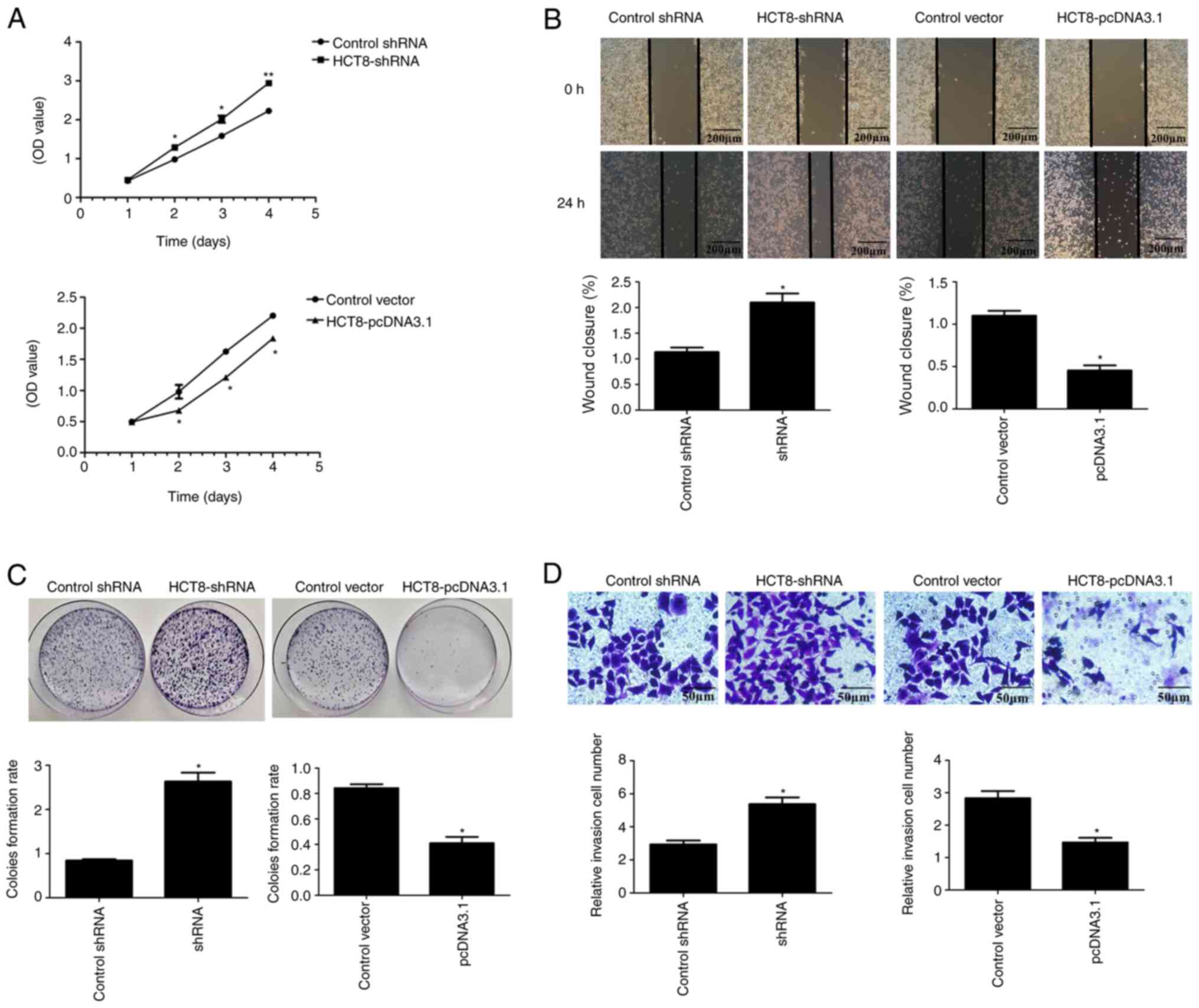
The data indicated that overexpression of ARL2
(pcDNA3.1) significantly inhibited the proliferation of the HCT8
and HCT116 cells, while ARL2 knockdown (shRNA) significantly
increased the proliferation of the HCT8 (Fig. 3A) and HCT116 (Fig. 4A) cells (P<0.05).
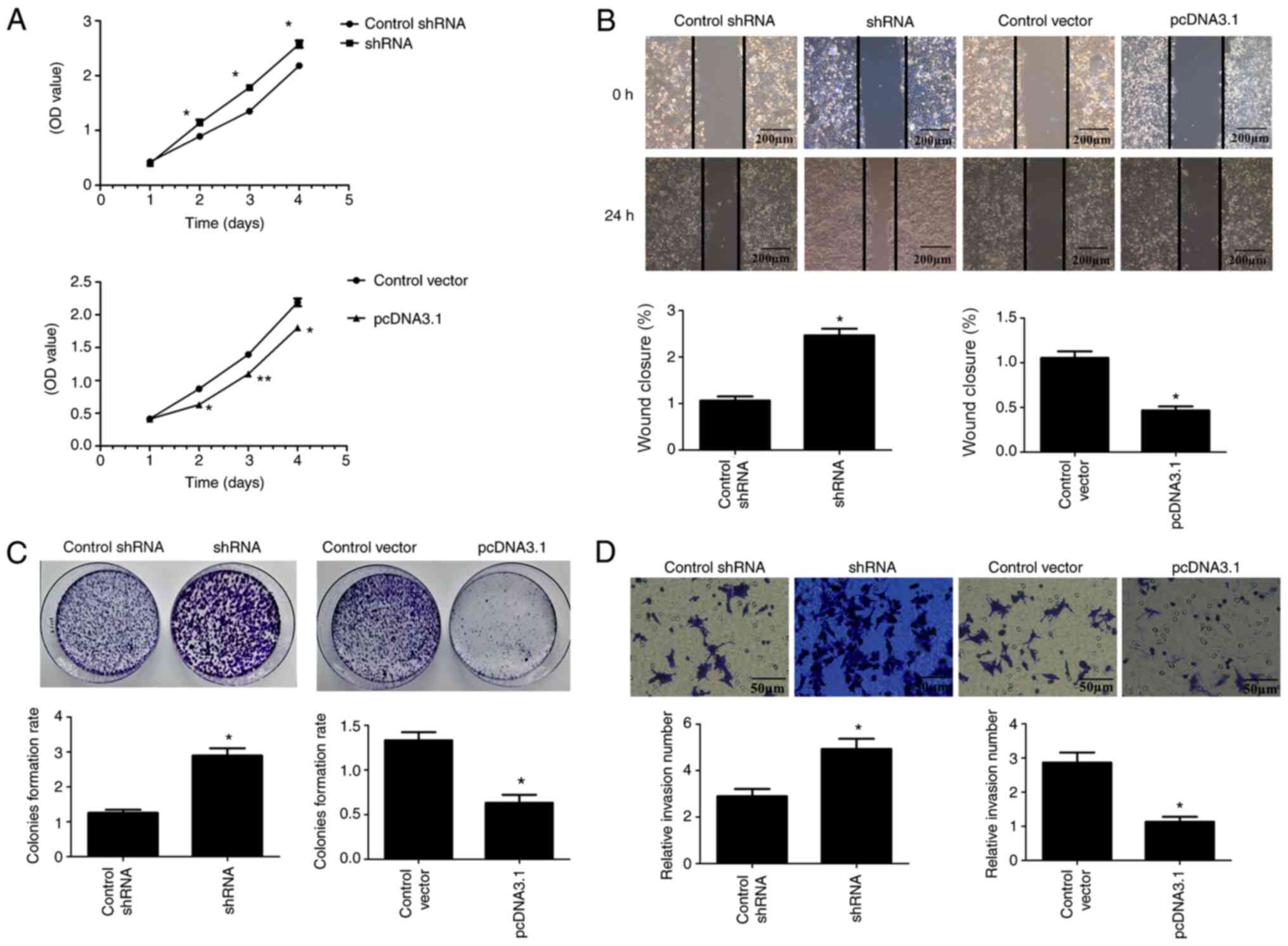 | Figure 4.ARL2 expression and HCT116
proliferation, migration and invasion. (A) Cell Counting Kit-8
assays were used to examine HCT116 cell proliferation in control,
ARL2 overexpressing and knockdown HCT8 cells. (B) Wound healing
assays were used to examine cell migration in control, ARL2
overexpressing and knockdown HCT116 cells. Scale bar, 200 µm. (C)
Colony formation assays were used to examine HCT116 cell
proliferation in control, ARL2 overexpressing and knockdown HCT116
cells. (D) Matrigel assay was used to examine cell invasion in
control, ARL2 overexpressing and knockdown HCT116 cells. Scale bar,
50 µm. The data are presented as the mean ± SD from three
independent experiments performed in triplicate. *P<0.05 vs
control vector or control shRNA, **P<0.001 vs. control vector or
control shRNA. ARL2, ADP ribosylation factor-like GTPase 2; sh,
short inhibiting; pcDNA3.1, overexpressing ARL2 plasmid; OD,
optical density. |
In addition, the colony formation assay was
performed to detect the colony-formation ability of the CRC cells.
As expected, colony formation of the HCT8 and HCT116 cells, with
ARL2 overexpression plasmid, was significantly decreased compared
with that in the control group. In contrast, the HCT8 and HCT116
cells with ARL2 shRNA sequences demonstrated significantly
increased colony formation ability compared with that in the
control group (Figs. 3C and 4C; P<0.05). These data indicated that
ARL2 reduced the proliferation of the HCT8 and HCT116 cells.
Overexpressed ARL2 inhibits the
migratory and invasive abilities of the CRC cells
The microtubule network has a critical role in
regulating cell migration and invasion (23); therefore, the wound healing and
Matrigel assays were used to examine whether overexpression or ARL2
knockdown inhibited or promoted the migration and invasion of the
CRC cells. As shown in Figs. 3B and
4B, the migration ability of the
HCT8 and HCT116 cells, with ARL2 overexpressing plasmid, was
significantly decreased compared with that in the control group,
while the migration ability of the HCT8 and HCT116 cells, with ARL2
knockdown plasmid was significantly increased compared with that in
the control group.
The Matrigel assay revealed similar results
demonstrating that ARL2 overexpressing cells (pcDNA3.1) had reduced
invasive abilities, whereas ARL2 knockdown (shRNA) cells exhibited
increased invasive abilities (Figs.
3D and 4D).
These results indicated that ARL2 reduced the
migratory and invasive abilities of the HCT8 and HCT116 cells.
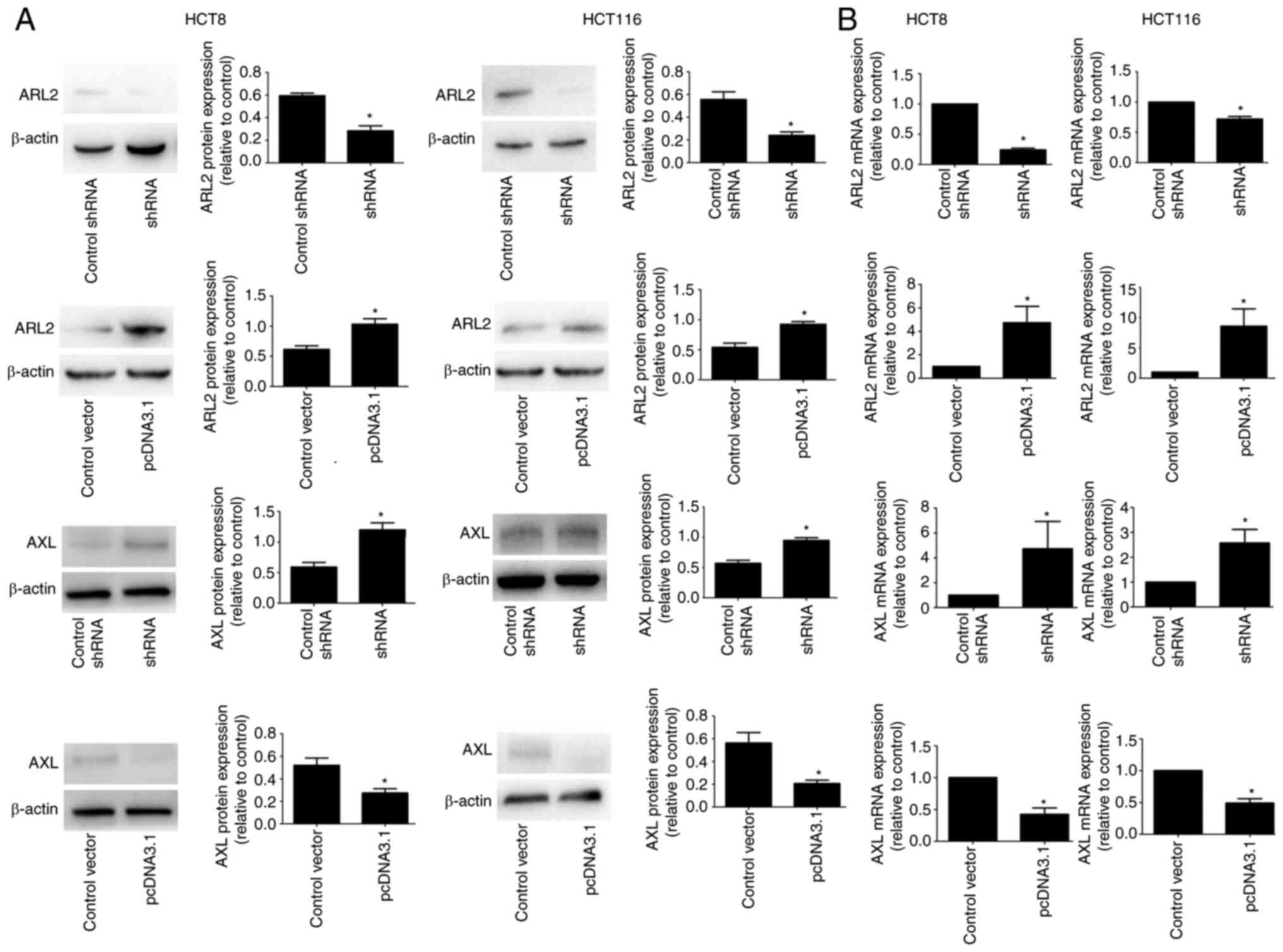
ARL2 reduces AXL expression in the CRC
cells
To investigate the functions of ARL2 in CRC, its
downstream target, AXL (11) was
studied in additional experiments. Previous studies confirmed that
AXL played a key role in the functional regulation of CRC cells
(16,17). On this basis, the effects of ARL2
expression on AXL were investigated in the HCT8 and HCT116 cells.
Initially, western blot analysis was used to detect the expression
levels of AXL in the HCT8 and HCT116 cells, following ARL2
overexpression and knockdown. The results indicated that ARL2
overexpression reduced AXL protein expression, whereas ARL2
knockdown increased AXL protein expression levels (Fig. 5A). In addition, RT-qPCR assays
demonstrated that ARL2 overexpression decreased AXL mRNA expression
level, whereas ARL2 knockdown increased AXL mRNA expression level
(Fig. 5B).
With increased ARL2 protein and mRNA expression
levels, there was a decrease in AXL protein and mRNA expression
levels, while the opposite results were found when ARL2 expression
was knocked down.
Discussion
Microtubule network dynamics is important to the
regulation of physiological processes, such as cell mitosis and
migration (24,25). For example, ARL2 has been associated
with the occurrence of specific malignant tumors, such as glioma,
breast cancer and cervical cancer (11,13,15).
However, the protein expression pattern and pathophysiological role
of ARL2 in different types of cancer is controversial, as the
expression of ARL2 is decreased in glioma and increased in
hepatocellular carcinoma (11,14), and
the role of ARL2 in CRC is unclear. In the present study, increased
protein expression levels of ARL2 were found in CRC tissues, which
is consistent with a previous study (26). ARL2 overexpression suppressed
migration, invasion and proliferation of two CRC cell lines (HCT8
and HCT116). In contrast, ARL2 knockdown enhanced migration,
invasion and proliferation. There was decreased AXL expression in
the CRC cells when ARL2 was overexpressed. The results of the
present study suggested that ARL2 may regulate the migration and
invasion of the CRC cells by negatively regulating AXL expression.
Therefore, ARL2 may be considered a protective factor against CRC
development.
An increasing number of studies have demonstrated
that ARL2 was associated with the development and progression of
various types of cancer. ARL2 has been reported to inhibit the
proliferation, migration and tumorigenicity of glioma cells and may
be considered a new prognostic and therapeutic target for glioma
(11). It was also reported that
ARL2 played an oncogenic role in cervical cancer (15). The current study demonstrated that
ARL2 protein expression level was increased in CRC tissues, while
its overexpression decreased cell migration, invasion and
proliferation, suggesting that it may be a protective factor
against CRC progression.
As a member of the TAM group of receptors, AXL acts
as an inhibitor of cytokine receptors mediates cell activation and
promotes apoptotic cell removal (27,28). AXL
gene silencing and drug inhibition suppressed proliferation,
migration and survival in CRC cells (16,17). In
the present study, the results indicated that ARL2 was inversely
associated with AXL expression in CRC cells. Furthermore, previous
studies have indicated that AXL functions as a tumor promoter in
cancer. For example, AXL induced the activation of downstream AKT,
and inhibited the proliferation and migration of pancreatic cancer
cells in vitro (29). In
addition, GAS6 promoted gastric cancer invasiveness by activating
AXL (30). Previous studies have
demonstrated the role of AXL in regulating cell growth, migration
and tumorigenesis of CRC cells (29,30). AXL
acted as a tumor promoter (31,32),
which may partially be suppressed by overexpressed ARL2 expression
in CRC cells. The present study provided the first preliminary
evidence of AXL was negatively associated with ARL2. To elucidate
the mechanism by which ARL2 and AXL expression in CRC cells,
western blot and RT-qPCR analyses were used to detect the protein
and mRNA expression levels of ARL2 and AXL in HCT8 and HCT116
cells, following overexpression or knockdown of ARL2. At both the
protein and mRNA expression, increased ARL2 led to decreased AXL
and decreased ARL2 led to increased AXL. The results revealed that
the increase in the levels of ARL2 was accompanied by decrease in
AXL expression level and may have reduced CRC cell progression.
Therefore, ARL2 may be negatively correlated with the expression
level of AXL at the gene level.
In conclusion, the present study described the
increased expression levels of ARL2 in clinical CRC samples. ARL2
could play a crucial role in CRC. In addition, significant evidence
was provided demonstrating that increased ARL2 expression in CRC
cells inhibited proliferation, colony formation, migration and
invasion. Furthermore, the results confirmed that ARL2 was
negatively associated with AXL in CRC cells and maybe a protective
factor in CRC. Taken together, the results suggested that ARL2
could play an important inhibitory role in the proliferation,
migration and tumorigenicity of CRC cells and ARL2 was negatively
associated with AXL expression. Therefore, ARL2 may be a new
predictive and therapeutic target for CRC.
Acknowledgements
The authors would like to thank Dr Yingying Cui and
Dr Kai Cao (Department of Pathology, The Affiliated Hospital of
Xuzhou Medical University) for discussions pertaining to the
present study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed in the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
SF designed the study. XP, YW, BM and WC performed
the experiments and analyzed the data. WC, YW and XP wrote the
manuscript. SF and BM revised the manuscript. All authors read and
approved the final version of the manuscript and agree to be
accountable all aspects of the study to ensure the accuracy or
integrity of all parts of the work.
Ethics approval and consent to
participate
This study was approved by the Institutional Ethics
Committee of the Affiliated Hospital of Xuzhou Medical University
(approval no. XYFY2019-KL157-01) and adhered to the principles of
the Declaration of Helsinki. Each patient provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mattiuzzi C, Sanchis-Gomar F and Lippi G:
Concise update on colorectal cancer epidemiology. Ann Transl Med.
7:609. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gu X, Zheng R, Xia C, Zeng H, Zhang S, Zou
X, Yang Z, Li H and Chen W: Interactions between life expectancy
and the incidence and mortality rates of cancer in China: A
population-based cluster analysis. Cancer Commun (Lond). 38:442018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zeng H, Chen W, Zheng R, Zhang S, Ji JS,
Zou X, Xia C, Sun K, Yang Z, Li H, et al: Changing cancer survival
in China during 2003-15: A pooled analysis of 17 population-based
cancer registries. Lancet Glob Health. 6:e555–e567. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dai W, Li Y, Mo S, Feng Y, Zhang L, Xu Y,
Li Q and Cai G: A robust gene signature for the prediction of early
relapse in stage I–III colon cancer. Mol Oncol. 12:463–475. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen K, Koe CT, Xing ZB, Tian X, Rossi F,
Wang C, Tang Q, Zong W, Hong WJ, Taneja R, et al: Arl2- and
Msps-dependent microtubule growth governs asymmetric division. J
Cell Biol. 212:661–676. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ozdemir ES, Jang H, Gursoy A, Keskin O and
Nussinov R: Arl2-mediated allosteric release of farnesylated KRas4B
from shuttling factor PDEδ. J Phys Chem B. 122:7503–7513. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stettin D, Waldmann A, Wolters M, Trunz B,
Schauder P and Hahn A: Infection with Helicobacter pylori-outcome
of a cross-sectional investigation. Dtsch Med Wochenschr.
132:2677–2682. 2007.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang F and Cheong JK: The renewed battle
against RAS-mutant cancers. Cell Mol Life Sci. 73:1845–1858. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cox AD, Fesik SW, Kimmelman AC, Luo J and
Der CJ: Drugging the undruggable RAS: Mission possible? Nat Rev
Drug Discov. 13:828–851. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stephen AG, Esposito D, Bagni RK and
McCormick F: Dragging ras back in the ring. Cancer Cell.
25:272–281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Guan G, Cheng W, Jiang Y, Shan F,
Wu A, Cheng P and Guo Z: ARL2 overexpression inhibits glioma
proliferation and tumorigenicity via down-regulating AXL. BMC
Cancer. 18:5992018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li HJ, Sun XM, Li ZK, Yin QW, Pang H, Pan
JJ, Li X and Chen W: LncRNA UCA1 promotes mitochondrial function of
bladder cancer via the MiR-195/ARL2 signaling pathway. Cell Physiol
Biochem. 43:2548–2561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Béghin A, Matera E, Brunet-Manquat S and
Dumontet C: Expression of Arl2 is associated with p53 localization
and chemosensitivity in a breast cancer cell line. Cell Cycle.
7:3074–3082. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hass HG, Vogel U, Scheurlen M and Jobst J:
Gene-expression analysis identifies specific patterns of
dysregulated molecular pathways and genetic subgroups of human
hepatocellular carcinoma. Anticancer Res. 36:5087–5096. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng R, Men J, Ma R, Wang Q, Wang Y, Sun Y
and Ren J: miR-214 down-regulates ARL2 and suppresses growth and
invasion of cervical cancer cells. Biochem Biophys Res Commun.
484:623–630. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Martinelli E, Martini G, Cardone C,
Troiani T, Liguori G, Vitagliano D, Napolitano S, Morgillo F,
Rinaldi B, Melillo RM, et al: AXL is an oncotarget in human
colorectal cancer. Oncotarget. 6:23281–23296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uribe DJ, Mandell EK, Watson A, Martinez
JD, Leighton JA, Ghosh S and Rothlin CV: The receptor tyrosine
kinase AXL promotes migration and invasion in colorectal cancer.
PLoS One. 12:e01799792017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bläker H: Grading of tumors in the tubular
digestive tract: Esophagus, stomach, colon and rectum. Pathologe.
37:293–298. 2016.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hou P, Li L, Chen F, Chen Y, Liu H, Li J,
Bai J and Zheng J: PTBP3-mediated regulation of ZEB1 mRNA stability
promotes epithelial-mesenchymal transition in breast cancer. Cancer
Res. 78:387–398. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qiu C, Bu X and Jiang Z: Protocadherin-10
acts as a tumor suppressor gene, and is frequently downregulated by
promoter methylation in pancreatic cancer cells. Oncol Rep.
36:383–389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tohidi F, Sadat SM, Bolhassani A and
Yaghobi R: Construction and production of HIV–VLP harboring MPER-V3
for potential vaccine study. Curr HIV Res. 15:434–439.
2017.PubMed/NCBI
|
|
23
|
Seetharaman S and Etienne-Manneville S:
Microtubules at focal adhesions-a double-edged sword. J Cell Sci.
132:jcs2328432019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
van Vuuren RJ, Botes M, Jurgens T, Joubert
AM and van den Bout I: Novel sulphamoylated 2-methoxy estradiol
derivatives inhibit breast cancer migration by disrupting
microtubule turnover and organization. Cancer Cell Int. 19:12019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Galmarini CM, Martin M, Bouchet BP,
Guillen-Navarro MJ, Martínez-Diez M, Martinez-Leal JF, Akhmanova A
and Aviles P: Plocabulin, a novel tubulin-binding agent, inhibits
angiogenesis by modulation of microtubule dynamics in endothelial
cells. BMC Cancer. 18:1642018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Long LM, He BF, Huang GQ, Guo YH, Liu YS
and Huo JR: microRNA-214 functions as a tumor suppressor in human
colon cancer via the suppression of ADP-ribosylation factor-like
protein 2. Oncol Lett. 9:645–650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou C, Cunningham L, Marcus AI, Li Y and
Kahn RA: Arl2 and Arl3 regulate different microtubule-dependent
processes. Mol Biol Cell. 17:2476–2487. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Newman LE, Schiavon CR, Zhou C and Kahn
RA: The abundance of the ARL2 GTPase and its GAP, ELMOD2, at
mitochondria are modulated by the fusogenic activity of mitofusins
and stressors. PLoS One. 12:e01751642017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song X, Akasaka H and Wang H,
Abbasgholizadeh R, Shin JH, Zang F, Chen J, Logsdon CD, Maitra A,
Bean AJ and Wang H: Hematopoietic progenitor kinase 1
down-regulates the oncogenic receptor tyrosine kinase AXL in
pancreatic cancer. J Biol Chem. 295:2348–2358. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bae CA, Ham IH, Oh HJ, Lee D, Woo J, Son
SY, Yoon JH, Lorens JB, Brekken RA, Kim TM, et al: Inhibiting the
GAS6/AXL axis suppresses tumor progression by blocking the
interaction between cancer-associated fibroblasts and cancer cells
in gastric carcinoma. Gastric Cancer. 23:824–836. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vajkoczy P, Knyazev P, Kunkel A, Capelle
HH, Behrndt S, von Tengg-Kobligk H, Kiessling F, Eichelsbacher U,
Essig M, Read TA, et al: Dominant-negative inhibition of the Axl
receptor tyrosine kinase suppresses brain tumor cell growth and
invasion and prolongs survival. Proc Natl Acad Sci USA.
103:5799–5804. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng P, Phillips E, Kim SH, Taylor D,
Hielscher T, Puccio L, Hjelmeland AB, Lichter P, Nakano I and
Goidts V: Kinome-wide shRNA screen identifies the receptor tyrosine
kinase AXL as a key regulator for mesenchymal glioblastoma
stem-like cells. Stem Cell Reports. 4:899–913. 2015. View Article : Google Scholar : PubMed/NCBI
|