Introduction
In the year 1915, Dr William Damshek first coined
the term ‘myeloproliferative disorders’ (MPD) to classify
phenotypically varied neoplasms that originate from a myeloid
progenitor cell, namely chronic myeloid leukemia (CML),
polycythaemia vera (PV), essential thrombocythemia (ET) and primary
myelofibrosis (PMF) (1). Fialkow
et al reported that MPD are clonal stem cell disorders
affecting both lymphoid and myeloid lineage (2). However, the identification of the
Philadelphia chromosome in CML delineated the classification of MPD
(3). The incidence rates for PV
(0.4–2.8/100,000/year) in European Union and US registries are
higher compared with ET (0.38–1.7/100,000/year) and PMF
(0.1–1/100,000/year) (4,5). The median survival of PV and ET is
longer (8–10 years) compared with PMF (2–5 years) (4,5).
With advancement of molecular techniques, in 2005
four different research groups published their findings regarding a
recurrent somatic mutation in the MPN other than CML (6–9). The
JAK2 gene was affected at codon 617 of exon 14, resulting in
a valine to phenylalanine substitution (9). This was followed by another JAK2
mutation identified in exon 12 that lead the World Health
Organization to include it as a diagnostic criteria for breakpoint
cluster region Abelson murine leukemia viral oncogene homolog 1
(BCR-ABL) negative MPNs (10,11).
JAK2 is a tyrosine kinase belonging to the Janus family of
proteins, which also includes JAK1, JAK3 and TYK2
(12,13). When interacting with cytokines, these
kinases are activated via phosphorylation and further trigger a
signal transduction cascade, which involves the STAT transcription
factors (STAT1-8) that translocate into the nucleus and
stimulate transcription of downstream targets (12,14).
Studies have demonstrated that the JAK-STAT signaling
pathway is utilized by a variety of cytokines for proliferation,
survival and differentiation of hematopoietic cells (15–17). The
role of JAK-STAT signaling in hematopoiesis has been
demonstrated in mice with a germline deletion of JAK2,
leading to embryonic lethal effects due to disrupted erythropoiesis
(18). Mutations in the pseudokinase
domain of JAK2 results in an uninhibited phosphorylation of
the tyrosine kinase and constitutive activation of the
JAK-STAT pathway, which is central to the pathogenesis of
MPNs that possess this gain-of-function mutation (19). JAK2 mutations are reported in
~95% of PVs, 50–60% of ETs and 50–60% of PMFs, which led to the
investigation of differing phenotypes resulting from the same
genetic aberration (7). MPNs without
the aforementioned genetic abnormalities are relegated to the
miscellaneous category and in the past few years, considerable work
has been done to characterize JAK2V617F and BCR-ABL
negative MPN (15,20,21). In
this context various mutations in genes involved in cell signaling
(including SH2B adaptor protein 3, E3 ubiquitin-protein ligase
CBL, NRAS, NF1 and FLT3), DNA methylation
[TET2, DNA methyltransferase (DNMT)3A], metabolism
[(NADP) cytoplasmic isocitrate dehydrogenase 1 and 2; IDH 1
and 2], histone modification (EZH2 and polycomb group
protein ASXL1) and RNA splicing (serine/arginine-rich
splicing factor 2 (SRSF2), splicing factor 3B subunit 1
(SF3B1) and splicing factor (U2AF1) have been
identified (15). These mutations
provide an insight into the disease progression but have not been
able to explain the pathogenesis of this MPN subtype. Therefore,
the present study aimed to investigate the microarray-based
expression profiles of bone marrow derived CD34(+) positive cells
from JAK2V617F positive vs. JAK2V617F negative
MPNs.
Materials and methods
Sample collection and sorting
Bone marrow samples were collected from seven
patients with MPN as per guidelines and approval by Institutional
Ethics Committee at St. John's Medical College and Hospital,
Bengaluru (approval no. 103/2016). The median age of patients was
39 years (32–47 years). The samples were collected between June
2016 to July 2018. Patients >18 years old and classified
according to the World Health Organization criteria for MPNs
(20) were included in the study.
CD34 cells were isolated from bone marrow samples using CD34
Microbead kit from Miltenyi Biotec, Inc., as per the manufacturer's
instructions.
Microarray analysis
RNA was isolated from CD34 cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
Whole transcriptome analysis of the seven samples was performed
using the Whole Transcript (WT) PLUS Reagent kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The assay workflow in
the order included first-stand cDNA synthesis, second-strand cDNA
synthesis, cRNA amplification, cRNA purification and
quantification, 2nd cycle ss-cDNA synthesis, template RNA removal,
ss-cDNA purification and quantification, fragmentation, terminal
labelling and hybridization to WT. All the steps were performed as
per manufacturer's instructions. In total, 100 ng RNA was used as
the input.
The quality check of CEL file was performed using
Transcriptome Analysis Console software version 4.0.1 (Thermo
Fisher Scientific, Inc.) and the files were normalized using the
Oligo-R-based Bioconductor package (22). The processing was done using RMA
method for R (23).
The normalized RMA files were checked for any
discrepancies using the top ten housekeeping gene and their
expression values across the sample. Principal Component Analysis
was performed to cluster the samples. The samples that were
clustered together based on the housekeeping genes were used for
further analysis.
The normalized text file was then subjected to the
AltAnalyze (http://www.altanalyze.org/) automated tool for
analysis. Differentially expressed genes in the JAK2V617F
negative vs. JAK2V617F positive samples were identified
using ±1.5 as the fold-change cut-off with a statistical
significance of <0.05. Prune ontology was calculated using the z
score (initial filtering 1.96 cut-off). The database used for
analysis was Ensembl (https://asia.ensembl.org/). The pathway network was
generated automatically using the AltAnalyze tool.
Bioinformatic analysis
Differentially expressed genes were calculated using
±1.5 fold-change and P≤0.05. Database for Annotation, Visualization
and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) tool version 6.8 was used
for Gene Ontology (GO), and functional and pathway enrichment
analyses (24,25). The protein-protein interaction (PPI)
network was constructed using STRING database version 11
(https://string-db.org/) using minimum confidence
0.4 (26). The genes which were
enriched for DNA modification and stemness properties based on GO
analysis were fed into StemChecker tool (http://stemchecker.sysbiolab.eu/) for analyzing
stemness markers and transcription factors for enriched genes
(27). Hierarchical clustering and
heatmaps were created using the ClustVis online tool (28).
Reverse transcription-quantitative
(RT-q)PCR
RNA was isolated from cells using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.). In total, 1 µg RNA
was converted into cDNA using the MMLV enzyme (Invitrogen; Thermo
Fisher Scientific, Inc.) as per the manufacturer's instructions.
Gene expression was evaluated by using the TB Green Premix Ex Taq
(Takara Bio, Inc.) as per manufacturer's instructions and qPCR
setup was run on a 7500 Fast Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). GUSB was used
as housekeeping gene. Primer sequences for selected genes are
presented in Table I. The relative
quantification was calculated using the 2−ΔΔCq method
(29). MPN mutation panel, including
CALR type I and II and myeloproliferative leukemia virus (MPL)
mutations (W515L, W515K, W515A and S505N), were analyzed using the
TRUPCR MPN panel kit (3B BlackBio Biotech India Ltd.) as per
manufacturer's instructions.
 | Table I.Primer sequences for selected
genes. |
Table I.
Primer sequences for selected
genes.
| Gene | Forward primer,
5′-3′ | Reverse primer,
5′-3′ |
|---|
| GUSB |
AGCCCATTATTCAGAGCGAG |
CCAAATGAGCTCTCCAACC |
| SUV39H1 |
TATGACTGCCCAAATCGTG |
TGATCTCTCCCACGTACTCC |
| MYB |
GCAGTGACGAGGATGATGAG |
CTGTTCCATTCTGTTCCACC |
Flow cytometry
The bone marrow cells post red blood cell lysis (150
mM ammonium chloride, 10 mM potassium bicarbonate, 0.1 mM
ethylenediaminetetraacetic acid) were labelled with CD34 FITC
(clone 581 cat. no. 555821; BD Biosciences) at room temperature for
30 min in dark and analyzed using a FACS Calibur instrument (BD
Biosciences). The results were analyzed using FCS4 Express software
version 6.06.0025 (De Novo Software).
Statistical analysis
The statistical significance for CD34(+) cells was
calculated using an unpaired student's t-test using SPSS v16 (IBM).
P<0.05 was considered to indicate a statistically significant
difference. All Q-PCR experiments were performed in triplicates
(n=3). The differential expression of genes was calculated using
one-way ANOVA and Benjamini and Hochberg false discovery rate
(FDR).
Results
Gene expression patterns in CD34(+)
cells isolated from JAK2V617F negative vs. positive neoplasms
The role of JAK2V617F mutation is well
established in driving a subset of the myeloproliferative neoplasms
(6–9); however, the etiology of the
JAK2V617F negative MPNs is unknown. To understand this,
JAK2V617F mutant positive and negative MPNs were compared
using microarray-based transcriptional analysis. The hematopoietic
cancer stem cells are determinant of the cellular hierarchy of
tumor progression in hematological malignancies; however, the
underlying mechanism is poorly understood in most cases, except CML
wherein the role of translocation and resultant chimeric protein is
a driving force in hematopoietic stem cells (HSCs) (30). To understand the changes that may be
driving the JAK2V617F negative MPNs, bone marrow derived
CD34(+) cells were targeted in the present study. CD34(+) fractions
were separated from patient samples and subjected to microarray
analysis. Tables II and III present the baseline characteristics
and mutation profile of the patients involved in the present study.
Patients who were JAK2V617F negative were also negative for
CALR type 1 and 2 mutations and MPL mutations (W515L,
W515K, W515A and S505N) (Table
III).
 | Table II.Baseline characteristics of the
patients. |
Table II.
Baseline characteristics of the
patients.
| ID | Age, years | Sex | Hb, g/dl | Total WBC counts,
×109/l | Platelet count,
×109/l | Janus kinase 2
V617F status |
|---|
| 1 | 39 | M | 19.1 | 8.43 | 297 | Absent |
| 2 | 41 | M | 16.9 | 11.81 | 286 | Present |
| 3 | 39 | M | 17.5 | 7.71 | 337 | Absent |
| 4 | 35 | M | 17.9 | 9.77 | 209 | Absent |
| 5 | 28 | M | 23.2 | 4.59 | 108 | Absent |
| 6 | 47 | M | 17.5 | 6.63 | 35 | Absent |
| 7 | 32 | M | 20.4 | 9.89 | 13 | Present |
 | Table III.Mutation profile of patients negative
for JAK2V617F. |
Table III.
Mutation profile of patients negative
for JAK2V617F.
| ID | JAK2V617F
status | MPL W515L
status | MPL W515K
status | MPL W515A
status | MPL S505N
status | CALR type 1,
p.L367fs*46 | CALR type 2,
K385fs*47 |
|---|
| 1 | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| 3 | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| 4 | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| 5 | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
| 6 | Absent | Absent | Absent | Absent | Absent | Absent | Absent |
The microarray analyses of CD34(+) cells isolated
from JAK2V617F negative and positive samples resulted in
21,448 gene signatures. The analysis resulted in 472 upregulated
and 202 downregulated genes. The maximally downregulated genes,
with >3-fold difference were 3-oxo-5-α-steroid 4-dehydrogenase 1
(SRD5A1), matrix metalloproteinase (MMP7), Wilms
tumor protein (WT1) and Ankyrin repeat and death
domain-containing protein 1B (ANKDD1B), while the maximally
upregulated genes with >3-fold difference were X-ray repair
cross-complementing protein (XRCC6), TUBB,
histone-lysine N-methyltransferase SUV39H1 and pantetheinase
VNN1 as shown in Table IV.
The expression of SUV39H1 and MYB was validated in
five samples used for microarray analysis as shown in Fig. S1 where in the expression of both the
genes was higher in JAK2V617F negative samples compared with
control samples.
 | Table IV.Fold change of selected genes in
JAK2V617F negative vs. positive samples. |
Table IV.
Fold change of selected genes in
JAK2V617F negative vs. positive samples.
| Gene name | Fold-change |
|---|
| SRD5A1 | −7.71 |
| ANKDD1B | −5.13 |
| MMP7 | −3.31 |
| WT1 | −3.54 |
| VNN1 | 22.39 |
| SUV39H1 | 5.03 |
| MYB | 4.64 |
| XRCC | 3.38 |
| TUBB | 4.43 |
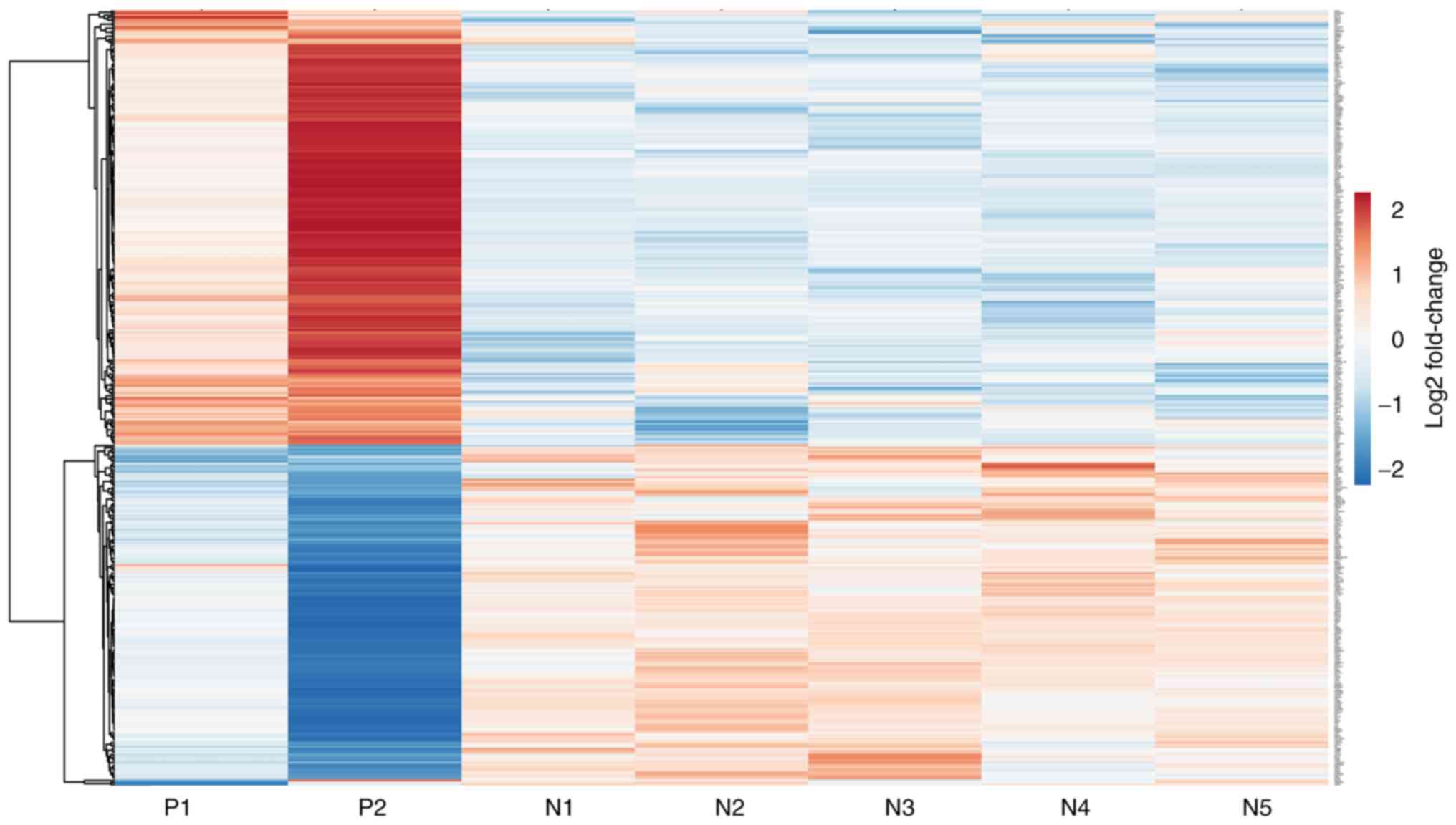
Hierarchical clustering of significantly upregulated
and downregulated genes was performed to understand the correlation
between samples (Fig. 1).
Hierarchical clustering separated seven samples into two different
groups consistent with the experimental design. Red bars indicated
a high level of expression, blue bars indicated a low level of
expression and white bars indicated no significant difference
(Fig. 1). There was a distinctive
difference in the expression of genes in JAK2V617F positive
and negative samples (Fig. 1). The
expression pattern of genes was homogeneous within the mutation
negative samples; however, there was heterogeneity within mutation
positive samples.
Gene Ontology and STRING analysis
reveals distinct cell cycle and epigenetic signatures in JAK2V617F
negative vs. positive samples
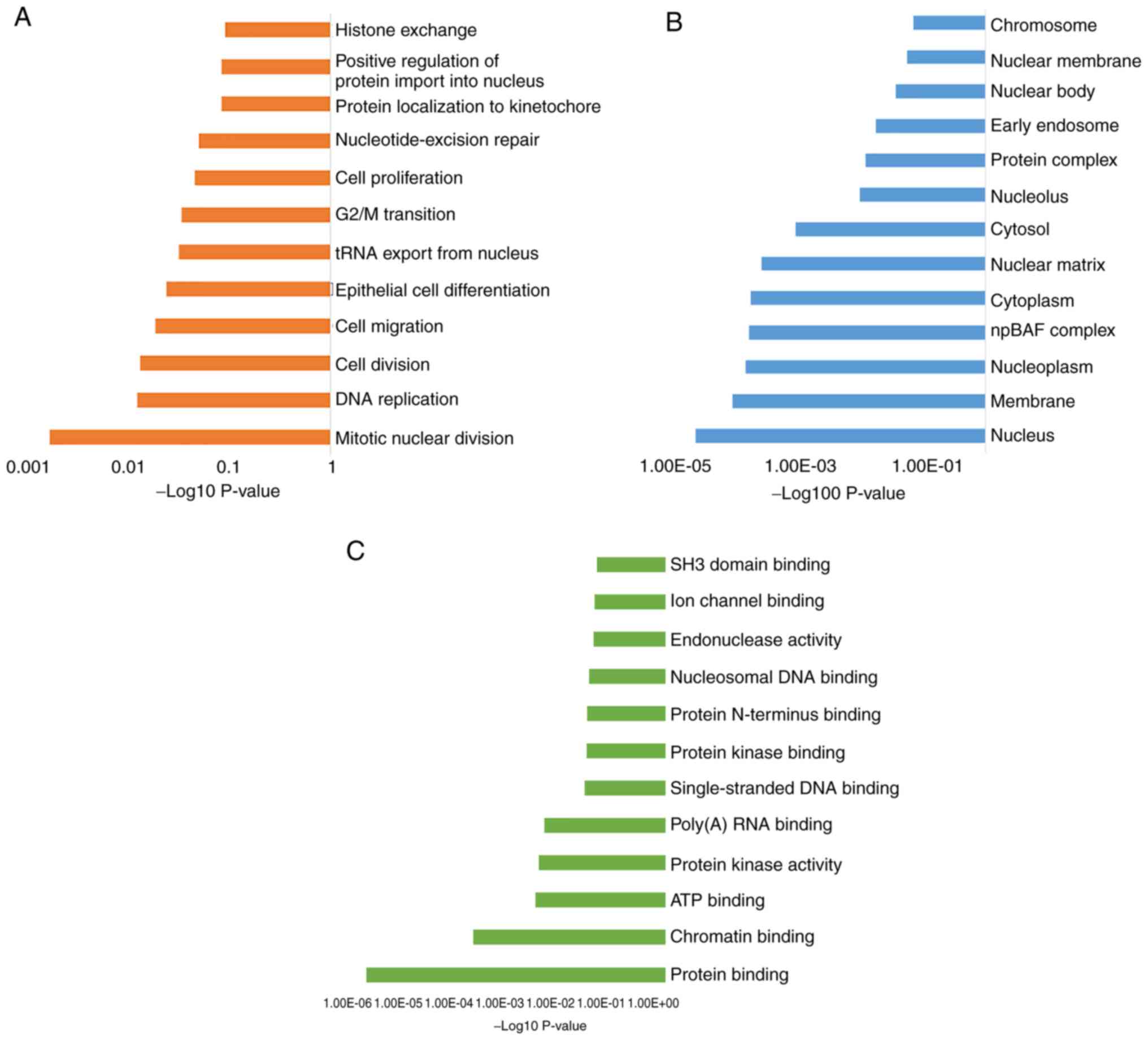
To understand the important biological processes
that determine the phenotypes in these two subsets of MPNs, GO was
performed. The enriched genes and common pathways were analyzed
using DAVID (Fig. 2). The
distribution of the transcripts in the GO terms revealed a distinct
set of biological processes (BP) (Fig.
2A), including ‘mitotic nuclear division’, ‘DNA replication’
and genes involved in ‘cell division, migration and proliferation’.
Notably, all the BPs that were identified were indicative of a
DNA-related processes, such as histone exchange and G2/M
transition. The cellular components category also indicated an
enrichment of genes associated with the nuclear compartment, such
as ‘nucleus’, ‘nucleoplasm’ and ‘nuclear matrix’ (Fig. 2B), suggesting alterations in events
occurring in the nuclear compartment. The most significant GO terms
describing molecular functions for the enriched genes were ‘protein
binding’, ‘chromatin binding’ and ‘ATP binding’ (Fig. 2C).
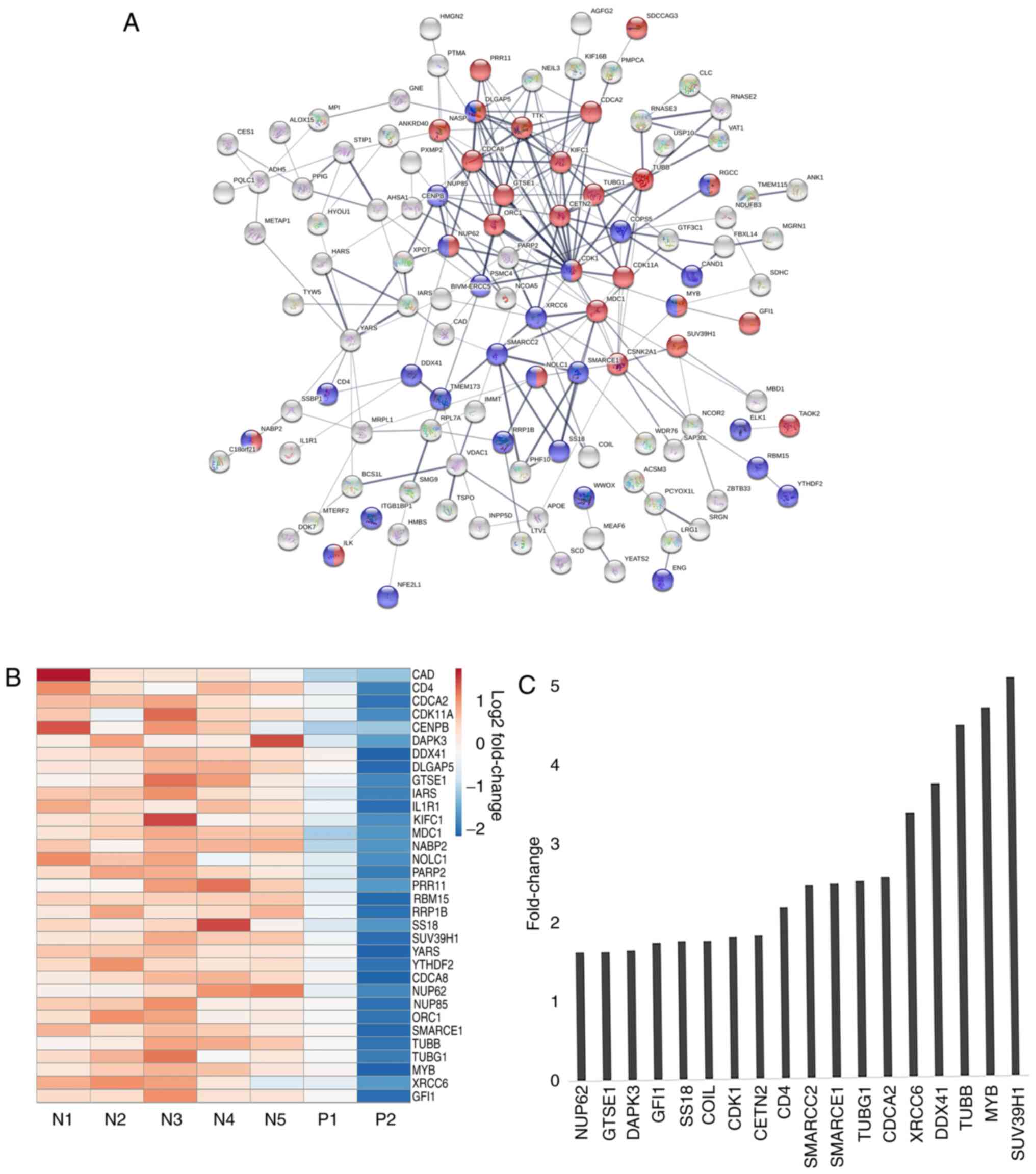
Following the aforementioned observations, the
interactions between the enriched proteins were analyzed using the
STRING database to understand the important molecular processes
that may be activated in the JAK2V617F (−) MPNs. The PPI
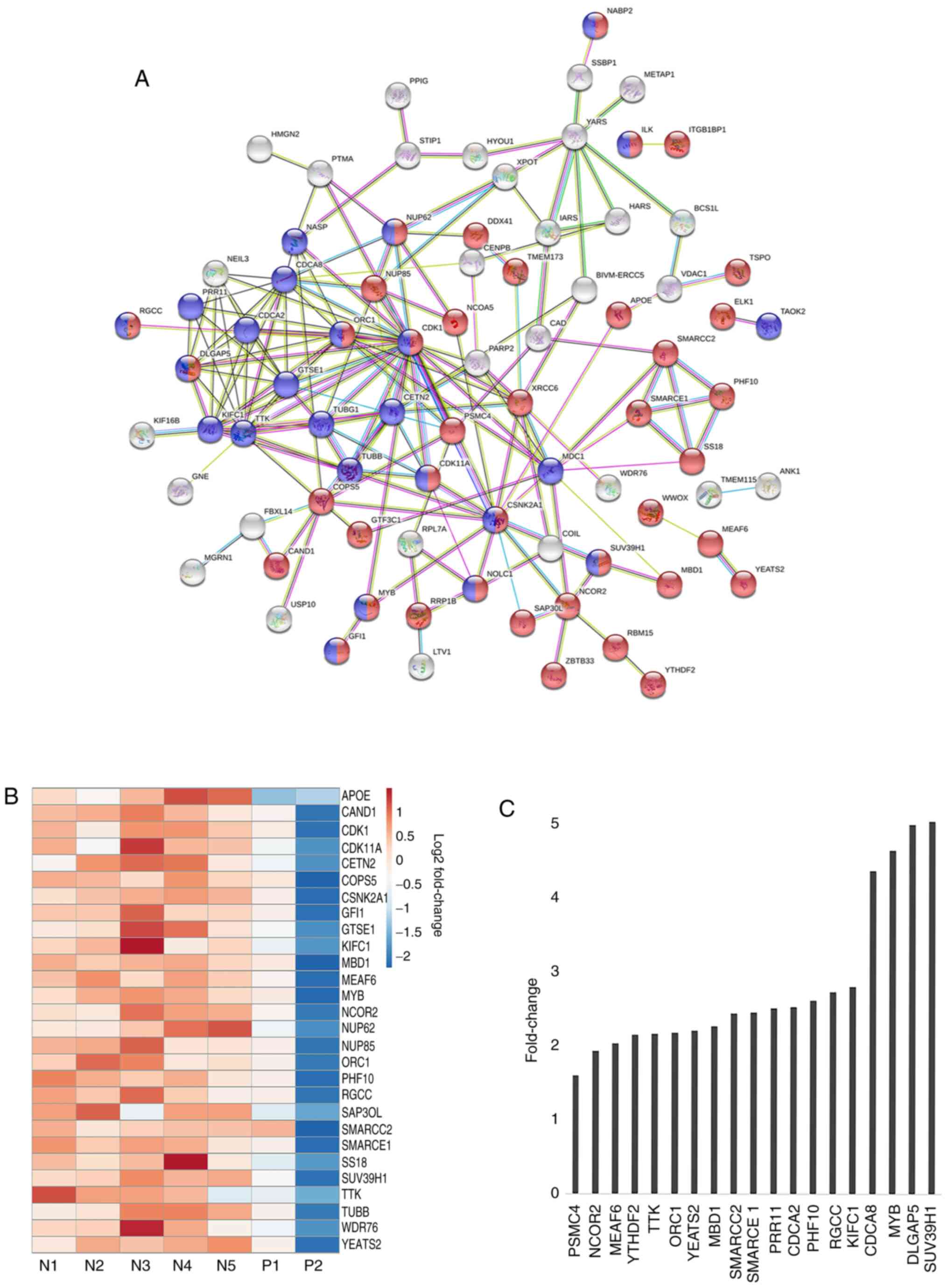
network constructed for the upregulated genes is shown in Fig. 3A. Notably, the STRING analysis
revealed the existence of a distinct network of genes represented
by tight network clusters. A detailed analysis of the network
indicated the involvement of genes in two main biological
processes: Cell cycle (red) and nucleobase modification (blue)
(Fig. 3A). The central tight network
cluster included genes involved in the cell cycle and its
regulation, such as CDK1, TUBB, G2 and S phase-expressed
protein (GTSE1), borealin (CDCA8), SUV39H1 and
centrin 2 (CETN2) (Fig. 3A).
Nucleobase modifying proteins, such as nuclear pore complex protein
NUP85, NUP62, MYB, SS18, nucleolar and coiled-body
phosphoprotein NOLC1, SWI/SNF complex subunit SMARCC2
and SMARCE1, were also linked in the network (Fig. 3A). SUV39H1 encodes a histone
methyltransferase that tri-methylates lysine 9 of histone H3, which
leads to transcriptional gene silencing (31). SMARCC1 and SMARCE1 are part of
the SWI/SNF chromatin remodeling complex involved in
transcriptional activation of target genes in an ATP-dependent
manner (32). The majority of the
proteins in the network were involved in acetylation and
phosphorylation (Fig. 3A). Proteins
such as hypoxia up-regulated protein HYOU1, Isoleucine-tRNA
ligase (IARS), tyrosine t-RNA synthetase YARS, dual
specificity protein kinase TTK, acyl-coenzyme A synthetase
ACSM3, CDK1, XRCC and carbamoyl-phosphate synthetase
CAD marked for nucleotide binding and acetylation indicating
towards the role of DNA modification in this subset of MPNs.
However, unlike other hematological malignancies, such as acute
myeloid leukemia (AML) (33), little
information is available on epigenetic modulations and their role
in regulating hematopoiesis and fibrosis in MPNs. A heatmap was
constructed to visualize the differences in gene expression between
samples for genes comprising the aforementioned tight cluster
(Fig. 3B). There was a categorical
difference between samples with the red bars indicating a high
expression of genes in mutant negative samples and blue bars
indicating decreased expression. A bar graph representing the
fold-change between mutant positive and negative samples shows the
significant relative difference in expression of selected few genes
involved in cell cycle and nucleotide modification, such as MYB,
SUV39H1, XRCC, SMARCC2, SMARCCE1 and CDK1 (Fig. 3C).
A network analysis was also performed for
downregulated genes, which highlighted the genes involved in
developmental process, response to stimuli, multicellular
organismal process and regulation of apoptosis (Fig. S2). Key genes involved in this
network included muscarinic acetylcholine receptor (CHRM3),
IL6, Kalirin (KALRN), MMP7 and ankyrin repeat
domain-containing protein ANKRD1.
Enrichment of chromatin modifiers in
JAK2V617F negative vs. positive samples
Analyses using the DAVID and STRING databases based
on GO and network enrichment for the upregulated genes suggested an
association between the significantly upregulated genes and
stem-like properties. Enriched genes which clustered into DNA
modification and stemness properties (based on DAVID GO analysis)
were further analyzed using StemChecker for evaluating the stemness
signatures and transcription factors associated with these genes.
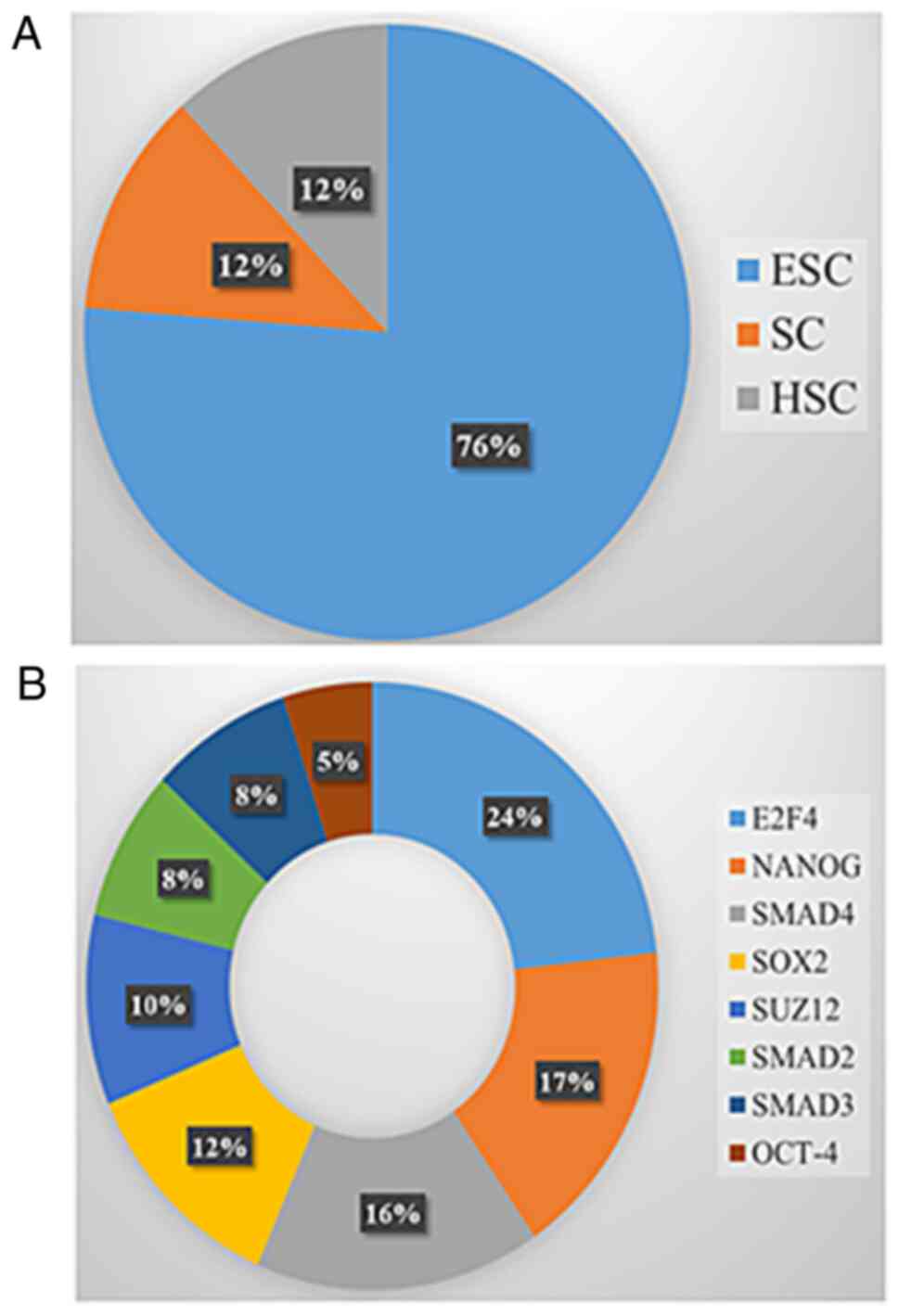
Seven genes (12%) were linked to the hematopoietic signature
(Fig. 4A) while forty-seven (76%)
genes were related to the embryonic signature (Fig. 4A). The abilities of self-renewal,
maintenance and differentiation of stem cells serve as a core
reservoir of cancer initiation and development and tumor growth
(34). Elevated numbers of embryonic
or lineage-specific stem cells can lead to deregulation and altered
differentiation of the progeny (34). The expansion of one specific lineage
in mutant negative MPNs might be attributed to an altered
hematopoietic stem cell. Notably, 76% of genes matched with
embryonic stem cells (ESC) (Fig.
4A). ESC belongs to the primitive class of stem cells and are
capable of differentiating into various cell types (35). Owing to their diverse nature, even a
minute perturbation in these cells can lead to notable changes in
cellular physiology (35). The
transcription factors (TFs) associated with stem cells were also
analyzed (Fig. 4B). Overexpression
of stem-cell specific TFs may contribute to the pathological
self-renewal characteristics of cancer stem cells (36). The majority of the genes mapped to
E2F4 and NANOG TF studies by Boyer et al
(37) (Figs. 4B and S3) which are involved in cell cycle and
pluripotency respectively (38,39). To
further evaluate the correlation between upregulated genes and
stemness and chromatin modification, a network was constructed
using STRING (Fig. 5A). The central
tight cluster mainly comprised of genes implicated in the cell
cycle, nucleobase modification and chromatin modification (Fig. 5A). These genes included MYB,
tubulin γ, probable ATP-dependent RNA helicase DDX41, TUBB,
IARS, SS18, CDCA8 and TTK (Fig. 5A). The heat map representing the
relative expression of genes involved in the cluster indicated a
higher expression of these genes in mutant negative samples
(Fig. 5B). There was heterogeneity
in mutant positive samples, but mutant negative samples had a
homogenous profile. The bar graph representing the fold-changes of
a select few key chromatin modifiers from the PPI network shows the
significant relative difference in expression between mutant
positive and negative samples such as chromatin modification
protein (YEATS2), MYST-associated factor (MEAF6),
methyl CpG binding protein (MBD1), nuclear receptor
corepressor (NCOR2), PHD finger protein (PHF10) and
SS18 (Fig. 5C).
SUV39H1 is a well-known epigenetic marker and has an
established role in MPN (40);
however, the role of MYB is unknown in this context. Therefore, the
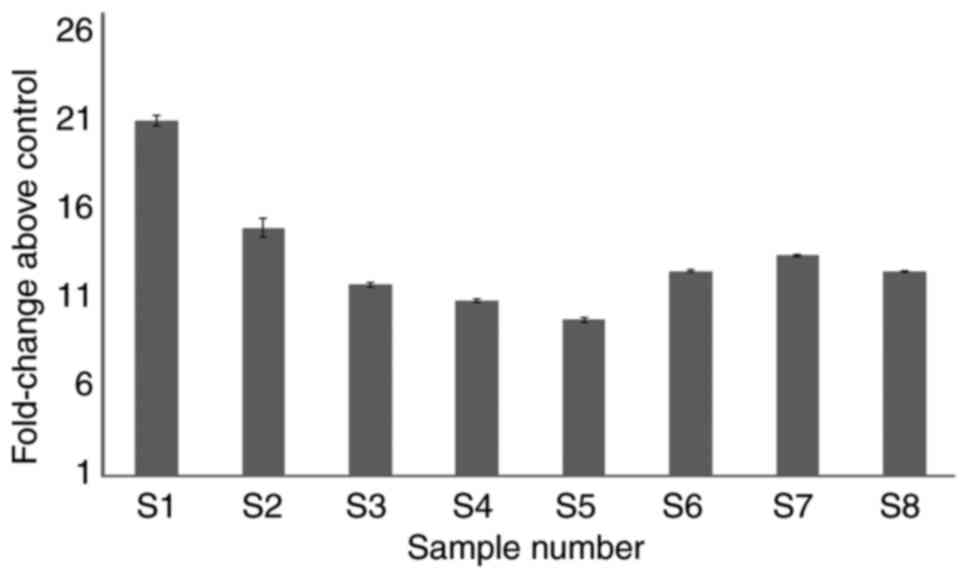
expression of MYB was evaluated in JAK2V617F negative
samples. There was a ~10-fold increase in MYB expression in
the patient samples compared with controls (Fig. 6).
On the basis of the aforementioned observations, the
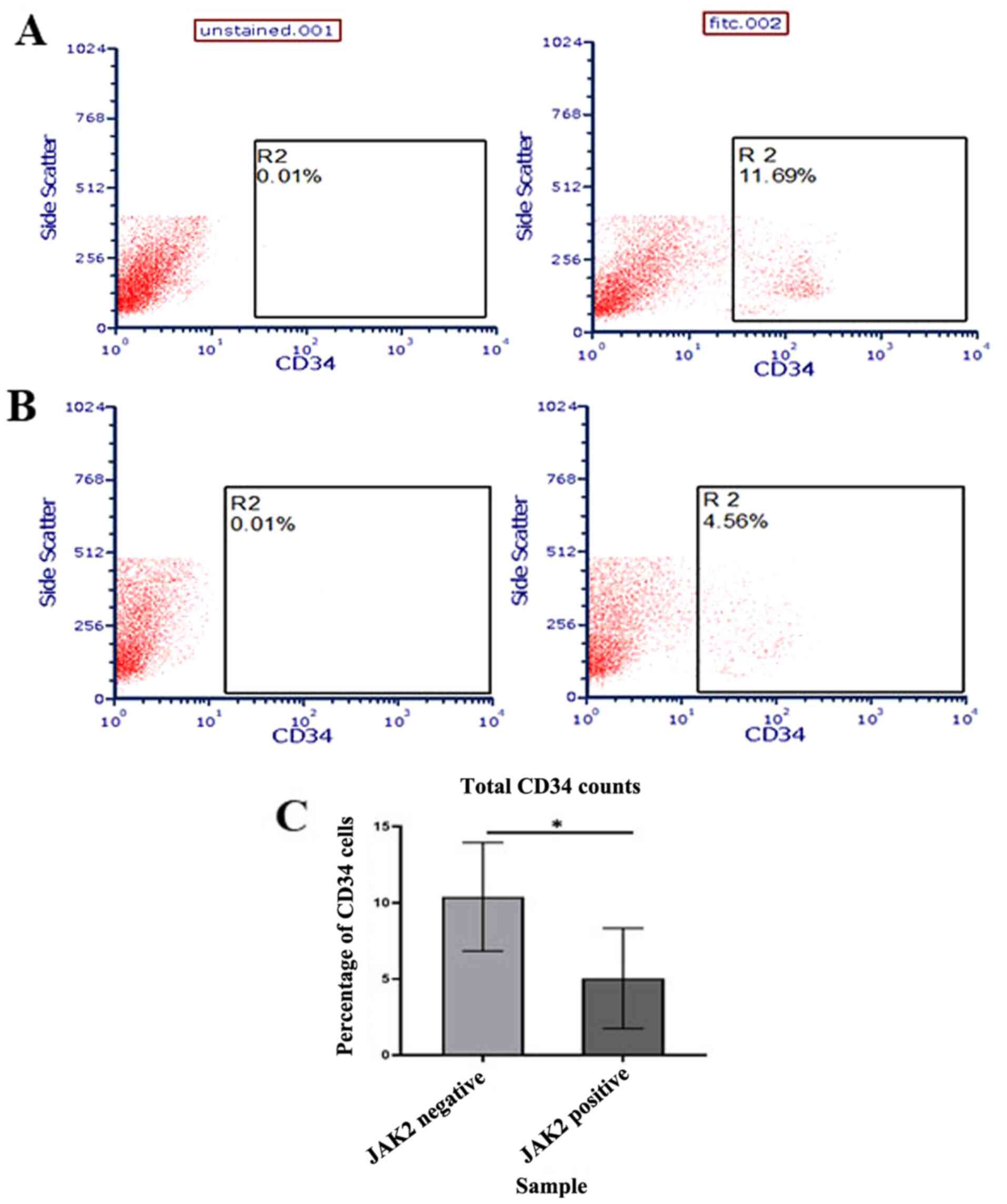
stem cells in patient samples were further evaluated. CD34 was
selected as the marker for interest as it is the most characterized
and reported HSC marker shown to be involved in various
hematological malignancies (41).
Using flow cytometry, it was demonstrated that there was a
significant increase in CD34 counts in JAK2V617F negative
samples compared with positive samples (P<0.05; Fig. 7C), which concurred with the
aforementioned bioinformatic analysis.
Discussion
The identification of the JAK2V671F mutation
distinguished BCR-ABL negative MPN into two broad categories
of JAK2 mutant positive and negative neoplasms (15,21). The
activation and role of the JAK-STAT pathway has been well
characterized in MPNs (19). It is
the mutation negative subset of MPN that has drawn considerable
interest in last few years. Seminal studies have identified
mutations in CALR, MPL, ASXL1 and TET2 genes as
drivers in certain a subset of JAK2V617F mutation negative
neoplasms (15,21,42). It
is important to identify the factors that drive this cohort of
mutant negative neoplasms. The MPN initiating cell is likely a
hematopoietic stem or progenitor cell that, under perturbed
conditions, deviates from the normal hematopoietic differentiation
(43). To understand the molecular
events that mediate tumor progression in mutant negative MPNs, the
present study compared the transcriptional profiles of CD34(+)
cells from JAK2 mutant positive and JAK2 mutant
negative samples. Microarray based transcriptional analysis
revealed the enrichment of a set of genes unique to the JAK2
mutant negative samples.
The present gene expression data revealed distinct
signatures in mutant negative and positive samples. The GO analysis
and STRING network of the upregulated genes identified two key
networks of proteins involved in the cell cycle and chromatin
modification. The present data revealed that there was an
association between genes that are involved in transcriptional
activation, DNA damage response and checkpoint regulation,
microtubule stabilization and spindle assembly. These results
conformed to previous findings reporting that dysregulation of
events in the mitotic cell cycle are involved in multiple cancer
types, including those of hematopoietic origin (44). CDK1 is involved in the
G1/S to G2/M transition by binding to M phase
cyclins. It is also involved in phosphorylation of key genes, such
as MYB, HIST1H3F, EZH2, histone H3-like centromeric protein
and nucleophosmin, that are further involved in downstream DNA
modification (45–50). GTSE1 accumulates in the
nucleus and binds the tumor suppressor protein p53, shuttling it
out of the nucleus and repressing its ability to induce apoptosis.
GTSE1 is only expressed in the S and G2 phase of
the cell cycle (51).
Notably, the other key network identified consisted
of genes involved in the modification of chromatin. Mutations in
genes involved in epigenetic regulation, such as TET2, ASXL1,
DNMT3a, EZH2 and IDH1, have been previously reported in
MPNs (52). Epigenetic changes
control the expression of DNA in numerous ways, including DNA
methylation, acetylation and modification of histones. These are
important epigenetic regulatory processes that control various
cellular events, including stemness maintenance and differentiation
(52). The covalent modifications of
histones at N-terminal lysine residues can lead to transcriptional
activation and repression. Histone methylation markers, such as
H3K4, are associated with transcription activation, while H3K9 and
others are associated with transcriptional repression (53). The role of individual histone
modifications within MPNs is currently unknown. Similarly,
polycomb-related proteins, such as PRC2 and ASXL1,
have been shown to be involved in chromatin remodeling in MPNs
(52,54). The present study reported the
enrichment of chromatin modifying enzymes, such as YEATS2,
MEAF6, SMARCE1, SMARCC2, NCoR2, SUV39H1 in JAK2 negative
samples. Reports suggest that these genes regulate the epigenetic
landscape of cells (31,32,55–59).
Similarly, SUV39H1, NCoR2 and CDK1 are involved in
acute promyelocytic leukemia pathogenesis and treatment response
(60). SUV39H1, CDK1 and
XRCC are involved in chromosome architecture and chromatin
assembly (31,45,61–63).
MBD1 bind to methylated DNA and repress transcription of
target genes (64). PHF10,
SMARCC2, SMARCE1 and SS18 form part of chromatin
remodeling complex altering the DNA-nucleosome biology (32,65,66).
MYB, a helix-turn-helix DNA-binding transcriptional
regulator, drives leukemia progression in AML and mixed lineage
leukemia-fusion leukemia. MYB targets genes involved in
myeloid differentiation, cellular proliferation, cell cycle,
apoptosis and cell signaling (67).
MYB activation by c-MYC via DNMT-1
upregulation promotes the progression of leukemia and lymphoma stem
cells (68). The present study
validated the expression of SUV39H1 and MYB in the
samples sent for array. Since the role of SUV39H1 has been
previously explored in MPN (40),
the expression of MYB in mutation negative samples was
evaluated. A >10-fold increase in MYB expression in
JAK2V671F negative samples was observed; however, the exact
function of MYB and the underlying molecular mechanisms in
the pathogenesis of mutation negative MPN need to be further
evaluated. Since there was an enrichment of genes involved in
epigenetic modification, the association between these genes and
stemness was investigated. Using StemChecker, 7 and 47 genes linked
to the hematopoietic and embryonic signatures were reported,
respectively. In total, 24 and 17% of these genes were under the
control of E2F4 and NANOG transcription factors,
respectively. JAK2 is involved in the self-renewal of mouse
ES cells by regulating the H3Y41 phosphorylation on the
NANOG promoter (69).
TET2 also regulates the differentiation of ESC by inducing
methylation at NANOG promoter (70). The increased level of the CD34
hematopoietic stem cell marker in mutation negative MPN was
correlated with the increase in stem-like properties, which may be
due to altered epigenetics in these samples. The exact mechanism
and chain of events that result in the gain of stemness and
epigenetic properties in these subsets of MPN are yet to be
investigated.
Due to an increasing number of studies
investigating the role of epigenetics in MPN, clinical trials have
already begun to supplement existing treatment regimens for MPN.
Histone deacetylase inhibitors LBH589 and ITF2357 are in phase I
and II clinical studies (71,72).
DNMT inhibitors azacytidine and decitabine are also in phase II
trials as a treatment approach in MPN (73,74).
The underlying causes regulating the initiation and
pathogenesis of JAK2 negative MPN are multidimensional. Over
the years several studies have identified driver mutations
(15,21), which has improved our understanding
of MPN and aided in diagnosis and treatment of the disease. The
present study focused on the epigenetics and the signaling between
the associated genes rather than mutations. In the absence of
driver mutations, the study highlighted the role of key
dysregulated events that might promote the pathogenesis of MPN. To
the best of our knowledge, the study is first of its kind to
observe a distinct epigenetic signature in JAK2V617F
negative patients in an Indian cohort. The altered expression of
genes involved in epigenetic regulation and a further crosstalk
between the epigenome and the transcription machinery may result in
aberrant hematopoiesis that further contributes to disease
progression. The limitation of the present study that should be
addressed in future research is the evaluation and comparison of
absolute CD34 counts in the bone marrow and circulating blood of
patients who are positive and negative for the JAK2V617F
mutation. Additional work needs to be conducted to resolve the
exact function of these genes in disease progression. A further
investigation into the MPN-specific epigenome will lead to an
improved characterization of JAK2V617F negative MPN and help
improve prognosis and treatment.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Sitalakshmi
Subramanian, (Department of Transfusion Medicine and
Immunohematology, St. John's Medical College and Hospital), for
support with use of the flow cytometer facility at St. John's
Medical College and Hospital.
Funding
The present study was funded by The Rajiv Gandhi
University of Health Sciences (grant no. M-73:2015-2016). Mugdha
Sharma is supported by fellowship from Council for Scientific and
Industrial Research, Government of India.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request. The additional datasets generated and/or
analyzed during the current study are available in the StemChecker
repository, [http://stemchecker.sysbiolab.eu/].
Authors' contributions
MS conceptualized the project, designed and
executed experiments, contributed to data analysis, manuscript and
figure preparation. CB contributed to the data analysis and
manuscript preparation. SBS performed the flow cytometry and
analyzed the data. JP contributed to the sample collection and
clinical data analysis. LY contributed to molecular data analysis.
CR conceptualized the project and contributed to the clinical data
analysis. SS conceptualized the project, contributed to
experimental design, data analysis and manuscript preparation. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Bone marrow samples were collected from patients as
per guidelines and approval by Institutional Ethics Committee at
St. John's Medical College and Hospital, Bengaluru in accordance
with Declaration of Helsinki (approval no. 103/2016). All patients
provided written informed consent for participation in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Glossary
Abbreviations
Abbreviations:
|
BCR
|
breakpoint cluster region
|
|
ABL
|
Abelson murine leukemia viral
oncogene homolog 1
|
|
AML
|
acute myeloid leukemia
|
|
DNMT
|
DNA methyltransferase
|
|
IL
|
interleukin
|
References
|
1
|
Dameshek W: Some speculations on the
myeloproliferative syndromes. Blood. 6:372–375. 1951. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fialkow PJ, Gartler SM and Yoshida A:
Clonal origin of chronic myelocytic leukemia in man. Proc Natl Acad
Sci USA. 58:1468–1471. 1967. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tefferi A: The history of
myeloproliferative disorders: Before and after dameshek. Leukemia.
22:3–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moulard O, Mehta J, Fryzek J, Olivares R,
Iqbal U and Mesa RA: Epidemiology of myelofibrosis, essential
thrombocythemia, and polycythemia vera in the European union. Eur J
Haematol. 92:289–297. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mehta J, Wang H, Iqbal SU and Mesa R:
Epidemiology of myeloproliferative neoplasms in the United States.
Leuk Lymphoma. 55:595–600. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baxter EJ, Scott LM, Campbell PJ, East C,
Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N,
et al: Acquired mutation of the tyrosine kinase JAK2 in human
myeloproliferative disorders. Lancet. 365:1054–1061. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Levine RL, Wadleigh M, Cools J, Ebert BL,
Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et
al: Activating mutation in the tyrosine kinase JAK2 in polycythemia
vera, essential thrombocythemia, and myeloid metaplasia with
myelofibrosis. Cancer Cell. 7:387–397. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
James C, Ugo V, Le Couedic JP, Staerk J,
Delhommeau F, Lacout C, Garçon L, Raslova H, Berger R,
Bennaceur-Griscelli A, et al: A unique clonal JAK2 mutation leading
to constitutive signalling causes polycythaemia vera. Nature.
434:1144–1148. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kralovics R, Passamonti F, Buser AS, Teo
SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M and Skoda RC: A
gain-of-function mutation of JAK2 in myeloproliferative disorders.
N Engl J Med. 352:1779–1790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Scott LM, Tong W, Levine RL, Scott MA,
Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison
CN, et al: JAK2 exon 12 mutations in polycythemia vera and
idiopathic erythrocytosis. N Engl J Med. 356:459–468. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Silver RT, Chow W, Orazi A, Arles SP and
Goldsmith SJ: Evaluation of WHO criteria for diagnosis of
polycythemia vera: A prospective analysis. Blood. 122:1881–1886.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bandaranayake RM, Ungureanu D, Shan Y,
Shaw DE, Silvennoinen O and Hubbard SR: Crystal structures of the
JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat
Struct Mol Biol. 19:754–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Saharinen P, Takaluoma K and Silvennoinen
O: Regulation of the Jak2 tyrosine kinase by its pseudokinase
domain. Mol Cell Biol. 20:3387–3395. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vainchenker W and Constantinescu SN:
JAK/STAT signaling in hematological malignancies. Oncogene.
32:2601–2613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vainchenker W and Kralovics R: Genetic
basis and molecular pathophysiology of classical myeloproliferative
neoplasms. Blood. 129:667–679. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Neubauer H, Cumano A, Muller M, Wu H,
Huffstadt U and Pfeffer K: Jak2 deficiency defines an essential
developmental checkpoint in definitive hematopoiesis. Cell.
93:397–409. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Staerk J and Constantinescu SN: The
JAK-STAT pathway and hematopoietic stem cells from the JAK2 V617F
perspective. JAKSTAT. 1:184–190. 2012.PubMed/NCBI
|
|
18
|
Parganas E, Wang D, Stravopodis D, Topham
DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van
Deursen JM, et al: Jak2 is essential for signaling through a
variety of cytokine receptors. Cell. 93:385–395. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Freitas RM and da Costa Maranduba CM:
Myeloproliferative neoplasms and the JAK/STAT signaling pathway: An
overview. Rev Bras Hematol Hemoter. 37:348–353. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barbui T, Thiele J, Gisslinger H,
Kvasnicka HM, Vannucchi AM, Guglielmelli P, Orazi A and Tefferi A:
The 2016 WHO classification and diagnostic criteria for
myeloproliferative neoplasms: Document summary and in-depth
discussion. Blood Cancer J. 8:152018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tefferi A: Novel mutations and their
functional and clinical relevance in myeloproliferative neoplasms:
JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 24:1128–1138.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Team RC: R: A Language and Environment for
Statistical Computing. R Foundation for Statistical Computing;
Vienna, Austria: 2012
|
|
24
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
26
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1):D607–D613. 2019.
View Article : Google Scholar
|
|
27
|
Pinto JP, Kalathur RK, Oliveira DV, Barata
T, Machado RS, Machado S, Pacheco-Leyva I, Duarte I and Futschik
ME: StemChecker: A web-based tool to discover and explore stemness
signatures in gene sets. Nucleic Acids Res. 43:W72–W77. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Metsalu T and Vilo J: ClustVis: A web tool
for visualizing clustering of multivariate data using principal
component analysis and heatmap. Nucleic Acids Res. 43:W566–W570.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cilloni D and Saglio G: Molecular
pathways: BCR-ABL. Clin Cancer Res. 18:930–937. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peters AH, O'Carroll D, Scherthan H,
Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner
M, Kohlmaier A, et al: Loss of the Suv39h histone
methyltransferases impairs mammalian heterochromatin and genome
stability. Cell. 107:323–337. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schaniel C, Ang YS, Ratnakumar K, Cormier
C, James T, Bernstein E, Lemischka IR and Paddison PJ:
Smarcc1/Baf155 couples self-renewal gene repression with changes in
chromatin structure in mouse embryonic stem cells. Stem Cells.
27:2979–2991. 2009.PubMed/NCBI
|
|
33
|
Goldman SL, Hassan C, Khunte M, Soldatenko
A, Jong Y, Afshinnekoo E and Mason CE: Epigenetic modifications in
acute myeloid leukemia: Prognosis, treatment, and heterogeneity.
Front Genet. 10:1332019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lim WF, Inoue-Yokoo T, Tan KS, Lai MI and
Sugiyama D: Hematopoietic cell differentiation from embryonic and
induced pluripotent stem cells. Stem Cell Res Ther. 4:712013.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blum B and Benvenisty N: The
tumorigenicity of human embryonic stem cells. Adv Cancer Res.
100:133–158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lambert M, Jambon S, Depauw S and
David-Cordonnier MH: Targeting transcription factors for cancer
treatment. Molecules. 23:14792018. View Article : Google Scholar
|
|
37
|
Boyer LA, Lee TI, Cole MF, Johnstone SE,
Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG,
et al: Core transcriptional regulatory circuitry in human embryonic
stem cells. Cell. 122:947–956. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hsu J and Sage J: Novel functions for the
transcription factor E2F4 in development and disease. Cell Cycle.
15:3183–3190. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gawlik-Rzemieniewska N and Bednarek I: The
role of NANOG transcriptional factor in the development of
malignant phenotype of cancer cells. Cancer Biol Ther. 17:1–10.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Son HJ, Kim JY, Hahn Y and Seo SB:
Negative regulation of JAK2 by H3K9 methyltransferase G9a in
leukemia. Mol Cell Biol. 32:3681–3694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sidney LE, Branch MJ, Dunphy SE, Dua HS
and Hopkinson A: Concise review: Evidence for CD34 as a common
marker for diverse progenitors. Stem Cells. 32:1380–1389. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shammo JM and Stein BL: Mutations in MPNs:
Prognostic implications, window to biology, and impact on treatment
decisions. Hematology Am Soc Hematol Educ Program. 2016:552–560.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mead AJ and Mullally A: Myeloproliferative
neoplasm stem cells. Blood. 129:1607–1616. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Aleem E and Arceci RJ: Targeting cell
cycle regulators in hematologic malignancies. Front Cell Dev Biol.
3:162015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bertoli C, Skotheim JM and de Bruin RA:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Werwein E, Cibis H, Hess D and Klempnauer
KH: Activation of the oncogenic transcription factor B-Myb via
multisite phosphorylation and prolyl cis/trans isomerization.
Nucleic Acids Res. 47:103–121. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Koseoglu MM, Dong J and Marzluff WF:
Coordinate regulation of histone mRNA metabolism and DNA
replication: Cyclin A/cdk1 is involved in inactivation of histone
mRNA metabolism and DNA replication at the end of S phase. Cell
Cycle. 9:3857–3863. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zeng X, Chen S and Huang H:
Phosphorylation of EZH2 by CDK1 and CDK2: A possible regulatory
mechanism of transmission of the H3K27me3 epigenetic mark through
cell divisions. Cell Cycle. 10:579–583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu Z, Zhou X, Wang W, Deng W, Fang J, Hu
H, Wang Z, Li S, Cui L, Shen J, et al: Dynamic phosphorylation of
CENP-A at Ser68 orchestrates its cell-cycle-dependent deposition at
centromeres. Dev Cell. 32:68–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jiang PS, Chang JH and Yung BY: Different
kinases phosphorylate nucleophosmin/B23 at different sites during
G(2) and M phases of the cell cycle. Cancer Lett. 153:151–160.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Monte M, Benetti R, Buscemi G, Sandy P,
Del Sal G and Schneider C: The cell cycle-regulated protein human
GTSE-1 controls DNA damage-induced apoptosis by affecting p53
function. J Biol Chem. 278:30356–30364. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
McPherson S, McMullin MF and Mills K:
Epigenetics in myeloproliferative neoplasms. J Cell Mol Med.
21:1660–1667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nielsen HM, Andersen CL, Westman M,
Kristensen LS, Asmar F, Kruse TA, Thomassen M, Larsen TS, Skov V,
Hansen LL, et al: Publisher correction: Epigenetic changes in
myelofibrosis: Distinct methylation changes in the myeloid
compartments and in cases with ASXL1 mutations. Sci Rep.
8:173112018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang
S, Andrews FH, Wang X, Gagea M, Wen H, et al: YEATS2 links histone
acetylation to tumorigenesis of non-small cell lung cancer. Nat
Commun. 8:10882017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Panagopoulos I, Micci F, Thorsen J,
Gorunova L, Eibak AM, Bjerkehagen B, Davidson B and Heim S: Novel
fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial
stromal sarcoma. PLoS One. 7:e393542012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Battaglia S, Maguire O and Campbell MJ:
Transcription factor co-repressors in cancer biology: Roles and
targeting. Int J Cancer. 126:2511–2519. 2010.PubMed/NCBI
|
|
58
|
Viiri KM, Korkeamaki H, Kukkonen MK,
Nieminen LK, Lindfors K, Peterson P, Mäki M, Kainulainen H and Lohi
O: SAP30L interacts with members of the Sin3A corepressor complex
and targets Sin3A to the nucleolus. Nucleic Acids Res.
34:3288–3298. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
DeVilbiss AW, Boyer ME and Bresnick EH:
Establishing a hematopoietic genetic network through locus-specific
integration of chromatin regulators. Proc Natl Acad Sci USA.
110:E3398–3407. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Arteaga MF, Mikesch JH, Fung TK and So CW:
Epigenetics in acute promyelocytic leukaemia pathogenesis and
treatment response: A TRAnsition to targeted therapies. Br J
Cancer. 112:413–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Crepaldi L, Policarpi C, Coatti A,
Sherlock WT, Jongbloets BC, Down TA and Riccio A: Binding of TFIIIC
to sine elements controls the relocation of activity-dependent
neuronal genes to transcription factories. PLoS Genet.
9:e10036992013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Malik P, Zuleger N, de las Heras JI,
Saiz-Ros N, Makarov AA, Lazou V, Meinke P, Waterfall M, Kelly DA
and Schirmer EC: NET23/STING promotes chromatin compaction from the
nuclear envelope. PLoS One. 9:e1118512014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Thacker J and Zdzienicka MZ: The XRCC
genes: Expanding roles in DNA double-strand break repair. DNA
Repair (Amst). 3:1081–1090. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Spruijt CG, Gnerlich F, Smits AH,
Pfaffeneder T, Jansen PW, Bauer C, Münzel M, Wagner M, Müller M,
Khan F, et al: Dynamic readers for 5-(hydroxy)methylcytosine and
its oxidized derivatives. Cell. 152:1146–1159. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Krasteva V, Crabtree GR and Lessard JA:
The BAF45a/PHF10 subunit of SWI/SNF-like chromatin remodeling
complexes is essential for hematopoietic stem cell maintenance. Exp
Hematol. 48:58–71 e15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kadoch C and Crabtree GR: Reversible
disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic
fusion in synovial sarcoma. Cell. 153:71–85. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Pattabiraman DR and Gonda TJ: Role and
potential for therapeutic targeting of MYB in leukemia. Leukemia.
27:269–277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Hu T, Chong Y, Cai B, Liu Y, Lu S and
Cowell JK: DNA methyltransferase 1-mediated CpG methylation of the
miR-150-5p promoter contributes to fibroblast growth factor
receptor 1-driven leukemogenesis. J Biol Chem. 294:18122–18130.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Griffiths DS, Li J, Dawson MA, Trotter MW,
Cheng YH, Smith AM, Mansfield W, Liu P, Kouzarides T, Nichols J, et
al: LIF-independent JAK signalling to chromatin in embryonic stem
cells uncovered from an adult stem cell disease. Nat Cell Biol.
13:13–21. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Langlois T, da Costa Reis Monte-Mor B,
Lenglet G, Droin N, Marty C, Le Couédic JP, Almire C, Auger N,
Mercher T, Delhommeau F, et al: TET2 deficiency inhibits mesoderm
and hematopoietic differentiation in human embryonic stem cells.
Stem Cells. 32:2084–2097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Mascarenhas J, Sandy L, Lu M, Yoon J,
Petersen B, Zhang D, Ye F, Newsom C, Najfeld V, Hochman T, et al: A
phase II study of panobinostat in patients with primary
myelofibrosis (PMF) and post-polycythemia vera/essential
thrombocythemia myelofibrosis (post-PV/ET MF). Leuk Res. 53:13–19.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rambaldi A, Dellacasa CM, Finazzi G,
Carobbio A, Ferrari ML, Guglielmelli P, Gattoni E, Salmoiraghi S,
Finazzi MC, Di Tollo S, et al: A pilot study of the
Histone-Deacetylase inhibitor Givinostat in patients with JAK2V617F
positive chronic myeloproliferative neoplasms. Br J Haematol.
150:446–455. 2010.PubMed/NCBI
|
|
73
|
Silverman LR, McKenzie DR, Peterson BL,
Holland JF, Backstrom JT, Beach CL and Larson RA; Cancer Leukemia
Group B, : Further analysis of trials with azacitidine in patients
with myelodysplastic syndrome: Studies 8421, 8921, and 9221 by the
cancer and leukemia Group B. J Clin Oncol. 24:3895–3903. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kantarjian HM, O'Brien S, Huang X,
Garcia-Manero G, Ravandi F, Cortes J, Shan J, Davisson J,
Bueso-Ramos CE and Issa JP: Survival advantage with decitabine
versus intensive chemotherapy in patients with higher risk
myelodysplastic syndrome: Comparison with historical experience.
Cancer. 109:1133–1137. 2007. View Article : Google Scholar : PubMed/NCBI
|