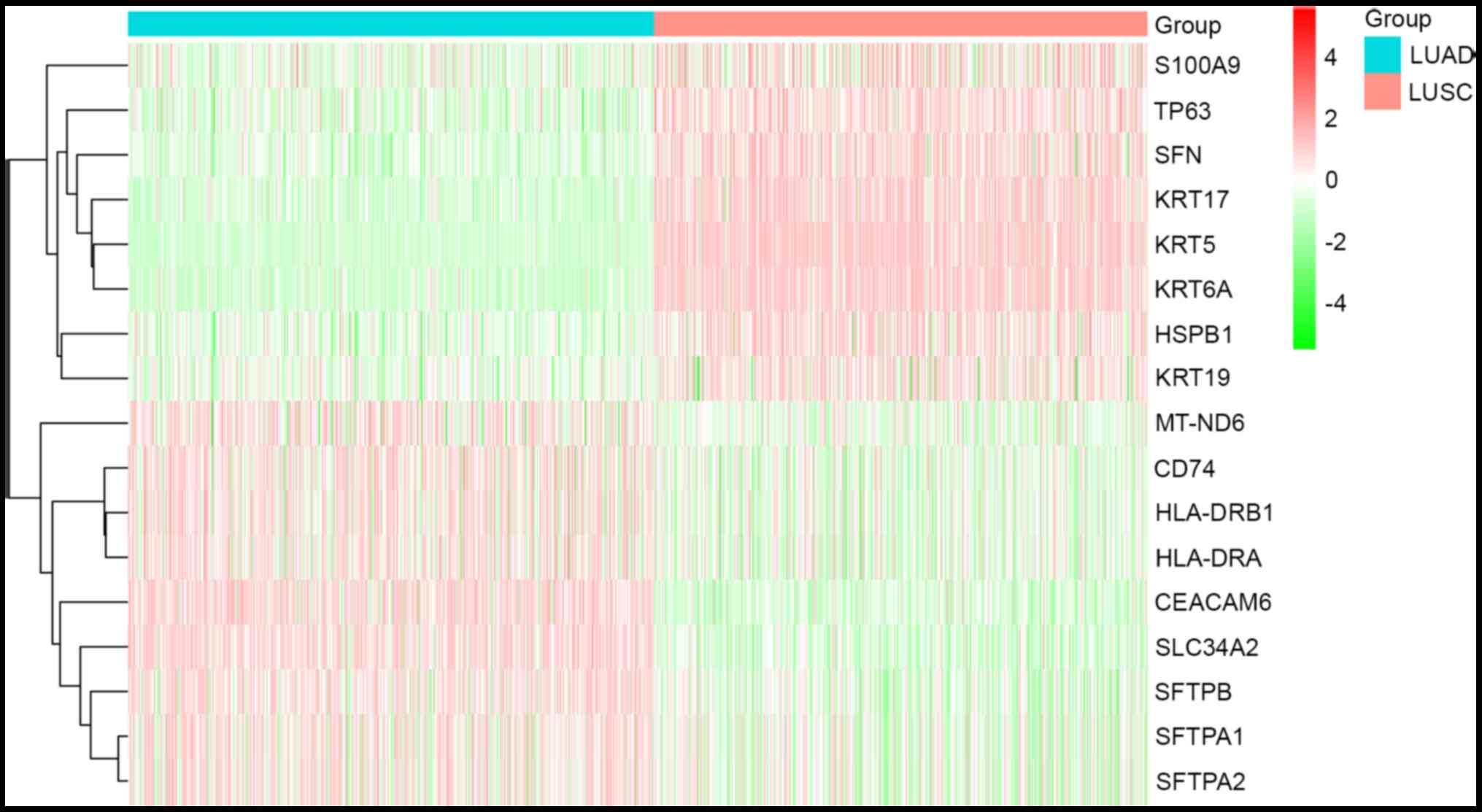
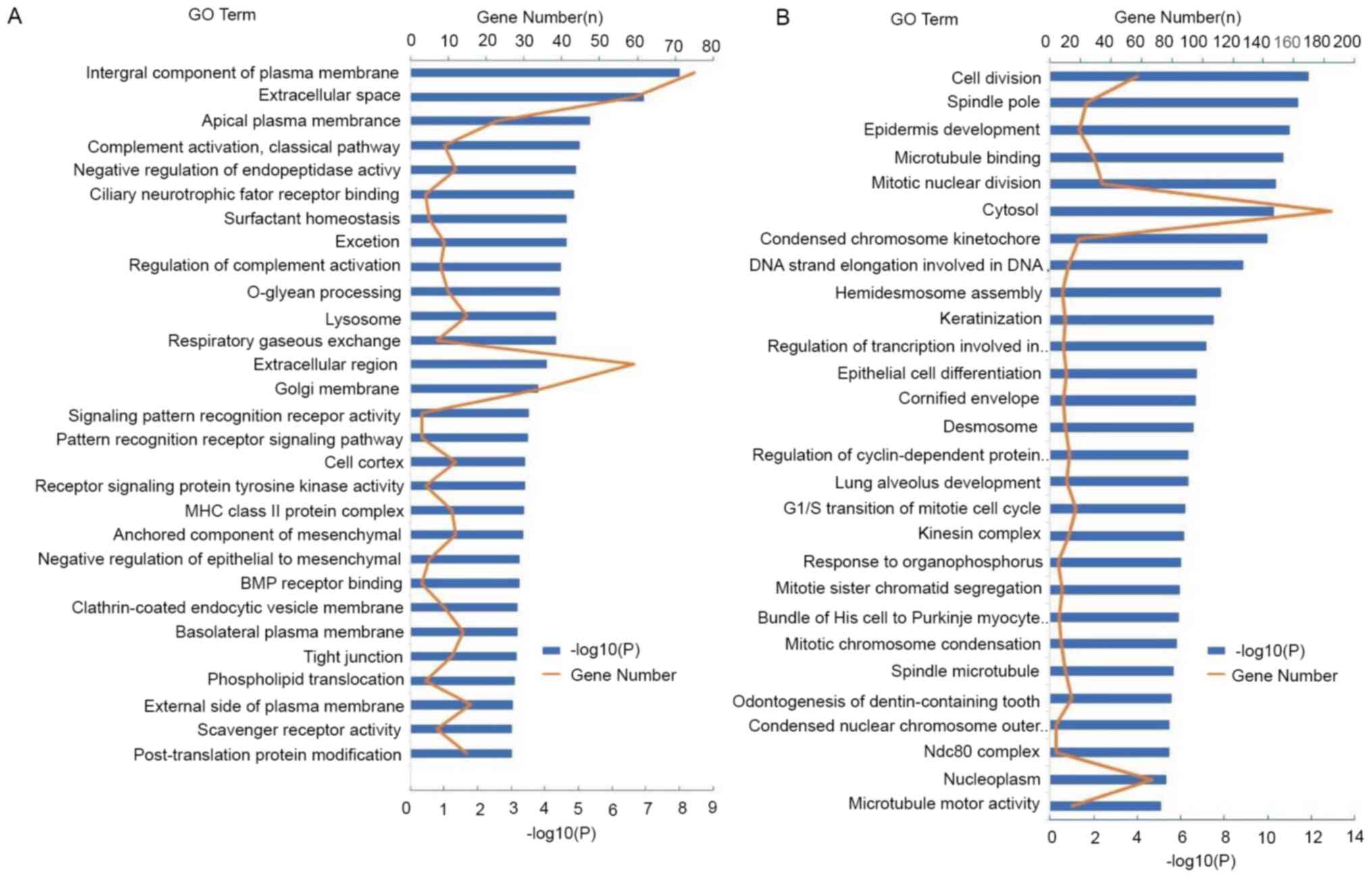
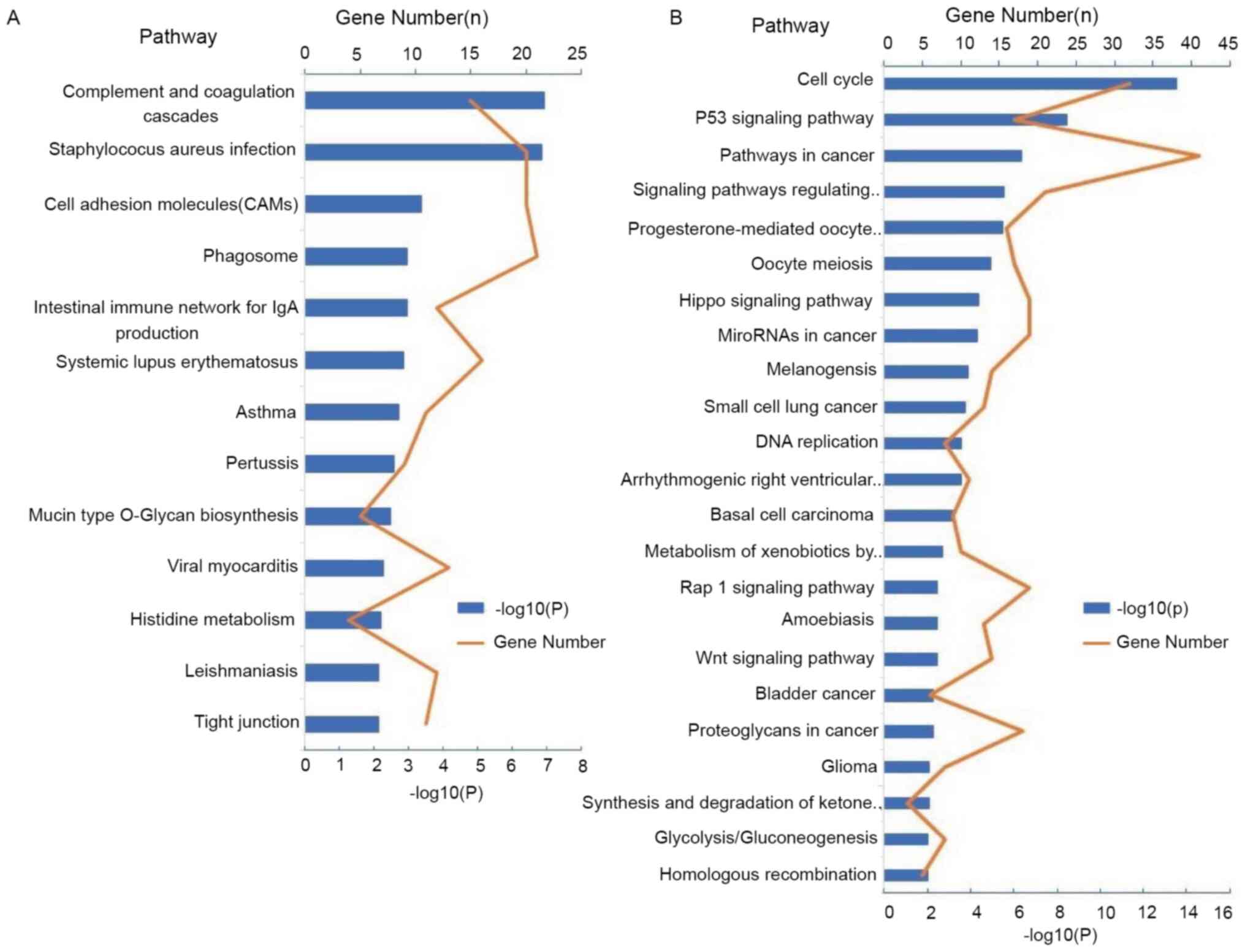
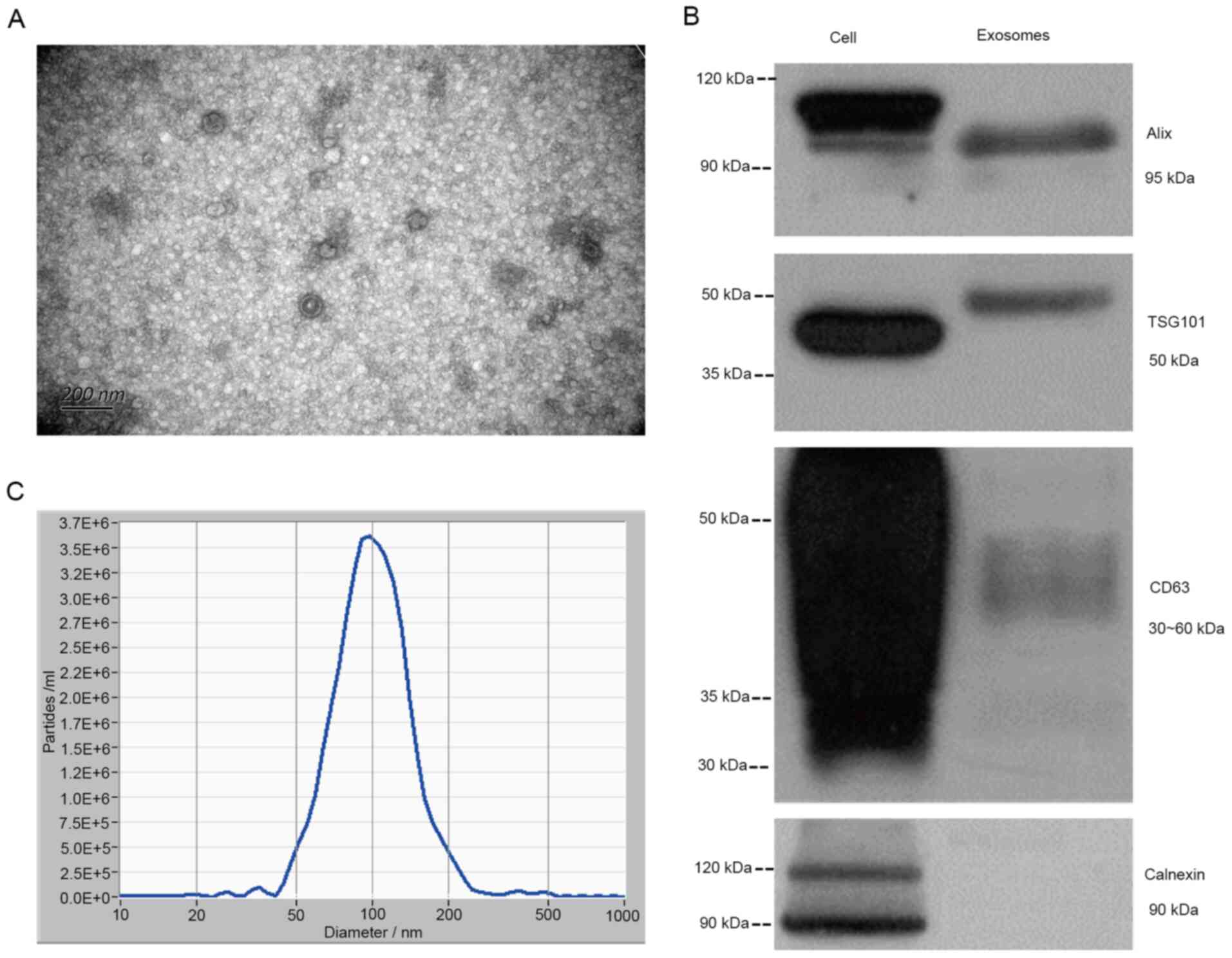
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Otsmane A, Kacimi G, Adane S, Cherbal F
and Aouichat Bouguerra S: Clinico-epidemiological profile and redox
imbalance of lung cancer patients in Algeria. J Med Life.
11:210–217. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang H, Lin Y and Liang Y: Treatment of
lung carcinosarcoma and other rare histologic subtypes of non-small
cell lung cancer. Curr Treat Options Oncol. 18:542017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim SL, Jia Z, Lu Y, Zhang H, Ng CT, Bay
BH, Shen HM and Ong CN: Metabolic signatures of four major
histological types of lung cancer cells. Metabolomics. 14:1182018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Björkqvist AM, Husgafvel-Pursiainen K,
Anttila S, Karjalainen A, Tammilehto L, Mattson K, Vainio H and
Knuutila S: DNA gains in 3q occur frequently in squamous cell
carcinoma of the lung, but not in adenocarcinoma. Genes Chromosomes
Cancer. 22:79–82. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cooper WA, Lam DC, O'toole SA and Minna
JD: Molecular biology of lung cancer. J Thorac Dis. 5 (Suppl
5):S479–S490. 2013.PubMed/NCBI
|
|
7
|
Davidson MR, Gazdar AF and Clarke BE: The
pivotal role of pathology in the management of lung cancer. J
Thorac Dis. 5 (Suppl 5):S463–S478. 2013.PubMed/NCBI
|
|
8
|
Langer CJ, Besse B, Gualberto A, Brambilla
E and Soria JC: The evolving role of histology in the management of
advanced non-small-cell lung cancer. J Clin Oncol. 28:5311–5320.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tomasini P, Khobta N, Greillier L and
Barlesi F: Ipilimumab: Its potential in non-small cell lung cancer.
Ther Adv Med Oncol. 4:43–50. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiao J, Lu X, Chen X, Zou Y, Liu A, Li W,
He B, He S and Chen Q: Eight potential biomarkers for
distinguishing between lung adenocarcinoma and squamous cell
carcinoma. Oncotarget. 8:71759–71771. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu F, Vermesh O, Mani V, Ge TJ, Madsen
SJ, Sabour A, Hsu EC, Gowrishankar G, Kanada M, Jokerst JV, et al:
The exosome total isolation chip. ACS Nano. 11:10712–10723. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fitts CA, Ji N, Li Y and Tan C: Exploiting
exosomes in cancer liquid biopsies and drug delivery. Adv Healthc
Mater. 8:e18012682019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carretero-González A, Otero I,
Carril-Ajuria L, de Velasco G and Manso L: Exosomes: Definition,
role in tumor development and clinical implications. Cancer
Microenviron. 11:13–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chuo ST, Chien JC and Lai CP: Imaging
extracellular vesicles: Current and emerging methods. J Biomed Sci.
25:912018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Revenfeld ALS Bæk R, Nielsen MH,
Stensballe A, Varming K and Jørgensen M: Diagnostic and prognostic
potential of extracellular vesicles in peripheral blood. Clin Ther.
36:830–846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang N, Song X, Liu L, Niu L, Wang X, Song
X and Xie L: Circulating exosomes contain protein biomarkers of
metastatic non-small-cell lung cancer. Cancer Sci. 109:1701–1709.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bustin SA: Quantification of mRNA using
real-time reverse transcription PCR (RT-PCR): Trends and problems.
J Mol Endocrinol. 29:23–39. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vandesompele J, De Preter K, Pattyn F,
Poppe B, Van Roy N, De Paepe A and Speleman F: Accurate
normalization of real-time quantitative RT-PCR data by geometric
averaging of multiple internal control genes. Genome Biol.
3:RESEARCH00342002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lourenço AP, Mackert A, Cristino ADS and
Simões ZLP: Validation of reference genes for gene expression
studies in the honey bee, Apis mellifera, by quantitative
real-time RT-PCR. Apidologie. 39:372–385. 2008. View Article : Google Scholar
|
|
23
|
Macabelli CH, Ferreira RM, Gimenes LU, de
Carvalho NA, Soares JG, Ayres H, Ferraz ML, Watanabe YF, Watanabe
OY, Sangalli JR, et al: Reference gene selection for gene
expression analysis of oocytes collected from dairy cattle and
buffaloes during winter and summer. PLoS One. 9:e932872014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong ZZ, Tang CH, Zhang Z, Zhou W, Zhao R,
Wang L, Xu JC, Wu YY, Wu J, Zhang X, et al: Simultaneous detection
of exosomal membrane protein and RNA by highly sensitive aptamer
assisted multiplex-PCR. ACS Appl Bio Mater. 3:2560–2567. 2020.
View Article : Google Scholar
|
|
25
|
Pan H, Yang XW, Bidne K, Hellmich RL,
Siegfried BD and Zhou XG: Selection of reference genes for RT-qPCR
analysis in the monarch butterfly, Danaus plexippus (L.), a
migrating bio-indicator. PLoS One. 10:e01294822015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang S, An S, Li Z, Wu F, Yang Q, Liu Y,
Cao J, Zhang H, Zhang Q and Liu X: Identification and validation of
reference genes for normalization of gene expression analysis using
qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae).
Gene. 555:393–402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Stat Soc B. 57:289–300. 1995.
|
|
28
|
Liu P, Wang H, Liang Y, Hu A, Xing R,
Jiang L, Yi L and Dong J: LINC00852 promotes lung adenocarcinoma
spinal metastasis by targeting S100A9. J Cancer. 9:4139–4149. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stewart PA, Welsh EA, Slebos RJC, Fang B,
Izumi V, Chambers M, Zhang G, Cen L, Pettersson F, Zhang Y, et al:
Proteogenomic landscape of squamous cell lung cancer. Nat Commun.
10:35782019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim Y, Shiba-Ishii A, Nakagawa T, Iemura
SI, Natsume T, Nakano N, Matsuoka R, Sakashita S, Lee S, Kawaguchi
A, et al: Stratifin regulates stabilization of receptor tyrosine
kinases via interaction with ubiquitin-specific protease 8 in lung
adenocarcinoma. Oncogene. 37:5387–5402. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Z, Yang MQ, Lei L, Fei LR, Zheng YW,
Huang WJ, Li ZH, Liu CC and Xu HT: Overexpression of KRT17 promotes
proliferation and invasion of non-small cell lung cancer and
indicates poor prognosis. Cancer Manag Res. 11:7485–7497. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Khayyata S, Yun S, Pasha T, Jian B,
McGrath C, Yu G, Gupta P and Baloch Z: Value of P63 and CK5/6 in
distinguishing squamous cell carcinoma from adenocarcinoma in lung
fine-needle aspiration specimens. Diagn Cytopathol. 37:178–183.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang B, Zhang W, Zhang M, Wang X, Peng S
and Zhang R: KRT6A promotes EMT and cancer stem cell transformation
in lung adenocarcinoma. Technol Cancer Res Treat.
19:15330338209212482020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Konda JD, Olivero M, Musiani D, Lamba S
and Di Renzo MF: Heat-shock protein 27 (HSP27, HSPB1) is synthetic
lethal to cells with oncogenic activation of MET, EGFR and BRAF.
Mol Oncol. 11:599–611. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ohtsuka T, Sakaguchi M, Yamamoto H, Tomida
S, Takata K, Shien K, Hashida S, Miyata-Takata T, Watanabe M,
Suzawa K, et al: Interaction of cytokeratin 19 head domain and HER2
in the cytoplasm leads to activation of HER2-Erk pathway. Sci Rep.
6:395572016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song Y, Sun Y, Lei Y, Yang K and Tang R:
YAP1 promotes multidrug resistance of small cell lung cancer by
CD74-related signaling pathways. Cancer Med. 9:259–268. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tokumoto H: Analysis of HLA-DRB1-related
alleles in Japanese patients with lung cancer-relationship to
genetic susceptibility and resistance to lung cancer. J Cancer Res
Clin Oncol. 124:511–516. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Martins I, Deshayes F, Baton F, Forget A,
Ciechomska I, Sylla K, Aoudjit F, Charron D, Al-Daccak R and
Alcaide-Loridan C: Pathologic expression of MHC class II is driven
by mitogen- activated protein kinases. Eur J Immunol. 37:788–797.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hong KP, Shin MH, Yoon S, Ji GY, Moon YR,
Lee OJ, Choi SY, Lee YM, Koo JH, Lee HC, et al: Therapeutic effect
of anti CEACAM6 monoclonal antibody against lung adenocarcinoma by
enhancing anoikis sensitivity. Biomaterials. 67:32–41. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Yang W, Pu Q, Yang Y, Ye S, Ma Q,
Ren J, Cao Z, Zhong G, Zhang X, et al: The effects and mechanisms
of SLC34A2 in tumorigenesis and progression of human non-small cell
lung cancer. J Biomed Sci. 22:522015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee S, Kim D, Kang J, Kim E, Kim W, Youn H
and Youn B: Surfactant protein B suppresses lung cancer progression
by inhibiting secretory phospholipase A2 activity and arachidonic
acid production. Cell Physiol Biochem. 42:1684–1700. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hasegawa Y, Takahashi M, Ariki S, Saito A,
Uehara Y, Takamiya R, Kuronuma K, Chiba H, Sakuma Y, Takahashi H
and Kuroki Y: Surfactant protein A down-regulates epidermal growth
factor receptor by mechanisms different from those of surfactant
protein D. J Biol Chem. 292:18565–18576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang Y, Kuan PJ, Xing C, Cronkhite JT,
Torres F, Rosenblatt RL, DiMaio JM, Kinch LN, Grishin NV and Garcia
CK: Genetic defects in surfactant protein A2 are associated with
pulmonary fibrosis and lung cancer. Am J Hum Genet. 84:52–59. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Min L, Zhu S, Chen L, Liu X, Wei R, Zhao
L, Yang Y, Zhang Z, Kong G, Li P and Zhang S: Evaluation of
circulating small extracellular vesicles derived miRNAs as
biomarkers of early colon cancer: A comparison with plasma total
miRNAs. J Extracell Vesicles. 8:16436702019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Yin Z, Fan J, Zhang S and Yang W:
The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal
Transduct Target Ther. 4:472019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li C, Li C, Zhi C, Liang W, Wang X, Chen
X, Lv T, Shen Q, Song Y, Lin D and Liu H: Clinical significance of
PD-L1 expression in serum-derived exosomes in NSCLC patients. J
Transl Med. 17:3552019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen R, Xu X, Qian Z, Zhang C, Niu Y, Wang
Z, Sun J, Zhang X and Yu Y: The biological functions and clinical
applications of exosomes in lung cancer. Cell Mol Life Sci.
76:4613–4633. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fu L, Wang H, Wei D, Wang B, Zhang C, Zhu
T, Ma Z, Li Z, Wu Y and Yu G: The value of CEP55 gene as a
diagnostic biomarker and independent prognostic factor in LUAD and
LUSC. PLoS One. 15:e02332832020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang C, Tan S, Liu WR, Lei Q, Qiao W, Wu
Y, Liu X, Cheng W, Wei YQ, Peng Y and Li W: RNA-Seq profiling of
circular RNA in human lung adenocarcinoma and squamous cell
carcinoma. Mol Cancer. 18:1342019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kowal J, Tkach M and Théry C: Biogenesis
and secretion of exosomes. Curr Opin Cell Biol. 29:116–125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Molina-Vila MA, Mayo-de-Las-Casas C,
Giménez-Capitán A, Jordana-Ariza N, Garzón M, Balada A, Villatoro
S, Teixidó C, García-Peláez B, Aguado C, et al: Liquid biopsy in
non-small cell lung cancer. Front Med (Lausanne).
3:692016.PubMed/NCBI
|
|
52
|
Yoshioka Y, Konishi Y, Kosaka N, Katsuda
T, Kato T and Ochiya T: Comparative marker analysis of
extracellular vesicles in different human cancer types. J Extracell
Vesicles. Jun 18–2013.(Epub ahead of print). do:
10.3402/jev.v2i0.20424 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Vlassov AV, Magdaleno S, Setterquist R and
Conrad R: Exosomes: Current knowledge of their composition,
biological functions, and diagnostic and therapeutic potentials.
Biochim Biophys Acta. 1820:940–948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang K, Chen R, Feng Z, Zhu YM, Sun XX,
Huang W and Chen ZN: Identification of differentially expressed
genes in non-small cell lung cancer. Aging (Albany NY).
11:11170–11185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mehta A, Sriramanakoppa NN, Agarwal P,
Viswakarma G, Vasudevan S, Panigrahi M, Kumar D, Saifi M, Chowdhary
I, Doval DC and Suryavanshi M: Predictive biomarkers in nonsmall
cell carcinoma and their clinico-pathological association. South
Asian J Cancer. 8:250–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Miyazono K, Kamiya Y and Morikawa M: Bone
morphogenetic protein receptors and signal transduction. J Biochem.
147:35–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Deng T, Lin D, Zhang M, Zhao Q, Li W,
Zhong B, Deng Y and Fu X: Differential expression of bone
morphogenetic protein 5 in human lung squamous cell carcinoma and
adenocarcinoma. Acta Biochim Biophys Sin (Shanghai). 47:557–563.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Powell DW: Barrier function of epithelia.
Am J Physiol. 241:G275–G288. 1981.PubMed/NCBI
|
|
59
|
Tsukita S, Furuse M and Itoh M:
Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol.
2:285–293. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hichino A, Okamoto M, Taga S, Akizuki R,
Endo S, Matsunaga T and Ikari A: Down-regulation of Claudin-2
expression and proliferation by epigenetic inhibitors in human lung
adenocarcinoma A549 cells. J Biol Chem. 292:2411–2421. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pai VC, Hsu CC, Chan TS, Liao WY, Chuu CP,
Chen WY, Li CR, Lin CY, Huang SP, Chen LT and Tsai KK: ASPM
promotes prostate cancer stemness and progression by augmenting
Wnt-Dvl-3-β-catenin signaling. Oncogene. 38:1340–1353. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gai M, Bianchi FT, Vagnoni C, Vernì F,
Bonaccorsi S, Pasquero S, Berto GE, Sgrò F, Chiotto AA, Annaratone
L, et al: ASPM and CITK regulate spindle orientation by affecting
the dynamics of astral microtubules. EMBO Rep. 18:18702017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yuan YJ, Sun Y, Gao R, Yin ZZ, Yuan ZY and
Xu LM: Abnormal spindle-like microcephaly-associated protein (ASPM)
contributes to the progression of lung squamous cell carcinoma
(LSCC) by regulating CDK4. J Cancer. 11:5413–5423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kemp CJ, Donehower LA, Bradley A and
Balmain A: Reduction of p53 gene dosage does not increase
initiation or promotion but enhances malignant progression of
chemically induced skin tumors. Cell. 74:813–822. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jackson EL, Olive KP, Tuveson DA, Bronson
R, Crowley D, Brown M and Jacks T: The differential effects of
mutant p53 alleles on advanced murine lung cancer. Cancer Res.
65:10280–10288. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Gibbons DL, Byers LA and Kurie JM:
Smoking, p53 mutation, and lung cancer. Mol Cancer Res. 12:3–13.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yang Z, Zhuan B, Yan Y, Jiang S and Wang
T: Integrated analyses of copy number variations and gene
differential expression in lung squamous-cell carcinoma. Biol Res.
48:472015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang S, Li M, Ji H and Fang Z: Landscape
of transcriptional deregulation in lung cancer. BMC Genomics.
19:4352018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cheng H, Yang X, Si H, Saleh AD, Xiao W,
Coupar J, Gollin SM, Ferris RL, Issaeva N, Yarbrough WG, et al:
Genomic and transcriptomic characterization links cell lines with
aggressive head and neck cancers. Cell Rep. 25:1332–1343.e5. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yoshida M, Yokota E, Sakuma T, Yamatsuji
T, Takigawa N, Ushijima T, Yamamoto T, Fukazawa T and Naomoto Y:
Development of an integrated CRISPRi targeting ΔNp63 for treatment
of squamous cell carcinoma. Oncotarget. 9:29220–29232. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li X, Shi G, Chu Q, Jiang W, Liu Y, Zhang
S, Zhang Z, Wei Z, He F, Guo Z and Qi L: A qualitative
transcriptional signature for the histological reclassification of
lung squamous cell carcinomas and adenocarcinomas. BMC Genomics.
20:8812019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bednarek K, Kostrzewska-Poczekaj M,
Szaumkessel M, Kiwerska K, Paczkowska J, Byzia E, Ustaszewski A,
Janiszewska J, Bartochowska A, Grenman R, et al: Downregulation of
CEACAM6 gene expression in laryngeal squamous cell carcinoma is an
effect of DNA hypermethylation and correlates with disease
progression. Am J Cancer Res. 8:1249–1261. 2018.PubMed/NCBI
|
|
73
|
Duxbury MS, Matros E, Clancy T, Bailey G,
Doff M, Zinner MJ, Ashley SW, Maitra A, Redston M and Whang EE:
CEACAM6 is a novel biomarker in pancreatic adenocarcinoma and PanIN
lesions. Ann Surg. 241:491–496. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zang M, Zhang Y, Zhang B, Hu L, Li J, Fan
Z, Wang H, Su L, Zhu Z, Li C, et al: CEACAM6 promotes tumor
angiogenesis and vasculogenic mimicry in gastric cancer via FAK
signaling. Biochim Biophys Acta. 1852:1020–1028. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Borczuk AC, Gorenstein L, Walter KL,
Assaad AA, Wang L and Powell CA: Non-small-cell lung cancer
molecular signatures recapitulate lung developmental pathways. Am J
Pathol. 163:1949–1960. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Cui S, Cheng Z, Qin W and Jiang L:
Exosomes as a liquid biopsy for lung cancer. Lung Cancer.
116:46–54. 2018. View Article : Google Scholar : PubMed/NCBI
|