Introduction
Renal cell carcinoma (RCC) accounts for >90% of
renal cancers (1). Among the RCC
subtypes, renal clear cell carcinoma (ccRCC) accounts for ~80% of
all cases (2). The effects of
radiotherapy and chemotherapy on RCC remain unsatisfactory
(3), and despite following
nephrectomy, the risk of recurrence or metastasis is up to 40%
(4,5). Thus, it remains essential to identify
novel therapeutic targets and potential prognostic biomarkers.
SIRT6 is a member of the silent information
regulatory protein family, and is a histone deacetylase and
ADP-ribosyltransferase protease that depends on nicotinamide
adenine dinucleotide (6).
SIRT6 plays an important role in several biological
processes, including transcriptional regulation, glucose/lipid
metabolism, DNA damage repair and life span regulation (6,7).
Increasing evidence suggests that SIRT6 expression is
closely associated with the occurrence and development of different
types of cancer (8,9). For example, SIRT6 plays a key
regulatory role in liver cancer (10), lung cancer (11), breast cancer (12), colorectal cancer (13) and reproductive system cancer
(14,15).
It has also been suggested that SIRT6 may
play a dual role in cancers (16).
For example, in non-small cell lung cancer (NSCLC), SIRT6
suppresses Twist1 expression and thereby inhibits the proliferation
of NSCLC cells (11). Conversely,
SIRT6 interacts with Ku70 in liver cancer, which promotes
its deacetylation to block Bax expression and thereby potentiates
its mitochondrial translocation to inhibit apoptotic cell death of
liver cancer cells (17). Currently,
the potential role of SIRT6 and its underlying molecular
mechanisms in renal cancer remain unknown. Therefore, in the
present study, the expression pattern, clinical significance and
biological function of SIRT6 in ccRCC was investigated.
Materials and methods
Human samples
A total of 60 pairs of ccRCC tissues and adjacent
normal tissues (≥2 cm away from the edge of the tumor site) used in
the present study were obtained from patients who were
pathologically diagnosed with ccRCC and who had partial (47 cases)
or radical nephrectomy (13 cases) between May 2018 and November
2019 at the First Hospital of China Medical University (Shenyang,
China). The average age of the patients was 67.6 years (age range,
28–80 years) and there were 37 males and 23 females. Tissue samples
were stored at −80°C until subsequent experimentation.
The proteins extracted from 20 pairs of tissue
samples were assessed via western blot analysis to detect
SIRT6 protein expression in ccRCC tissues and adjacent
normal tissues. According to the renal cancer stage defined in the
eighth edition of American Joint Committee on Cancer (18), the remaining 40 patients with ccRCC
were divided into two groups, TNM (n=28; I–II) or TNM (n=12;
III–IV). The RNAs extracted from 40 pairs of tissue samples were
assessed via reverse transcription-quantitative (RT-q)PCR analysis,
and the association between SIRT6 expression and the
clinicopathological characteristics of patients with ccRCC was
assessed using the χ2 test. The present study was
approved by the Ethics Committee of the First Hospital of China
Medical University (Institutional review board no. 2018-64-2;
Shenyang, China), and performed in accordance with the Declaration
of Helsinki (19). Written informed
consent was provided by all patients prior to the study start.
Cell culture
ccRCC-derived 769-P and 786-O cells were purchased
from Shanghai Institute for Biological Sciences, Chinese Academy of
Sciences. Cells were maintained in RPMI-1640 medium supplemented
with 10% heat-inactivated fetal bovine serum (FBS, both purchased
from Gibco; Thermo Fisher Scientific, Inc.), 100 units/ml
penicillin and 100 µg/ml streptomycin (Thermo Fisher Scientific,
Inc.), at 37°C with 5% CO2.
Cell transfection
Small interfering (si)RNA targeting two different
sites of SIRT6 (siRNA-SIRT6#1 and siRNA-SIRT6#2) and its negative
control (siRNA-NC) were purchased from Shanghai GenePharma Co.,
Ltd. siRNA was transfected into the cells at a final concentration
of 40 pM, according to the manufacturer's instructions. ccRCC cells
were transfected using Lipofectamine® RNAiMAX
(Invitrogen; Thermo Fisher Scientific, Inc.) and opti-MEM (Gibco;
Thermo Fisher Scientific, Inc.). After transfection, the cells were
incubated at 37°C for 48 h for subsequent cell experiments. The
sequences of the siRNAs are listed in Table I.
 | Table I.Sequences of siRNAs and shRNAs, and
the primers used for quantitative PCR. |
Table I.
Sequences of siRNAs and shRNAs, and
the primers used for quantitative PCR.
| A, siRNA names and
sequences |
|---|
| siRNA | Sequence
(5′-3′) |
| si-NC forward |
UUCUCCGAACGUGUCACGUTT |
| si-NC reverse |
ACGUGACACGUUCGGAGAATT |
| si-SIRT6#1
forward |
GUGGAAGAAUGUGCCAAGUTT |
| si-SIRT6#1
reverse |
ACUUGGCACAUUCUUCCACTT |
| si-SIRT6#2
forward |
GAAGAAUGUGCCAAGUGUATT |
| si-SIRT6#2
reverse |
UACACUUGGCACAUUCUUCTT |
|
| B, shRNA names
and sequences |
|
| shRNA | Sequence
(5′-3′) |
| sh-NC |
TTCTCCGAACGTGTCACGT |
| sh-SIRT6 |
GAAGAATGTGCCAAGTGTA |
|
| C, Primer names
and sequences |
|
| Primer | Sequence
(5′-3′) |
| SIRT6 forward |
CCATCCTAGACTGGGAGGACT |
| SIRT6 reverse |
GGATCTGCAGCGATGTACCC |
| β-actin
forward |
CATGCCATCCTGCGTCTGGAC |
| β-actin
reverse |
CAGGCAGCTCGTAGCTCTTCTCC |
B-cell lymphoma 2 (Bcl-2) overexpression vectors
(pcDNA3.0-Bcl-2) and empty vectors (pcDNA3.0) were purchased from
Obio Technology Co., Ltd., and ccRCC cells were transfected using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and opti-MEM (Gibco; Thermo Fisher Scientific,
Inc.). Transfection efficiency was verified by western blot
analysis.
In the rescue experiment, the 769-P cells were
transfected with si-SIRT6#1 and incubated at 37°C with 5%
CO2 for 24 h, then transfected with Bcl-2 overexpression
vectors and incubated under the same conditions. Transfection
efficiency was verified using western blot analysis 24 h later,
then the Cell Counting Kit-8 experiment was performed.
Lentiviral short hairpin (sh)RNA
vector construction and infection
To construct the ccRCC cells with stable
SIRT6 knockdown in in vivo animal studies, a
lentiviral shRNA vector targeting the human SIRT6 gene
(pLenti-CMV-shSIRT6-PGK-Puro) was constructed by Obio Technology
Co., Ltd. The sequences used to construct the targeting
sh-SIRT6 and sh-NC are listed in Table I.
293T cells were purchased from the Cell Bank of Type
Culture Collection of Chinese Academy of Sciences and maintained in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS, at 37°C with 5% CO2. Following incubation for 24 h
at 37°C, the cells were transfected with 0.5 µg sh-SIRT6 or sh-NC
using FuGENE™ HD transfection reagent (Promega Corporation) and the
Packaging Plasmid Mix (Sigma-Aldrich; Merck KGaA). Following
incubation overnight at 37°C, the media were replaced with 10 ml
fresh medium and the virus-containing supernatants (sh-NC and
sh-SIRT6) were collected after 48 h.
769-P cells were transfected with sh-NC and
sh-SIRT6, respectively, at 37°C for 72 h. To establish
stable cell lines, 2 µg/ml puromycin (cat. no. A1113803; Thermo
Fisher Scientific, Inc.) was added to the medium for 1 week
following transfection with the lentiviral vectors. Knockdown
efficiency was verified via RT-qPCR and western blot analyses.
RT-qPCR
Total RNA was extracted from tissues using
TRIzol® reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The concentration and
purity of the RNA solution were detected using a NanoDrop 2000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using the
PrimeScript™ RT Master Mix (cat. no. RR036A; Takara Biotechnology
Co., Ltd.), at 37°C for 15 min and 85°C for 5 sec, and was held at
4°C until further use. qPCR was subsequently performed using the
SYBR premix ExTaq™ kit (cat. no. RR420A; Takara Biotechnology Co.,
Ltd.). Relative expression levels were detected using a
LightCycler™480 II system (Roche Diagnostics) and normalized to the
internal reference gene β-actin. The relative expression levels
were calculated using the 2−ΔΔCq method (20). The following thermocycling conditions
were used: Initial denaturation at 95°C for 5 min, 95°C for 10 sec
and 60°C for 30 sec for 45 cycles, with a final cycle of 95°C for
15 sec, 60°C for 1 min and 40°C for 30 sec. The primer sequences
used for qPCR are listed in Table
I.
For the expression of SIRT6 in patients with ccRCC
in different clinical stages, β-actin was used as internal controls
for expression data normalization. The expression of tumor tissue
was determined using the 2−ΔΔCq method by comparing with
the adjacent normal tissue.
Western blot analysis
Total protein was extracted from the 769-P and 786-O
cells using RIPA lysis buffer (Sigma-Aldrich; Merck KGaA). Protein
concentration was determined using the BCA method (Beijing Solarbio
Science & Technology Co., Ltd.) and 40 µg protein/lane was
separated by 10% SDS-PAGE. The separated proteins were subsequently
transferred onto PVDF membranes (EMD Millipore) and blocked with 5%
skimmed milk at room temperature for 30 min. Subsequently,
appropriate primary antibody dilutions were prepared and incubated
with the membranes overnight at 4°C. The antibodies included rabbit
anti-SIRT6 (cat. no. 2590s; Cell Signaling Technology,
Inc.), rabbit anti-Bcl-2 (cat. no. ab182858; Abcam), rabbit
anti-Bax (cat. no. ab32503; Abcam) and rabbit anti-β-actin (cat.
no. BS0061; BIOSS) (all 1:1,000). After washing with PBS three
times, the membranes were incubated with HRP-conjugated goat
anti-rabbit IgG (1:5,000; cat. no. sc2357; Santa Cruz
Biotechnology, Inc.) for 1 h at room temperature. And protein bands
were visualized using the Pierce™ ECL Plus western blotting
substrate (Thermo Fisher Scientific, Inc.). To determine the
expression intensity of SIRT6 relative to normal adjacent tissues
more accurately, the relative expression levels were detected using
ImageJ v1.51 software (National Institutes of Health) and
normalized to the internal reference gene β-actin. Subsequently,
the ratio of the tumor tissue compared to the adjacent normal
tissue was calculated as the relative expression level.
CCK-8 assay
The effect of SIRT6 on cell proliferation was
assessed via the CCK-8 assay. Cells were collected 24 h
post-transfection and seeded into a 96-well plate at a density of
1,500 cells/well. Following incubation for 4 h at 37°C, with 5%
CO2, 10 µl CCK-8 reagent (Vazyme Biotech Co., Ltd.) was
added to each well and the reaction mixtures were incubated for an
additional 3 h at 37°C, with 5% CO2. Cell proliferation
was subsequently analyzed at a wavelength of 450 nm, using a
microplate reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
The effect of SIRT6 on cell proliferation and
the association between SIRT6 and cisplatin sensitivity of ccRCC
were assessed using the colony formation assay. Cells were
collected 24 h post-transfection and seeded into 6-well plates at a
density of 1,500 cells/well. Following incubation for 10 days at
37°C with 5% CO2, cells were fixed with 4%
paraformaldehyde solution for 30 min at room temperature, washed
three times with PBS and subsequently stained with 0.1% crystal
violet (Sigma-Aldrich; Merck KGaA) for 30 min at room temperature.
Cells were extensively re-washed with PBS and images were captured
using a light microscope (magnification, ×40). The number of viable
colonies was defined as >50 cells/colony. The results were
quantified using ImageJ v1.51 software (National Institutes of
Health).
For the cisplatin sensitivity assay, the 769-P cells
were collected 24 h post-transfection and seeded into 6-well plates
at a density of 2,000 cells/well, then incubated with DMSO or 2.5
µM cisplatin at 37°C with 5% CO2 for 2 weeks.
Subsequently, the cells were fixed, stained and counted according
to the aforementioned method.
Transwell assay
The Transwell assay was performed to assess the
effect of SIRT6 on the migratory and invasive abilities of
ccRCC cells. Cells were collected 24 h post-transfection. Briefly,
1×105 cells were plated in the upper chambers of
Transwell plates (Corning, Inc.) in 200 µl serum-free RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.). For the invasion
assay, Transwell membranes were precoated with 55 µl Matrigel (1:7
dilution, Corning, Inc.) for 30 min at 37°C, RPMI-1640 medium (600
µl) supplemented with 10% FBS was plated in the lower chambers.
Following incubation for 24 h at 37°C and 5% CO2, the
cells were fixed with 4% formaldehyde at room temperature for 30
min and subsequently stained with 0.1% crystal violet at room
temperature for 30 min. Cell were washed three times with PBS and
the unmigrated cells were removed using cotton swabs. Images were
captured using a Leica DM3000 microscope (Leica Microsystems GmbH;
magnification, ×40 and ×100). The numbers of cells were counted in
≥5 independent fields of view, using ImageJ v1.51 software
(National Institutes of Health).
In vivo animal studies
A total of 42 male BALB/c nude mice (body weight,
18–20 g; aged 4–6 weeks) were purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. Of these, 30 mice were used
for pre-experiments to determine the ccRCC cell lines (769-P or
786-O), whether to use Matrigel and different inoculated cell
numbers. The remaining 12 mice were used for the in vivo
tumor formation experiment. All animal experiments were approved by
the Ethics Committee of Medical Experimental Animal Welfare of
China Medical University (approval no. 2019227; Shenyang, China),
and performed at the Experimental Animal Department of China
Medical University.
The experimental mice were classified into two
groups (n=6 mice/group), sh-SIRT6 and sh-NC. All mice
received subcutaneous lateral injection of 769-P cells
(5×106 cells). sh-SIRT6 or sh-NC were suspended
in 200 µl serum-free RPMI-1640 medium/Matrigel (1:1 mixture). All
mice were housed and maintained under specific pathogen-free
conditions in clear cages with free access to food and water, at
room temperature (22–25°C), with 50% humidity and 12-h light/dark
cycles. Tumor formation was monitored daily. All mice were
anaesthetized with an intraperitoneal injection of 50 mg/kg
pentobarbital sodium (Sigma-Aldrich; Merck KGaA) 2 weeks
post-injection, and cervical vertebrae were dislocated. Following
euthanasia, lack of heartbeat was used to verify mortality. The
maximum diameter of the observed tumors was 14 mm. No mice had
multiple tumors. The tumors were removed and weighed, and whole
tissue lysates were extracted from each tumor for western blot
analysis.
Bioinformatics analysis
To determine the expression pattern and clinical
characteristics of SIRT6, a public dataset [The Cancer
Genome Atlas (TCGA)-KIRC cohort] was analyzed in the UALCAN
database (http://ualcan.path.uab.edu). TCGA
database (https://portal.gdc.cancer.gov) was used to download
clinical data of patients with ccRCC and SIRT6 expression
data. The association between SIRT6 expression level and
survival in patients with ccRCC was assessed using GraphPad Prism
v8 software (GraphPad Software, Inc.). The association between
SIRT6 expression and survival in patients with pan-cancer was
assessed using Kaplan-Meier plotter database (http://kmplot.com/analysis). The Kaplan-Meier plot was
created in the following forms: i) All cases (based on the median
SIRT6 expression value) and ii) Stage I and II/Stage III and IV
(based on the best grouping with the smallest P-value). P<0.05
was considered to indicate a statistically significant difference
(only statistical results are presented). The Human Protein Atlas
database (https://www.proteinatlas.org) was used to compare
SIRT6 protein expression in ccRCC tissues and normal
tissues.
Statistical analysis
Statistical analysis was performed using SPSS 21.0
software (IBM Corp.). All experiments were performed in triplicate
and data are presented as the mean ± standard deviation. Survival
analysis was performed using the Kaplan-Meier method and log-rank
test. The χ2 test was used to assess the association
between SIRT6 expression and the clinicopathological
characteristics of patients with ccRCC. Paired Student's t-test was
used to compare SIRT6 expression in tumor tissues (stage
I/II or III/IV) and matched adjacent normal tissues. One-way ANOVA
followed by Tukey's post hoc test were used to compare differences
between multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
SIRT6 expression is associated with
poor prognosis of patients with ccRCC
To determine the role of SIRT6 in ccRCC,
mRNA-seq data within the UALCAN database was used to determine
SIRT6 expression in ccRCC. The results demonstrated that
SIRT6 expression was significantly higher in ccRCC tissues
compared with normal tissues, and its high expression was dependent
on cancer stage (Fig. 1A). In
addition, ccRCC tissues at all stages highly expressed SIRT6
compared with normal tissues (P<0.001). To determine the
association between SIRT6 expression and the prognosis of
patients with ccRCC, Kaplan-Meier survival analysis was performed
using the TCGA-KIRC dataset. A total of 342 cases were classified
into two groups, based on the median SIRT6 expression value,
with 171 cases in the high SIRT6 group and 171 cases in the
low SIRT6 group. As presented in Fig. 1B, high SIRT6 expression was
closely associated with poor prognosis of patients with ccRCC
(log-rank, P=0.0009). In addition, the survival rates of patients
with high SIRT6 expression were lower compared with those
with low SIRT6 expression level, regardless of the disease
stage (P=0.0595 and P=0.0127, respectively; Fig. 1C and D). The association between
SIRT6 expression and the clinicopathological characteristics
of patients in the dataset was assessed to determine the clinical
significance of SIRT6 in ccRCC. The results demonstrated
that SIRT6 expression was positively associated with TNM
stage and distant metastasis (P<0.05; Table II). The patient information used to
perform Kaplan-Meier survival analysis is presented in Table SI. The Kaplan-Meier plotter database
was used to determine the survival rates of patients with cancer in
TCGA database. As presented in Fig.
S1, SIRT6 expression was associated with poor prognosis
of patients with ccRCC and hepatocellular carcinoma. Conversely,
SIRT6 expression was associated with favorable prognosis in
patients with bladder carcinoma, cervical squamous cell carcinoma,
head and neck squamous cell carcinoma, pancreatic adenocarcinoma,
stomach adenocarcinoma and uterine corpus endometrial
carcinoma.
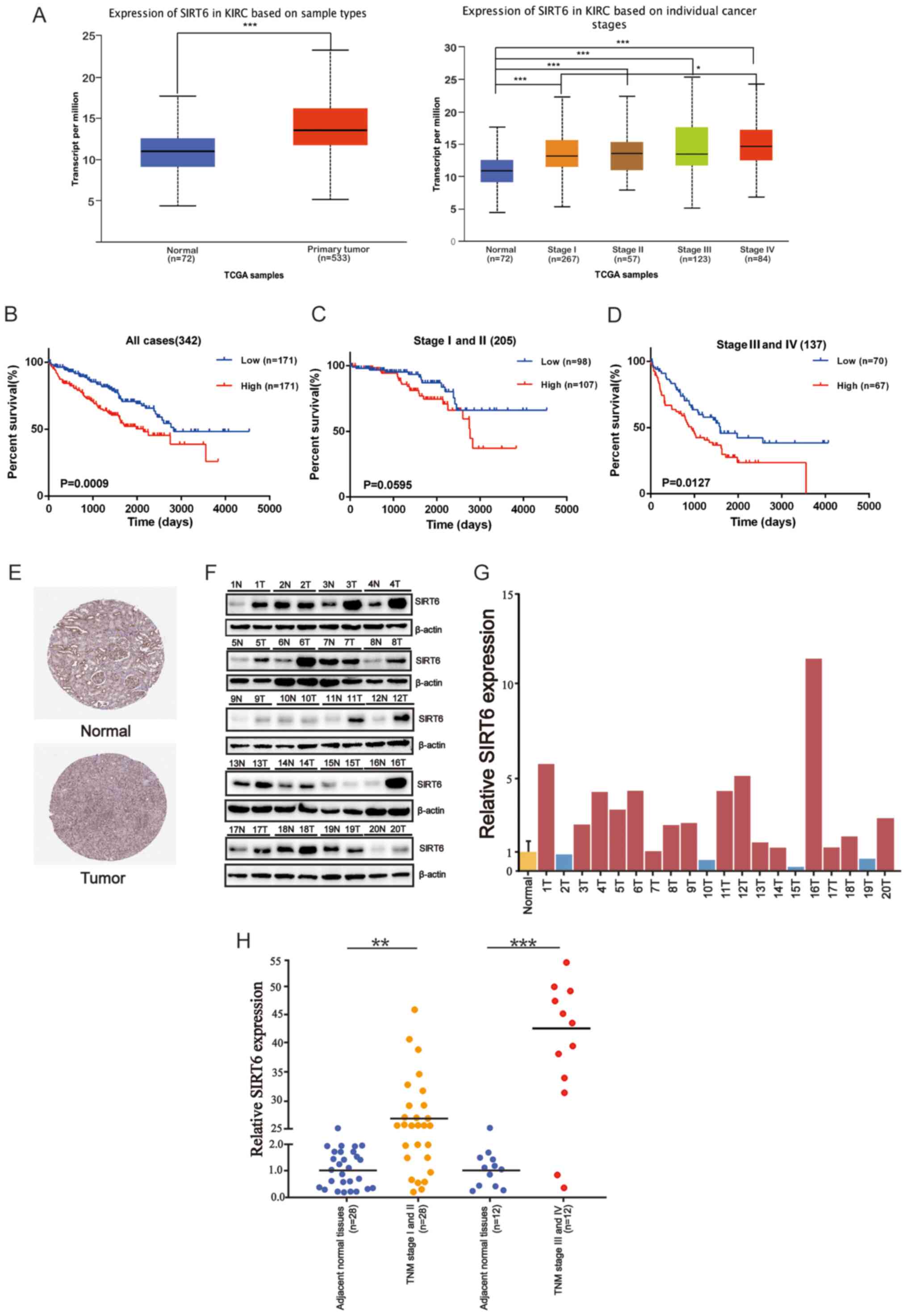 | Figure 1.Upregulation of SIRT6
expression is associated with poor prognosis of ccRCC. (A)
Differential expression of SIRT6 between ccRCC tissues and
normal tissues. (B) Kaplan-Meier plot and log-rank test were used
to assess overall survival time in patients with ccRCC, based on
SIRT6 expression in TCGA databased and the clinical
information of the patients with ccRCC. (C) Kaplan-Meier plot
presenting the survival of the patients at stages I and II. (D)
Kaplan-Meier plot presenting the survival of patients at stages III
and IV. (E) Representative images of immunohistochemistry staining
of SIRT6 in paired cancer tissues and adjacent normal
tissues via the Human Protein Atlas. (F) Western blot analysis was
performed to detect SIRT6 protein expression in ccRCC tissues and
adjacent normal tissues. β-actin was used as the loading control.
(G) Densitometric quantitative analysis of SIRT6 protein expression
in 20 pairs of ccRCC tissues and adjacent normal tissues. (H)
SIRT6 mRNA expression in 40 pairs of cancer tissues and
adjacent normal tissues. Control refers to normal kidney tissues,
and SIRT6 expression was normalized to the control.
SIRT6 mRNA expression was calculated using the comparative
Ct method. Relative expression intensity values were calculated
using the 2−∆∆Cq method. *P<0.05, **P<0.01,
***P<0.001. SIRT6, sirtuin 6; ccRCC, clear cell renal cell
carcinoma; TCGA, The Cancer Genome Atlas; KIRC, Kidney Renal Clear
Cell Carcinoma; N, normal; T, tumor; TNM,
tumor-node-metastasis. |
 | Table II.Association between SIRT6 expression
and the clinicopathological characteristics in patients with clear
cell renal cell carcinoma (n=342). |
Table II.
Association between SIRT6 expression
and the clinicopathological characteristics in patients with clear
cell renal cell carcinoma (n=342).
|
| SIRT6 expression
(TCGA) |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | <Median
(n=171) | ≥Median
(n=171) | Total number of
patients, n | P-value |
|---|
| Age, years |
|
|
| 0.4479 |
|
<60 | 76 | 83 | 159 |
|
|
≥60 | 95 | 88 | 183 |
|
| Sex |
|
|
| 0.3069 |
|
Male | 116 | 107 | 223 |
|
|
Female | 55 | 64 | 119 |
|
| TNM stage |
|
|
| 0.0058a |
|
I–II | 115 | 90 | 205 |
|
|
III–IV | 56 | 81 | 137 |
|
| Distant
metastasis |
|
|
| 0.0122b |
|
Negative | 144 | 125 | 269 |
|
|
Positive | 27 | 46 | 73 |
|
The Human Protein Atlas database was used to compare
SIRT6 protein expression between ccRCC tissues and normal
tissues. As presented in Fig. 1E,
SIRT6 immunohistochemical staining was substantially
stronger in ccRCC tissues compared with normal tissues. Western
blot analysis was subsequently performed to verify these results,
using tissue samples obtained from 20 patients with ccRCC. As
presented in Fig. 1F, SIRT6
protein expression was significantly higher in ccRCC tissues
compared with normal tissues. Quantitative densitometer analysis
demonstrated that SIRT6 was highly expressed in 16/20 pairs of
ccRCC tissues (P<0.01 or P<0.001; Fig. 1G). In addition, RT-qPCR analysis was
performed to detect SIRT6 expression in 40 pairs of ccRCC
tissues and adjacent normal tissues. The results demonstrated that
SIRT6 expression was upregulated 27-fold on average in the TNM
(I–II) stages of ccRCC and 43-fold on average in the TNM (III–IV)
stages (P<0.01 or P<0.001; Fig.
1H). To assess the significance of SIRT6 expression in ccRCC,
the association between SIRT6 expression and the
clinicopathological characteristics were assessed using ccRCC
tissues. The results demonstrated that high SIRT6 expression was
significantly associated with higher tumor TNM stage and distant
metastasis in ccRCC (Table III).
However, no significant differences were observed between SIRT6
expression and age or sex. These results suggest that SIRT6
expression is upregulated, and significantly associated with TNM
stage and distant metastasis in ccRCC. This is consistent with the
results from TCGA database (Table
II). Taken together, these results suggest that SIRT6 acts as a
proto-oncogene in ccRCC.
 | Table III.Association between SIRT6 expression
and the clinicopathological characteristics of patient with ccRCC
(n=40). |
Table III.
Association between SIRT6 expression
and the clinicopathological characteristics of patient with ccRCC
(n=40).
|
| SIRT6
expression |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | ≥Median <Median
(n=20) | Total number
(n=20) | of patients, n | P-value |
|---|
| Age, years |
|
|
| 0.5073 |
|
<60 | 14 | 12 | 26 |
|
|
≥60 | 6 | 8 | 14 |
|
| Sex |
|
|
| 0.5186 |
|
Male | 13 | 11 | 24 |
|
|
Female | 7 | 9 | 16 |
|
| TNM stage |
|
|
| 0.0058b |
|
I–II | 18 | 10 | 28 |
|
|
III–IV | 2 | 10 | 12 |
|
| Distant
metastasis |
|
|
| 0.0177a |
|
Negative | 19 | 13 | 32 |
|
|
Positive | 1 | 7 | 8 |
|
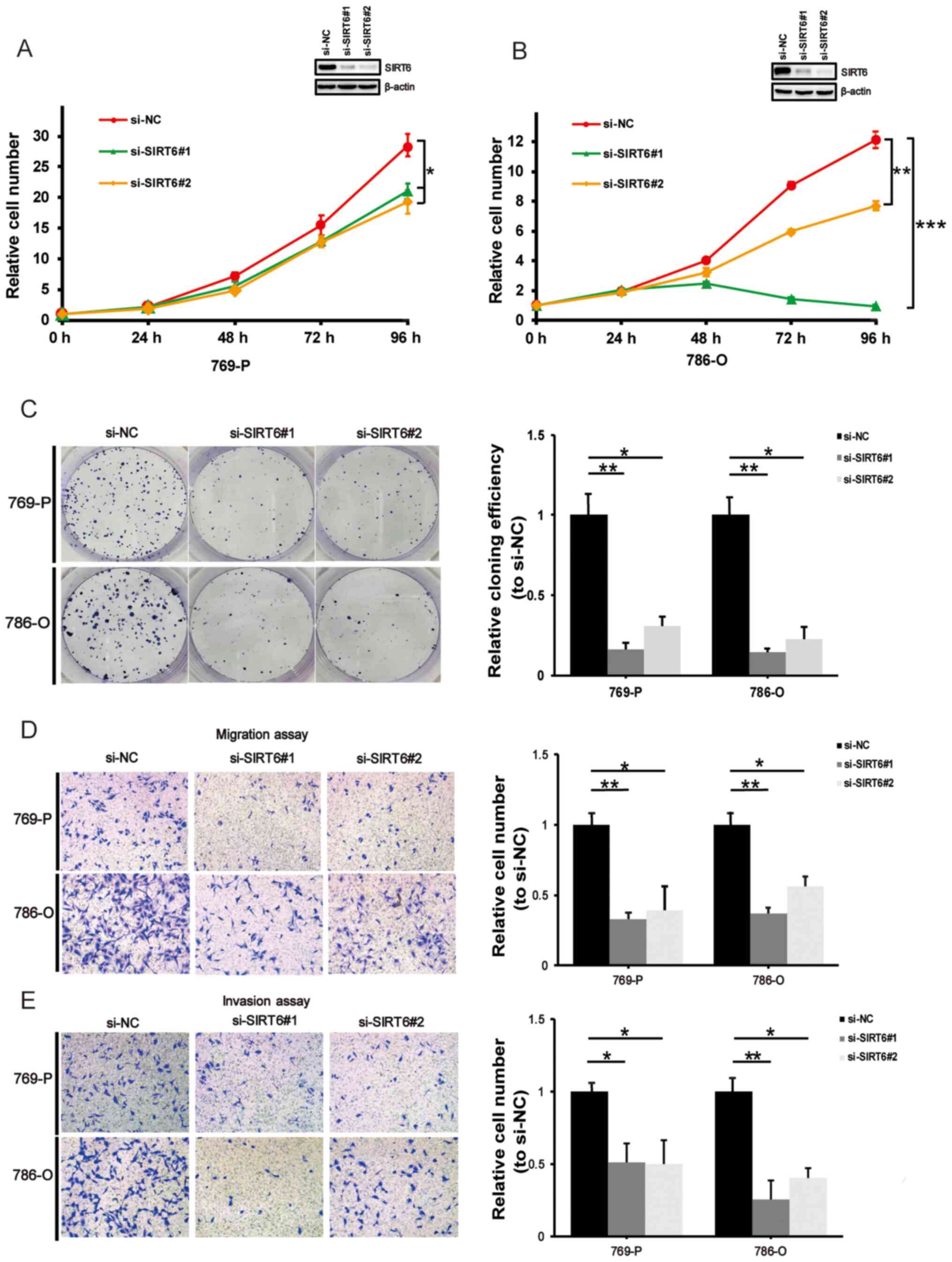
SIRT6 knockdown inhibits ccRCC cell
proliferation, migration and invasion
The effect of SIRT6 on cell proliferation,
migration and invasion in ccRCC was assessed. ccRCC-derived 769-P
and 786-O cells were transfected with si-NC or si-SIRT6. As
expected, SIRT6 knockdown significantly decreased the
proliferative rate (P<0.05; Fig.
2A and P<0.01 or P<0.001; Fig.
2B) and colony formation ability (P<0.05 or P<0.01;
Fig. 2C) of 769-P and 786-O cells.
Consistent with these results, the migration and the invasion
assays demonstrated that SIRT6 knockdown attenuated the cell
migratory and invasive abilities (P<0.05 or P<0.01) (Fig. 2D and E).
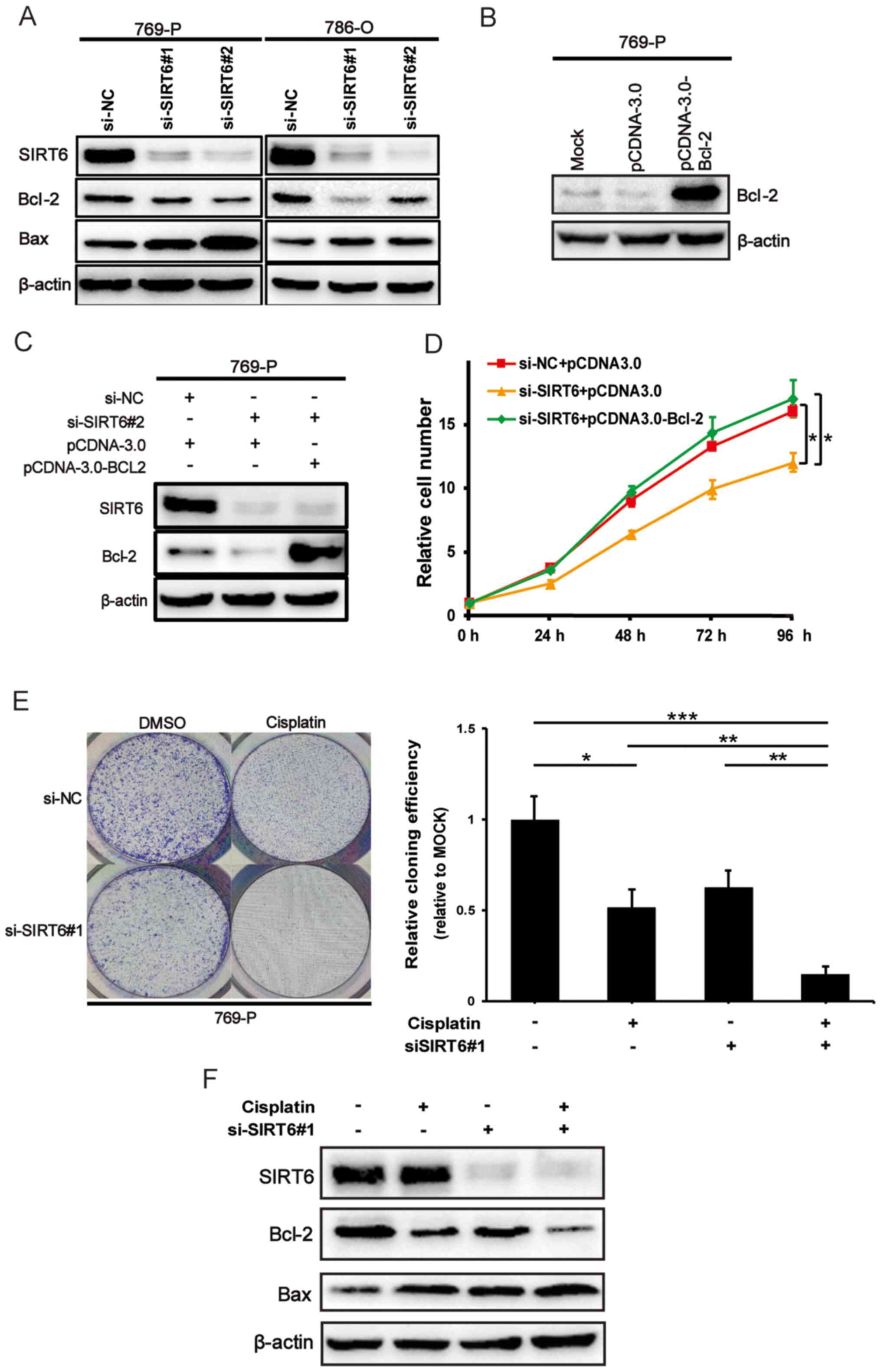
SIRT6 promotes ccRCC cell
proliferation via Bcl-2
It has been demonstrated that SIRT6 prevents
the mitochondrial translocation of Bax in liver cancer cells, and
SIRT6-mediated deacetylation Ku70 attenuates apoptotic cell
death of hepatocellular carcinoma (HCC) cells (17), suggesting that SIRT6 may
participate in the pro-survival Bcl-2 pathway. To confirm this
hypothesis, 769-P and 786-O cells were transfected with the si-NC
or si-SIRT6 in the present study, and western blot analysis
was performed to detect the protein expression levels of Bcl-2 and
Bax. As presented in Fig. 3A, Bcl-2
expression notably decreased, while Bax expression notably
increased following SIRT6 knockdown in ccRCC cells. Bcl-2
expression was subsequently overexpressed in 769-P cells, and the
transfection efficiency is presented in Fig. 3B. Bcl-2 expression was overexpressed
following SIRT6 knockdown. Notably, the results demonstrated
that SIRT6 depletion-mediated decrease in the proliferative
ability of 769-P cells was restored following overexpression of
Bcl-2 (P<0.05) (Fig. 3C and D).
Taken together, these results suggest that SIRT6 promotes
ccRCC cell proliferation, at least in part, through regulation of
the pro-survival Bcl-2 pathway.
SIRT6 knockdown enhances cisplatin
sensitivity of ccRCC cells
The results of the present study suggest that
SIRT6 acts as a proto-oncogene in ccRCC. Thus, it was
assessed whether SIRT6 affects the chemosensitivity of ccRCC
cells via the colony formation assay. SIRT6-depleted 769-P
cells were treated with or without 2.5 µM cisplatin. After 2 weeks
of treatment, viable cells colonies were stained with crystal
violet and observed under a microscope. As presented in Fig. 3E (P<0.05 or P<0.01 or
P<0.0001), the number of cell colonies in cisplatin-exposed
SIRT6-knockdown cells were significantly smaller compared
with the control cells treated with cisplatin. In addition, western
blot analysis was performed to detect the related protein levels in
these four groups. As presented in Fig.
3F, Bcl-2 protein expression significantly decreased in
SIRT6 knockdown cells exposed to cisplatin, while Bax
protein expression significantly increased. Collectively, these
results suggest that SIRT6 knockdown enhances the
sensitivity of ccRCC cells to cisplatin.
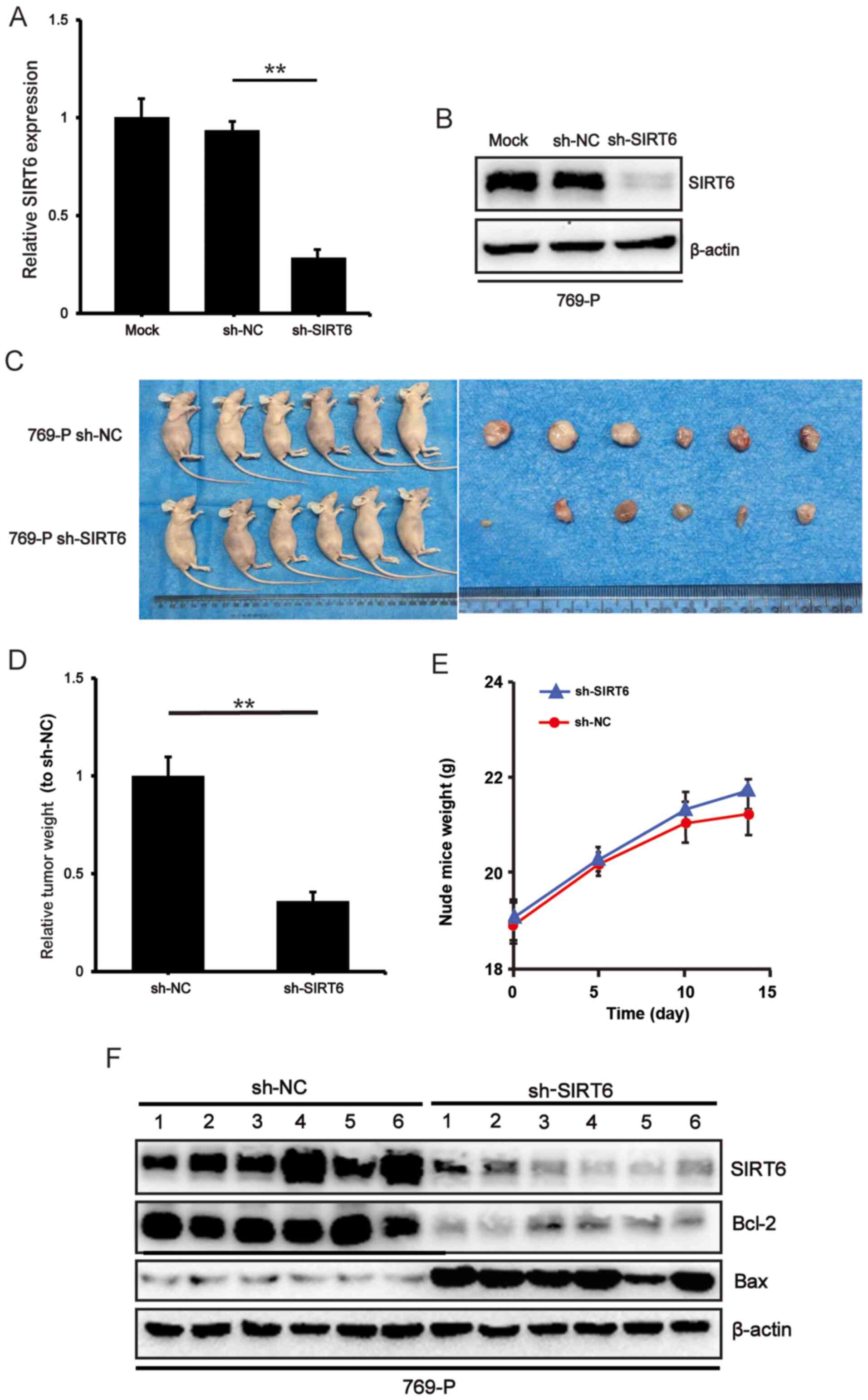
SIRT6 knockdown inhibits tumor
proliferation in vivo
To determine the potential effect of SIRT6
knockdown on tumor growth in vivo, RT-qPCR and western blot
analyses were performed to verify the efficiency of SIRT6 knockdown
on stable 769-P cells (P<0.01) (Fig.
4A and B). A BALB/c xenograft model was subsequently
established via subcutaneous injection of SIRT6-depleted
769-P cells. The representative images of tumors in the control and
experimental groups were taken 2 weeks after injection. The results
demonstrated that the tumor weight was significantly lower in the
sh-SIRT6 group compared with the sh-NC group (P<0.01)
(Fig. 4C and D), suggesting that
SIRT6 knockdown notably decreases the tumor growth rate
arising from 769-P cells in vivo. The average body weight of
both groups of mice was recorded over a 2-week period and the
changes in body weight were plotted (Fig. 4E). In the last 3 days of the
experiment, a small number of mice lost weight; however, no
statistically significant differences were observed between the two
groups. Based on the conclusion that SIRT6 plays a role as an
oncogene in ccRCC, If the tumor continued to grow in the mice for a
week, the weight of mice may drop >10% of the starting weight,
and mice in the sh-NC group may experience more significant weight
loss compared with that in mice in the sh-SIRT6 group. Western blot
analysis demonstrated that the protein expression levels of Bcl-2
and Bax were downregulated and upregulated in SIRT6-depleted
tumors compared with the control tumors, respectively (Fig. 4F). Taken together, these results
suggest that SIRT6 prevents the tumor forming ability of
ccRCC cells by suppressing the pro-survival Bcl-2 pathway.
Discussion
To the best of our knowledge, the present study was
the first to demonstrate that SIRT6 participates in the
acquisition and/or promotion of the malignant properties of ccRCC
through potentiation of the pro-survival Bcl-2 pathway. Taken
together, the results of the present study suggest that
SIRT6 acts as a proto-oncogene in ccRCC, and may be a
potential therapeutic target of ccRCC.
Increasing evidence suggests that SIRT6 acts
as a tumor suppressor in lung, breast and pancreatic cancers
(11,12,21).
Conversely, it has also been demonstrated that SIRT6 acts as
a proto-oncogene in other types of cancer. For example, Ming et
al (22) reported that UV
exposure-mediated induction of SIRT6 stimulates
cyclooxygenase 2 expression through inhibition of the
adenylate-activated protein kinase pathway, thereby promoting the
proliferation, as well as the survival, of the epidermal layer in
skin. Notably, skin tumor formation was significantly attenuated in
SIRT6-knockout mice. In addition, Bauer et al
(23) demonstrated that
SIRT6-induced cytokine secretion and cell motility is
promoted by activation of calcium channels in pancreatic cancer
cells. Khongkow et al (24)
reported that SIRT6 is highly expressed in paclitaxel- and
epirubicin-resistant MCF-7 breast cancer cells compared with their
parental cells. According to their results, gene silencing and
overexpression of SIRT6 increases and decreases the
sensitivity to paclitaxel and epirubicin, respectively. In
addition, although renal cell carcinoma is not sensitive to
radiotherapy and chemotherapy, cisplatin is one of the most
extensive and effective chemotherapeutic agents for several types
of human cancer, including testicular, bladder, ovarian,
colorectal, lung and head and neck cancers (25–27).
Cisplatin acts by directly binding to DNA to produce a
cisplatin-DNA adduct. If cisplatin-DNA adducts are not efficiently
processed by the cellular repair mechanism, the programmed cell
death pathway is initiated. Molecular mechanisms that disrupt these
pre-apoptotic signals are thought to be responsible for tumor
resistance to chemotherapy (28).
Thus, cisplatin has been the standard experimental drug to study
the molecular mechanism of chemosensitivity in renal cell carcinoma
(29–31). In accordance with these observations,
the results of the present study demonstrated that high
SIRT6 expression was associated with poor prognosis of
patients with ccRCC, and SIRT6 knockdown decreased the
proliferative, migratory and invasive abilities of ccRCC cells, and
enhanced their sensitivity to cisplatin. Furthermore, SIRT6
knockdown suppressed tumor growth in vivo, suggesting that
SIRT6 acts as a proto-oncogene in ccRCC.
Notably, it has been demonstrated that SIRT6
prohibits cancer cell apoptosis (32–34). In
prostate cancer cells, SIRT6 knockdown decreases the
expression levels of pro-survival/anti-apoptotic Bcl-2, and
enhances their chemosensitivity (15). In liver cancer cells, SIRT6
blocks the mitochondrial translocation of pro-apoptotic Bax,
thereby decreases the apoptotic rate of HCC cells via deacetylation
of Ku70 (17). Based on the results
of the present study, SIRT6 knockdown in ccRCC cells
downregulated and upregulated Bcl-2 and Bax expression,
respectively, which enhanced their sensitivity to cisplatin. In
addition, overexpression of Bcl-2 in ccRCC cells restored
SIRT6 depletion-mediated decrease of their proliferative
rate. Bcl-2 is one of the most representative anti-apoptotic gene
products, whereby aberrant Bcl-2 expression in cancer cells is
associated with their histological types, poor prognosis of
patients, and resistance to radiotherapy and chemotherapy (35–37).
Elevated Bcl-2 expression dissociates Bax-Bax homo-dimers, and the
resultant free Bax and Bcl-2 form is more stable than Bax-Bcl-2
hetero-dimers, thereby preventing Bax-Bax homo-dimer-induced
apoptotic cell death (38,39). SIRT6-mediated differential
regulation of Bcl-2 and Bax disrupts the balance between them,
thereby suppressing mitochondrial apoptotic cell death (17).
The present study was not without limitations. For
example, downstream pathway components of SIRT6 promoting
cancer, including TRPM2 and TNF1 α, and the in vivo
verification of the effect of SIRT6 on cisplatin sensitivity
were not investigated.
In conclusion, the results of the present study
suggest that SIRT6 regulates Bcl-2 to acquire and/or promote
the malignant properties of ccRCC, and thus may act as an
attractive therapeutic target of ccRCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Haotian Xing
(Department of Urology, The First Hospital of China Medical
University, Shenyang, China) for his guidance on the bioinformatics
analysis.
Funding
The present study was supported in part by the
National Natural Science Foundation of China (grant nos. 81672523
and 81472404).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding authors upon reasonable
request.
Authors' contributions
YZ and MY designed and supervised the present
study. JA, JY, KL and ZZ performed the in vitro experiments
and collected the data, while JY and YY performed the in
vivo experiments and collected the data. JA and JY analyzed and
interpreted the results. JA, MY and YZ drafted the initial
manuscript and made substantial revisions. A, JY and YZ confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript. J
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Hospital of China Medical University
(Institutional review board no. 2018-64-2; Shenyang, China) and
performed in accordance with the Declaration of Helsinki (19). Written informed consent was provided
by all patients prior to the study start. All animal experiments
were approved by The Ethics Committee of Medical Experimental
Animal Welfare of China Medical University (approval no. 2019227;
Shenyang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gray R, Hentschke R, Isaac S, Mead R,
Ozturk A, Rieley P, Smale K and Stern R: Sampling variation of
reported results. Nature. 234:230–231. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen YL, Ge GJ, Qi C, Wang H, Wang HL, Li
LY, Li GH and Xia LQ: A five-gene signature may predict sunitinib
sensitivity and serve as prognostic biomarkers for renal cell
carcinoma. J Cell Physiol. 233:6649–6660. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Escudier B, Porta C, Schmidinger M,
Rioux-Leclercq N, Bex A, Khoo V, Gruenvald V and Horwich A: Renal
cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 27:v58–v68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lam JS, Shvarts O, Leppert JT, Pantuck AJ,
Figlin RA and Belldegrun AS: Postoperative surveillance protocol
for patients with localized and locally advanced renal cell
carcinoma based on a validated prognostic nomogram and risk group
stratification system. J Urol. 174:466–472. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, He J, Liao M, Hu M, Li W, Ouyang
H, Wang X, Ye T, Zhang Y and Ouyang L: An overview of Sirtuins as
potential therapeutic target: Structure, function and modulators.
Eur J Med Chem. 161:48–77. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haigis MC and Sinclair DA: Mammalian
sirtuins: Biological insights and disease relevance. Ann Rev
Pathol. 5:253–295. 2010. View Article : Google Scholar
|
|
8
|
Sebastián C, Zwaans BM, Silberman DM,
Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber
D, et al: The histone deacetylase SIRT6 is a tumor suppressor that
controls cancer metabolism. Cell. 151:1185–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Marquardt JU, Fischer K, Baus K, Kashyap
A, Ma S, Krupp M, Linke M, Teufel A, Zechner U, Strand D, et al:
Sirtuin-6-dependent genetic and epigenetic alterations are
associated with poor clinical outcome in hepatocellular carcinoma
patients. Hepatology. 58:1054–1064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang C, Yu Y, Huang Q and Tang K: SIRT6
regulates the proliferation and apoptosis of hepatocellular
carcinoma via the ERK1/2 signaling pathway. Mol Med Rep.
20:1575–1582. 2019.PubMed/NCBI
|
|
11
|
Han Z, Liu L, Liu Y and Li S: Sirtuin
SIRT6 suppresses cell proliferation through inhibition of Twist1
expression in non-small cell lung cancer. Int J Clin Exp Pathol.
7:4774–4781. 2014.PubMed/NCBI
|
|
12
|
Khongkow M, Olmos Y, Gong C, Gomes AR,
Monteiro LJ, Yagüe E, Cavaco TB, Khongkow P, Man EP, Laohasinnarong
S, et al: SIRT6 modulates paclitaxel and epirubicin resistance and
survival in breast cancer. Carcinogenesis. 34:1476–1486. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi W, Fitchev PS, Cornwell ML, Greenberg
J, Cabe M, Weber CR, Roy HK, Crawford SE and Savkovic SD: FOXO3
growth inhibition of colonic cells is dependent on intraepithelial
lipid droplet density. J Biol Chem. 288:16274–16281. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Colas E, Perez C, Cabrera S, Pedrola N,
Monge M, Castellvi J, Eyzaguirre F, Gregorio J, Ruiz A, Llaurado M,
et al: Molecular markers of endometrial carcinoma detected in
uterine aspirates. Int J Cancer. 129:2435–2444. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Xie QR, Wang B, Shao J, Zhang T,
Liu T, Huang G and Xia W: Inhibition of SIRT6 in prostate cancer
reduces cell viability and increases sensitivity to
chemotherapeutics. Protein Cell. 4:702–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Desantis V, Lamanuzzi A and Vacca A: The
role of SIRT6 in tumors. Haematologica. 103:1–4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tao NN, Ren JH, Tang H, Ran LK, Zhou HZ,
Liu B, Huang AL and Chen J: Deacetylation of Ku70 by SIRT6
attenuates Bax-mediated apoptosis in hepatocellular carcinoma.
Biochem Biophys Res Commun. 485:713–719. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC cancer staging manual. (8th edition).
Springer International Publishing; pp. 739–747. 2017
|
|
19
|
World Medical Association: World medical
association declaration of Helsinki: Ethical principles for medical
research involving human subjects. JAMA. 310:2191–2194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kugel S, Sebastián C, Fitamant J, Ross KN,
Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al:
SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell.
165:1401–1415. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ming M, Han W, Zhao B, Sundaresan NR, Deng
CX, Gupta MP and He YY: SIRT6 promotes COX-2 expression and acts as
an oncogene in skin cancer. Cancer Res. 74:5925–5933. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bauer I, Grozio A, Lasigliè D, Basile G,
Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A, et al:
The NAD+-dependent histone deacetylase SIRT6 promotes cytokine
production and migration in pancreatic cancer cells by regulating
Ca2+ responses. J Biol Chemistry. 287:40924–40937. 2012.
View Article : Google Scholar
|
|
24
|
Khongkow P, Gomes AR, Gong C, Man EP,
Tsang JW, Zhao F, Monteiro LJ, Coombes RC, Medema RH, Khoo US and
Lam EW: Paclitaxel targets FOXM1 to regulate KIF20A in mitotic
catastrophe and breast cancer paclitaxel resistance. Oncogene.
35:990–1002. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Prestayko AW, D'Aoust JC, Issell BF and
Crooke ST: Cisplatin (cis-diamminedichloroplatinum II).
Cancer Treat Rev. 6:17–39. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lebwohl D and Canetta R: Clinical
development of platinum complexes in cancer therapy: An historical
perspective and an update. Eur J Cancer. 34:1522–1534. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Galanski M: Recent developments in the
field of anticancer platinum complexes. Recent Pat Anticancer Drug
Discov. 1:285–295. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Siddik ZH: Cisplatin: Mode of cytotoxic
action and molecular basis of resistance. Oncogene. 22:7265–7279.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hueber PA, Waters P, Clark P, Clarke P,
Eccles M and Goodyer P: PAX2 inactivation enhances
cisplatin-induced apoptosis in renal carcinoma cells. Kidney Int.
69:1139–1145. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu W, Chen H, Wong N, Haynes W, Baker CM
and Wang X: Pseudohypoxia induced by miR-126 deactivation promotes
migration and therapeutic resistance in renal cell carcinoma.
Cancer Lett. 394:65–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yan L, Ding B, Liu H, Zhang Y, Zeng J, Hu
J, Yao W, Yu G, An R, Chen Z, et al: Inhibition of SMYD2 suppresses
tumor progression by down-regulating microRNA-125b and attenuates
multi-drug resistance in renal cell carcinoma. Theranostics.
9:8377–8391. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bai L, Lin G, Sun L, Liu Y, Huang X, Cao
C, Guo Y and Xie C: Upregulation of SIRT6 predicts poor prognosis
and promotes metastasis of non-small cell lung cancer via the
ERK1/2/MMP9 pathway. Oncotarget. 7:40377–40386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Azuma Y, Yokobori T, Mogi A, Altan B,
Yajima T, Kosaka T, Onozato R, Yamaki E, Asao T, Nishiyama M and
Kuwano H: SIRT6 expression is associated with poor prognosis and
chemosensitivity in patients with non-small cell lung cancer. J
Surg Oncol. 112:231–237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Strub T, Ghiraldini FG, Carcamo S, Li M,
Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ,
et al: SIRT6 haploinsufficiency induces BRAF melanoma cell
resistance to MAPK inhibitors via IGF signalling. Nat Commun.
9:34402018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu L, Lin X, Zheng Y and Zhou H: Silencing
of heat shock protein 27 increases the radiosensitivity of
non-small cell lung carcinoma cells. Mol Med Rep. 20:613–621.
2019.PubMed/NCBI
|
|
36
|
Chrysovergis A, Papanikolaou VS, Tsiambas
E, Ragos V, Peschos D and Kyrodimos E: Digital Analysis of BCL2
expression in laryngeal squamous cell carcinoma. Anticancer Res.
39:1253–1257. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li H, Wang H, Deng K, Han W, Hong B and
Lin W: The ratio of Bcl-2/Bim as a predictor of cisplatin response
provides a rational combination of ABT-263 with cisplatin or
radiation in small cell lung cancer. Cancer Biomark. 24:51–59.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zha H, Aimé-Sempé C, Sato T and Reed JC:
Proapoptotic protein Bax heterodimerizes with Bcl-2 and
homodimerizes with Bax via a novel domain (BH3) distinct from BH1
and BH2. J Biol Chemistry. 271:7440–7444. 1996. View Article : Google Scholar
|
|
39
|
Reed JC, Zha H, Aime-Sempe C, Takayama S
and Wang HG: Structure-function analysis of Bcl-2 family proteins.
Regulators of programmed cell death. Adv Exp Med Biol. 406:99–112.
1996. View Article : Google Scholar : PubMed/NCBI
|