Introduction
Breast cancer is one of the most widespread types of
cancer among women worldwide and the most common cause of
cancer-related deaths (11.6 1% of the total cancer deaths)
according to GLOBCAN 2018 (1,2). It was
estimated that in 2019, >300,000 new cases of breast cancer
would be diagnosed in the USA, and ~41,760 women will die from this
disease (3). Hence, identification
of novel prognostic markers and therapeutic targets are urgently
needed for breast cancer. It has been reported that microRNA
(miRNA/miR)-203 is abnormally expressed in several sub-types of
breast cancer, such as circulating tumor cells positive metastatic
breast cancer and triple-negative breast cancer suggesting that
miR-203 may be used as potential prognostic marker (4,5). In
addition, a study demonstrated that miR-203 could attenuate breast
cancer cell viability and migration ability in vitro via
reducing the abundance of Runt-related transcription factor 2 in
MDA-MB-231 cells (6).
The role of miR-203 in cancer has been investigated
in some studies (7–9). For example, miR-203 induced mesenchymal
to epithelial transition via repressing Snail family
transcriptional repressor 2 in prostate cells (7). In addition, miR-203 affected the
proliferation of ovarian cancer cells via the JAK-STAT pathway and
regulated the proliferation and apoptosis of ovarian cancer cells
by targeting the inhibition of SOCS3 expression (8). miR-203 also suppressed bladder cancer
cell growth by targeting Twist family bHLH transcription factor 1
(9). However, the mechanism of
action of miR-203 and target genes in breast cancer remain elusive.
Hence, identifying the targets and key pathways of miR-203 in
breast cancer is of great importance.
High-throughput transcriptome technology provides
insight into the expression profile of hundreds to tens of
thousands of genes (10). In
addition, pathway analysis technologies allow the mapping of gene
expression data as pathway maps based on their respective
functional annotation and known molecular interactions (11). Hence, the current study aimed to
investigate the mechanism and target genes of miR-203 in breast
cancer by combining bioinformatics analysis and in vitro
experiments, which may help to improve the treatment and outcome of
patients with breast cancer. The present study aimed to identify
potential drugs by conducting a virtual screening analysis for
miR-203 targets, which may provide a faster and cheaper strategy
for expanding the arsenal of approved breast cancer drugs.
Materials and methods
Cell culture and miRNA
transfection
The cell line SUM159 breast cancer was obtained from
the Chinese Academy of Sciences Cell Bank, and cells were
propagated in a 51% CO2 atmosphere at 37°C and
maintained in Ham's F-12 Nutrient Mixture (F12; cat. no. 21700075;
Thermo Fisher Scientific, Inc.) with 101% (vol/vol) fetal bovine
serum (FBS; cat. no. 10270-106; Gibco; Thermo Fisher Scientific,
Inc.) and 11% (vol/vol) penicillin/streptomycin (cat. no. 10378016,
Thermo Fisher Scientific, Inc.). The miR-203 mimics and scrambled
controls (miR-203 NC) were synthesized by Shanghai GenePharma Co.,
Ltd. The sequences were: miR-203 mimics,
5′-TGCTTTGGCCACTGACTGTCC-3′; miR-203 negative control (NC),
5′TCGCCACATGATCGCCTAAGT-3′. Cells were seeded into 6-well plates at
a density of 1×105 cells/well and cultured in fresh F12
medium 101% FBS and without antibiotics 24 h prior to transfection.
Subsequently, cells were transfected with miR-203 mimics or miR-203
NC using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at a final concentration of 100 nmol/l in each
group, according to the manufacturer's instructions. Transfected
cells were cultured at 37°C for 6 h prior to the replacement of
complete medium. pCMV6-XL5-FKBP5 vector for the overexpression of
FKBP5 and empty control vector pCMV6-XL5 were purchased from
OriGene Technologies Inc. (cat. no. SC117569, PCMV6XL5). Cells were
separately transfected with miR-203 mimics (miR-203),
pCMV6-XL5-FKBP5 and miR-203 mimics (FKBP5) or Cntrl (empty control
vector pCMV6-XL5 and miR-203 NC) using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Transfected cells were cultured at
37°C for 6 h prior to the replacement of complete medium. Cells
were harvested for subsequent experiments 24 h following
transfection and all experiments were performed in triplicate.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA from SUM159 cells transfected with miR-203
mimics or miR-203 NC was extracted using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific Inc.) and cDNA was
synthesized using iScript™ cDNA Synthesis Kit (Biorad Laboratories
Inc.) following the manufacturer's instructions. iQ™ SYBR Green
supermix (Biorad Laboratories Inc.) was used to perform RT-qPCR. A
7500 HT Fast Real-Time PCR system (Applied Biosystems; Thermo
Fisher Scientific Inc.) was used to cycle and quantify reactions.
The thermocycling conditions were as follows: denaturation at 95°C
for 20 sec, followed by 40 cycles of 95°C for 10 sec, 60°C for 20
sec and 70°C for 10 sec. Relative gene expression levels of miR-203
were evaluated. U6 was used as the miRNA endogenous normalization
control. The relative expression levels of FKBP5 were evaluated
relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The
primer sequences used were as follows: miR-203 forward
5′-GGGGTGAAATGTTTAGGAC-3; reverse 5′-CAGTGCGTGTCGTGGAGT-3′; U6
forward 5′-CTCGCTTCGGCAGCACA-3′, reverse
5′-AACGCTTCACGAATTTGCGT-3′; FKBP5 forward 5′-GGGGTGAAATGTTTAGGAC-3;
reverse 5′-CAGTGCGTGTCGTGGAGT-3′; and GAPDH forward
5′-TCAAGAAGGTGGTGAAGCAG-3′, reverse 5′-CGCTGTTGAAGTCAGAGGAG-3′. The
relative expression of each target amplicon was calculated using
the 2−ΔΔCq method (12).
Data preprocessing and analysis of
differentially expressed genes (DEGs)
To investigate the downstream molecular pathways of
miR-203, potentially mediating its effects in breast cancer, gene
expression arrays were carried out in SUM159 cells transfected with
miR-203 mimics or miR-203 NC. The changes in the miRNA expression
levels were detected in total RNA isolated by a TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific Inc.). Reverse
transcription and hybridization were performed using the Human
Genome U133 Plus 2.0 Array (cat. no. 902482; Affymetrix, Inc.).
This microarray had 9,921 probe sets representing ~6,500 genes and
interrogated >47,000 transcripts. The raw data was submitted to
National Microbiology Data Center (SUB1610765979725, http://nmdc.cn/resource/attachment/detail/NMDCX0000106).
The original expression profiles were transformed into a matrix
using Affy package in R v. 3.6.1 (13). Subsequently, the Limma package v.
3.40.2 (14) was utilized to
identify DEGs. The threshold was set at P<0.05 and |log(fold
change)|>1. A volcano plot and heatmap of DEGs were constructed
using the pheatmap v.1.0.12 package in R v. 3.6.1 (https://cran.r-project.org/web/packages/pheatmap/index.html).
Pathway enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG,
http://www.genome.jp/kegg/) is a
database used for the systematic analysis of gene function via
linking genomic information with high-level systemic function
(15). After obtaining the DEGs, the
ClusterProfiler 3.12.0 package (16)
was utilized to perform KEGG enrichment analyses, with a cut-off
value of P<0.05. Simultaneously, Gene Set Enrichment Analysis
(GSEA) based on KEGG gene sets in the MSigDB database (17) was carried out to identify the
enriched pathways between SUM159 cells transfected with or without
miR-203, with cut-off values of false discovery rate (FDR) <251%
and P<0.05, which were recommended by GSEA v.4.0.3 (17).
Predicting the targets of miR-203,
protein-protein interaction network (PPI) and core genes in the PPI
network
The potential targets of miRNA-203 were predicted
using TargetScan 7.2 (18).
Subsequently, the intersection of genes identified by Targetscan
7.2 database and downregulated DEGs were then regarded as candidate
targets of miR-203. Additionally, the human PPI network was
retrieved from the Search Tool for the Retrieval of Interacting
Genes (STRING) database (19) and
constructed using both TargetScan and downregulated DEGs. Hub genes
are considered important biomarkers (10); therefore, the EcCentricity,
Betweenness and Stress algorithms were utilized to identify the hub
genes by cytoHubba plugin of Cytoscape v.3.7.1 (20). In addition, the prognostic value of
the hub genes was validated with the Kaplan Meier plotter (KMplot)
database (21).
Validation of transcriptome analyses
by western blot analysis
SUM159 cells transfected with miR-203 mimics or
miR-203 NC were lysed at 4°C in buffer (1 mM β-glycerophosphate,
2.5 mM sodium pyrophosphate, 20 mM Tris-HCl (pH 7.4), 1% Triton
X-100, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA and 1 mM
Na3VO4) supplemented with protease inhibitor
cocktail (1:1,000; cat. no. P8340; Merck KGaA). The protein
concentration was determined with a bicinchoninic acid (BCA)
protein assay kit (cat. no. 23225; Thermo Fisher Scientific Inc.).
Protein samples (20 µg) were separated by 121% SDS-PAGE and then
transferred onto a PVDF membrane. Following blocking with 51% (w/v)
skimmed milk in wash buffer (TBS and 0.051% Tween-20) for 1 h at
room temperature, the membranes were incubated 4°C with the
following primary antibodies: anti-GAPDH (1:10,000; cat. no. G9545;
Sigma-Aldrich; Merck KGaA) and anti-FK506 binding protein 5 (FKBP5;
1:1,000; cat. no. sc-271547; Santa Cruz Biotechnology, Inc.).
Subsequently, the membranes were incubated with a corresponding
HRP-conjugated IgG secondary antibody (1:5,000; cat. nos. ab6721
and ab6789; Abcam.) at room temperature for 1 h. The immunoreactive
signals were scanned and quantified using the ImageQuant LAS 4000
software v.1.2 (Cytiva).
Wound healing motility assay
Cells were harvested for scratch assay at 24 h
following transfection and all experiments were performed in
triplicate. Briefly, SUM159 cells were seeded into a 12-well plate
at a density of 1×105 cells/well. The wound was
generated when cells reached 90–951% confluency by scratching the
surface of the plates with a pipette tip. Subsequently, the cells
were washed thrice with PBS to remove cell debris and incubated for
15 h with fresh F12 medium 11% FBS and without antibiotics.
Finally, the migrated cells were imaged under a light
phase-contrast microscope (magnification, ×40). The migration
distance in each group was measured using ImageJ v.1.8.0 (National
Institutes of Health) (22) and
statistically analyzed.
Identifying potential drug repurposing
candidates
FKBP5 3D structure was downloaded from the Protein
Data Bank (PDB; 6SAF; http://www.rcsb.org/) and its biding sites were
identified by Maestro 2019-1 software (Schrödinger, LLC) (23). Subsequently, a library of 2,106 Food
and Drug Administration (FDA)-approved drugs obtained from the
ZINC15 database (24) was
constructed. Finally, virtual screening and molecular docking was
performed using the Maestro 2019-1 (Schrödinger, LLC) software to
identify potential drug-repurposing candidates. The best potential
drug was determined based on the glide score (25).
Free fatty acid content assay
SUM159 cells were transfected with miR-203 or
miR-203 NC as described earlier. Then, SUM159 cells were seeded in
6-well plates at a density of 1×105 cells/well with
complete medium. Subsequently cells were harvested for fatty acid
content assay after 24 h. Fatty acids were extracted with a Free
Fatty Acid Content assay kit (cat. no. D799794, Sangon Biotech,
Co., Ltd.) according to the manufacturer's instructions. As the
manufacturer's protocol, copper ions could combine free fatty acid
to form fatty acid copper salt. Its content is in direct proportion
to free fatty acids. The content of fatty acid can be calculated by
measuring the content of copper ions with copper reagent by
colorimetry.
Statistical analysis
All experiments were repeated at least 3 times.
Quantitative data are expressed as the mean ± standard error of the
mean (SEM). All statistical analyses were carried out using the
SPSS 18.0 software (SPSS, Inc.). The figures were generated using
tGraph Prism 7.0 software (GraphPad Software, Inc.). All data were
checked for normal distribution and homogeneity of variance using
Kolmogorov-Smirnov one-sample and Levene's tests. Statistical
evaluation of the data was performed by using the unpaired
Student's t-test or Kruskal Wallis non-parametric test with a post
hoc Dunn's test. Survival analysis was performed using KMplot with
an auto-selected best cutoff and log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Results
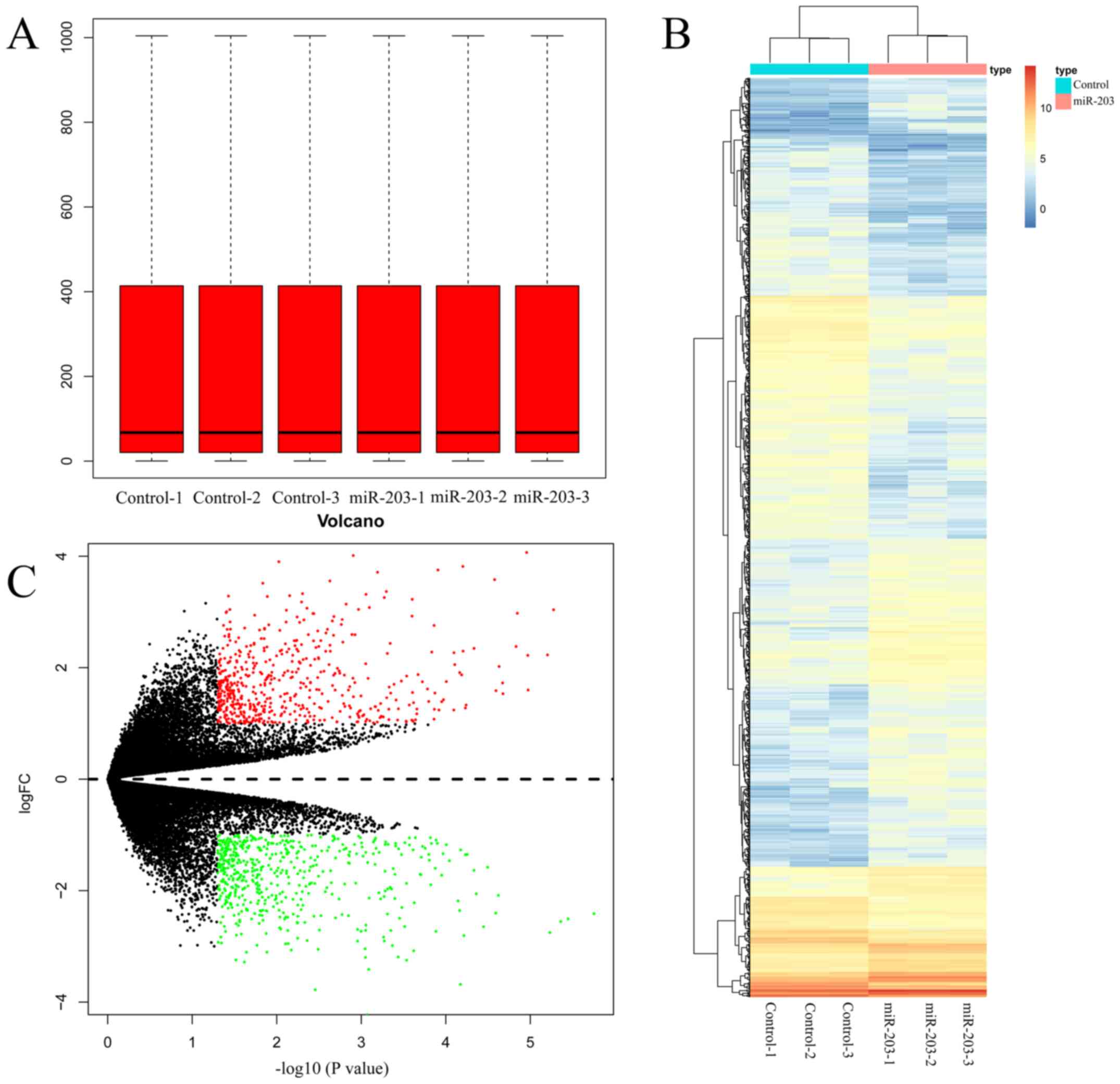
Identification of DEGs
To assess the transfection efficiency of miR-203 in
SUM159 cells, RT-qPCR was performed and the results demonstrated
that the expression of miR-203 was significantly increased by
2.6-fold compared with the control group (SUM159 cells transfected
with miR-203 NC) (P<0.05; Fig.
S1). The box plots for the expression values for all genes in
each sample following normalization by R package Limma 3.40.2 are
represented in Fig. 1A. On comparing
miR-203-overexpressing SUM159 cells with Cntrl (SUM159 cells
transfected with miR-203 NC) counterparts, 1,101 DEGs were
identified with a threshold of P<0.05 and |logFC| >1
(Table SI). In addition, a heatmap
of DEGs was constructed to visualize their expression levels in
different samples (Fig. 1B). Volcano
plots of DEGs drawn by pheatmap 1.0.12 for the two treatment groups
were presented in Fig. 1C. The
significantly downregulated genes are indicated by green dots, and
the significantly upregulated ones by red dots. Black dots indicate
genes with no significant differences in gene expression.
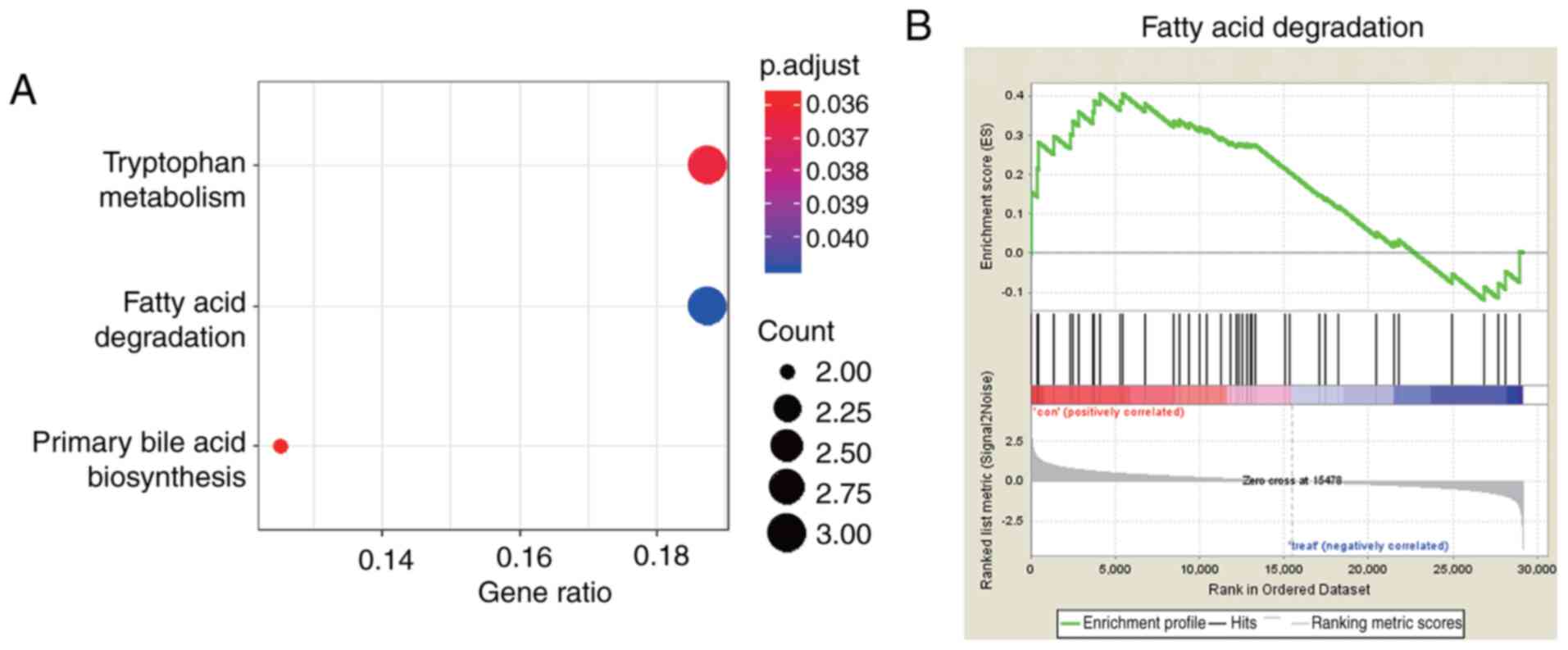
Identification of key pathways between
KEGG and GSEA
To further investigate the function of the
aforementioned DEGs, KEGG pathway enrichment analysis was carried
out. Pathways which were significantly enriched (P<0.05) were
identified using the ClusterProfiler package. Based on the KEGG
database, DEGs were significantly enriched in the ‘tryptophan
metabolism’, ‘fatty acid degradation’ and ‘primary bile acid
biosynthesis’ pathways (Fig. 2A).
GSEA was carried out to summarize genome-wide gene expression
changes into gene sets, other than DEGs, between Cntrl (SUM159
cells transfected with miR-203 NC) and treatment groups (SUM159
cells transfected with miR-203). A total of 25 pathways enriched in
the Cntrl group (P<0.05 and FDR<25%, Table SII), and 24 in the treatment group
were predicted by applying GSEA (P<0.05 and FDR<25%; Table SIII). Between KEGG pathway
enrichment analysis and GSEA, one overlapping pathway was obtained,
namely the ‘fatty acid degradation’ pathway (Fig. 2A and B). Hence, the fatty acid
content was evaluated using the free fatty acid content assay kit
and the results demonstrated that the levels of fatty acids were
significantly reduced in the miR-203 group compared with the
control group (SUM159 cells transfected with miR-203, P<0.05;
Fig. S2).
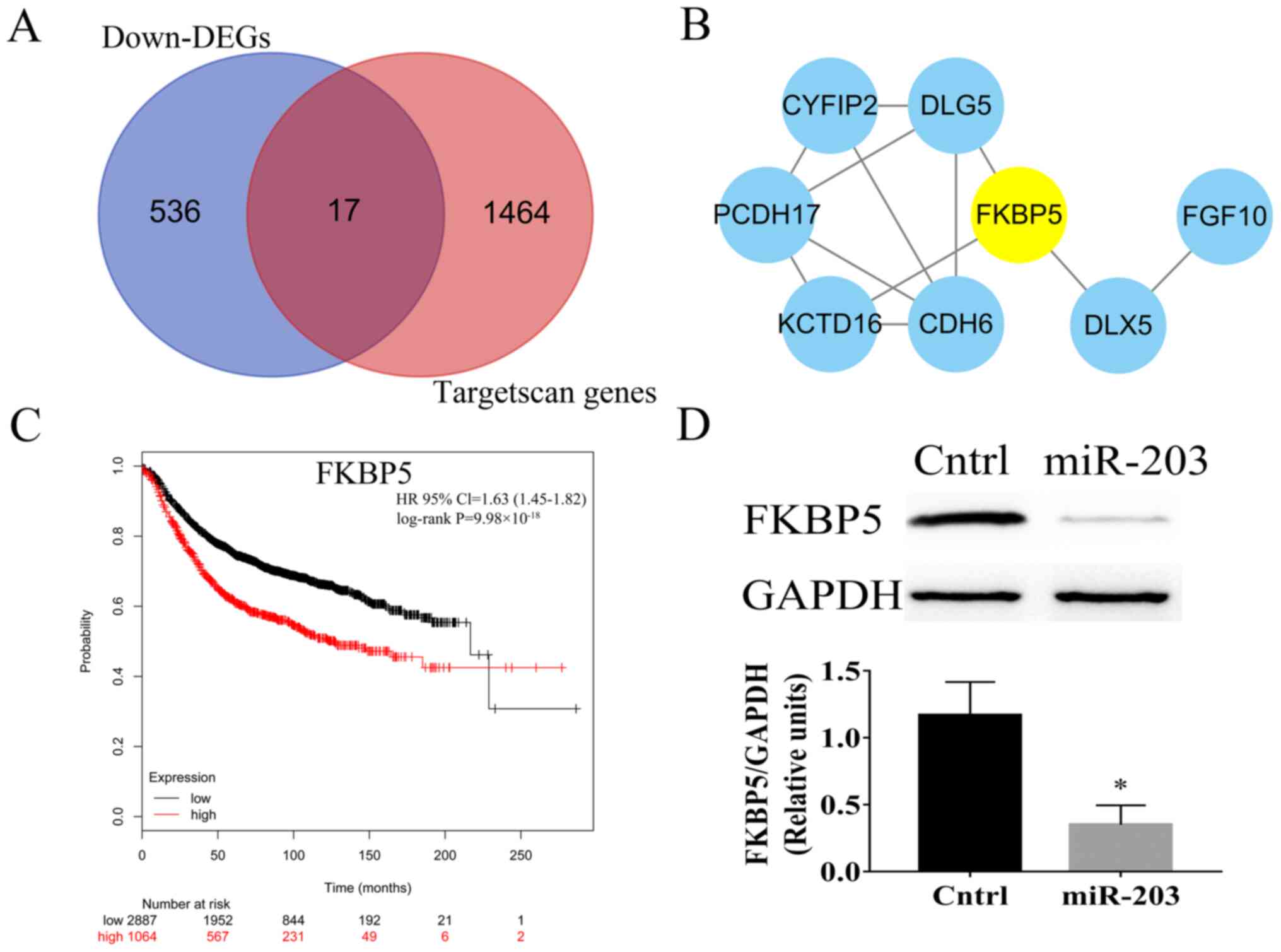
Predicted target genes of miR-203, PPI network, and
core genes in the PPI network. A total of 1481 target genes of
miR-203 were predicted using the Targetscan database. Among them,
17 genes were downregulated DEGs (Fig.
3A). Subsequently, to reveal the interactions among target
genes, the genes were subjected to screening in the STRING
database. Only interactions with a combined score of >0.15 were
considered significant (19). The
PPI network was comprised of 17 nodes (8 nodes with combined score
>0.15) and 12 edges (Fig. 3B).
FKBP5 was identified as the top overlapping gene among 3 ranking
methods (EcCentricity, Betweenness and Stress), using the cytoHubba
plugin (Table SIV). In addition,
overall survival analysis based on the Kmplot database with an
auto-selected best cutoff and log-rank test was performed to
further verify the role of the FKBP5 hub gene. The analysis
revealed that FKBP5 was significantly associated with breast cancer
(log-rank P<0.05 and HR=1.63; Fig.
3C). This finding was further verified by detecting the
expression levels of FKBP5 using western blot analysis. Compared
with the Cntrl group (SUM159 cells transfected with miR-NC), the
protein expression levels of FKBP5 were significantly downregulated
in the miR-203 group (P<0.05; Fig.
3D). The western blotting results were in line with those
observed in DEGs.
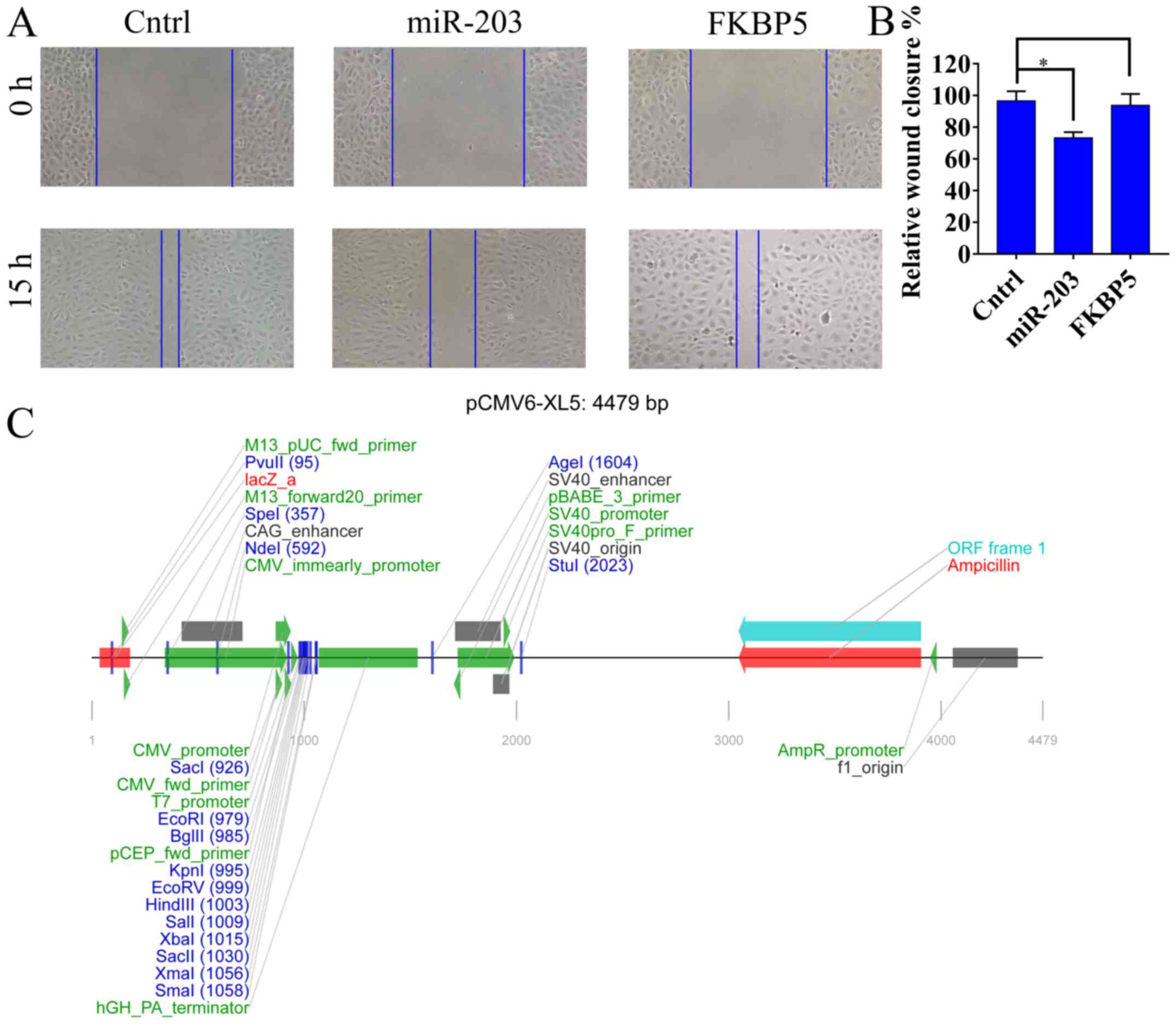
FKBP5 upregulation restores the
migration ability of miR-203 overexpressing SUM159 cells
To assess the transfection efficiency of miR-203 and
FKBP5 in SUM159 cells, RT-qPCR was performed. RT-qPCR results
demonstrated that miR-203 and FKBP5 expression significantly
increased by 2.1-fold and 1.8-fold compared with the Cntrl group
(SUM159 cells transfected with miR-NC and empty control vector
pCMV6-XL5), respectively (P<0.05; Fig S3). In addition, to further verify
that miR-203 could directly target FKBP5, scratch wound healing
assays were performed in SUM159 (Cntrl, SUM159 cells transfected
with miR-NC and empty control vector pCMV6-XL5), SUM159 miR-203
overexpressing (SUM159 cells transfected with miR-203), and SUM159
FKBP5 overexpressing (FKBP5, SUM159 cells transfected with miR-203
and pCMV6-XL5-FKBP5) breast cancer cells. As demonstrated in
Fig. 4A, after 15 h the wound
closure was almost the same between the Cntrl and FKBP5 groups.
However, in the miR-203 group the SUM159 cell migration was slower
compared with the Cntrl group (P<0.05; Figs. 4A and B). The liner map of empty
vector pCMV6-XL5 was shown in Fig.
4C.
ZINC000003944422 is considered as a
potential drug repurposing candidate of FKBP5
Virtual screening (Maestro 2019-1) was utilized to
identify potential drug repurposing candidates for FKBP5. Hence,
the 3D protein structure of FKBP5 was downloaded from PDB (6SAF).
According to the glide scores, ZINC000003944422 was the top 1 hit
obtained from the structure-based virtual screening process
(Table SV). The 3D structure of
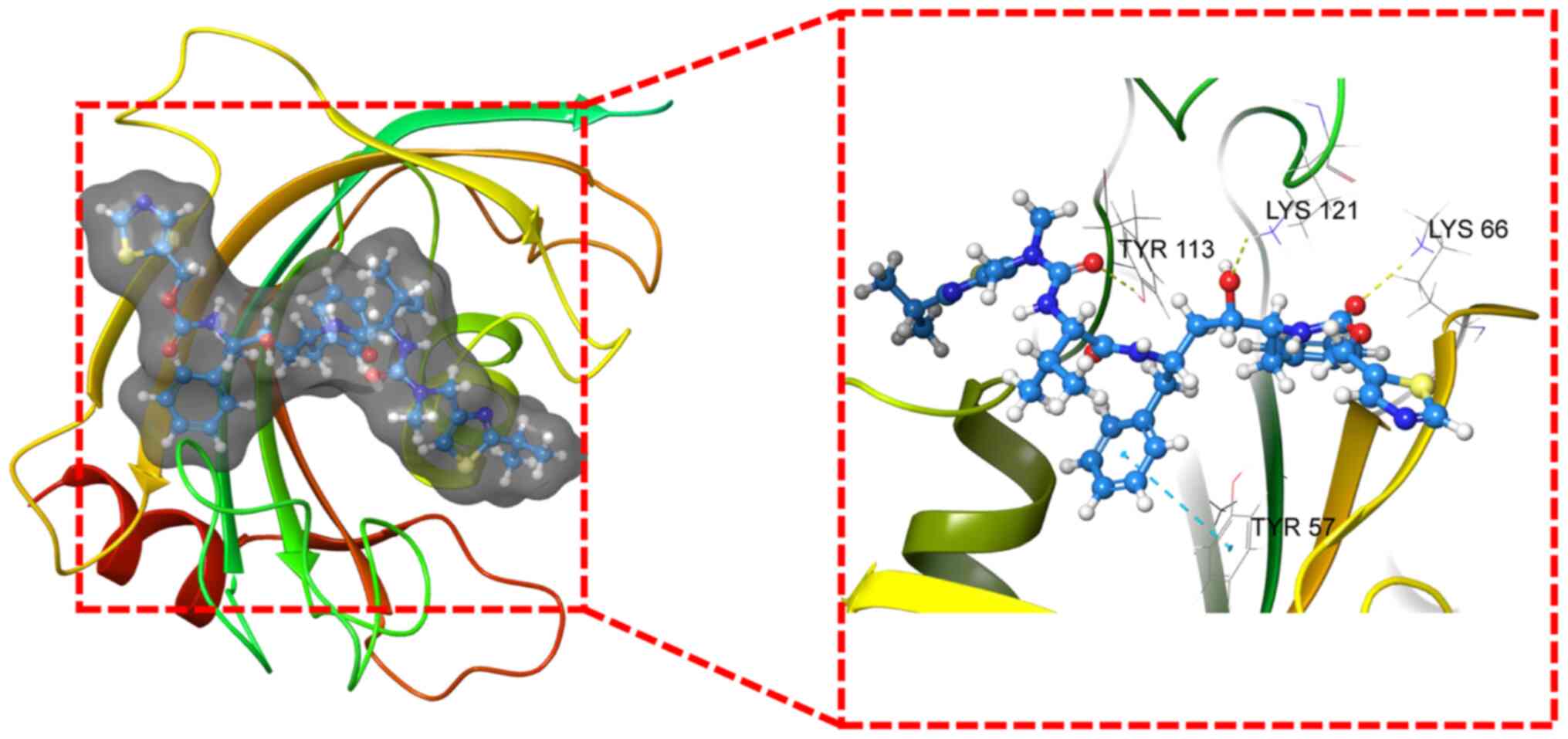
ZINC000003944422 is presented in Fig.
5. Additionally, hydrogen bonds and Pi interactions were
identified in the ligand-protein complex between FKBP5 and
ZINC000003944422 (Fig. 5). In
addition, ZINC000003944422 is also known as Norvir.
Discussion
In 2019, breast cancer was the most common type of
cancer and the second leading cause of cancer-related death among
women in the USA after lung cancer (3). Recently, several studies have
elucidated the key roles of miRNAs in regulating gene expression
during cancer development. For example, high expression of miR-190
suppressed breast cancer metastasis (26). miR-21 promoted breast cancer
proliferation by targeting Leucine zipper transcription factor-like
1 (LZTF1) (27). Emerging evidence
has suggested that miR-203 may serve an import role in cancer
(7–9). However, the specific underlying
molecular mechanisms of miR-203 in breast cancer remain unclear.
The present study demonstrated that miR-203 can directly target
FKBP5 and is involved in the ‘fatty acid degradation’ pathway in
breast cancer. In addition, in the present study potential drugs
that inhibit FKBP5 were identified using virtual screening
analysis.
To explore the underlying mechanism of miR-203 in
breast cancer in the present study, SUM159 were cells were
transfected. According to RT-qPCR results, the transfection
efficiency in the present study was like previous studies (28,29). A
microarray analysis based on the SUM159 cells transfected with
miR-203 mimics or miR-203 NC was conducted in the present study.
According to the results of microarray, the ‘fatty acid
degradation’ pathway was the most commonly enriched pathway in the
KEGG pathway enrichment and GSEA results. In the present study,
GSEA was carried out to summarize genome-wide gene expression
changes in gene sets other than DEGs between Cntrl and treatment
groups. Fatty acid contents were significantly decreased in SUM159
cells transfected with miR-203 mimics compared with SUM159 cells
transfected with miR-203 NC. Fatty acids are fundamental substrates
required for energy storage, synthesis of membranes, generation of
signaling molecules and formation of lipid droplets in cancer cells
(30). Overactivation of fatty acid
metabolism is one of the most aberrant metabolic alterations in
cancer cells, which promotes cancer cell survival and helps
maintain their invasive ability (31). The results of the current study were
in line with a previous one, suggesting that miR-203 inhibits lung
cancer cell metastasis via targeting fatty acid binding protein 4
(FABP4) (32). In addition, another
study demonstrated that fatty acid degradation may serve an
important role in breast cancer cell survival and proliferation via
regulating oxidative stress (33).
Taken together, the results of the present study were in accordance
with the ones obtained from the previous aforementioned studies.
Hence, we hypothesized that overexpression of miR-203 may reduce
the contents of fatty acids which in turn may inhibit cancer cell
growth and metastasis.
In the present study, one hub target gene, FKBP5,
was identified by PPI network and target analysis. In addition,
wound healing assays indicated that miR-203 overexpression induced
inhibition of migration of SUM159 cells were reversed by FKBP5
overexpression. These results suggested that miR-203 may directly
target FKBP5 in breast cancer. It has been reported that FKBP5 is
involved in breast and ovarian cancer (34,35).
Another study demonstrated that FKBP5 may act as a tumor suppressor
and affect cell response to chemotherapy (36). The present study further supported
the findings of previous studies, indicating that FKBP5 may be
directly targeted by miR-203 in breast cancer.
In the present study, the potential drug repurposing
candidates were identified using virtual screening analysis based
on 2,106 FDA-approved drugs. The analysis revealed that FKBP5 may
be a potential therapeutic target in breast cancer. In addition,
ZINC000003944422 was found as a potential target-drug for FKBP5.
Screening in the ZINC15 database in the present study also
demonstrated that ZINC000003944422 was Norvir. It has been reported
that Norvir inhibits the proliferation of renal cancer (37) and bladder cancer cells (38). In line with previous studies, the
findings of the present study suggested that Norvir may improve the
outcome of patients with breast cancer. Hence, drug development
could be accelerated to improve the outcome of patients with breast
cancer. However, further experiments should be carried out to
verify the results of the current study.
The present study had limitations. The results in
the present study were mainly based on SUM159 cells and the
potential drug was identified by virtual screening (instead of
performing laboratory experiments). Further studies based on
different breast cancer cell lines are needed to verify the results
of the present study. In addition, well-designed laboratory
experiments are required to prove that ZINC000003944422 is a
potential drug for FKBP5.
Overall, in the present study 1,101 DEGs were
identified between Cntrl and breast cancer tissues. In addition,
KEGG pathway enrichment and GSEA revealed that miR-203 was enriched
in the ‘fatty acid degradation’ pathway in breast cancer. In
addition, a total of 17 target genes of miR-203 were identified in
the downregulated DEGs and Targetscan 7.2. To further detect the
hub genes among target genes, hub gene analysis was performed using
the cytoHubba plugin. In addition, virtual screening analysis
predicted that ZINC000003944422 may be a potential target-drug for
FKBP5. Taken together, the aforementioned findings indicated that
miR-203 may directly target FKBP5 in breast cancer via fatty acid
degradation and potential drugs, hence providing a novel treatment
approach for breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. Publicly available datasets
were analyzed in this study. The data can be found in the ZINC15
(http://zinc15.docking.org/) and Kmplot
(https://kmplot.com/analysis/)
databases.
Authors' contributions
DY and JY contributed to the study design, data
collection, data interpretation and manuscript preparation. BX and
YF contributed to data collection, data analysis and statistical
analysis. DY and JY confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
DEGs
|
differentially expressed genes
|
|
PPI
|
protein-protein interaction
network
|
|
Cntrl
|
control
|
|
miRNA/miR
|
microRNA
|
References
|
1
|
Telli ML, Gradishar WJ and Ward JH: NCCN
Guidelines Updates: Breast Cancer. J Natl Compr Canc Netw.
17:552–555. 2019.PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Madhavan D, Zucknick M, Wallwiener M, Cuk
K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R,
et al: Circulating miRNAs as surrogate markers for circulating
tumor cells and prognostic markers in metastatic breast cancer.
Clin Cancer Res. 18:5972–5982. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taipaleenmäki H, Browne G, Akech J, Zustin
J, van Wijnen AJ, Stein JL, Hesse E, Stein GS and Lian JB:
Targeting of Runx2 by miR-135 and miR-203 Impairs Progression of
Breast Cancer and Metastatic Bone Disease. Cancer Res.
75:1433–1444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Qu Y, Li WC, Hellem MR, Rostad K, Popa M,
McCormack E, Oyan AM, Kalland KH and Ke XS: miR-182 and miR-203
induce mesenchymal to epithelial transition and self-sufficiency of
growth signals via repressing SNAI2 in prostate cells. Int J
Cancer. 133:544–555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu HP, Zhang Y, Liu ZT, Qi H, Zheng XM,
Qi LH and Wang JY: miR-203 regulates proliferation and apoptosis of
ovarian cancer cells by targeting SOCS3. Eur Rev Med Pharmacol Sci.
23:9286–9294. 2019.PubMed/NCBI
|
|
9
|
Shen J, Zhang J, Xiao M, Yang J and Zhang
N: miR-203 Suppresses Bladder Cancer Cell Growth and Targets
Twist1. Oncol Res. 26:1155–1165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang F, Qiu W, Li R, Hu J, Luo S, Zhang T,
He X and Zheng C: Genome-wide identification of the interactions
between key genes and pathways provide new insights into the
toxicity of bisphenol F and S during early development in
zebrafish. Chemosphere. 213:559–567. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res 47D. D590–D595. 2019. View Article : Google Scholar
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy - analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smyth GK, Michaud J and Scott HS: Use of
within-array replicate spots for assessing differential expression
in microarray experiments. Bioinformatics. 21:2067–2075. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res 40D. D109–D114.
2012. View Article : Google Scholar
|
|
16
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
19
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res 47D. D607–D613. 2019.
View Article : Google Scholar
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res Treat. 160:439–446.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rueden CT, Schindelin J, Hiner MC, DeZonia
BE, Walter AE, Arena ET and Eliceiri KW: ImageJ2: ImageJ for the
next generation of scientific image data. BMC Bioinformatics.
18:5292017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salam NK, Nuti R and Sherman W: Novel
method for generating structure-based pharmacophores using
energetic analysis. J Chem Inf Model. 49:2356–2368. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Irwin JJ, Sterling T, Mysinger MM, Bolstad
ES and Coleman RG: ZINC: A free tool to discover chemistry for
biology. J Chem Inf Model. 52:1757–1768. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Wang X, Xiong Y, Li CD, Xu Q, Shen
L, Chandra Kaushik A and Wei DQ: An Integrated Pan-Cancer Analysis
and Structure-Based Virtual Screening of GPR15. Int J Mol Sci.
20:62262019. View Article : Google Scholar
|
|
26
|
Yu Y, Luo W, Yang ZJ, Chi JR, Li YR, Ding
Y, Ge J, Wang X and Cao XC: miR-190 suppresses breast cancer
metastasis by regulation of TGF-β-induced epithelial-mesenchymal
transition. Mol Cancer. 17:702018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang H, Tan Z, Hu H, Liu H, Wu T, Zheng C,
Wang X, Luo Z, Wang J, Liu S, et al: microRNA-21 promotes breast
cancer proliferation and metastasis by targeting LZTFL1. BMC
Cancer. 19:7382019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia Y, Wang Y, Wang Q, Ghaffar M, Wang Y,
Sheng W and Zhang F: Increased miR-203-3p and reduced miR-21-5p
synergistically inhibit proliferation, migration, and invasion in
esophageal cancer cells. Anticancer Drugs. 30:38–45. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang W, Wu Y, Cheng D and He Z: Mechanism
of epithelial mesenchymal transition inhibited by miR 203 in non
small cell lung cancer. Oncol Rep. 43:437–446. 2020.PubMed/NCBI
|
|
30
|
Currie E, Schulze A, Zechner R, Walther TC
and Farese RV Jr: Cellular fatty acid metabolism and cancer. Cell
Metab. 18:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen TT and Li H: Fatty acid metabolism
and prospects for targeted therapy of cancer. Eur J Lipid Sci
Technol. 119:16003662017. View Article : Google Scholar
|
|
32
|
Chen JC and Wu X: [miR-203 inhibits lung
cancer cell metastasis by targeting fatty acid binding protein 4].
Nan Fang Yi Ke Da Xue Xue Bao. 38:578–583. 2018.(In Chinese).
PubMed/NCBI
|
|
33
|
Mikalayeva V, Ceslevičienė I, Sarapinienė
I, Žvikas V, Skeberdis VA, Jakštas V and Bordel S: Fatty Acid
Synthesis and Degradation Interplay to Regulate the Oxidative
Stress in Cancer Cells. Int J Mol Sci. 20:13482019. View Article : Google Scholar
|
|
34
|
Moore NL, Edwards DP and Weigel NL: Cyclin
A2 and its associated kinase activity are required for optimal
induction of progesterone receptor target genes in breast cancer
cells. J Steroid Biochem Mol Biol 144B. 471–482. 2014. View Article : Google Scholar
|
|
35
|
Sun NK, Huang SL, Chang PY, Lu HP and Chao
CC: Transcriptomic profiling of taxol-resistant ovarian cancer
cells identifies FKBP5 and the androgen receptor as critical
markers of chemotherapeutic response. Oncotarget. 5:11939–11956.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hou J and Wang L: FKBP5 as a selection
biomarker for gemcitabine and Akt inhibitors in treatment of
pancreatic cancer. PLoS One. 7:e362522012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sato A and Asano T, Ito K and Asano T:
17-Allylamino-17-demethoxygeldanamycin and ritonavir inhibit renal
cancer growth by inhibiting the expression of heat shock factor-1.
Int J Oncol. 41:46–52. 2012.PubMed/NCBI
|
|
38
|
Sato A and Asano T, Okubo K, Isono M and
Asano T: Nelfinavir and Ritonavir Kill Bladder Cancer Cells
Synergistically by Inducing Endoplasmic Reticulum Stress. Oncol
Res. 26:323–332. 2018. View Article : Google Scholar : PubMed/NCBI
|