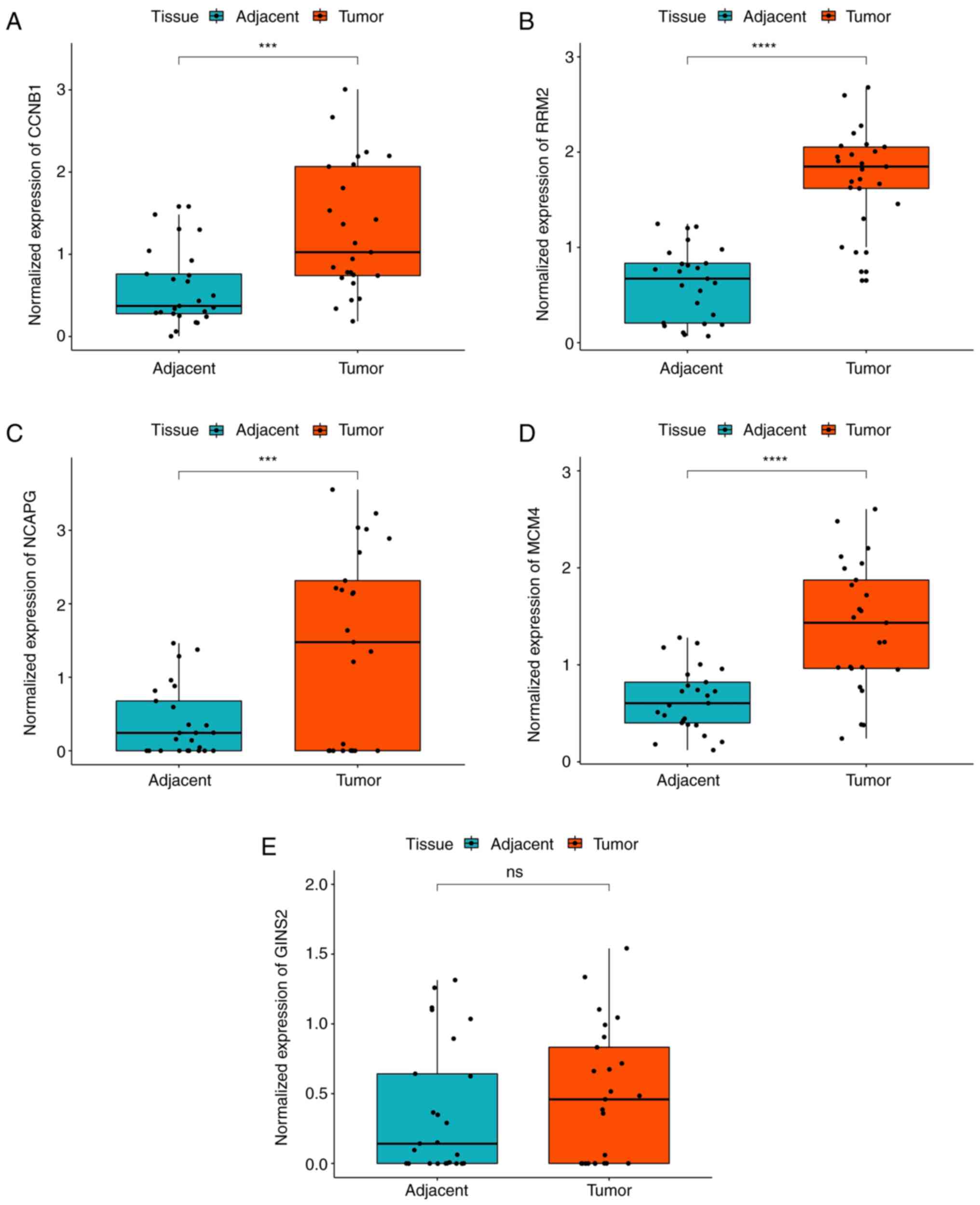
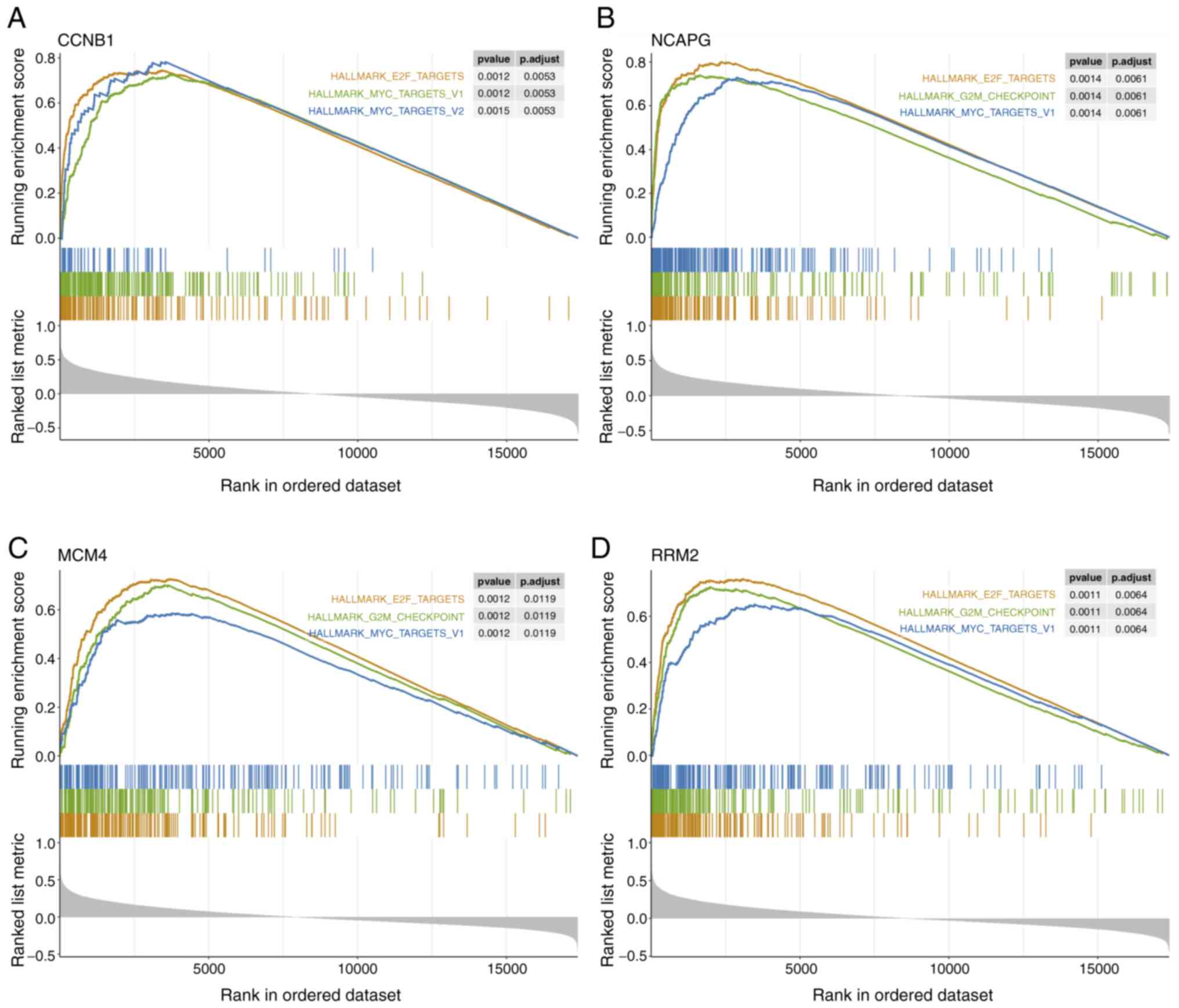
|
1
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T,
Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, et
al: Global, regional, and national cancer incidence, mortality,
years of life lost, years lived with disability, and
disability-adjusted life-years for 29 cancer groups, 1990 to 2016:
A systematic analysis for the global burden of disease study. JAMA
Oncol. 4:1553–1568. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hwang KT, Kim J, Jung J, Chang JH, Chai
YJ, Oh SW, Oh S, Kim YA, Park SB and Hwang KR: Impact of breast
cancer subtypes on prognosis of women with operable invasive breast
cancer: A population-based study using SEER database. Clin Cancer
Res. 25:1970–1979. 2019.PubMed/NCBI
|
|
3
|
Bauer KR, Brown M, Cress RD, Parise CA and
Caggiano V: Descriptive analysis of estrogen receptor
(ER)-negative, progesterone receptor (PR)-negative, and
HER2-negative invasive breast cancer, the so-called triple-negative
phenotype: A population-based study from the California cancer
registry. Cancer. 109:1721–1728. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carey L, Winer E, Viale G, Cameron D and
Gianni L: Triple-negative breast cancer: Disease entity or title of
convenience? Nat Rev Clin Oncol. 7:683–692. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hwang SY, Park S and Kwon Y: Recent
therapeutic trends and promising targets in triple negative breast
cancer. Pharmacol Ther. 199:30–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li MX, Jin LT, Wang TJ, Feng YJ, Pan CP,
Zhao DM and Shao J: Identification of potential core genes in
triple negative breast cancer using bioinformatics analysis. Onco
Targets Ther. 11:4105–4112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lv X, He M, Zhao Y, Zhang L, Zhu W, Jiang
L, Yan Y, Fan Y, Zhao H, Zhou S, et al: Identification of potential
key genes and pathways predicting pathogenesis and prognosis for
triple-negative breast cancer. Cancer Cell Int. 19:1722019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Komatsu M, Yoshimaru T, Matsuo T, Kiyotani
K, Miyoshi Y, Tanahashi T, Rokutan K, Yamaguchi R, Saito A, Imoto
S, et al: Molecular features of triple negative breast cancer cells
by genome-wide gene expression profiling analysis. Int J Oncol.
42:478–506. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maire V, Némati F, Richardson M,
Vincent-Salomon A, Tesson B, Rigaill G, Gravier E, Marty-Prouvost
B, De Koning L, Lang G, et al: Polo-like kinase 1: A potential
therapeutic option in combination with conventional chemotherapy
for the management of patients with triple-negative breast cancer.
Cancer Res. 73:813–823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41((Database Issue)):
D991–D995. 2013.PubMed/NCBI
|
|
12
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: Protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47D:D607–D613. 2019.
View Article : Google Scholar
|
|
13
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doncheva NT, Assenov Y, Domingues FS and
Albrecht M: Topological analysis and interactive visualization of
biological networks and protein structures. Nat Protoc. 7:670–685.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bader GD and Hogue CWV: An automated
method for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liberzon A, Birger C, Thorvaldsdóttir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Freeman LC: Centrality in social networks
conceptual clarification. Soc Netw. 1:215–239. 1978. View Article : Google Scholar
|
|
22
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hartwell LH and Weinert TA: Checkpoints:
Controls that ensure the order of cell cycle events. Science.
246:629–634. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang BN, Bueno Venegas A, Hickson ID and
Chu WK: DNA replication stress and its impact on chromosome
segregation and tumorigenesis. Semin Cancer Biol. 55:61–69. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Joerger AC and Fersht AR: The p53 pathway:
Origins, inactivation in cancer, and emerging therapeutic
approaches. Annu Rev Biochem. 85:375–404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Beberok A, Wrześniok D, Rok J, Rzepka Z,
Respondek M and Buszman E: Ciprofloxacin triggers the apoptosis of
human triple-negative breast cancer MDA-MB-231 cells via the
p53/Bax/Bcl-2 signaling pathway. Int J Oncol. 52:1727–1737.
2018.PubMed/NCBI
|
|
28
|
Zhu X, Wang K, Zhang K, Zhang T, Yin Y and
Xu F: Ziyuglycoside I inhibits the proliferation of MDA-MB-231
breast carcinoma cells through inducing p53-mediated G2/M cell
cycle arrest and intrinsic/extrinsic apoptosis. Int J Mol Sci.
17:19032016. View Article : Google Scholar
|
|
29
|
Singletary KW and Gapstur SM: Alcohol and
breast cancer: Review of epidemiologic and experimental evidence
and potential mechanisms. JAMA. 286:2143–2151. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao M, Howard EW, Parris AB, Guo Z, Zhao
Q and Yang X: Alcohol promotes migration and invasion of
triple-negative breast cancer cells through activation of p38 MAPK
and JNK. Mol Carcinog. 56:849–862. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shekhar MP, Pauley R and Heppner G: Host
microenvironment in breast cancer development: Extracellular
matrix-stromal cell contribution to neoplastic phenotype of
epithelial cells in the breast. Breast Cancer Res. 5:130–135. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu S, Zhang X, Shi C, Lin J, Chen G, Wu
B, Wu L, Shi H, Yuan Y, Zhou W, et al: Altered microRNAs expression
profiling in cumulus cells from patients with polycystic ovary
syndrome. J Transl Med. 13:2382015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kuznetsov VA, Tang Z and Ivshina AV:
Identification of common oncogenic and early developmental pathways
in the ovarian carcinomas controlling by distinct prognostically
significant microRNA subsets. BMC Genomics. 18 (Suppl 6):S6922017.
View Article : Google Scholar
|
|
34
|
Cheng L, Yang H, Zhao H, Pei X, Shi H, Sun
J, Zhang Y, Wang Z and Zhou M: MetSigDis: A manually curated
resource for the metabolic signatures of diseases. Brief Bioinform.
20:203–209. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18:7222017. View Article : Google Scholar
|
|
36
|
Zhao X, Sun S, Zeng X and Cui L:
Expression profiles analysis identifies a novel three-mRNA
signature to predict overall survival in oral squamous cell
carcinoma. Am J Cancer Res. 8:450–461. 2018.PubMed/NCBI
|
|
37
|
Luo Y, Shen D, Chen L, Wang G, Liu X, Qian
K, Xiao Y, Wang X and Ju L: Identification of 9 key genes and small
molecule drugs in clear cell renal cell carcinoma. Aging (Albany
NY). 11:6029–6052. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gavet O and Pines J: Progressive
activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell.
18:533–543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Agarwal R, Gonzalez-Angulo AM, Myhre S,
Carey M, Lee JS, Overgaard J, Alsner J, Stemke-Hale K, Lluch A,
Neve RM, et al: Integrative analysis of cyclin protein levels
identifies cyclin b1 as a classifier and predictor of outcomes in
breast cancer. Clin Cancer Res. 15:3654–3662. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sparano JA, Gray RJ, Makower DF, Pritchard
KI, Albain KS, Hayes DF, Geyer CE Jr, Dees EC, Perez EA, Olson JA
Jr, et al: Prospective validation of a 21-gene expression assay in
breast cancer. N Engl J Med. 373:2005–2014. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang QQ, Chen J, Zhou DL, Duan YF, Qi CL,
Li JC, He XD, Zhang M, Yang YX and Wang L: Dipalmitoylphosphatidic
acid inhibits tumor growth in triple-negative breast cancer. Int J
Biol Sci. 13:471–479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tian S, Chen Y, Yang B, Lou C, Zhu R, Zhao
Y and Zhao H: F1012-2 inhibits the growth of triple negative breast
cancer through induction of cell cycle arrest, apoptosis, and
autophagy. Phytother Res. 32:908–922. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Thadani R, Uhlmann F and Heeger S:
Condensin, chromatin crossbarring and chromosome condensation. Curr
Biol. 22:R1012–R1021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Q, Su R, Shan C, Gao C and Wu P:
Non-SMC condensin I complex, subunit G (NCAPG) is a novel mitotic
gene required for hepatocellular cancer cell proliferation and
migration. Oncol Res. 26:269–276. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen J, Qian X, He Y, Han X and Pan Y:
Novel key genes in triple-negative breast cancer identified by
weighted gene co-expression network analysis. J Cell Biochem.
120:16900–16912. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tye BK: MCM proteins in DNA replication.
Annu Rev Biochem. 68:649–686. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kwok HF, Zhang SD, McCrudden CM, Yuen HF,
Ting KP, Wen Q, Khoo US and Chan KY: Prognostic significance of
minichromosome maintenance proteins in breast cancer. Am J Cancer
Res. 5:52–71. 2014.PubMed/NCBI
|
|
48
|
Issac MSM, Yousef E, Tahir MR and Gaboury
LA: MCM2, MCM4, and MCM6 in breast cancer: Clinical utility in
diagnosis and prognosis. Neoplasia. 21:1015–1035. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qiu WG, Polotskaia A, Xiao G, Di L, Zhao
Y, Hu W, Philip J, Hendrickson RC and Bargonetti J: Identification,
validation, and targeting of the mutant p53-PARP-MCM chromatin axis
in triple negative breast cancer. NPJ Breast Cancer. 3:12017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Aye Y, Li M, Long MJ and Weiss RS:
Ribonucleotide reductase and cancer: Biological mechanisms and
targeted therapies. Oncogene. 34:2011–2021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen WX, Yang LG, Xu LY, Cheng L, Qian Q,
Sun L and Zhu YL: Bioinformatics analysis revealing prognostic
significance of RRM2 gene in breast cancer. Biosci Rep.
39:BSR201820622019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shah KN, Wilson EA, Malla R, Elford HL and
Faridi JS: Targeting ribonucleotide reductase M2 and NF-κB
activation with didox to circumvent tamoxifen resistance in breast
cancer. Mol Cancer Ther. 14:2411–2421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liang WH, Li N, Yuan ZQ, Qian XL and Wang
ZH: DSCAM-AS1 promotes tumor growth of breast cancer by reducing
miR-204-5p and up-regulating RRM2. Mol Carcinog. 58:461–473. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Jones DT, Lechertier T, Mitter R, Herbert
JM, Bicknell R, Jones JL, Li JL, Buffa F, Harris AL and
Hodivala-Dilke K: Gene expression analysis in human breast cancer
associated blood vessels. PLoS One. 7:e442942012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Labib K and Gambus A: A key role for the
GINS complex at DNA replication forks. Trends Cell Biol.
17:271–278. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zheng M, Zhou Y, Yang X, Tang J, Wei D,
Zhang Y, Jiang J, Chen Z and Zhu P: High GINS2 transcript level
predicts poor prognosis and correlates with high histological grade
and endocrine therapy resistance through mammary cancer stem cells
in breast cancer patients. Breast Cancer Res Treat. 148:423–436.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Peng L, Song Z, Chen D, Linghu R, Wang Y,
Zhang X, Kou X, Yang J and Jiao S: GINS2 regulates matrix
metallopeptidase 9 expression and cancer stem cell property in
human triple negative Breast cancer. Biomed Pharmacother.
84:1568–1574. 2016. View Article : Google Scholar : PubMed/NCBI
|