Introduction
RNA helicases are a class of enzymes that regulate
the structure and function of RNAs by utilizing the energy of
nucleoside triphosphate (NTP) to unwind the secondary structure of
RNAs, which interferes with protein interactions in certain cases
(1). The amino acid sequence of RNA
helicases is characterized by the existence of two typical
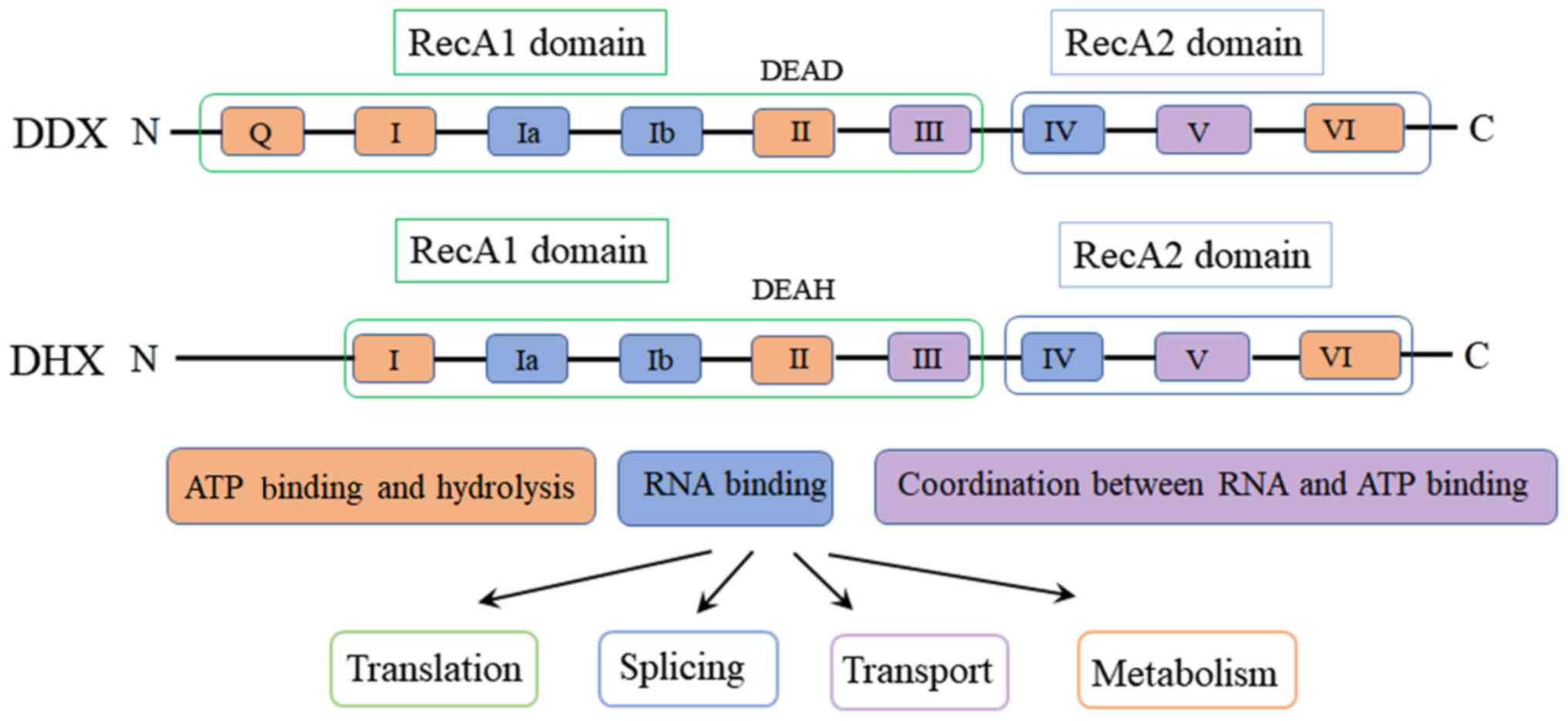
RecA-like helicase domains, the RecA1 domain and the RecA2 domain
(Fig. 1). The conserved motifs I,
Ia, Ib and III are located in the RecA1 domain, whereas the
conserved motifs IV, V and VI are located in the RecA2 domain
(2). The non-conserved N-terminal
domain (NTR) and C-terminal domain (CTR) are located at both ends
of the catalytic core domain. The NTR and CTR domains determine the
expression pattern, subcellular localization and substrate
specificity of the enzyme (3,4).
According to the amino acid sequence and structure,
RNA helicases can be divided into six super-families (SF) 1–6,
among which SF2 contains the most members (5). SF2 contains 8–9 highly conserved
motifs, including motifs Q, I, Ia, Ib, II, III, IV, V and VI, each
of which display a distinct function. Motifs I, II, VI and Q bind
to NTP and catalyze hydrolysis; motifs Ia, Ib and IV primarily bind
to RNA substrates; and motifs III and V connect the NTP binding and
RNA recognition sites, promoting NTP-dependent RNA unwinding
(6,7). Based on the amino acid sequence of
conserved motif II, SF2 are divided into two subfamilies: DEAD-box
helicase and DEAH-box helicase (DEAH), which are named after the
amino acid sequence Asp(D)-Glu(E)-Ala(A)-Asp(D)/His(H) of motif II,
referred to as DDX and DHX, respectively (8). Non-conserved domains and conserved
motifs 8–9 determine several important biological properties of RNA
helicases, including splicing, transportation, translation
initiation, RNA degradation and ribosome synthesis (Fig. 1) (9).
Emerging evidence suggests that numerous RNA
helicases are dysregulated in tumor tissues: DDX1 is highly
expressed in neuroblastoma and retinoblastoma (10) and breast cancer (11); DDX2A is highly expressed in melanoma
(12) and hepatocellular carcinoma
(13); DDX3 is upregulated in breast
(14) and liver cancer (15); DDX48 is upregulated in gastric cancer
(16); DDX43 is upregulated in acute
myeloid leukemia (17) and lung
cancer (18); and DDX5 and DDX6 are
upregulated in colorectal cancer (19,20). In
contrast, DDX3 is downregulated in certain types of liver and lung
cancer (21). Moreover, DHX32 is
downregulated in acute lymphocytic leukemia, but upregulated in
colorectal and breast cancer (Table
I) (10,12,13,17–36). The
aforementioned studies indicate that the abnormal expression of RNA
helicases is significantly associated with cancer initiation and
development.
 | Table I.Association between RNA helicases and
tumors. |
Table I.
Association between RNA helicases and
tumors.
| RNA helicases | Associated
tumor | Expression
level | (Refs.) |
|---|
| DDX1 | Neuroblastoma,
retinoblastoma | Upregulated | (10) |
| DDX2A | Melanoma,
hepatocellular carcinoma | Upregulated | (12,13) |
| DDX4 | Ovarian cancer,
blood-derived cancer | Upregulated | (22,23) |
| DDX3 | Breast, liver
cancer/liver, lung cancer |
Up/downregulated | (21) |
| DDX5 | Colorectal,
prostate, breast cancer | Upregulated | (19,24) |
| DDX6 | Gastric cancer,
colorectal cancer | Upregulated | (20,25) |
| DDX11 | Invasive melanoma,
lung adenocarcinoma | Upregulated | (26,27) |
| DDX17 | Colorectal
cancer | Upregulated | (24) |
| DDX39 | Hepatocellular
carcinoma, lung cancer | Upregulated | (29,30) |
| DDX43 | Acute myeloid
leukemia, lung cancer | Upregulated | (17,18) |
| DDX48 | Gastric cancer,
vaginal carcinoma | Upregulated | (31,32) |
| DDX53 | Gastric, cervical
and lung cancer | Upregulated | (33,34)) |
| DHX9 | Lung cancer | Upregulated | (35) |
| DHX15 | Glioma | Upregulated | (36) |
| DHX32 | Colorectal
cancer/acute lymphocytic leukemia |
Up/downregulated | (37,52) |
Discovery of RNA helicase DHX32
DHX32 is a helicase of the DHX family that was
discovered in 2002 by Abdelhaleem (37) based on the similarity to the amino
acid sequence of the DDX15 helicase domain. The potentially encoded
sequence FLJ10889 (accession no. XM 045832) was identified by
performing a National Center for Biotechnology Information
non-redundant protein database search, which was then used to
search the expressed sequence tag database to obtain the
overlapping expressed sequence tags (AL599197) with 5′ ends. The
full-length sequence was confirmed via reverse transcription-PCR
amplification and sequencing, and DHX32 was identified as the DEAH
helicase homolog. At the same time, a variant transcript (DHX32∆5)
with a 243-bp deletion in exon 5 was also identified. Both DHX32
and DHX32∆5 were confirmed by cloning from HL-60 bone marrow
leukemia cells (37,38).
Structure of DHX32 gene
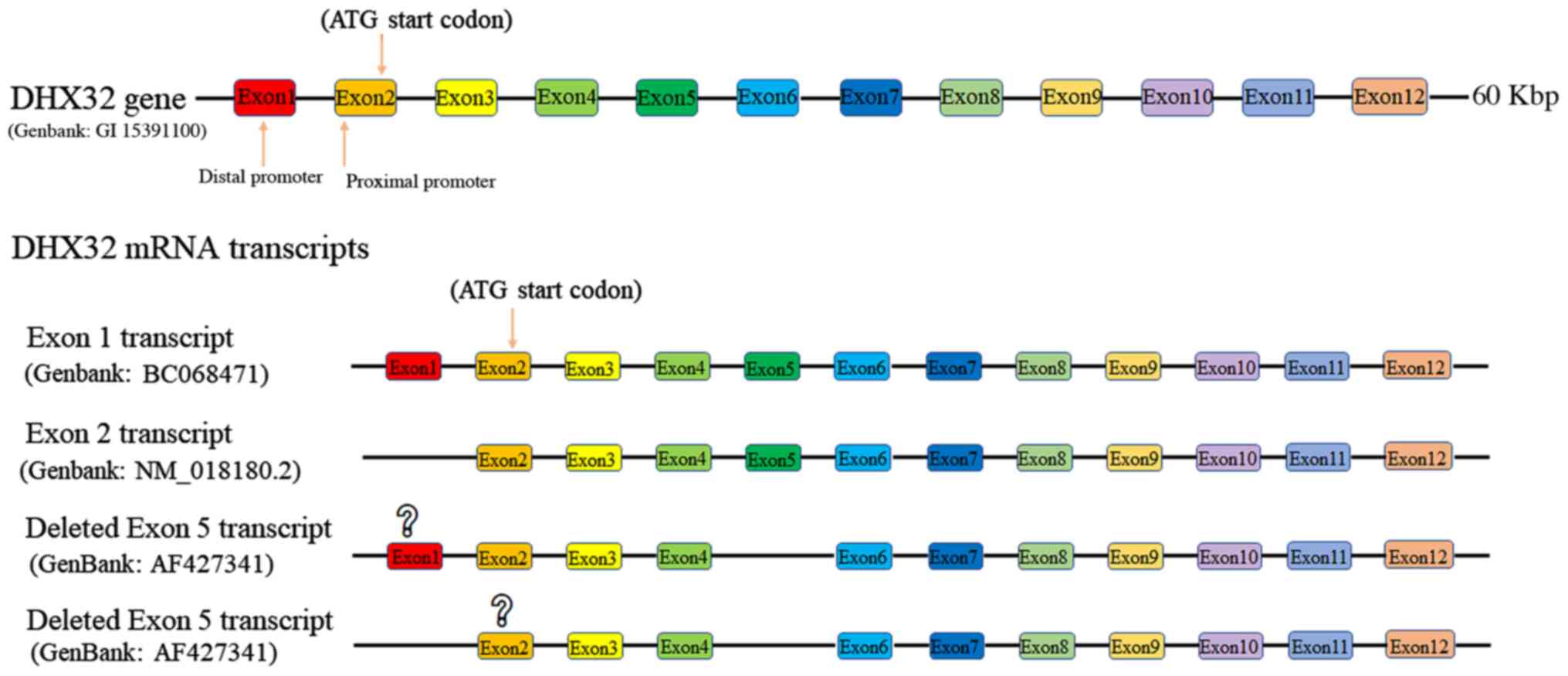
Human DHX32 coding sequence (Gene ID: 55760) is
located on chromosome 10q26.2. The gene contains 12 exons and the
full-length mRNA (Genbank: AF427340) is 3071 bp in length (Fig. 2). Exon 1 contains a CpG island, which
serves as a potential distant promoter. Exon 2 contains the
proximal promoter, which contains the TATA box sequence. DHX32 RNA
in the thymus and viscera are transcribed from both the distal
(Genbank: XM 017016404.1) and proximal promoters (Genbank: NM
018180.2). DHX32 transcripts in the bone marrow are only
transcribed from the proximal promoter (Genbank: NM 018180.2)
(38). Although the two set of DHX32
RNA contain different 5′-untranslated regions, they share the same
translational initiation ATG codon. The use of different promoters
represents a layer of regulation for the tissue- and developmental
stage-specific gene expression of DHX32 (39).
The coding sequence of DHX32 encompasses 11 exons.
DHX32 protein contains 743 amino acids, with a molecular weight of
84 kDa (Genbank: NM_018180.2). In addition, alternative splicing of
DHX32 mRNA yields the DHX32∆5 variant, which has a 243-bp deletion
in exon 5 (Genbank: AF427341). The corresponding protein has a
deletion of 81 residues (284aa-364aa) and a molecular weight of 75
kDa. However, which promoter is used for this variant mRNA has not
yet been identified (Fig. 2)
(38).
Helicase activity of DHX32
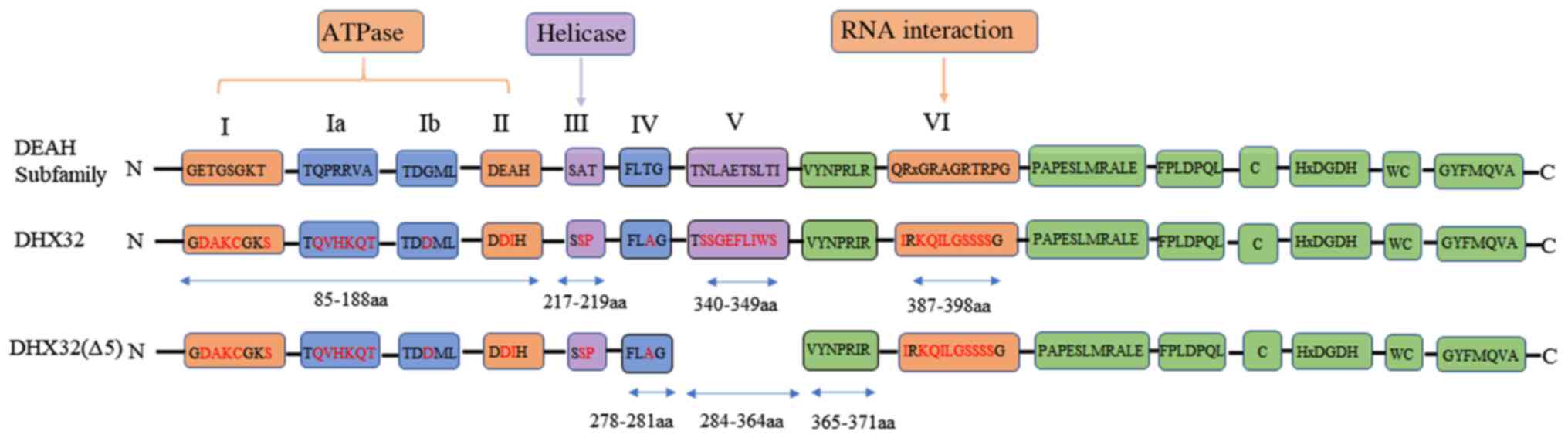
Compared with other members of the DEAH family,
DHX32 has a unique helicase domain sequence that is characterized
by the presence of amino acid substitutions in all eight motifs
(Fig. 3) (37). Motifs I and II are the ATP-binding
sites. Structural-based analyses demonstrate that not only the
lysine in motif I, but also the aspartate, glutamine and histidine
residues in motif II are critical for enzymatic activity (40). Except for the glutamine residue in
motif II, which is replaced by aspartate, all other key amino acids
are conserved. Therefore, it is likely that DHX32 is ATPase active.
However, compared with the traditional DEAH family, the
serine-alanine-threonine (SAT) domain in motif III of the DHX32 is
replaced by the serine-serine-proline (SSP) domain (Fig. 3), which is involved in ATP hydrolysis
and RNA unwinding (41). Several key
residues in the QRxGRxGR sequence of motif VI are not conserved in
DHX32. Since the QRxGRxGR domain is the key site for nucleic acid
substrate binding (42), it is
likely that DHX32 may not display substrate binding and thus, will
not display helicase activity. In addition to containing a SSP
motif III, DHX32∆5 also has a deletion of motif V (Fig. 3), which connects the ATP binding and
RNA recognition sites required for the unwinding of RNA (7). Therefore, it is speculated that DHX32∆5
displays a lower helicase activity compared with DHX32.
Subcellular location of DHX32
DHX32 is widely distributed in human tissues,
including the bone marrow, thymus, spleen, rectum and breast.
Confocal microscopy revealed that DHX32 is localized in the nucleus
and mitochondria of HeLa cells (43). RNA helicases that regulate mRNA
transcription and splicing are located in the nucleus, whereas
those that regulate translation are located in the cytoplasm
(44). The location of DHX32 in the
nucleus and mitochondria reflects the diversity of its functions.
Electron microscope immunocytochemistry revealed that DHX32 is
localized in the mitochondria of mouse hepatocytes (45), and is localized in the nucleus and
the inner mitochondrial membrane in HL-60 leukemia cells (43). Iborra et al (46) demonstrated that newly synthesized RNA
molecules of mitochondria accumulate along the inner membrane. Alli
et al (43) double stained
cells with anti-bromouridine and anti-DHX32 antibodies,
demonstrating DHX32 mitochondria localization, which was similar to
the position of newly synthesized RNA in mitochondria.
Collectively, the aforementioned studies indicate that DHX32 is
involved in regulating mitochondrial gene expression. Heat shock
protein 60 (Hsp60), an important molecular chaperone, is primarily
distributed in the mitochondria in Jurkat cells. Chen et al
(47) reported that when DHX32 is
highly expressed, Hsp60 was transferred from the mitochondria to
the cytoplasm in Jurkat cells. Therefore, DHX32 may also regulate
the redistribution of mitochondrial-associated proteins, and then,
may regulate gene expression in both the nucleus and
mitochondria.
DHX32 and cell differentiation
DHX32 expression is different in normal lymphoid
tissues. In lymphoid follicles, DHX32 expression is higher in
lymphocytes at the germinal center compared with the mantle region.
In the spleen, lymphocytes of red pulp express higher levels of
DHX32 compared with the white pulp. In the thymus, DHX32 expression
is higher in the thymic medulla compared with the thymic cortex
(38). The aforementioned results
indicate that DHX32 expression in normal lymphoid tissues is
associated with lymphocyte activation and differentiation status.
Further studies demonstrate that DHX32 expression is low in
precursor T lymphoblastic lymphoma derived from precursor cells of
acute lymphoblastic leukemia, and high in large B-cell lymphoma
derived from mature lymphocytes. Furthermore, DHX32 expression in
CD4 and CD8 double-negative cells and double-positive cells is
significantly lower compared with single-positive cells, indicating
that DHX32 expression is positively correlated with lymphocyte
proliferation and differentiation (48). Overall, DHX32 expression is closely
associated with cell proliferation, differentiation and
apoptosis.
DHX32 and cancer
DHX32 and hematological tumors
In 2002, by using RNA hybridization, Abdelhaeem
(37) demonstrated that DHX32 was
downregulated in acute lymphocytic leukemia cells and patients'
cancer cells. T cell lines, precursor B cell lines and precursor B
cell lymphocytic leukemia samples from patients also displayed
downregulation of DHX32 expression, indicating that DHX32
downregulation is not restricted to a specific lymphocyte lineage.
Interestingly, there were no significant differences in other DEAH
expression levels, including DDX15 and DDX9, in the myeloid and
lymphoid lines (37), suggesting
that downregulated RNA helicase expression is DHX32-specific. In
2005, immunohistochemical staining demonstrated that DHX32
expression was low in follicular lymphoma and Burkitt lymphoma, but
is high in mantle cell lymphoma, large B-cell lymphoma and
Reed-Steenberg cells of nodular sclerotic Hodgkin lymphoma with a
higher degree of malignancy (38).
The aforementioned finding provided evidence that DHX32 expression
is dysregulated in lymphoma, and the expression level is
significantly different in various types of tumor tissues. The
higher the malignancy, the more significant the alterations in
DHX32 expression levels are, which suggests the potential of DHX32
to serve as a novel biomarker for lymphoma prognosis. A study
recruiting 28 patients with primary chemotherapy-resistance
leukemia was conducted by McNeer et al (49). The aforementioned study reported that
DHX32 gene deletions were the cause of chemotherapy-resistance in
pediatrics with acute myeloid leukemia. Moreover, the frequency of
DHX32 mutant allele was increased from 14 to 39% after
chemotherapy. The cBioPortal of TCGA pan-cancer repository (RRID:
SCR_014555, URL: http://www.cbioportal.org/) was used to explore DHX32
functionality in big data. There were 31 studies (12,845 samples)
in myeloid leukemia and lymphoid leukemia. These data were analyzed
by using a log-rank test (R package survival) and found that the
DHX32 missense mutations are associated with the poor prognosis of
patients in myeloid leukemia and lymphocytic leukemia. These
findings confirm that DHX32 is abnormally expressed in
hematological tumors and is associated with the malignancy of the
tumor, further demonstrating the potential of DHX32 as a prognostic
biomarker in leukemia.
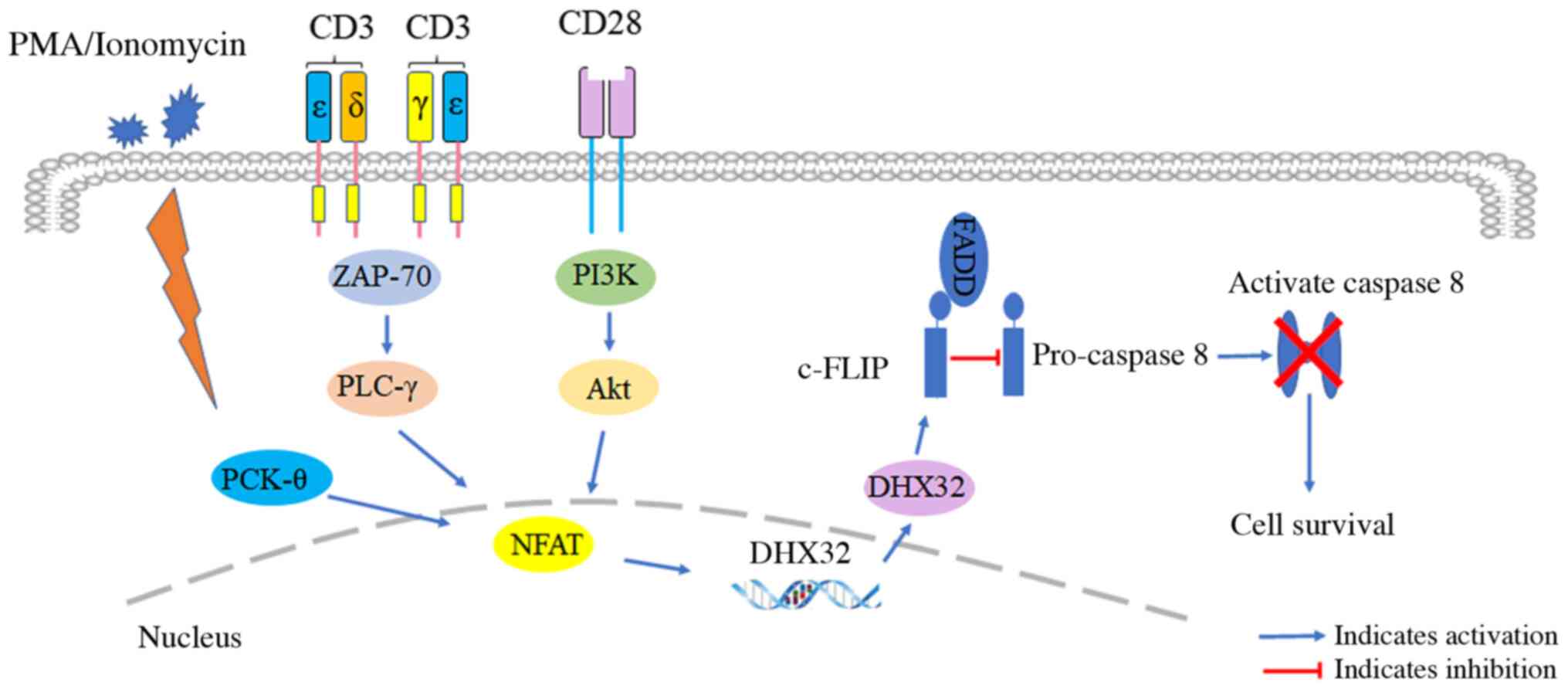
In 2006, it was reported that DHX32 expression was
significantly upregulated when co-stimulated by CD3 and CD28
antibodies, or stimulated by phorbol 12-myristate 13-acetate, or
ionomycin alone in Jurkat cells with low background DHX32
expression (50). Noteworthy, only
the mRNA isoform transcribed from the proximal promoter was
expressed in activated Jurkat cells. The proximal promoter of DHX32
has a binding site for the nuclear transcription factor nuclear
factor of activated T-cells (NFAT) (50). It was speculated that DHX32
expression could be regulated by NFAT during T cell activation.
Forced expression of DHX32 in Jurkat cells did not affect cell
proliferation and the response to chemotherapeutic agents,
including actinomycin D and etoposide. However, forced DHX32
expression downregulated the expression of antiapoptotic protein
c-FLIP was associated with the Fas signaling pathway and promoted
apoptosis. In normal peripheral blood lymphocytes, co-stimulation
with phytohemagglutinin, and CD3 and CD28 antibodies upregulated
the expression of DHX32 and c-FLIP, and inhibited apoptosis
(51). Therefore, it has been
proposed that DHX32 overexpression can increase the response to
apoptosis associated with Fas signaling by regulating the
expression of c-FLP protein, contributing to tumorigenesis
(Fig. 4).
DHX32 and solid tumors
In 2009, by using mRNA differential display PCR to
screen the progression and metastasis-specific markers of
colorectal cancer, Huang et al (52) reported that DHX32 expression in
colorectal cancer was higher compared with adjacent non-cancerous
tissues, and the expression was significantly associated with tumor
thrombus formation, lymph node metastasis, histological grade and
Dukes stage of the cancer, indicating that abnormal DHX32
expression may be involved in colorectal cancer initiation and
progression. Furthermore, Lin et al (53) discovered that DHX32 promoted
colorectal cell proliferation, invasion and migration. The
quantitative PCR analyses identified that genes involved in cell
proliferation, apoptosis, invasion and migration were regulated by
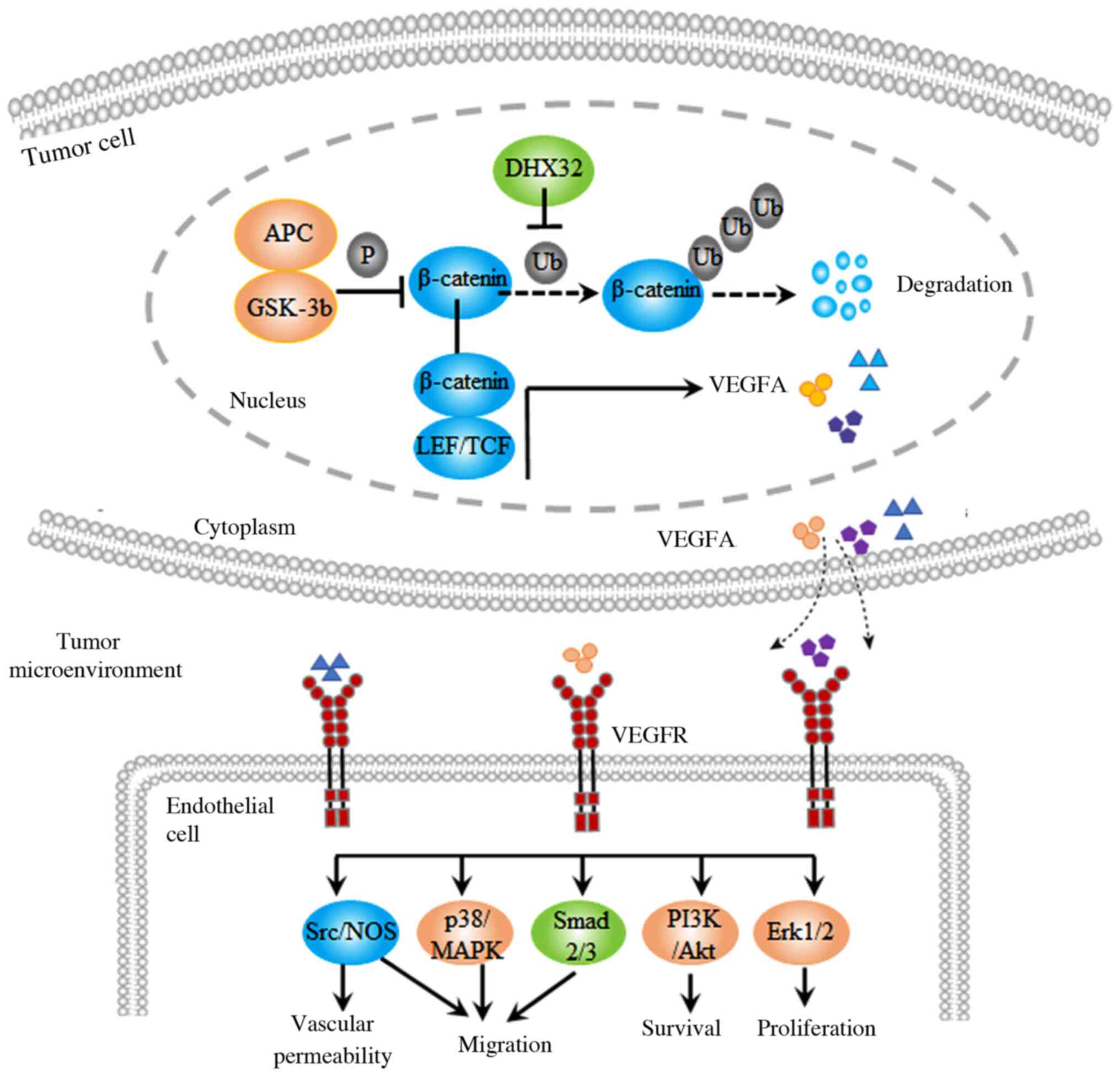
DHX32, which included VEGFA. Further research demonstrated that
DHX32 increased the transcriptional regulatory activity of
β-catenin, which is an upstream regulator of the VEGFA gene. DHX32
upregulation inhibited β-catenin ubiquitination, prolonged the
half-life of β-catenin and improved the stability of β-catenin,
thus promoting VEGFA expression. DHX32 knockdown significantly
inhibited colorectal cancer xenograft growth and angiogenesis in
nude mice (54). The results
indicated that DHX32 induces VEGFA expression by augmenting
β-catenin signaling, thereby promoting angiogenesis in colorectal
cancer (Fig. 5).
In solid tumors, DHX32 expression has been evaluated
in pathological samples. In colorectal cancer, the association
between DHX32 expression levels and clinical pathology was analyzed
in 139 colorectal cancer tissues and 93 corresponding adjacent
non-cancerous tissues via immunohistochemistry. The results
demonstrated that DHX32 was upregulated in colorectal cancer
tissues compared with adjacent tissues, and DHX32 expression was
associated with tumor microvascular density, degree of
differentiation and poor patient prognosis (54). Similarly, in a cohort of 193 patients
with breast cancer, DHX32 mRNA and protein expression levels were
increased compared with adjacent non-cancerous tissues, and there
was statistical significance between DHX32 expression and breast
cancer clinical stage, histological grade, lymph node metastasis
and proliferation marker Ki-67. Kaplan-Meier survival analysis
revealed that increased DHX32 expression was associated with poor
prognosis in breast cancer. Moreover, the Cox proportional hazard
model suggested that DHX32 expression was an independent prognostic
factor for low survival and disease-free survival in breast cancer
(55). In summary, DHX32 expression
is upregulated in colorectal and breast cancer, and its expression
is significantly associated with the occurrence, development and
poor prognosis of the cancer, indicating that DHX32 might serve as
a novel biomarker for colorectal and breast cancer.
Perspective and future direction
DHX32 has a unique helicase domain that contains
eight conserved motifs that are different from other RNA helicases.
The variation of these conserved amino acid residues does not
affect the ATPase activity, but does alter the unwinding activity.
The function of RNA helicase does not simultaneously depend on
ATPase activity and unwinding activity. Alternative splicing is a
post-transcriptional modification process that allows a gene to
encode multiple proteins that have similar structures but different
functions. Abnormal splicing is closely associated with cancer
initiation and progression. Some splicing variants are specifically
expressed in tumor tissues and can be used as biomarkers for tumor
diagnosis and targets for cancer treatments (56). DHX32 has two isomers; however,
whether their ATPase and helicase activities are similar, whether
they share same expression pattern in cancer and how they affect
cancer cell growth, differentiation and survival are not completely
understood and require further investigation.
DHX32 expression is significantly upregulated in
colorectal and breast cancer. In addition, the abundance of DHX32
in colorectal and breast cancer is associated with the clinical and
pathological features of the patients. DHX32 does not have a signal
peptide and is an intracellular protein. However, several
non-secretory proteins, including DDX48 helicase (16) and hsp70 (57,58), can
be detected in the circulation and serve as biomarkers for cancer,
since overexpressed proteins in cancer cells are often discharged
into the circulation. Thus, whether DHX32 proteins can be detected
in peripheral blood of the patients requires further investigation.
In addition, additional bioinformatics analyses are required to
establish the association between the abundance of DHX32, and
clinical and pathological features of samples. Therefore,
developing a novel method for assessing DHX32 protein in peripheral
blood and liquid biopsy samples, and determining whether DHX32 can
serve as a biomarker for screening, diagnosis and prognosis of
colorectal and breast cancer are important for translational
medicine.
DHX32 expression is dysregulated in cancer cells,
indicating its potential as a target for anticancer treatment.
However, this is an under-explored area of research as the majority
of current efforts to develop drugs targeting helicase are focused
on DDX3 (59,60). RK-33 is a small molecule inhibitor
that binds to the ATP-binding site of DDX3, inhibiting its enzyme
activity. Preclinical studies have reported that RK-33 is a
promising drug for treating breast cancer. It was hypothesized that
DHX32 may serve as a target for controlling the Wnt signaling
pathway, thus could be used for the treatment of colorectal and
breast cancer. However, the structure of DHX32 has not been
previously reported. Therefore, the molecular structure of DHX32
should be identified to facilitate the development of drugs to
target DHX32.
DHX32 is a multifunctional protein that regulates
ribosome biosynthesis, transcription, splicing and translation of
mRNA, as well as numerous other biological activities. DHX32 serves
important roles in cell proliferation, differentiation and
apoptosis, promoting cancer initiation and progression, as well as
tumor angiogenesis. Interestingly, in human acute lymphoblastic
leukemia, DHX32 expression is downregulated, but in colorectal and
breast cancer, DHX32 expression is significantly upregulated. The
mechanism underlying abnormal DHX32 expression in tumors is not
completely understood; therefore, further investigation is
required.
Acknowledgements
Not applicable.
Funding
This work was supported by the Youth Foundation
Project of Fujian Provincial Health Department (grant no.
2014-2-69); Fujian Provincial Health and Family Planning
Commission, Fujian, China (grant no. 2016J01626).
Availability of data and materials
Not applicable.
Authors' contributions
QW researched the topic literation, designed the
study and wrote the initial manuscript. JG, YC, HL, JW, ZF and FW
revised for important intellectual content, ZZ revised for
important intellectual content and supervised. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jankowsky E, Gross CH, Shuman S and Pyle
AM: Active disruption of an RNA-protein interaction by a DExH/D RNA
helicase. Science. 291:121–125. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fairman-Williams ME, Guenther UP and
Jankowsky E: SF1 and SF2 helicases: Family matters. Curr Opin
Struct Biol. 20:313–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He Y, Andersen GR and Nielsen KH:
Structural basis for the function of DEAH helicases. EMBO Rep.
11:180–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith WA, Schurter BT, Wong-Staal F and
David M: Arginine methylation of RNA helicase a determines its
subcellular localization. J Biol Chem. 279:22795–22798. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Singleton MR, Dillingham MS and Wigley DB:
Structure and mechanism of helicases and nucleic acid translocases.
Annu Rev Biochem. 76:23–50. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marchat LA, Arzola-Rodríguez SI,
Hernandez-de la Cruz O, Lopez-Rosas I and Lopez-Camarillo C:
DEAD/DExH-Box RNA Helicases in Selected Human Parasites. Korean J
Parasitol. 53:583–595. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du Pont KE, Davidson RB, McCullagh M and
Geiss BJ: Motif V regulates energy transduction between the
flavivirus NS3 ATPase and RNA-binding cleft. J Biol Chem.
295:1551–1564. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Robert F and Pelletier J: Perturbations of
RNA helicases in cancer. Wiley Interdiscip Rev RNA. 4:333–349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abdelhaleem M: RNA helicases: Regulators
of differentiation. Clin Biochem. 38:499–503. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Godbout R, Li L, Liu RZ and Roy K: Role of
DEAD box 1 in retinoblastoma and neuroblastoma. Future Oncol.
3:575–587. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Taunk NK, Goyal S, Wu H, Moran MS, Chen S
and Haffty BG: DEAD box 1 (DDX1) expression predicts for local
control and overall survival in early stage, node-negative breast
cancer. Cancer. 118:888–898. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Eberle J, Krasagakis K and Orfanos CE:
Translation initiation factor eIF-4A1 mRNA is consistently
overexpressed in human melanoma cells in vitro. Int J Cancer.
71:396–401. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shuda M, Kondoh N, Tanaka K, Ryo A,
Wakatsuki T, Hada A, Goseki N, Igari T, Hatsuse K, Aihara T, et al:
Enhanced expression of translation factor mRNAs in hepatocellular
carcinoma. Anticancer Res. 20:2489–2494. 2000.PubMed/NCBI
|
|
14
|
Heerma van Voss MR, Schrijver WAME, Ter
Hoeve ND, Hoefnagel LD, Manson QF, van der Wall E, Raman V and van
Diest PJ; Dutch Distant Breast Cancer Metastases Consortium, : The
prognostic effect of DDX3 upregulation in distant breast cancer
metastases. Clin Exp Metastasis. 34:85–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Shen GH, Xie JM, Li B and Gao QG:
Rottlerin upregulates DDX3 expression in hepatocellular carcinoma.
Biochem Biophys Res Commun. 495:1503–1509. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia Q, Kong XT, Zhang GA, Hou XJ, Qiang H
and Zhong RQ: Proteomics-based identification of DEAD-box protein
48 as a novel autoantigen, a prospective serum marker for
pancreatic cancer. Biochem Biophys Res Commun. 330:526–532. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin J, Chen Q, Yang J, Qian J, Deng ZQ,
Qian W, Chen XX, Ma JC, Xiong DS, Ma YJ, et al: DDX43 promoter is
frequently hypomethylated and may predict a favorable outcome in
acute myeloid leukemia. Leuk Res. 38:601–607. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma N, Xu HE, Luo Z, Zhou J, Zhou Y and Liu
M: Expression and significance of DDX43 in lung adenocarcinoma. Pak
J Pharm Sci. 30 (Suppl 4):1491–1496. 2017.PubMed/NCBI
|
|
19
|
Dai L, Pan G, Liu X, Huang J, Jiang Z, Zhu
X, Gan X, Xu Q and Tan N: High expression of ALDOA and DDX5 are
associated with poor prognosis in human colorectal cancer. Cancer
Manag Res. 10:1799–1806. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taniguchi K, Sugito N, Kumazaki M,
Shinohara H, Yamada N, Matsuhashi N, Futamura M, Ito Y, Otsuki Y,
Yoshida K, et al: Positive feedback of DDX6/c-Myc/PTB1 regulated by
miR-124 contributes to maintenance of the Warburg effect in colon
cancer cells. Biochim Biophys Acta. 1852:1971–1980. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He Y, Zhang D, Yang Y, Wang X, Zhao X,
Zhang P, Zhu H, Xu N and Liang S: A double-edged function of DDX3,
as an oncogene or tumor suppressor, in cancer progression (Review).
Oncol Rep. 39:883–892. 2018.PubMed/NCBI
|
|
22
|
Kim KH, Kang YJ, Jo JO, Ock MS, Moon SH,
Suh DS, Yoon MS, Park ES, Jeong N, Eo WK, et al: DDX4 (DEAD box
polypeptide 4) colocalizes with cancer stem cell marker CD133 in
ovarian cancers. Biochem Biophys Res Commun. 447:315–322. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schudrowitz N, Takagi S, Wessel GM and
Yajima M: Germline factor DDX4 functions in blood-derived cancer
cell phenotypes. Cancer Sci. 108:1612–1619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Janknecht R: Multi-talented DEAD-box
proteins and potential tumor promoters: p68 RNA helicase (DDX5) and
its paralog, p72 RNA helicase (DDX17). Am J Transl Res. 2:223–234.
2010.PubMed/NCBI
|
|
25
|
Taniguchi K, Iwatsuki A, Sugito N,
Shinohara H, Kuranaga Y, Oshikawa Y, Tajirika T, Futamura M,
Yoshida K, Uchiyama K, et al: Oncogene RNA helicase DDX6 promotes
the process of c-Myc expression in gastric cancer cells. Mol
Carcinog. 57:579–589. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bhattacharya C, Wang X and Becker D: The
DEAD/DEAH box helicase, DDX11, is essential for the survival of
advanced melanomas. Mol Cancer. 11:822012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Liu L, Liu X, Xu P, Hu Q and Yu Y:
The role of upregulated DDX11 as a potential prognostic and
diagnostic biomarker in lung adenocarcinoma. J Cancer.
10:4208–4216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vychytilova-Faltejskova P, Svobodova
Kovarikova A, Grolich T, Prochazka V, Slaba K, Machackova T,
Halamkova J, Svoboda M, Kala Z, Kiss I, et al: MicroRNA biogenesis
pathway genes are deregulated in colorectal cancer. Int J Mol Sci.
20:202019. View Article : Google Scholar
|
|
29
|
Zhang T, Ma Z, Liu L, Sun J, Tang H, Zhang
B, Zou Y and Li H: DDX39 promotes hepatocellular carcinoma growth
and metastasis through activating Wnt/β-catenin pathway. Cell Death
Dis. 9:6752018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sugiura T, Nagano Y and Noguchi Y: DDX39,
upregulated in lung squamous cell cancer, displays RNA helicase
activities and promotes cancer cell growth. Cancer Biol Ther.
6:957–964. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee S, Baek M, Yang H, Bang YJ, Kim WH, Ha
JH, Kim DK and Jeoung DI: Identification of genes differentially
expressed between gastric cancers and normal gastric mucosa with
cDNA microarrays. Cancer Lett. 184:197–206. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hellman K, Alaiya AA, Becker S, Lomnytska
M, Schedvins K, Steinberg W, Hellström AC, Andersson S, Hellman U
and Auer G: Differential tissue-specific protein markers of vaginal
carcinoma. Br J Cancer. 100:1303–1314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cho B, Lee H, Jeong S, Bang YJ, Lee HJ,
Hwang KS, Kim HY, Lee YS, Kang GH and Jeoung DI: Promoter
hypomethylation of a novel cancer/testis antigen gene CAGE is
correlated with its aberrant expression and is seen in premalignant
stage of gastric carcinoma. Biochem Biophys Res Commun. 307:52–63.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park SY, Kim WJ, Byun JH, Lee JJ, Jeoung
D, Park ST and Kim Y: Role of DDX53 in taxol-resistance of cervix
cancer cells in vitro. Biochem Biophys Res Commun. 506:641–647.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao S, Sun R, Wang W, Meng X, Zhang Y,
Zhang N and Yang S: RNA helicase DHX9 may be a therapeutic target
in lung cancer and inhibited by enoxacin. Am J Transl Res.
9:674–682. 2017.PubMed/NCBI
|
|
36
|
Ito S and Koso H: RNA helicase DHX15 acts
as a tumor suppressor in glioma. Cancer Science. Wiley; Hoboken,
NJ, USA: 109. pp. 782018
|
|
37
|
Abdelhaleem M: The novel helicase
homologue DDX32 is down-regulated in acute lymphoblastic leukemia.
Leuk Res. 26:945–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alli Z, Ho M and Abdelhaleem M: Expression
of DHX32 in lymphoid tissues. Exp Mol Pathol. 79:219–223. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Landry JR, Mager DL and Wilhelm BT:
Complex controls: The role of alternative promoters in mammalian
genomes. Trends Genet. 19:640–648. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tai CL, Pan WC, Liaw SH, Yang UC, Hwang LH
and Chen DS: Structure-based mutational analysis of the hepatitis C
virus NS3 helicase. J Virol. 75:8289–8297. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Caruthers JM and McKay DB: Helicase
structure and mechanism. Curr Opin Struct Biol. 12:123–133. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schneider S, Hotz HR and Schwer B:
Characterization of dominant-negative mutants of the DEAH-box
splicing factors Prp22 and Prp16. J Biol Chem. 277:15452–15458.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Alli Z, Ackerley C, Chen Y, Al-Saud B and
Abdelhaleem M: Nuclear and mitochondrial localization of the
putative RNA helicase DHX32. Exp Mol Pathol. 81:245–248. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Di Liegro CM, Schiera G and Di Liegro I:
Regulation of mRNA transport, localization and translation in the
nervous system of mammals (Review). Int J Mol Med. 33:747–762.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Alli Z, Ho M, Ackerley C and Abdelhaleem
M: Characterization of murine Dhx32. Exp Mol Pathol. 83:115–118.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Iborra FJ, Kimura H and Cook PR: The
functional organization of mitochondrial genomes in human cells.
BMC Biol. 2:92004. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen Y, Alli Z, Ackerley C, Al-Saud B and
Abdelhaleem M: Altered distribution of heat shock protein 60
(Hsp60) with dysregulated expression of DHX32. Exp Mol Pathol.
82:256–261. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Abdelhaleem M, Sun TH and Ho M: DHX32
expression suggests a role in lymphocyte differentiation.
Anticancer Res. 25:2645–2648. 2005.PubMed/NCBI
|
|
49
|
McNeer NA, Philip J, Geiger H, Ries RE,
Lavallée VP, Walsh M, Shah M, Arora K, Emde AK, Robine N, et al:
Genetic mechanisms of primary chemotherapy resistance in pediatric
acute myeloid leukemia. Leukemia. 33:1934–1943. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Alli Z, Nam EH, Beimnet K and Abdelhaleem
M: The activation-induced expression of DHX32 in Jurkat T cells is
specific and involves calcium and nuclear factor of activated T
cells. Cell Immunol. 237:141–146. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Alli Z, Chen Y, Abdul Wajid S, Al-Saud B
and Abdelhaleem M: A role for DHX32 in regulating T-cell apoptosis.
Anticancer Res. 27:373–377. 2007.PubMed/NCBI
|
|
52
|
Huang C, Liang X, Huang R and Zhang Z:
Up-regulation and clinical relevance of novel helicase homologue
DHX32 in colorectal cancer. Journal of experimental & clinical
cancer research. CR (East Lansing Mich). 28:112009.
|
|
53
|
Lin H, Liu W, Fang Z, Liang X, Li J, Bai
Y, Lin L, You H, Pei Y, Wang F, et al: Overexpression of DHX32
contributes to the growth and metastasis of colorectal cancer. Sci
Rep. 5:92472015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lin H, Fang Z, Su Y, Li P, Wang J, Liao H,
Hu Q, Ye C, Fang Y, Luo Q, et al: DHX32 promotes angiogenesis in
colorectal cancer through augmenting β-catenin signaling to induce
expression of VEGFA. EBioMedicine. 18:62–72. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang M, Zhang G, Wang Y, Ma R, Zhang L, Lv
H, Fang F and Kang X: DHX32 expression is an indicator of poor
breast cancer prognosis. Oncol Lett. 13:942–948. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Pajares MJ, Ezponda T, Catena R, Calvo A,
Pio R and Montuenga LM: Alternative splicing: An emerging topic in
molecular and clinical oncology. Lancet Oncol. 8:349–357. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhan R, Leng X, Liu X, Wang X, Gong J, Yan
L, Wang L, Wang Y, Wang X and Qian LJ: Heat shock protein 70 is
secreted from endothelial cells by a non-classical pathway
involving exosomes. Biochem Biophys Res Commun. 387:229–233. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lancaster GI and Febbraio MA:
Exosome-dependent trafficking of HSP70: A novel secretory pathway
for cellular stress proteins. J Biol Chem. 280:23349–23355. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bol GM, Khan R, Heerma van Voss MR,
Tantravedi S, Korz D, Kato Y and Raman V: PLGA nanoparticle
formulation of RK-33: An RNA helicase inhibitor against DDX3.
Cancer Chemother Pharmacol. 76:821–827. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Heerma van Voss MR, Vesuna F, Bol GM,
Afzal J, Tantravedi S, Bergman Y, Kammers K, Lehar M, Malek R,
Ballew M, et al: Targeting mitochondrial translation by inhibiting
DDX3: A novel radiosensitization strategy for cancer treatment.
Oncogene. 37:63–74. 2018. View Article : Google Scholar : PubMed/NCBI
|