Introduction
Glioma (GM) is the most common type of malignant
brain tumor, with high recurrence, accounting for 74.6% of all
malignant central nervous system tumors in America between 2009 and
2013 (1). The morbidity of GM is
higher in men (56.3%) compared with in women (43.7%) (2). Even with aggressive treatments, such as
neurosurgical resection followed by radical combined
radiochemotherapies, these notoriously infiltrative tumors
invariably recur, and there has been no significant improvement in
the overall survival rate of patients with GM in recent decades
(3). Therefore, understanding the
complex biological characteristics of GM and identifying suitable
prognostic biomarkers are urgently needed.
Circular RNAs (circRNAs), a novel group of conserved
RNAs, exert pivotal functions in mammalian cells (4). The present evidence demonstrates that
circRNAs regulate the levels of miRNAs as sponges for miRNAs or
alter gene transcription by binding to RNA-binding proteins
(5,6). In recent years, circRNAs have been
reported to be closely associated with several diseases, such as
cancer and diabetes (7–9). Moreover, they are essential for the
development of GM. For example, circ-HIPK3 acts as a prognostic
biomarker for GM and promotes cell aggressiveness by targeting
miR-654/insulin-like growth factor 2 mRNA-binding protein 3
signaling (10). Another study
indicated that circPTN sponges miR-145-5p/miR-330-5p to promote
proliferation and stemness in GM (11). Androgen receptor (AR) is a
transcription factor that regulates eukaryotic gene expression and
affect cellular proliferation and differentiation in target tissues
(12). Suppressor of cytokine
signaling 2-antisense RNA 1 (SOCS2-AS1) is a type of long
non-coding RNA (lncRNA) that acts as oncogenic function in human
cancer progression (13). Homo
sapien (hsa)circRNA_104634 is spliced from the
aspartyl/asparaginyl β-hydroxylase (ASPH) gene
(chr8:62593526-62596747) and the spliced length is 264 nucleotides.
The current research aimed to identify the functions and possible
mechanisms of hsa_circRNA_104634 (circ-ASPH) in regulating GM cell
progression.
Materials and methods
Patients and tissues
In total, 60 fresh GM/non-tumor samples were
obtained during surgical resection from the patients with GM at the
Qiqihar Hospital Affiliated with Southern Medical University
(Qiqihar, China) from January 2014 to January 2016. The cohort
consists of 60 patients (age range: 17–67 years old; 39 male, 21
female). The inclusion criteria are as following: i) The patients
tissues were examined by at least two experienced pathologists; ii)
the patients underwent radical resection with a clear surgical
margin and the adjacent non-tumorous tissues were at least 1-cm
away from the tumor edge; iii) the patients with available
follow-up information (over the phone); iv) none of the patients
received anticancer treatment before the surgery and no history of
other types of cancer. The exclusion criteria are that the patients
with serious diseases or severe chronic diseases. Overall survival
time was determined as survival time after surgery. The specimens
were frozen in liquid nitrogen immediately after surgery and stored
at liquid nitrogen (−196°C). The project was authorized by The
Ethics Committee of Qiqihar Hospital Affiliated with Southern
Medical University and written informed consent was obtained from
all patients. The patients were divided into high- and
low-circ-ASPH groups according to the median expression level of
circ-ASPH in cancer tissues.
Cell culture and transfection
GM cell lines including U251, U87MG (glioblastoma of
unknown origin), LN229 and normal human astrocyte cells (NHAs) were
acquired from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences or The American Type Culture
Collection. The cells were cultivated in RPMI-1640 medium (HyClone;
Cyvita) containing 10% fetal bovine serum (FBS) (Gibco; Thermo
Fisher Scientific, Inc.) in a 37°C, 5% CO2
incubator.
MicroRNA (miR)-599 mimics, miR-599 inhibitor and
scramble oligonucleotide were obtained from Shanghai GenePharma
Co., Ltd. circ-ASPH/AR small interfering (si)RNAs and
overexpression vectors were obtained from Guangzhou RiboBio Co.,
Ltd. Transfection was conducted when cell confluence reached 60–80%
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
LN229 cells were used for knockdown experiments (transfection with
si-circ-ASPH-1/-2 or si-NC) because it has the highest expression
of circ-ASPH. U87MG cells were used for overexpression due to its
lowest circ-ASPH expression (transfection with circ-ASPH vector or
empty vector). The targeted sequences of siRNA-circ-ASPH are listed
as follows: si-circ-ASPH-1, 5′-AGTTTTATTAGAGACAAAGCA-3′ And
si-circ-ASPH-2, 5′-CCAAAGTTTTATTAGAGACAA-3′. si-AR sequence was
5′-AAGAUAAUAACUCAGUUCUUATT-3′. si-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. miR-599 mimics sequence was
5′-GUUGUGUCAGUUUAUCAAAC-3′. miR-599 inhibitor sequence was
5′-GUUUGAUAAACUGACACAAC-3′. Mimics-NC sequence was
5′-UUCUCCGAACGUGUCACGU-3′. Inhibitor-NC sequence was
5′-CAGUACUUUUGUGUAGUACAA-3′. In total, 5 µl of siRNA (20 µM) or 2.5
µg of plasmid vector with 5 µl of P3000™ reagent was diluted in 125
µl serum-free medium. After 5 min of incubation at 22–25°C, the
reagents in the two tubes were combined. After 15–20 min, the
mixtures were added into a six-well plate with serum-free medium.
Following 8 h of incubation at room temperature, the medium was
r-eplaced with medium containing 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). Total RNA isolation was performed at 48 h after
transfection. Protein extraction was conducted at 72 h after
transfection.
circRNA sequencing
Total RNA from 4 pairs of tumor/adjacent nontumorous
tissues was extracted using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's instructions.
Non-circular RNAs were digested by ribonuclease R. circRNAs were
amplified and transcribed into fluorescent cRNA by the random
priming method (14). Arrays were
scanned using the Illumina sequencing platform (Illumina,
Inc.).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA from GM specimens and cells was obtained
using TRIzol reagent. Reverse transcription assay was performed
using SuperScript RT kit in accordance with the manufacturer's
protocol (Invitrogen; Thermo Fisher Scientific, Inc.). The reaction
was set at 25°C for 10 min, 42°C for 60 min, 85°C for 5 min.
SYBR-Green qPCR master mix (Invitrogen; Thermo Fisher Scientific,
Inc.) was applied to conduct RT-qPCR. The reaction volume was 10
µl. The thermocycling conditions were as follows: 90°C For 5 min,
then 90°C for 15 sec, 60°C for 30 sec for 45 cycles. The
2−ΔΔCq method was employed to analyze gene expression
(15). For circRNA and mRNA
quantification, GAPDH was used as the internal reference. For miRNA
(small non-coding RNA) quantification, U6 was used as the internal
control. The primers for circ-ASPH, GAPDH and U6 were as follows:
circ-ASPH: 5′-AACTTATCAGAGGTGCTTCAAGG-3′ (Forward) and
5′-GAAGTTCCTGAGAGTCCGCC-3′ (reverse), ASPH:
5′-CATGGAGGACACAAGAATGGG-3′ (forward) and
5′-CCAAACGACAGCTACAGATGT-3′ (reverse), AR:
5′-GACGACCAGATGGCTGTCATT-3′ (forward) and 5′-GGGCGAAGTAGAGCATCCT-3′
(reverse), SOCS2-AS1: 5′-CCATACAGGTCAACTTTTCCACCAC-3′ (forward) and
5′-CCAACCTCAGCTCTGCTCTCTT-3′ (reverse), GAPDH:
5′-GGGAGCCAAAAGGGTCAT-3′ (forward) and 5′-GAGTCCTTCCACGATACCAA-3′
(reverse), miR-599: 5′-GTTGTGTCAGTTTATCAAAC-3′ (forward) and
5′-%CTCCATATCGCACTTTAATCTCTAACT-3′ (reverse), U6:
5′-ATTGGAACGATACAGAGAAGATT-3′ (forward) and
5′-GGAACGCTTCACGAATTTG-3′ (reverse).
Treatment of Actinomycin D
After cell inoculation in six-well plates
(2.5×105/well), the transcriptional inhibitor
actinomycin D (EMD Millipore) was added to the culture medium at 2
mg/ml for respective 0, 4, 8, 12 and 24 h. Then, total RNA was
extracted and RT-qPCR was used to detect the expression of
circ-ASPH and linear ASPH mRNA as aforementioned. The detection
methods used for RNA isolation and RT-qPCR were identical to
aforementioned.
Subcellular fractionation test
A PARIS kit (cat. no. AM1921; Thermo Fisher
Scientific, Inc.) was used to separate RNAs in the cytoplasmic and
nuclear fractions in accordance with the manufacturer's
instructions, followed by RT-qPCR with U6 and GAPDH as the nuclear
and cytoplasmic controls, respectively.
RNA immunoprecipitation (RIP)
A RIP assay was conducted using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore). GM
cells were transfected for 48 h. The adherent cells were cultured
until 80–90% confluency in dishes. In total, 2.0×107
cells were lysed using 200 µl complete RIP lysis buffer (EMD
Millipore). The cell lysates were incubated with 50 µl magnetic
beads protein A/G conjugated with 5 µg antibodies against Argonaute
2 (anti-Ago2; 1:30; cat. no. ab186733; Abcam) or IgG (anti-IgG;
1:50; cat. no. PP64B; EMD Millipore) for 30 min at room
temperature. Then 900 µl of RIP immunoprecipitation buffer was
added to each tube, which were incubated with rotation for 6 h at
4°C. The beads were then washed and incubated with 150 µl
proteinase K buffer for RNA purification at 55°C for 30 min with
shaking to digest the protein. After purification, enriched
circ-ASPH was quantified using RT-qPCR as aforementioned.
RNA pull-down assay
In total, 100 pmol biotin-labeled circ-ASPH and
oligonucleotide probes were incubated with 50 µl of magnetic
streptavidin beads (cat. no. 20164; Thermo Fisher Scientific, Inc.)
for 2 h at 22–25°C. LN229 and U87MG cells were lysed using Pierce
IP Lysis Buffer (Thermo Fisher Scientific, Inc.) according to the
manufacturer's manual. RNAs wer purified with TRIzol and the lysed
samples were sonicated. The sonication procedure is five series of
30 sec on (20 kHz) and 30 sec off at 4°C. Immediately after
sonication, samples were centrifuged for 5 min at 12,000 × g at
4°C. After adding hybridization buffer to the supernatants, 20 µl
of samples were then incubated overnight with the magnetic bead
mixture at 4°C. The supernatant was discarded and beads were washed
with 900 µl of wash buffer. In total, 95 µl of proteinease K buffer
and 5 µl of proteinase-K were added to the samples. The samples
were incubated for 45 min at 50°C then 10 min at 95°C. Samples were
chilled on ice for 3 min before separating the beads from RNAs.
RNAs were purified and DNA were removed using an RNA Purification
kit (cat. no. 12183555; Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's instructions. After
purification, enriched circ-ASPH and miRNAs were quantified by
RT-qPCR. A negative control was also performed using scrambled
oligonucleotide probes.
Dual-luciferase reporter assay
CircRNA Interactome (https://circinteractome.nia.nih.gov) and starBase 2.0
(http://starbase.sysu.edu.cn) were used
to predict the target miRNAs of circ-ASPH (16). The target genes of miR-599 were
predicted using the target gene prediction software, starBase 2.0
(17). Data from The Cancer Genome
Atlas (TCGA) datasets were analyzed using starBase 2.0. Mutant
(mut) or wild-type (wt) circ-ASPH and the 3′-untranslated region
(UTR) of AR containing the predicted binding site for miR-599 were
amplified using PCR and cloned into the pmiR-Repot vector (YouBio).
Wt or mut plasmids (50 ng) were cotransfected with scrambled
mimics-negative control (NC) or miR-599 mimics (20 nM; Shanghai
GenePharma Co., Ltd.) into 293T (Type Culture Collection of The
Chinese Academy of Sciences, Shanghai, China) cells using
Lipofectamine 3000. A luciferase assay was conducted using a dual
luciferase reporter assay kit (Promega Corporation) after 36 h of
transfection according to the manufacturer's instructions. The
specific target activity was expressed as the relative activity
ratio of firefly luciferase to Renilla luciferase.
Immunoblotting assay
Cell lysates harvested at 72 h post-transfection
were prepared using RIPA reagent (Beyotime Institute of
Biotechnology). A BCA assay kit (Beyotime Institute of
Biotechnology) was used to measure the concentration of proteins.
After that, all samples were incubated in boiling water for 10 min
to achieve protein denaturation. Then, 40 µg per lane of protein
from each sample was subjected to 10% SDS-PAGE electrophoresis to
separate proteins with different molecular weights. Proteins were
transferred to PVDF membranes, followed by blocking with non-fat
milk at room temperature for 1 h. The blocked membranes were then
incubated with GAPDH (1:10,000; cat. no. ab181602) and AR (1:1,000;
cat. no. ab198394) rabbit primary antibodies (both Abcam) for 12 h
at 4°C, followed by incubation with goat anti-rabbit IgG (HRP)
secondary antibody (1:5,000; cat. no. ab6721; Abcam) at room
temperature for 2 h. ECL™ Blotting Reagent (Sigma-Aldrich; Merck
KGaA) was used to develop the signals of each band.
Cell Counting Kit (CCK)-8 and
clone-forming assays
Cells were seeded onto 96-well plates at 2,000
cells/well after 48 h of transfection. The culture medium was
replaced with 90 µl of fresh medium and 10 µl of CCK-8 reagent
(Shanghai Yeasen Biotechnology Co., Ltd.) at the indicated time
points (0, 24, 48, 72 and 96 h). Absorption was detected at 450 nm
using a microplate reader (Thermo Fisher Scientific, Inc.). For the
clone-forming assay, 800 transfected cells were suspended in
RPMI-1640 medium and added to six-well plates. After culturing for
12 days, the cells were fixed with paraformaldehyde for 20 min at
room temperature and stained with crystal violet for another 20 min
at room temperature (Beyotime Institute of Biotechnology). Finally,
the colonies were manually counted.
Scratched wound assay
A scratched wound assay was conducted as described
previously (18). Briefly, LN229 and
U87MG cells were seeded in a six-well plate. The sample cells were
continuously cultured until the cell fusion rate reached >90%,
and then medium with a low concentration of serum (1%) was used to
replace the culture medium. The scraper was aligned at the lower
part of the plate and pushed up slightly to form a scratch. Images
were captured at preset time points (0 and 36 h for LN229 cells; 0
and 24 h for U87MG cells), and cell migration rates were then
calculated for each group. A light microscope was used for
visualizing and capturing the images. ImageJ version 1.50i software
(National Institutes of Health) was used to analyze the distance of
cell migrated.
Transwell experiments
A Transwell chamber (BD Biosciences) was employed to
determine cell migratory and invasive abilities. For the invasion
assay, the Transwell compartment was coated with Matrigel
(precooled at 4°C overnight) and placed in an incubator at 37°C for
4 h to form a reconstructed basement membrane. In total, 200 µl of
serum-free medium with the cells and 600 µl of corresponding medium
with 10% FBS were used for the upper and lower chambers,
respectively. After 24 h culturing at 37°C in a cell incubator, the
cells in the upper chamber were discarded, the cells on the lower
side of membrane were fixed with paraformaldehyde for 20 min at
room temperature and stained with crystal violet for another 20 min
at room temperature. The migratory or invasive cells were assessed
using a light microscope.
Statistical analysis
Data analyses were performed using SPSS 22.0
software (IBM, Corp.). For group comparisons, the differences
between two groups and multiple groups were analyzed via unpaired
Student's t-tests and one-way ANOVA with Tukey's test,
respectively. For comparisons between cancerous and adjacent normal
tissues, paired t-tests were used. Pearson's correlation analysis
was carried out to examine the correlation between miR-599,
circ-ASPH and AR. Survival curves were estimated by applying
Kaplan-Meier method and log-rank tests. P<0.05 was considered to
indicate a statistically significant difference.
Results
circ-ASPH expression is elevated in GM
and associated with an unfavorable prognosis
circRNA sequencing analysis revealed 128 circRNAs
(fold-change >2; P<0.05) that were differentially expressed
between four pairs of GM and adjacent normal controls. The
hierarchical clustering of the top 25 upregulated/downregulated
circRNAs is shown in Fig. 1A. GM and
non-cancerous tissues from the same four patients to verify the
most upregulated circRNAs, including hsa_circRNA_102211, _101314,
_104666, _001678, _100017 and _104634. Among these recruited
candidates, hsa_circRNA_104634 was the most upregulated circRNA
(Fig. 1B). hsa_circRNA_104634 was
spliced from the ASPH gene (chr8:62593526-62596747) and the spliced
length was 264 nucleotides. Thus, hsa_circRNA_104634 was named
circ-ASPH (Fig. 1C). The
differential expression of circ-ASPH in GM was then explored in a
large cohort of patients with GM (n=60). The data revealed that the
relative expression of circ-ASPH was significantly higher in GM
specimens compared with in non-tumorous tissues (P<0.01;
Fig. 1D). In addition, total RNA was
isolated, and RT-qPCR was used to measure circ-ASPH and linear ASPH
mRNA expression after treatment with actinomycin D at different
time points (0, 4, 8, 12 and 24 h). Linear ASPH mRNA had a shorter
half-life compared with circ-ASPH, suggesting the stability of
circ-ASPH (Fig. 1E). To further
confirm the clinical relevance of circ-ASPH, Kaplan-Meier analysis
was applied. The 60 patients with GM were divided into high- and
low-circ-ASPH groups in accordance with the median expression level
of circ-ASPH in cancer tissues. As a result, we found that
upregulation of circ-ASPH in tissue samples was associated with
lower overall survival rate for patients with GM after surgery
(Fig. 1F). The expression of
circ-ASPH in GM cell lines and NHAs was measured using RT-qPCR. The
expression levels of circ-ASPH were significantly higher in U251
and LN229 cells compared with in NHAs (P<0.01; Fig. 1G). Two siRNAs targeting the
back-spliced junction of circ-ASPH were used to silence circ-ASPH
expression in LN229 cells (Fig.
1H).
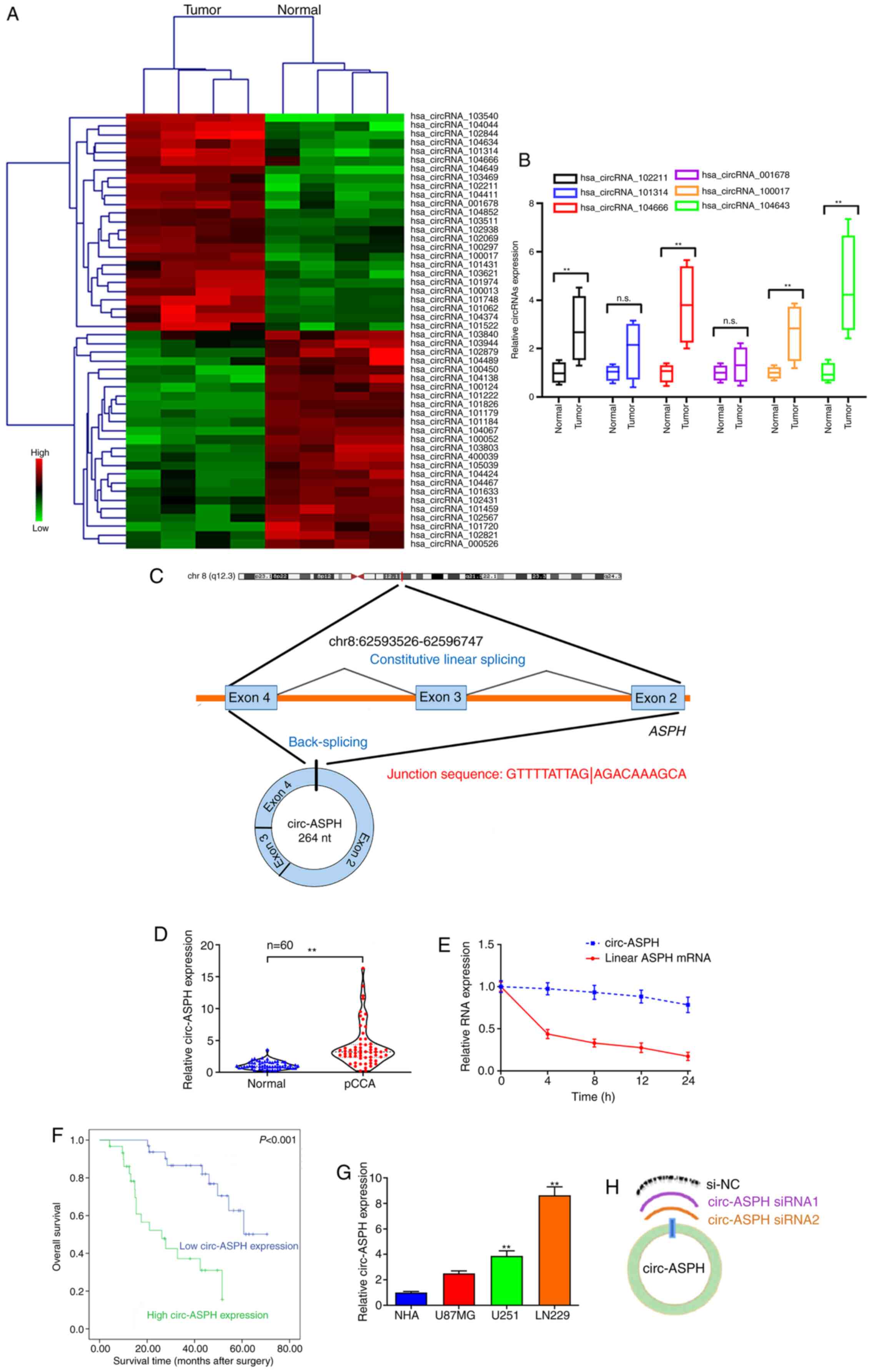 | Figure 1.circ-ASPH expression in GM tissues
and cells and its clinical value. (A) Clustered heatmap showing
tissue-specific circRNAs. (B) RT-qPCR for hsa_circRNA_102211,
_101314, _104666, _001678, _100017 and _104634 expression in
GM/non-cancerous specimens. (C) Schematic representation of
circ-ASPH formation. (D) circ-ASPH expression in 60 pairs of
GM/non-cancerous specimens by RT-qPCR. (E) Relative circ-ASPH and
linear ASPH mRNA expression at different time points. (F)
Kaplan-Meier analysis plots with log-rank test for overall survival
in patients with GM according to circ-ASPH expression. (G) Relative
expression of circ-ASPH in GM and NHA cells by RT-qPCR. (H)
Schematic for two circ-ASPH siRNAs targeting to the back-spliced
junction site of circ-ASPH. **P<0.01 vs. respective control. GM,
glioma; circ, circular; hsa, Homo sapien; RT-q, reverse
transcription-quantitative; NHA, normal human astrocyte; ASPH,
aspartyl/asparaginyl β-hydroxylase. |
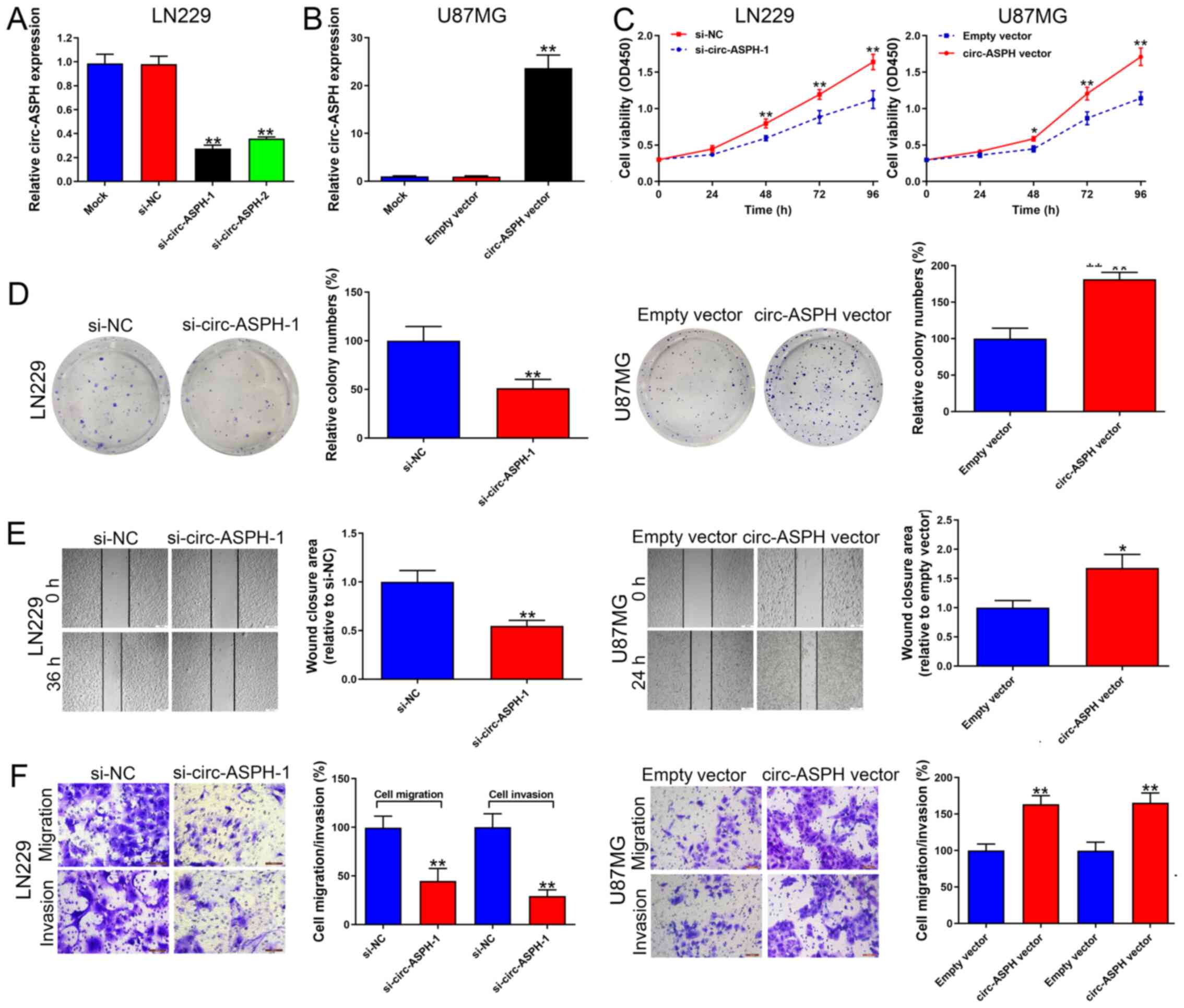
circ-ASPH accelerates GM cell
proliferation and aggressiveness
LN229 cells transfected with si-circ-ASPH-1 and
si-circ-ASPH-2 both strongly downregulated the expression of
circ-ASPH (vs. si-NC, both P<0.01; Fig. 2A). si-circ-ASPH-1 was chosen for
further functional study because it has a stronger knockdown
efficiency compared with si-circ-ASPH-2. Additionally, circ-ASPH
expression was upregulated in U87MG cells due to its low circ-ASPH
expression among the GM cell lines used in the present study. As a
result, circ-ASPH expression was significantly upregulated after
transfection with the circ-ASPH vector (vs. empty vector,
P<0.01; Fig. 2B). The CCK-8 and
colony formation results suggested that cell viability was
repressed by circ-ASPH-silencing in LN229 cells (Fig. 2C and D). In contrast,
circ-ASPH-overexpression resulted in higher cell viability and more
colonies formed in U87MG cells, suggesting the cell viability and
proliferation-promoting role of circ-ASPH (Fig. 2C and D). Depletion of circ-ASPH
significantly induced cell migratory potential inhibition in LN229
cells analyzed by wound healing and Transwell migration assays
(both P<0.01; Fig. 2E and F).
circ-ASPH elevation increased U87MG cell migration compared with
the empty vector group (P<0.05 and P<0.001; Fig. 2E and F). Afterwards, cell invasion
capacity was analyzed using a Matrigel invasion assay. It was found
that cell invasion was markedly attenuated in cells transfected
with si-circ-ASPH-1 compared with cells transfected with si-NC
(P<0.01; Fig. 2F). Moreover,
increased circ-ASPH levels led to more U87MG cells passing through
the Transwell membrane coated with Matrigel (P<0.01; Fig. 2F).
circ-ASPH sponges miR-599 to regulate
AR expression in GM cells
circ-ASPH was primarily localized to the cytoplasm
of LN229 and U87MG cell lines, as analyzed by a subcellular
distribution assay, indicating that circ-ASPH functions at the
post-transcriptional level (Fig.
3A). The potential interaction between circ-ASPH and miRNAs was
predicted using starBase 2.0 and circular RNA interactome online
databases. Only one miRNA, miR-599, was included in both databases
(Fig. 3B). circ-ASPH was
significantly enriched in the anti-Ago2 immunoprecipitated pool
compared with the anti-IgG pool (P<0.01). After knockdown of
circ-ASPH, the enrichment was partially decreased in both LN229 and
U87MG cells (anti-Ago2 si-NC vs. si-circ-ASPH-1; Fig. 3C). Additionally, overexpressed
circ-ASPH elevated the efficiency of pull-down using biotin-labeled
circ-ASPH probes (Fig. 3D).
Moreover, miR-599 was enriched in the circ-ASPH pull-downs from
LN229 and U87MG cells compared with the oligo probe group (both
P<0.01; Fig. 3E). Further
dual-luciferase reporter assays indicated that miR-599 mimics
markedly inhibited the luciferase signal of circ-ASPH wt but not
circ-ASPH mut, compared with mimics-NC (Fig. 3F and G). Bioinformatics analysis
predicted that AR is a potential target gene of miR-599. Moreover,
the results suggested that AR mRNA expression was significantly
elevated in GM cancerous tissue samples (P<0.01; Fig. 3H). Data from TCGA indicated that high
expression of AR in GM tissues is associated with worse overall
survival (Fig. 3I). Correlations
between circ-ASPH and AR mRNA/miR-599 expression were further
evaluated. A significant positive correlation between circ-ASPH and
AR mRNA expression was identified (R2=0.309, P=0.011;
Fig. 3J). However, the data failed
to demonstrate the negative correlation between circ-ASPH and
miR-599 expression (Fig. 3K). Next,
upregulation of AR in GM cell lines was identified (Fig. 3L). AR downregulation induced by
miR-599 mimics was also validated. Conversely, cell transfection
with the miR-599 inhibitor resulted in AR upregulation in U87MG
cells compared with the inhibitor-NC (P<0.01; Fig. 3M). A dual-luciferase reporter assay
demonstrated an inhibition of the luciferase activity of AR mRNA
3′-UTR wt, but did not change the luciferase intensity of AR mRNA
3′-UTR mut, confirming that AR mRNA 3′-UTR interacted with miR-599
(Fig. 3N and O).
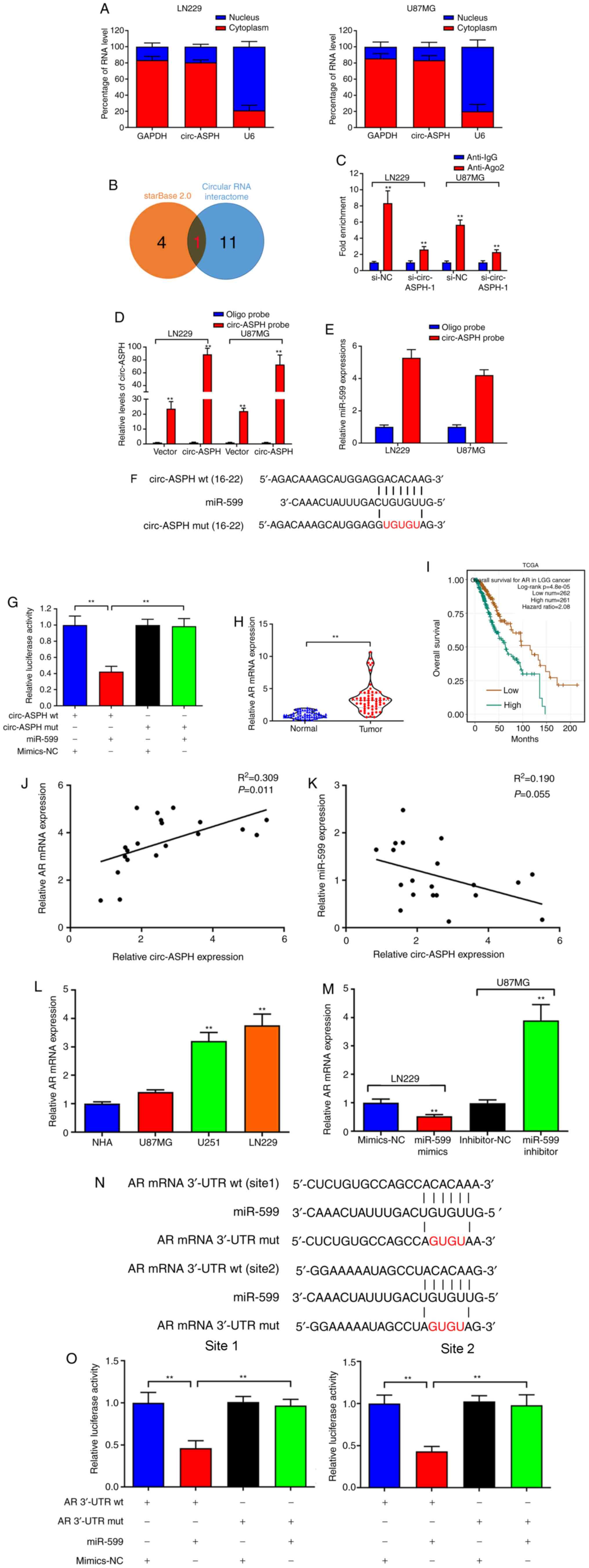 | Figure 3.circ-ASPH sponges miR-599 to
upregulate AR expression in GM. (A) RT-qPCR detection of the
percentage of circ-ASPH in the cytoplasmic and nuclear fractions of
LN229 and U87MG cells. (B) miRNAs that may be sponged by circ-ASPH
were predicted using the starBase 2.0 and circRNA interactome
databases. (C) Ago2-RNA immunoprecipitation assay for circ-ASPH
levels in LN229 and U87MG cells after transfection. (D) Lysates
prepared from LN229 and U87MG cells after transfection were
subjected to RNA pull-down assay. (E) RT-qPCR for miR-599
expression in LN229 and U87MG cell lysates. (F) Schematic
illustration of circ-ASPH-wt and circ-ASPH-mut luciferase reporter
vectors. (G) Binding ability between circ-ASPH and miR-599 was
detected by dual-luciferase reporter assay in 293T cells. (H)
Relative AR expression in GM/non-cancerous tissues analyzed using a
TCGA dataset. (I) Kaplan-Meier analysis for overall survival in
patients with GM according to AR expression in a TCGA dataset. (J
and K) Correlation analysis of circ-ASPH, miR-599 and AR mRNA
expression was explored in GM tissue samples. (L) Relative AR mRNA
expression in GM and NHA cells was detected by RT-qPCR. (M)
Relative AR mRNA expression was detected after transfection in
LN229 and U87MG cells by RT-qPCR. (N and O) Binding ability between
AR mRNA 3′-UTR and miR-599 was detected by dual-luciferase reporter
assay in 293T cells. **P<0.01 vs. respective control. GM,
glioma; circ, circular; RT-q, reverse transcription-quantitative;
miRNA/miR, microRNA; TCGA, The Cancer Genome Atlas; NHA, normal
human astrocytes; UTR, untranslated region; wt, wild-type; mut,
mutant; ASPH, ASPH, aspartyl/asparaginyl β-hydroxylase. |
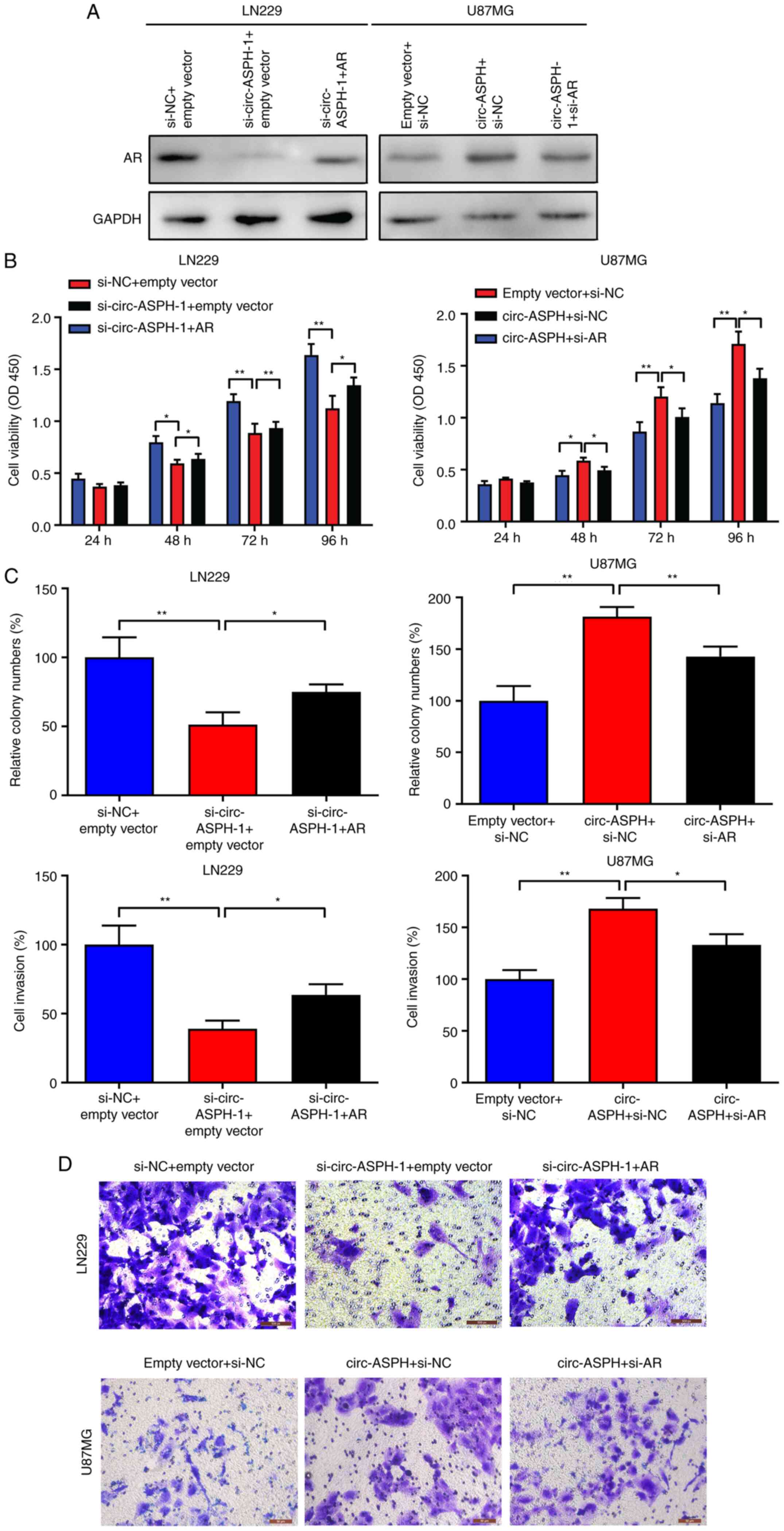
circ-ASPH facilitates cell
proliferation and invasion by regulating AR expression
To further verify whether the oncogenic function of
circ-ASPH is partly attributed to its regulation of AR, a rescue
assay was performed. As Fig. 4A
indicates, silencing circ-ASPH attenuated AR expression levels
compared with si-NC. However, this effect could be partly reversed
by cotransfection with si-circ-ASPH-1 and AR vector. Moreover,
overexpression of circ-ASPH led to enhanced expression of AR.
Further cotransfection with the circ-ASPH vector and si-AR partly
inhibited AR expression (Fig. 4A).
For the functional assay, CCK-8, clone-forming and Transwell
invasion assays showed that silencing of circ-ASPH attenuated cell
proliferation, clone formation and cell invasion in LN229 cells.
Meanwhile, cotransfection with si-circ-ASPH-1 and AR vector partly
promoted cell proliferation, colony formation and cell invasive
capacity (Fig. 4B-D). For U87MG
cells, overexpression of circ-ASPH contributed to cell
proliferation, clone forming and cell invasion. After
cotransfection with circ-ASPH vector and si-AR, the cell viability,
colony formation ability and invaded cells were partially decreased
(Fig. 4B-D).
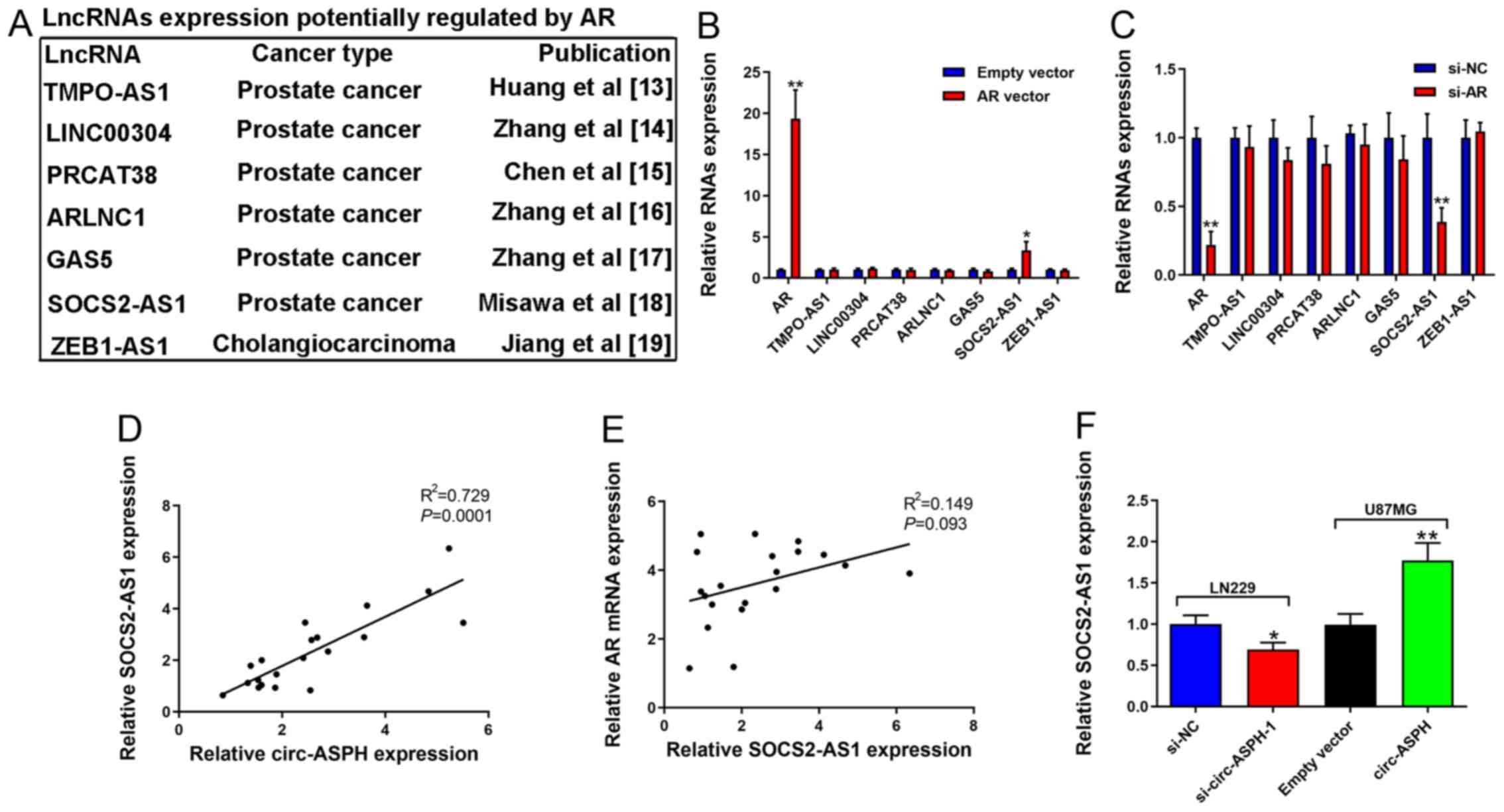
AR activates SOCS2-AS1 expression in
GM cells
AR functions as a steroid hormone-activated
transcription factor (19). A
previous study reported its downstream targets in various
malignancies, such as prostate cancer (20). The present study focused on lncRNAs
that have been reported as targets of AR in other cancer types,
including TMPO-AS1 (21), LINC00304
(22), PRCAT38 (23), ARLNC1 (24), GAS5 (25), SOCS2-AS1 (13), and ZEB1-AS1 (26) (Fig.
5A). It was identified that only the expression of SOCS2-AS1
was increased after AR overexpression (Fig. 5B). Consistent with expectations,
decreased AR markedly inhibited SOCS2-AS1 expression (Fig. 5C). There was a positive correlation
between circ-ASPH and SOCS2-AS1 expression (R2=0.729;
Fig. 5D). However, there was no
correlation between AR mRNA and SOCS2-AS1 expression (Fig. 5E). SOCS2-AS1 downregulation induced
by si-circ-ASPH-1 was validated. Similarly, elevated circ-ASPH led
to enhanced expression of SOCS2-AS1 (P<0.01; Fig. 5F).
Discussion
Recently, circRNAs have been clarified to regulate a
number important processes, such as metastasis, differentiation and
metabolism, leading to the progression of several diseases
including malignant tumors (27). In
previous studies, several circRNAs such as circ-FBXW7, circ-KIF4A
and circ-TTBK2 have been proven to play essential functions in GM
tumorigenesis (28–30). In the present study, a circRNA
microarray was conducted to study the expression profile in GM. A
total of 128 circRNAs with different expression levels were
identified. The top six upregulated circRNAs, namely
hsa_circRNA_102211, _101314, _104666, _001678, _100017 and _104634,
were analyzed using RT-qPCR. Among these circRNAs,
hsa_circRNA_104634 was the most upregulated. hsa_circRNA_104634 is
spliced from exons 2–4 of the ASPH gene (31). Kaplan-Meier analysis with the
log-rank test identified circ-ASPH as a prognostic indicator for
patients with GM. Nevertheless, the independent prognostic role of
circ-ASPH was not investigated because the clinical characteristics
of several patients were incomplete.
After confirming the expression levels of circ-ASPH
and its clinical relevance, a series of functional assays were
performed to determine its functions in GM cells. LN229 and U87MG
cells were selected for further study. The data illustrated that
decreased circ-ASPH inhibited GM cell proliferation, migration and
invasiveness and vice versa. The localization of circRNAs suggests
how they exert their functions (32). circ-ASPH is primarily localized to
the cytoplasm, which suggests that it has mechanisms in
post-transcriptional gene regulation (33). Several studies have determined the
importance of miR-599 in suppressing cancer progression, such as
gastric cancer and esophageal squamous cell carcinoma (34,35).
Moreover, miR-599 functions as a tumor suppressor by targeting
Ras-related protein Rab-27B and periostin in glioma (36,37). The
present study demonstrated that circ-ASPH counteracts
miR-599-mediated AR suppression by acting as a sponge for miR-599,
which broadens the understanding of the functions of miR-599. AR is
a steroid hormone-activated transcription factor (19). Upon binding the hormone ligand, the
receptor dissociates from accessory proteins, translocates into the
nucleus, dimerizes and then stimulates transcription of androgen
responsive genes (20). The current
study identified that the oncogenic role of circ-ASPH is partially
dependent on its regulation of AR. lncRNAs, including TMPO-AS1
(21), LINC00304 (22), PRCAT38 (23), ARLNC1 (24), GAS5 (25), SOCS2-AS1 (13), and ZEB1-AS1 (26), have been identified as direct targets
of AR in other malignancies, including prostate cancer (21–25) and
cholangiocarcinoma (26). Among
these lncRNAs, only SOCS2-AS1 expression was regulated by AR in GM,
suggesting a tissue-specific mechanism of AR. SOCS2-AS1 is a
well-known lncRNA that promotes prostate cancer cell development
and progression (13). Furthermore,
SOCS2-AS1 was also positively regulated by circ-ASPH in the present
study. Therefore, circ-ASPH/miR-599/AR/SOCS2-AS1 signaling
functions in GM progression.
In conclusion, circ-ASPH counteracted
miR-599-mediated AR suppression by acting as a sponge for miR-599.
Furthermore, AR positively regulated lncRNA SOCS2-AS1 expression
levels in GM. Taken together, these data suggested that the
circ-ASPH/miR-599/AR/SOCS2-AS1 axis may be a promising molecular
target for GM.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the study are
included in this published article. The additional datasets
generated and/or analyzed during the current study are available in
The Cancer Genome Atlas repository, (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga).
Authors' contributions
YQ conceived and designed the study. YQ and JZ
analyzed and interpreted the data. YQ, LQ, and LH performed the
in vitro assays. YQ and LQ were major contributors in
drafting the manuscript. YQ and LQ confirmed the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was authorized by the Ethics Committee of
Qiqihar Hospital Affiliated to Southern Medical University and
written informed consent was acquired from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ostrom QT, Gittleman H, Xu J, Kromer C,
Wolinsky Y, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical
report: Primary brain and other central nervous system tumors
diagnosed in the United States in 2009–2013. Neuro Oncol. 18
(Suppl_5):v1–v75. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ostrom QT, Cote DJ, Ascha M, Kruchko C and
Barnholtz-Sloan JS: Adult glioma incidence and survival by race or
ethnicity in the United States from 2000 to 2014. JAMA Oncol.
4:1254–1262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jiang T, Nam DH, Ram Z, Poon WS, Wang J,
Boldbaatar D, Mao Y, Ma W, Mao Q, You Y, et al: CGCG clinical
practice guidelines for the management of adult diffuse gliomas.
Cancer Lett. 375:263–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bolha L, Ravnik-Glavač M and Glavač D:
Circular RNAs: Biogenesis, function, and a role as possible cancer
biomarkers. Int J Genomics. 2017:62183532017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu Y, Yao Y, Zhong X, Leng K, Qin W, Qu L,
Cui Y and Jiang X: Downregulated circular RNA hsa_circ_0001649
regulates proliferation, migration and invasion in
cholangiocarcinoma cells. Biochem Biophys Res Commun. 49:455–461.
2018. View Article : Google Scholar
|
|
8
|
Zhang SJ, Chen X, Li CP, Li XM, Liu C, Liu
BH, Shan K, Jiang Q, Zhao C and Yan B: Identification and
characterization of circular RNAs as a new class of putative
biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci.
58:6500–6509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Wang Y, Piao H, Li B, Huang M, Zhu
Z, Li D, Wang T, Xu R and Liu K: Circular RNAs as potential
biomarkers and therapeutics for cardiovascular disease. PeerJ.
7:e68312019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin P, Huang Y, Zhu P, Zou Y, Shao T and
Wang O: CircRNA circHIPK3 serves as a prognostic marker to promote
glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem
Biophys Res Commun. 503:1570–1574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Chen T, Zhu Y, Li Y, Zhang Y, Wang
Y, Li X, Xie X, Wang J, Huang M, et al: circPTN sponges
miR-145-5p/miR-330-5p to promote proliferation and stemness in
glioma. J Exp Clin Cancer Res. 38:3982019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rahim B and O'Regan R: AR Signaling in
breast cancer. Cancers (Basel). 9:212017. View Article : Google Scholar
|
|
13
|
Misawa A, Takayama K, Urano T and Inoue S:
Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes
cell growth and inhibits apoptosis in prostate cancer cells. J Biol
Chem. 291:17861–17880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Su H, Lin F, Deng X, Shen L, Fang Y, Fei
Z, Zhao L, Zhang X, Pan H, Xie D, et al: Profiling and
bioinformatics analyses reveal differential circular RNA expression
in radioresistant esophageal cancer cells. J Transl Med.
14:2252016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu Y, Zhu J, Liu J and Qi L: Circular RNA
circ_0079593 indicates a poor prognosis and facilitates cell growth
and invasion by sponging miR-182 and miR-433 in glioma. J Cell
Biochem. 120:18005–18013. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gucalp A and Traina TA: The Androgen
Receptor: Is it a promising target? Ann Surg Oncol. 24:2876–2880.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schweizer MT and Yu EY: AR-signaling in
human malignancies: Prostate cancer and beyond. Cancers (Basel).
9:72017. View Article : Google Scholar
|
|
21
|
Huang W, Su X, Yan W, Kong Z, Wang D,
Huang Y, Zhai Q, Zhang X, Wu H, Li Y, et al: Overexpression of
AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and
poor prognosis in prostate cancer. Prostate. 78:1248–1261. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang P, Lu Y, Kong Z, Zhang Y, Fu F, Su
X, Huang Y, Wan X and Li Y: Androgen-responsive lncRNA LINC00304
ppromotes cell cycle and proliferation via regulating CCNA1.
Prostate. 79:994–1006. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z, Song X, Li Q, Xie L, Guo T, Su T,
Tang C, Chang X, Liang B and Huang D: Androgen receptor-activated
enhancers simultaneously regulate oncogene TMPRSS2 and lncRNA
PRCAT38 in prostate cancer. Cells. 8:8642019. View Article : Google Scholar
|
|
24
|
Zhang Y, Pitchiaya S, Cieślik M, Niknafs
YS, Tien JC, Hosono Y, Iyer MK, Yazdani S, Subramaniam S, Shukla
SK, et al: Analysis of the androgen receptor-regulated lncRNA
landscape identifies a role for ARLNC1 in prostate cancer
progression. Nat Genet. 50:814–824. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Su X, Kong Z, Fu F, Zhang P, Wang
D, Wu H, Wan X and Li Y: An androgen reduced transcript of lncRNA
GAS5 promoted prostate cancer proliferation. PLoS One.
12:e01823052017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang X, Li J, Wang W, Hu Z, Guan C, Zhao
Y, Li W and Cui Y: AR-induced ZEB1-AS1 represents poor prognosis in
cholangiocarcinoma and facilitates tumor stemness, proliferation
and invasion through mediating miR-133b/HOXB8. Aging (Albany NY).
12:1237–1255. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao ZG, Yang P, Huang J and Ding YQ:
CircFBXW7 alleviates glioma progression through regulating
miR-23a-3p/PTEN axis. Anat Rec (Hoboken). 304:279–290. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huo LW, Wang YF, Bai XB, Zheng HL and Wang
MD: circKIF4A promotes tumorogenesis of glioma by targeting
miR-139-3p to activate Wnt5a signaling. Mol Med. 26:292020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang HY, Zhang BW, Zhang ZB and Deng QJ:
Circular RNA TTBK2 regulates cell proliferation, invasion and
ferroptosis via miR-761/ITGB8 axis in glioma. Eur Rev Med Pharmacol
Sci. 24:2585–2600. 2020.PubMed/NCBI
|
|
31
|
Glažar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang Q, Li F, He AT and Yang BB: Circular
RNAs: Expression, localization, and therapeutic potentials. Mol
Ther. Jan 21–2021.(Epub ahead of print). doi:
10.1016/j.ymthe.2021.01.018. View Article : Google Scholar
|
|
33
|
Mumtaz PT, Taban Q, Dar MA, Mir S, Haq ZU,
Zargar SM, Shah RA and Ahmad SM: Deep insights in circular RNAs:
From biogenesis to therapeutics. Biol Proced. 22:102020. View Article : Google Scholar
|
|
34
|
Li C, Tian Y, Liang Y and Li Q:
Circ_0008035 contributes to cell proliferation and inhibits
apoptosis and ferroptosis in gastric cancer via miR-599/EIF4A1
axis. Cancer Cell Int. 20:842020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ba Y, Liu Y, Li C, Zhu Y and Xing W: HIPK3
promotes growth and metastasis of esophageal squamous cell
carcinoma via regulation of miR-599/c-MYC axis. Onco Targets Ther.
13:1967–1978. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang Y, Wang X, Zhang J and Lai R:
MicroRNA-599 suppresses glioma progression by targeting RAB27B.
Oncol Lett. 16:1243–1252. 2018.PubMed/NCBI
|
|
37
|
Zhang T, Ma G, Zhang Y, Huo H and Zhao Y:
miR-599 inhibits proliferation and invasion of glioma by targeting
periostin. Biotechnol Lett. 39:1325–1333. 2017. View Article : Google Scholar : PubMed/NCBI
|