|
1
|
Haugan K, Marcussen N, Kjølbye A, Nielsen
M, Hennan J and Petersen J: Treatment with the gap junction
modifier rotigaptide (ZP123) reduces infarct size in rats with
chronic myocardial infarction. J Cardiovasc Pharmacol. 47:236–242.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tsuchida S, Arai Y, Kishida T, Takahashi
KA, Honjo K, Terauchi R, Inoue H, Oda R, Mazda O and Kubo T:
Silencing the expression of connexin 43 decreases inflammation and
joint destruction in experimental arthritis. J Orthop Res.
31:525–530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
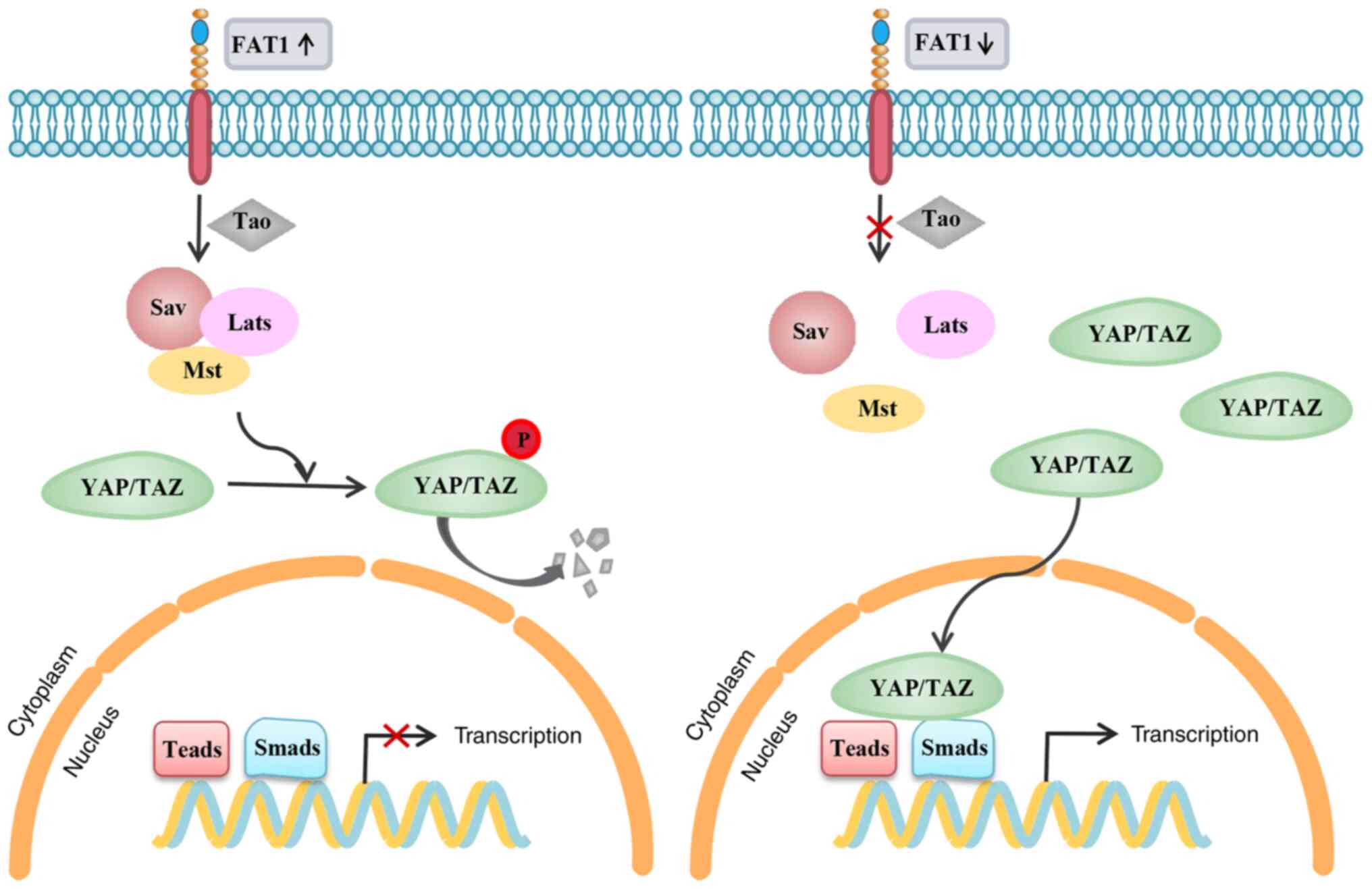
|
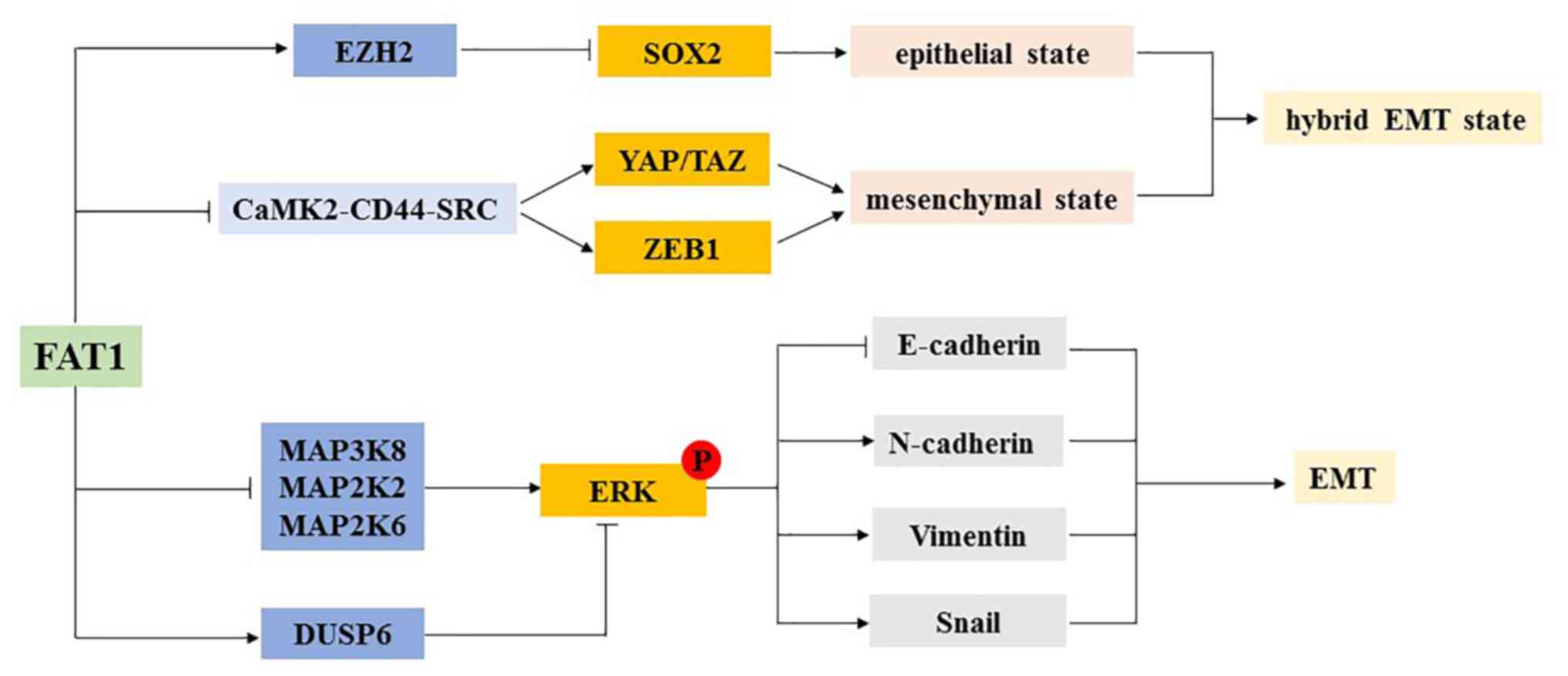
El-Bahrawy M, Talbot I, Poulsom R, Jeffery
R and Alison M: The expression of E-cadherin and catenins in
colorectal tumours from familial adenomatous polyposis patients. J
Pathol. 198:69–76. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nollet F, Kools P and van Roy F:
Phylogenetic analysis of the cadherin superfamily allows
identification of six major subfamilies besides several solitary
members. J Mol Biol. 299:551–572. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tanoue T and Takeichi M: New insights into
Fat cadherins. J Cell Sci. 118:2347–2353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bryant PJ, Huettner B, Held LI Jr, Ryerse
J and Szidonya J: Mutations at the fat locus interfere with cell
proliferation control and epithelial morphogenesis in
Drosophila. Dev Biol. 129:541–554. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sadeqzadeh E, de Bock C and Thorne R:
Sleeping giants: Emerging roles for the fat cadherins in health and
disease. Med Res Rev. 34:190–221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bennett F and Harvey K: Fat cadherin
modulates organ size in Drosophila via the
Salvador/Warts/Hippo signaling pathway. Curr Biol. 16:2101–2110.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silva E, Tsatskis Y, Gardano L, Tapon N
and McNeill H: The tumor-suppressor gene fat controls tissue growth
upstream of expanded in the hippo signaling pathway. Curr Biol.
16:2081–2089. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oh H and Irvine K: In vivo regulation of
Yorkie phosphorylation and localization. Development.
135:1081–1088. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang C, Axelrod J and Simon M: Regulation
of Frizzled by fat-like cadherins during planar polarity signaling
in the Drosophila compound eye. Cell. 108:675–688. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hill E, Broadbent I, Chothia C and Pettitt
J: Cadherin superfamily proteins in caenorhabditis elegans and
Drosophila melanogaster. J Mol Biol. 305:1011–1024. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
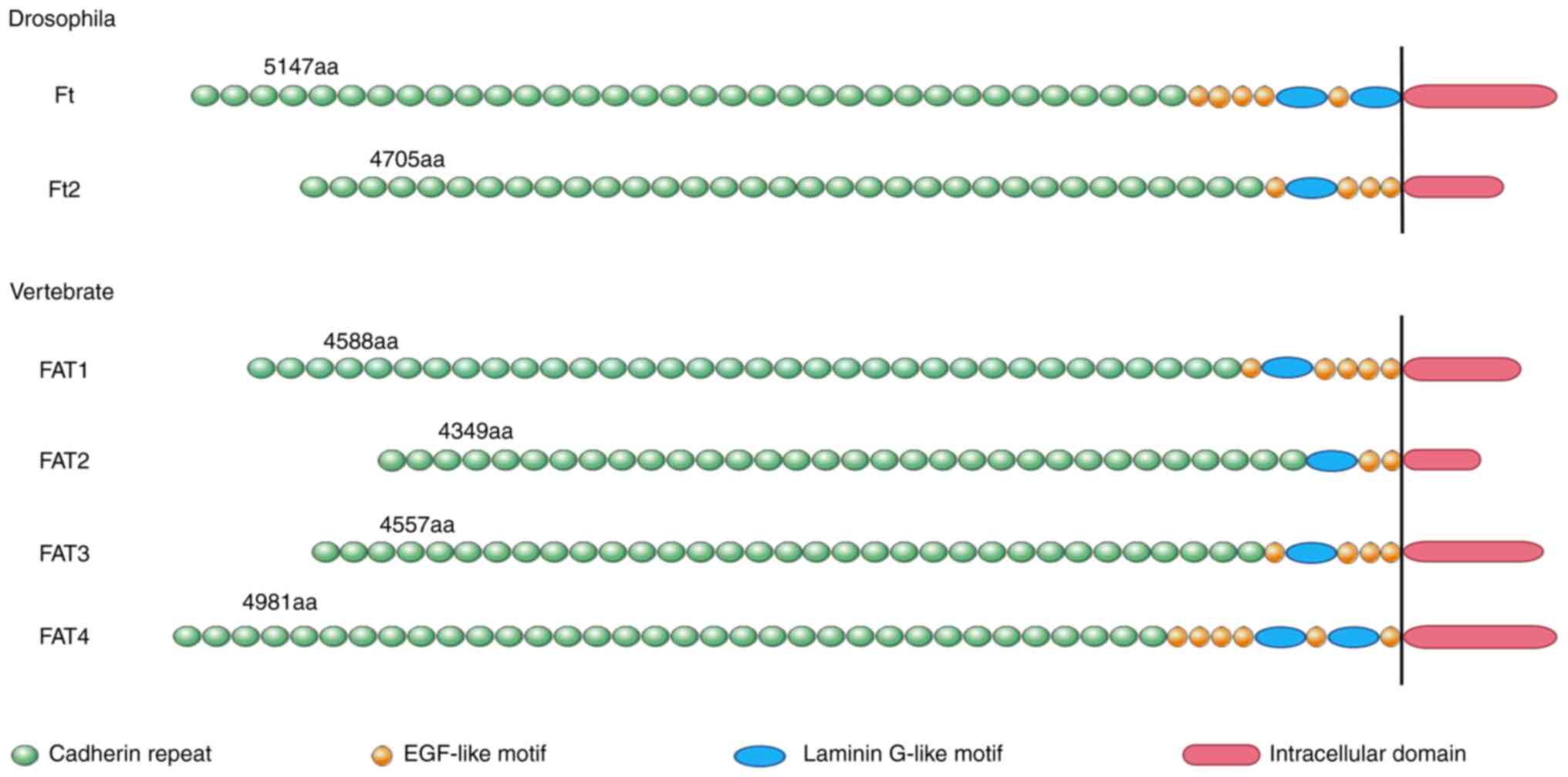
13
|
Down M, Power M, Smith SI, Ralston K,
Spanevello M, Burns GF and Boyd AW: Cloning and expression of the
large zebrafish protocadherin gene, Fat. Gene Expr Patterns.
5:483–490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Liu J, Liang X, Chen J, Hong J,
Li L, He Q and Cai X: History and progression of Fat cadherins in
health and disease. Onco Targets Ther. 9:7337–7343. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dunne J, Hanby AM, Poulsom R, Jones TA,
Sheer D, Chin WG, Da SM, Zhao Q, Beverley PC and Owen MJ: Molecular
cloning and tissue expression of FAT, the human homologue of the
Drosophila fat gene that is located on chromosome 4q34-q35
and encodes a putative adhesion molecule. Genomics. 30:207–223.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Valletta D, Czech B, Spruss T, Ikenberg K,
Wild P, Hartmann A, Weiss TS, Oefner PJ, Müller M, Bosserhoff AK
and Hellerbrand C: Regulation and function of the atypical cadherin
FAT1 in hepatocellular carcinoma. Carcinogenesis. 35:1407–1415.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sadeqzadeh E, de Bock CE, Zhang XD,
Shipman KL, Scott NM, Song C, Yeadon T, Oliveira CS, Jin B, Hersey
P, et al: Dual processing of FAT1 cadherin protein by human
melanoma cells generates distinct protein products. J Biol Chem.
286:28181–28191. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schreiner D, Müller K and Hofer H: The
intracellular domain of the human protocadherin hFat1 interacts
with Homer signalling scaffolding proteins. FEBS Lett.
580:5295–5300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morris LGT, Kaufman AM, Gong Y, Ramaswami
D, Walsh LA, Turcan S, Eng S, Kannan K, Zou Y, Peng L, et al:
Recurrent somatic mutation of FAT1 in multiple human cancers leads
to aberrant Wnt activation. Nat Genet. 45:253–261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ahmed AF, de Bock CE, Lincz LF, Pundavela
J, Zouikr I, Sontag E, Hondermarck H and Thorne RF: FAT1 cadherin
acts upstream of Hippo signalling through TAZ to regulate neuronal
differentiation. Cell Mol Life Sci. 72:4653–4669. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang
J, Qian Y, Ma Y, Wang F, Li H, et al: FAT1 prevents epithelial
mesenchymal transition (EMT) via MAPK/ERK signaling pathway in
esophageal squamous cell cancer. Cancer Lett. 397:83–93. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pastushenko I, Mauri F, Song Y, de Cock F,
Meeusen B, Swedlund B, Impens F, Van Haver D, Opitz M, Thery M, et
al: Fat1 deletion promotes hybrid EMT state, tumour stemness and
metastasis. Nature. 589:448–455. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mariot V, Roche S, Hourdé C, Portilho D,
Sacconi S, Puppo F, Duguez S, Rameau P, Caruso N, Delezoide AL, et
al: Correlation between low FAT1 expression and early affected
muscle in facioscapulohumeral muscular dystrophy. Ann Neurol.
78:387–400. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Blair IP, Chetcuti AF, Badenhop RF,
Scimone A, Moses MJ, Adams LJ, Craddock N, Green E, Kirov G, Owen
MJ, et al: Positional cloning, association analysis and expression
studies provide convergent evidence that the cadherin gene FAT
contains a bipolar disorder susceptibility allele. Mol Psychiatry.
11:372–383. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krause M, Bear J, Loureiro J and Gertler
F: The Ena/VASP enigma. J Cell Sci. 115:4721–4726. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Krause M, Leslie JD, Stewart M, Lafuente
EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H,
Way M, et al: Lamellipodin, an Ena/VASP ligand, is implicated in
the regulation of lamellipodial dynamics. Dev Cell. 7:571–583.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lafuente EM, van Puijenbroek AFL, Krause
M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer
TA, Gertler FB and Boussiotis VA: RIAM, an Ena/VASP and profilin
ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion.
Dev Cell. 7:585–595. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Renfranz P and Beckerle M: Doing
(F/L)PPPPs: EVH1 domains and their proline-rich partners in cell
polarity and migration. Curr Opin Cell Biol. 14:88–103. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moeller MJ, Soofi A, Braun GS, Li X, Watzl
C, Kriz W and Holzman LB: Protocadherin FAT1 binds Ena/VASP
proteins and is necessary for actin dynamics and cell polarization.
EMBO J. 23:3769–3779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tanoue T and Takeichi M: Mammalian Fat1
cadherin regulates actin dynamics and cell-cell contact. J Cell
Biol. 165:517–528. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nobes CD and Hall A: Rho GTPases control
polarity, protrusion, and adhesion during cell movement. J Cell
Biol. 144:1235–1244. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang P, Yan R, Zhang X, Wang L, Ke X and
Qu Y: Activating Wnt/β-catenin signaling pathway for disease
therapy: Challenges and opportunities. Pharmacol Ther. 196:79–90.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Perugorria MJ, Olaizola P, Labiano I,
Esparza-Baquer A, Marzioni M, Marin JJG, Bujanda L and Banales JM:
Wnt-β-catenin signalling in liver development, health and disease.
Nat Rev Gastroenterol Hepatol. 16:121–136. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Morin PJ, Sparks AB, Korinek V, Barker N,
Clevers H, Vogelstein B and Kinzler KW: Activation of
beta-catenin-Tcf signaling in colon cancer by mutations in
beta-catenin or APC. Science. 275:1787–1790. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hou R, Liu L, Anees S, Hiroyasu S and
Sibinga NES: The Fat1 cadherin integrates vascular smooth muscle
cell growth and migration signals. J Cell Biol. 173:417–429. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kaplan D, Meigs T and Casey P: Distinct
regions of the cadherin cytoplasmic domain are essential for
functional interaction with Galpha 12 and beta-catenin. J Biol
Chem. 276:44037–44043. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rockman SP, Currie SA, Ciavarella M,
Vincan E, Dow C, Thomas RJ and Phillips WA: Id2 is a target of the
beta-catenin/T cell factor pathway in colon carcinoma. J Biol Chem.
276:45113–45119. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kolligs FT, Nieman MT, Winer I, Hu G, Van
Mater D, Feng Y, Smith IM, Wu R, Zhai Y, Cho KR and Fearon ER:
ITF-2, a downstream target of the Wnt/TCF pathway, is activated in
human cancers with beta-catenin defects and promotes neoplastic
transformation. Cancer Cell. 1:145–155. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He TC, Sparks AB, Rago C, Hermeking H,
Zawel L, da Costa LT, Morin PJ, Vogelstein B and Kinzler KW:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cancer Genome Atlas Research Network, .
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Staley B and Irvine K: Hippo signaling in
Drosophila: Recent advances and insights. Dev Dyn. 241:3–15.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pan D: The hippo signaling pathway in
development and cancer. Dev Cell. 19:491–505. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Oh H and Irvine K: Yorkie: The final
destination of Hippo signaling. Trends Cell Biol. 20:410–417. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wei X, Shimizu T and Lai Z: Mob as tumor
suppressor is activated by Hippo kinase for growth inhibition in
Drosophila. EMBO J. 26:1772–1781. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
McCartney BM, Kulikauskas RM, LaJeunesse
DR and Fehon RG: The neurofibromatosis-2 homologue, Merlin, and the
tumor suppressor expanded function together in Drosophila to
regulate cell proliferation and differentiation. Development.
127:1315–1324. 2000.PubMed/NCBI
|
|
50
|
Camargo FD, Gokhale S, Johnnidis JB, Fu D,
Bell GW, Jaenisch R and Brummelkamp TR: YAP1 increases organ size
and expands undifferentiated progenitor cells. Curr Biol.
17:2054–2060. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dong J, Feldmann G, Huang J, Wu S, Zhang
N, Comerford SA, Gayyed MF, Anders RA, Maitra A and Pan D:
Elucidation of a universal size-control mechanism in
Drosophila and mammals. Cell. 130:1120–1133. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao B, Li L, Lei Q and Guan K: The
Hippo-YAP pathway in organ size control and tumorigenesis: An
updated version. Genes Dev. 24:862–874. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Beyer TA, Weiss A, Khomchuk Y, Huang K,
Ogunjimi AA, Varelas X and Wrana JL: Switch enhancers interpret
TGF-β and Hippo signaling to control cell fate in human embryonic
stem cells. Cell Rep. 5:1611–1624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wrighton K, Dai F and Feng X: A new kid on
the TGFbeta block: TAZ controls Smad nucleocytoplasmic shuttling.
Dev Cell. 15:8–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nishioka N, Inoue KI, Adachi K, Kiyonari
H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki
N, et al: The Hippo signaling pathway components Lats and Yap
pattern Tead4 activity to distinguish mouse trophectoderm from
inner cell mass. Dev Cell. 16:398–410. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ahmed A, de Bock C, Sontag E, Hondermarck
H, Lincz L and Thorne R: FAT1 cadherin controls neuritogenesis
during NTera2 cell differentiation. Biochem Biophys Res Commun.
514:625–631. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Skouloudaki K, Puetz M, Simons M, Courbard
JR, Boehlke C, Hartleben B, Engel C, Moeller MJ, Englert C, Bollig
F, et al: Scribble participates in Hippo signaling and is required
for normal zebrafish pronephros development. Proc Natl Acad Sci
USA. 106:8579–8584. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ye CY, Zheng CP, Ying WW and Weng SS:
Up-regulation of microRNA-497 inhibits the proliferation, migration
and invasion but increases the apoptosis of multiple myeloma cells
through the MAPK/ERK signaling pathway by targeting Raf-1. Cell
Cycle. 17:2666–2683. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yan P, Zhu H, Yin L, Wang L, Xie P, Ye J,
Jiang X and He X: Integrin αvβ6 promotes lung cancer proliferation
and metastasis through upregulation of IL-8-mediated MAPK/ERK
signaling. Transl Oncol. 11:619–627. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang WM, Xu Y, Wang YH, Sun HX, Sun YF, He
YF, Zhu QF, Hu B, Zhang X, Xia JL, et al: HOXB7 promotes tumor
progression via bFGF-induced activation of MAPK/ERK pathway and
indicated poor prognosis in hepatocellular carcinoma. Oncotarget.
8:47121–47135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Miller MB, Bi WL, Ramkissoon LA, Kang YJ,
Abedalthagafi M, Knoff DS, Agarwalla PK, Wen PY, Reardon DA,
Alexander BM, et al: MAPK activation and HRAS mutation identified
in pituitary spindle cell oncocytoma. Oncotarget. 7:37054–37063.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang Y, Wang G, Ma Y, Teng J, Wang Y, Cui
Y, Dong Y, Shao S, Zhan Q and Liu X: FAT1, a direct transcriptional
target of E2F1, suppresses cell proliferation, migration and
invasion in esophageal squamous cell carcinoma. Chin J Cancer Res.
31:609–619. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Saxena K, Jolly M and Balamurugan K:
Hypoxia, partial EMT and collective migration: Emerging culprits in
metastasis. Transl Oncol. 13:1008452020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Meng P, Zhang YF, Zhang W, Chen X, Xu T,
Hu S, Liang X, Feng M, Yang X and Ho M: Identification of the
atypical cadherin FAT1 as a novel glypican-3 interacting protein in
liver cancer cells. Sci Rep. 11:402021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Srivastava C, Irshad K, Dikshit B,
Chattopadhyay P, Sarkar C, Gupta DK, Sinha S and Chosdol K: FAT1
modulates EMT and stemness genes expression in hypoxic
glioblastoma. Int J Cancer. 142:805–812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pastushenko I, Brisebarre A, Sifrim A,
Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D,
Moers V, Lemaire S, et al: Identification of the tumour transition
states occurring during EMT. Nature. 556:463–468. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ponassi M, Jacques TS, Ciani L and ffrench
Constant C: Expression of the rat homologue of the
Drosophila fat tumour suppressor gene. Mech Dev. 80:207–212.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ciani L, Patel A, Allen N and
ffrench-Constant C: Mice lacking the giant protocadherin mFAT1
exhibit renal slit junction abnormalities and a partially penetrant
cyclopia and anophthalmia phenotype. Mol Cell Biol. 23:3575–3582.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Inoue T, Yaoita E, Kurihara H, Shimizu F,
Sakai T, Kobayashi T, Ohshiro K, Kawachi H, Okada H, Suzuki H, et
al: FAT is a component of glomerular slit diaphragms. Kidney Int.
59:1003–1012. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yaoita E, Kurihara H, Yoshida Y, Inoue T,
Matsuki A, Sakai T and Yamamoto T: Role of Fat1 in cell-cell
contact formation of podocytes in puromycin aminonucleoside
nephrosis and neonatal kidney. Kidney Int. 68:542–551. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Uglow EB, Slater S, Sala-Newby GB,
Aguilera-Garcia CM, Angelini GD, Newby AC and George SJ:
Dismantling of cadherin-mediated cell-cell contacts modulates
smooth muscle cell proliferation. Circ Res. 92:1314–1321. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lahrouchi N, George A, Ratbi I, Schneider
R, Elalaoui SC, Moosa S, Bharti S, Sharma R, Abu-Asab M, Onojafe F,
et al: Homozygous frameshift mutations in FAT1 cause a syndrome
characterized by colobomatous-microphthalmia, ptosis, nephropathy
and syndactyly. Nat Commun. 10:11802019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Puppo F, Dionnet E, Gaillard MC, Gaildrat
P, Castro C, Vovan C, Bertaux K, Bernard R, Attarian S, Goto K, et
al: Identification of variants in the 4q35 gene FAT1 in patients
with a facioscapulohumeral dystrophy-like phenotype. Hum Mutat.
36:443–453. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Park HJ, Lee W, Kim SH, Lee JH, Shin HY,
Kim SM, Park KD, Lee JH and Choi YC: FAT1 gene alteration in
facioscapulohumeral muscular dystrophy type 1. Yonsei Med J.
59:337–340. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Caruso N, Herberth B, Bartoli M, Puppo F,
Dumonceaux J, Zimmermann A, Denadai S, Lebossé M, Roche S, Geng L,
et al: Deregulation of the protocadherin gene FAT1 alters muscle
shapes: implications for the pathogenesis of facioscapulohumeral
dystrophy. PLoS Genet. 9:e10035502013. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Stahl EA, Breen G, Forstner AJ, McQuillin
A, Ripke S, Trubetskoy V, Mattheisen M, Wang Y, Coleman JRI, Gaspar
HA, et al: Genome-wide association study identifies 30 loci
associated with bipolar disorder. Nat Genet. 51:793–803. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bipolar Disorder and Schizophrenia Working
Group of the Psychiatric Genomics Consortium. Electronic address:
douglas.ruderfer@vanderbilt.edu; Bipolar Disorder and Schizophrenia
Working Group of the Psychiatric Genomics Consortium: Genomic
dissection of bipolar disorder and schizophrenia, including 28
subphenotypes, . Cell. 173:1705–1715.e1716. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Psychiatric GWAS Consortium Bipolar
Disorder Working Group, : Large-scale genome-wide association
analysis of bipolar disorder identifies a new susceptibility locus
near ODZ4. Nat Genet. 43:977–983. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Abou Jamra R, Becker T, Georgi A, Feulner
T, Schumacher J, Stromaier J, Schirmbeck F, Schulze TG, Propping P,
Rietschel M, et al: Genetic variation of the FAT gene at 4q35 is
associated with bipolar affective disorder. Mol Psychiatry.
13:277–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Neumann M, Seehawer M, Schlee C, Vosberg
S, Heesch S, von der Heide EK, Graf A, Krebs S, Blum H, Gökbuget N,
et al: FAT1 expression and mutations in adult acute lymphoblastic
leukemia. Blood Cancer J. 4:e2242014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
de Bock CE, Ardjmand A, Molloy TJ, Bone
SM, Johnstone D, Campbell DM, Shipman KL, Yeadon TM, Holst J,
Spanevello MD, et al: The Fat1 cadherin is overexpressed and an
independent prognostic factor for survival in paired
diagnosis-relapse samples of precursor B-cell acute lymphoblastic
leukemia. Leukemia. 26:918–926. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
GBD 2015 Mortality and Causes of Death
Collaborators, . Global, regional, and national life expectancy,
all-cause mortality, and cause-specific mortality for 249 causes of
death, 1980–2015: A systematic analysis for the global burden of
disease study 2015. Lancet. 388:1459–1544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Shariff MI, Cox IJ, Gomaa AI, Khan SA,
Gedroyc W and Taylor-Robinson SD: Hepatocellular carcinoma: Current
trends in worldwide epidemiology, risk factors, diagnosis and
therapeutics. Expert Rev Gastroenterol Hepatol. 3:353–367. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Dhanasekaran R, Limaye A and Cabrera R:
Hepatocellular carcinoma: Current trends in worldwide epidemiology,
risk factors, diagnosis, and therapeutics. Hepat Med. 4:19–37.
2012.PubMed/NCBI
|
|
85
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Bataller R and Brenner D: Liver fibrosis.
J Clin Invest. 115:209–218. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Farazi P and DePinho R: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Rosmorduc O and Housset C: Hypoxia: A link
between fibrogenesis, angiogenesis, and carcinogenesis in liver
disease. Semin Liver Dis. 30:258–270. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Argiris A, Karamouzis M, Raben D and
Ferris R: Head and neck cancer. Lancet. 371:1695–1709. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Pai SI and Westra WH: Molecular pathology
of head and neck cancer: Implications for diagnosis, prognosis, and
treatment. Annu Rev Pathol. 4:49–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kim KT, Kim BS and Kim JH: Association
between FAT1 mutation and overall survival in patients with human
papillomavirus-negative head and neck squamous cell carcinoma. Head
Neck. 38 (Suppl 1):S2021–S2029. 2016. View Article : Google Scholar
|
|
93
|
Gupta S, Kong W, Peng Y, Miao Q and
Mackillop W: Temporal trends in the incidence and survival of
cancers of the upper aerodigestive tract in Ontario and the United
States. Int J Cancer. 125:2159–2165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Lin SC, Lin LH, Yu SY, Kao SY, Chang KW,
Cheng HW and Liu CJ: FAT1 somatic mutations in head and neck
carcinoma are associated with tumor progression and survival.
Carcinogenesis. 39:1320–1330. 2018.PubMed/NCBI
|
|
95
|
Katoh Y and Katoh M: Comparative
integromics on FAT1, FAT2, FAT3 and FAT4. Int J Mol Med.
18:523–528. 2006.PubMed/NCBI
|
|
96
|
Liu CJ, Liu TY, Kuo LT, Cheng HW, Chu TH,
Chang KW and Lin SC: Differential gene expression signature between
primary and metastatic head and neck squamous cell carcinoma. J
Pathol. 214:489–497. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Martin D, Degese MS, Vitale-Cross L,
Iglesias-Bartolome R, Valera JLC, Wang Z, Feng X, Yeerna H, Vadmal
V, Moroishi T, et al: Assembly and activation of the Hippo
signalome by FAT1 tumor suppressor. Nat Commun. 9:23722018.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Pennathur A, Gibson M, Jobe B and Luketich
J: Oesophageal carcinoma. Lancet. 381:400–412. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey
SM, Dong ZW, Mark SD, Qiao YL and Taylor PR: Prospective study of
risk factors for esophageal and gastric cancers in the Linxian
general population trial cohort in China. Int J Cancer.
113:456–463. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Li WQ, Park Y, Wu JW, Ren JS, Goldstein
AM, Taylor PR, Hollenbeck AR, Freedman ND and Abnet CC: Index-based
dietary patterns and risk of esophageal and gastric cancer in a
large cohort study. Clin Gastroenterol Hepatol. 11:1130–1136.e1132.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang L, Zhou Y, Cheng C, Cui H, Cheng L,
Kong P, Wang J, Li Y, Chen W, Song B, et al: Genomic analyses
reveal mutational signatures and frequently altered genes in
esophageal squamous cell carcinoma. Am J Hum Genet. 96:597–611.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Sauer AG, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Britt KL, Cuzick J and Phillips KA: Key
steps for effective breast cancer prevention. Nat Rev Cancer.
20:417–436. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang L, Lyu S, Wang S, Shen H, Niu F, Liu
X, Liu J and Niu Y: Loss of FAT1 during the progression from DCIS
to IDC and predict poor clinical outcome in breast cancer. Exp Mol
Pathol. 100:177–183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lee S, Stewart S, Nagtegaal I, Luo J, Wu
Y, Colditz G, Medina D and Allred DC: Differentially expressed
genes regulating the progression of ductal carcinoma in situ to
invasive breast cancer. Cancer Res. 72:4574–4586. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Guo L, Qi J, Wang H, Jiang X and Liu Y:
Getting under the skin: The role of CDK4/6 in melanomas. Eur J Med
Chem. 204:1125312020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Salvador-Barbero B, Álvarez-Fernández M,
Zapatero-Solana E, El Bakkali A, Menéndez MDC, López-Casas PP, Di
Domenico T, Xie T, VanArsdale T, Shields DJ, et al: CDK4/6
inhibitors impair recovery from cytotoxic chemotherapy in
pancreatic adenocarcinoma. Cancer Cell. 37:340–353.e346. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Li Z, Razavi P, Li Q, Toy W, Liu B, Ping
C, Hsieh W, Sanchez-Vega F, Brown DN, Da Cruz Paula AF, et al: Loss
of the FAT1 tumor suppressor promotes resistance to CDK4/6
inhibitors via the hippo pathway. Cancer Cell. 34:893–905.e898.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Kang MH, Jeong GS, Smoot DT, Ashktorab H,
Hwang CM, Kim BS, Kim HS and Park YY: Verteporfin inhibits gastric
cancer cell growth by suppressing adhesion molecule FAT1.
Oncotarget. 8:98887–98897. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Döhner H, Bloomfield C, Frizzera G,
Frestedt J and Arthur D: Recurring chromosome abnormalities in
Hodgkin's disease. Genes Chromosomes Cancer. 5:392–398. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yu J and Li H: The expression of FAT1 is
associated with overall survival in children with medulloblastoma.
Tumori. 103:44–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Polascik TJ, Cairns P, Chang WY,
Schoenberg MP and Sidransky D: Distinct regions of allelic loss on
chromosome 4 in human primary bladder carcinoma. Cancer Res.
55:5396–5399. 1995.PubMed/NCBI
|
|
116
|
Shivapurkar N, Virmani AK, Wistuba II,
Milchgrub S, Mackay B, Minna JD and Gazdar AF: Deletions of
chromosome 4 at multiple sites are frequent in malignant
mesothelioma and small cell lung carcinoma. Clin Cancer Res.
5:17–23. 1999.PubMed/NCBI
|
|
117
|
Shivapurkar N, Maitra A, Milchgrub S and
Gazdar AF: Deletions of chromosome 4 occur early during the
pathogenesis of colorectal carcinoma. Hum Pathol. 32:169–177. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Chen M, Sun X, Wang Y, Ling K, Chen C, Cai
X, Liang X and Liang Z: FAT1 inhibits the proliferation and
metastasis of cervical cancer cells by binding β-catenin. Int J
Clin Exp Pathol. 12:3807–3818. 2019.PubMed/NCBI
|
|
119
|
Zhang H, Ramakrishnan SK, Triner D,
Centofanti B, Maitra D, Győrffy B, Sebolt-Leopold JS, Dame MK,
Varani J, Brenner DE, et al: Tumor-selective proteotoxicity of
verteporfin inhibits colon cancer progression independently of
YAP1. Sci Signal. 8:ra982015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Xu J, Wang B, Liu ZT, Lai MC, Zhang ML and
Zheng SS: miR-223-3p regulating the occurrence and development of
liver cancer cells by targeting FAT1 gene. Math Biosci Eng.
17:1534–1547. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Fan L, Campagnoli S, Wu H, Grandi A, Parri
M, De Camilli E, Grandi G, Viale G, Pileri P, Grifantini R, et al:
Negatively charged AuNP modified with monoclonal antibody against
novel tumor antigen FAT1 for tumor targeting. J Exp Clin Cancer
Res. 34:1032015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Katoh M: Function and cancer genomics of
FAT family genes (review). Int J Oncol. 41:1913–1918. 2012.
View Article : Google Scholar : PubMed/NCBI
|