Introduction
Lung cancer is a disease with the fastest increasing
morbidity and mortality rates, and is a notable public health issue
(1). In 2019, there were ~228,150
new cases and 142,670 deaths of lung cancer in the United States
(1). It is categorised into small
cell lung cancer and non-small cell lung cancer (NSCLC), with the
latter composing ~85% of all lung cancer cases in the United States
in 2015 (2). Despite the development
of therapeutic technology, the effect of lung cancer treatment
remains unsatisfactory, with a low survival rate of only 15%
(3,4). Therefore, it is important to elucidate
the pathogenesis of lung cancer occurrence and progression.
Long non-coding (lnc)RNAs have been found to be
involved in tumour initiation and tumorigenesis (5,6).
Furthermore, studies have shown that numerous lncRNAs, such as
lncRNA cancer susceptibility 15, forkhead box protein C2 and JHDM1D
antisense 1, promote the onset and progression of NSCLC (7–9).
LINC00473 is a novel oncogenic lncRNA and is upregulated in various
types of human cancer (10).
Reportedly, silencing LINC00473 can block the progression of
pancreatic cancer by strengthening miR-195-5p-targeted
downregulation of PD-L1 (11). Niu
et al (12) also reported
that LINC00473 regulates the expression of MAPK1 in breast cancer
by interacting with miR-198 (12).
However, there have been few reports on the effects of LINC00473 in
NSCLC.
microRNAs (miRNAs/miRs) are elements that can
inhibit the expression of target genes by hindering mRNA
translation, where imbalance can result in tumour suppressor gene
dysfunction, leading to the occurrence of tumours (13). It has been reported that lncRNA can
act as a sponge of miRNAs, reducing their regulatory effects on
target mRNAs (14). The present
study aimed to elucidate the mechanism by which LINC00473 affects
the progression of NSCLC. Using bioinformatics tools, the miRNAs
that bind to LINC00473 were predicted, and the miR-497-5p binding
site was revealed. Previously miRNA miR-497-5p has been found to
inhibit NSCLC cell proliferation and invasion as a tumour
suppressor gene, therefore it was speculated that LINC00473 may
influence the progression of NSCLC by targeting miR-497-5p and
affecting its activity.
Materials and methods
Sample collection
In total, 58 NSCLC tissues and normal adjacent
tissues (~5 cm away from the cancerous tissues; 39 males and 19
females; age range, 29–81 years; median age, 56 years) were
obtained from patients who underwent resection surgery at The
Second Hospital of Shandong University (Ji'nan, China) between July
2017 and August 2018. All samples were verified after a
histopathological double-blind assessment by two independent
pathologists and immediately stored at −80°C. Five-year survival
rates were calculated by the percentage of patients who survived
>60 months after resection surgery. Each patient signed an
informed consent form. The protocol of the research was approved by
The Ethics Committee of The Second Hospital of Shandong University
(Ji'nan, China).
Cell culture
NSCLC cell lines A549 and H1299 and normal human
bronchial epithelial cell line (HBE) were obtained from the Cell
Bank of the Type Culture Collection of the Chinese Academy of
Sciences and cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% bovine fetal serum (FBS), 100 U/ml
penicillin and 100 µg/ml streptomycin sulphate (both HyClone;
Cytiva) at 37°C in a humidified incubator with 5% CO2
atmosphere.
Cell transfection
Once cells reached 70–80% confluence, si-LINC00473
(5 nM; 5′-GCGCCGGGAGAUGCAUCACGAUGAA-3′), pcDNA3.1-LINC00473
(Shanghai GenePharma Co., Ltd.), miR-497-5p mimics (50 nM;
5′-CAGCAGCACACUGUGGUUUGU-3′) and miR-497-5p inhibitors (100 nM;
5′-ACAAACCACAGUGUGCUGCUG-3′) (Guangzhou RiboBio Co., Ltd.) were
transfected into A549 and H1299 cells, respectively, using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. As
a control, si-NC (5 nM; 5′-CACUGAUUUCAAAUGGUGCUAUU-3′),
pcDNA3.1-NC, mimics NC (50 nM; 5′-UUCUCCGAACGUGUCACGUTT-3′) and
inhibitors NC (100 nM; 5′-CAGUACUUUUGUGUAGUACAA-3′) were also
transfected into A549 and H1299 cells and were non-targeting. After
48 h at 37°C, cells were collected for subsequent analyses.
Luciferase reporter assay
LINC00473-wild-type (WT) and LINC00473-mutant (MUT)
inserts were ligated into pGL3 reporter vectors (Promega
Corporation) and transfected into A549 and H1299 cells
(5×105), respectively, in 24-well plates and incubated
for 24 h at 37°C. In parallel, miR-497-5p mimics or mimics NC
LINC00473-WT or LINC00473-MUT were respectively transfected into
A549 and H1299 cells using Lipofectamine 2000 according to the
manufacturer's instructions. Luciferase activity was detected using
the Dual-Luciferase Reporter Assay system (Promega Corporation).
Renilla luciferase activity was used for normalization.
Moreover, the binding site of LINC00473 and miR-497-5p was
predicted using DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/).
RNA immunoprecipitation
A549 and H1299 cells were lysed in complete RIP
lysis buffer (EMD Millipore) and then incubated with magnetic beads
(EMD Millipore) containing human anti-SNRNP70 antibody or IgG (EMD
Millipore; cat. nos. CS203216 and CS200621, respectively) overnight
at 4°C. After incubation, tubes were placed on a magnetic separator
(EMD Millipore) and supernatants were discarded. After washing with
RIP washing buffer, the samples were subsequently incubated with
proteinase K buffer in a heating block at 55°C for 30 min. Target
RNA was extracted using chloroform, purified using two salt
solutions (EMD Millipore), a precipitate enhancer (EMD Millipore)
and absolute ethanol, and detected by reverse
transcription-quantitative (RT-q)PCR analysis.
RT-qPCR
Total RNA was isolated from both tissues and cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed using PrimeScript™ RT
reagent kit (Takara Bio, Inc.) according to the manufacturer's
protocol. The relative gene expression of LINC00473 and miR-497-5p
was analysed using a SYBR® Premix Ex Taq™ kit (Takara
Bio, Inc.) and an ABI PRISM® 7500 Two-step Sequence
Detection system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The PCR conditions were 95°C for 3 min, followed by 40
cycles at 95°C for 5 sec and 60°C for 30 sec. The 2−ΔΔCq
method (15) was used for
quantification of LINC00473 and miR-497-5p expression. GAPDH and U6
expression levels were used as reference controls for LINC00473 and
miR-497-5p levels, respectively. Primers used for amplification are
listed in Table I.
 | Table I.Primer sequences for quantitative
expression analysis. |
Table I.
Primer sequences for quantitative
expression analysis.
| Gene | Sequence of
oligonucleotides, 5-3 |
|---|
| LINC00473 |
|
|
Forward |
GATGGAAAGGAGGGAAGG |
|
Reverse |
CACAGTGGGTCCAGGGTT |
|
microRNA-497-5p |
|
|
Forward |
CCTTCAGCAGCACACTGTGG |
|
Reverse |
CAGTGCAGGGTCCGAGGTAT |
| GAPDH |
|
|
Forward |
AGAAGGCTGGGGCTCATTTG |
|
Reverse | AGGGGCCATC
CACAGTCTTC |
| U6 |
|
|
Forward |
CTCGCTTCGGCAGCACA |
|
Reverse |
AACGCTTCACGAATTTGCGT |
Colony formation assay
A total of 1,000 transfected A549 and H1299 cells
were placed in 6-well plates and cultured in RPMI-1640 medium.
After 14 days of continuous incubation at 37°C, cells were stained
with crystal violet for 10 min at room temperature (Beyotime
Institute of Biotechnology) and observed manually under a light
microscope (magnification, ×40) for the number of colonies to be
counted (>50 cells were considered as a colony).
MTT assay
The transfected cells were resuspended in RPMI-1640
with 10% FBS, seeded in a 96-well plate (5×103/well),
and incubated at room temperature for 24–72 h. MTT solution was
then added, followed by incubation for 4 h and addition of 150 µl
DMSO to each well. Optical density at 490 nm was measured using a
microplate reader, and data were expressed as absorbance
values.
Apoptosis analysis
Apoptosis analysis was performed using an Annexin V
FITC Apoptosis kit (BD Biosciences; cat. no. 556420). Cells
transfected with si-LINC00473, pcDNA3.1-LINC00473 and miR-497-5p
inhibitors were cultured for 48 h, then incubated with
FITC-labelled Annexin V and PI in the dark at 25°C for 20 min.
Thereafter, the apoptosis rate of the cells was measured by flow
cytometry using a flow cytometer (BD FACSCanto II; BD Biosciences).
FlowJo version 10 software (FlowJo LLC) was used to analyse
apoptosis.
Transwell migration and invasion
assays
After 48 h of incubation, transfected A549 and H1299
cells were trypsinised and resuspended. For invasion assays, the
upper chamber surface of the Transwell insert was coated with 50
mg/l Matrigel (1:8) and air-dried at 4°C. For both migration and
invasion assays, 2×105 cells were added to the upper
chamber containing serum-free RPMI-1640 medium, and the lower
chamber was filled with RPMI-1640 medium supplemented with 5% FBS.
After 24 h of incubation, the cells were stained with crystal
violet for 20 min at room temperature. Finally, the filters were
washed with PBS and observed under a light microscope
(magnification, ×200). Four visual fields were randomly selected
and images were captured, and the visible cells were counted.
Western blotting
After 48 h incubation of transfected cells, samples
from each group were collected, and lysed with ice cold RIPA buffer
(Sigma-Aldrich; Merck KGaA) supplemented with a protease inhibitor
cocktail (Roche Diagnostics). The protein concentration was
measured using a BCA kit (Takara Bio, Inc.). Proteins (10 ng/lane)
were separated via 10% SDS-PAGE, followed by transfer to a PVDF
membrane. After 2.5 h of blocking with a BSA (neoFroxx GmbH)-TBST
solution (1× TBS, 1% Tween-20 and 5% w/v BSA) at room temperature,
the PVDF membrane was incubated with the following primary
antibodies: Bax (1:1,000; cat. no. 14796), Bcl-2 (1:1,000; cat. no.
4223), Matrix metalloproteinase (MMP)-2 (1:1,000; cat. no. 40994),
MMP-9 (1:1,000; cat. no. 15561), p44/42 (ERK1/2; 1:1,000; cat. no.
4695), phosphorylated-p44/42 (p-ERK1/2; Thr202/Tyr204; 1:1,000;
cat. no. 4370), p38 (1:1,000; cat. no. 14451) and
phosphorylated-p38 (Thr180/Tyr182; 1:1,000; cat. no. 4511) (all
Cell Signalling Technology, Inc.) at 4°C overnight. The following
day, the samples were incubated with HRP-conjugated secondary
antibodies (1:2,000; cat. no. 7074; Cell Signalling Technology,
Inc.) at room temperature for 2 h. The protein blots on the
membrane were imaged using a chemiluminescence kit (Pierce; Thermo
Fisher Scientific, Inc.). Data were quantified using ImageJ
software v1.41 (National Institutes of Health).
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiments and were analysed using SPSS 19.0
(IBM Corp). Paired t-tests were used to determine the statistical
differences between two groups. The overall survival rate was
analysed by the Kaplan-Meier method and differences were tested
using the log-rank test. The data difference among >2 groups
were determined using ANOVA and Bonferroni's post-hoc test. The
correlation between LINC00473 and miR-497-5p was analysed using
Pearson's correlation coefficient. P<0.05 was considered to
indicate a statistically significant difference.
Results
LINC00473 expression is upregulated in
NSCLC tissues and cells
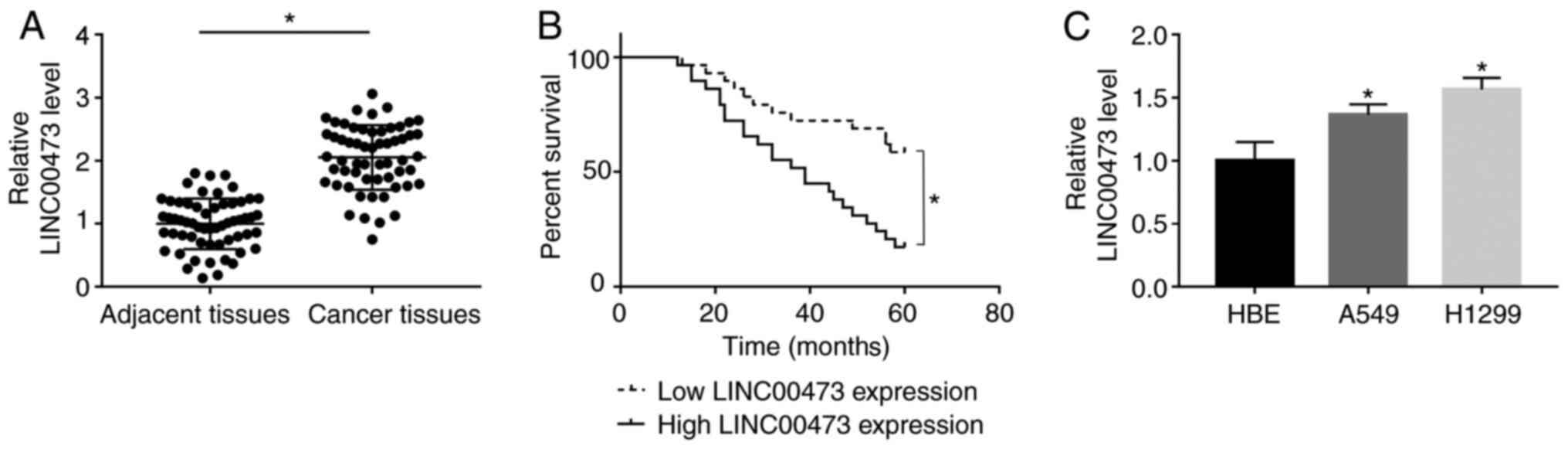
RT-qPCR results indicated that LINC00473 was
significantly upregulated in NSCLC tissues when compared with
adjacent tissues (P< 0.05; Fig.
1A). Furthermore, survival analysis indicated that patients
with a higher expression of LINC00473 had a significantly lower
5-year survival rate compared with patients with low expression of
LINC00473 (log-rank P<0.05; Fig.
1B). Additionally, significantly increased expression of
LINC00473 was observed in both A549 and H1299 cells compared with
HBE cells (P<0.05; Fig. 1C). The
present results indicated that LINC00473 expression was upregulated
in both NSCLC tissues and cells.
LINC00473 promotes proliferation and
inhibits apoptosis of NSCLC cells
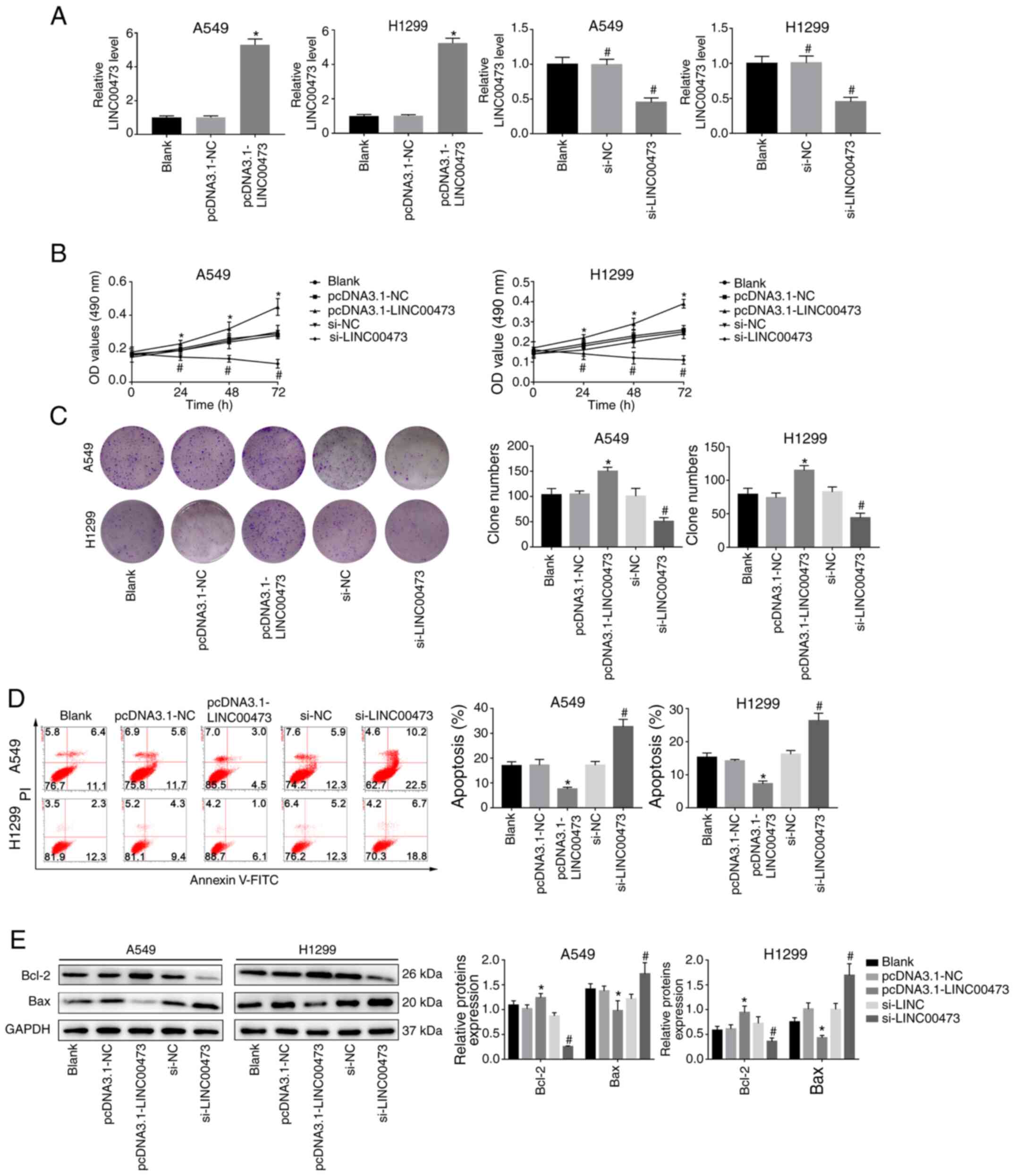
As indicated in Fig.
2A, LINC00473 was significantly upregulated in A549 and H1299
cells transfected with pcDNA3.1-LINC00473, compared with those
transfected with pcDNA3.1-NC (both P<0.05). Furthermore, the
expression of LINC00473 in A549 and H1299 cells was significantly
decreased after transfection with si-LINC00473 when compared with
the si-NC group (both P<0.05). The results of the MTT and colony
formation assays indicated that overexpression of LINC00473
promoted proliferation, whereas its downregulation inhibited the
proliferation of A549 and H1299 cells compared with their NC groups
(all P<0.05; Fig. 2B and C).
Moreover, the apoptosis rate (Annexin V+/PI+
cells) significantly decreased when LINC00473 was overexpressed and
significantly increased when LINC00473 was knocked down (both
P<0.05; Fig. 2D). Meanwhile, it
was observed that expression levels of Bax decreased while Bcl-2
increased with LINC00473-overexpression, and the opposite was
observed when LINC00473 was knocked down, with an increase in Bax
and decrease in Bcl-2 expression (P<0.05; Fig. 2E). These results suggested that
LINC00473 served a promoting role in proliferation and an
inhibitory role in apoptosis of NSCLC cells in vitro.
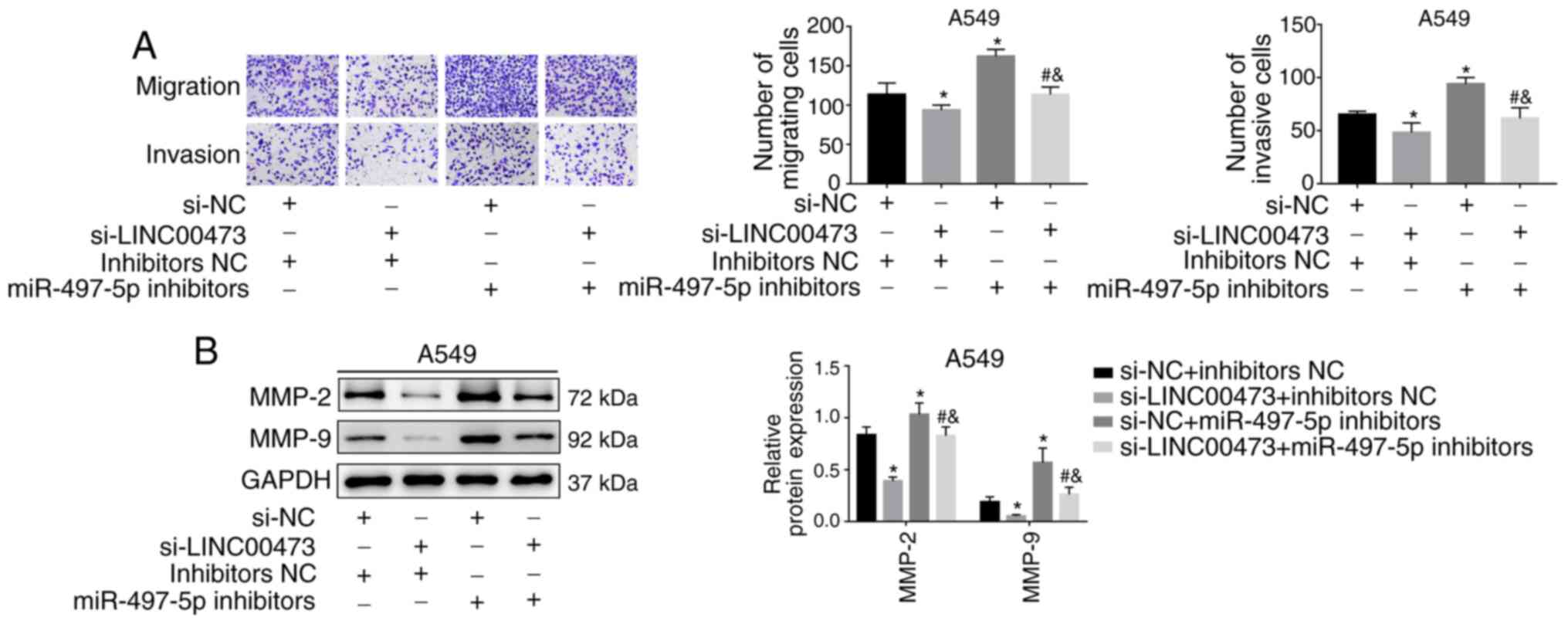
LINC00473 promotes the migration and
invasion of NSCLC cells
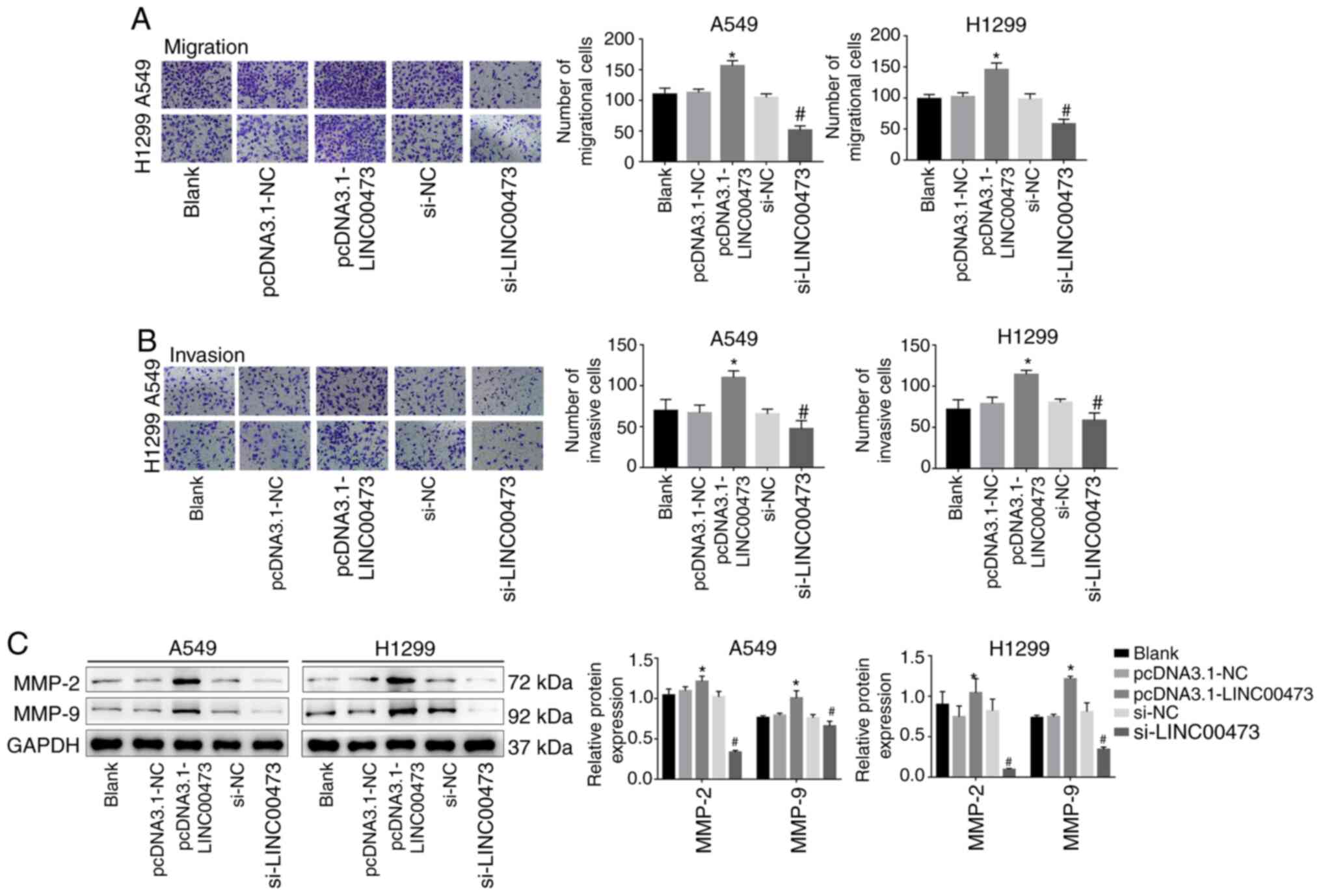
A Transwell migration assay demonstrated that the
overexpression of LINC00473 significantly enhanced the migration
and invasion activities of NSCLC cells (P<0.05; Fig. 3A and B). Conversely, migration was
significantly inhibited by LINC00473-knockdown (P<0.05; Fig. 3A and B). In addition,
LINC00473-overexpression significantly increased the expression of
the metastasis-associated proteins MMP-2 and MMP-9; however,
LINC00473-knockdown significantly decreased their expression (all
P<0.05; Fig. 3C). The results
demonstrated that LINC00473 promoted the migration and invasion of
NSCLC cells in vitro.
LINC00473 competitively binds to
miR-497-5p
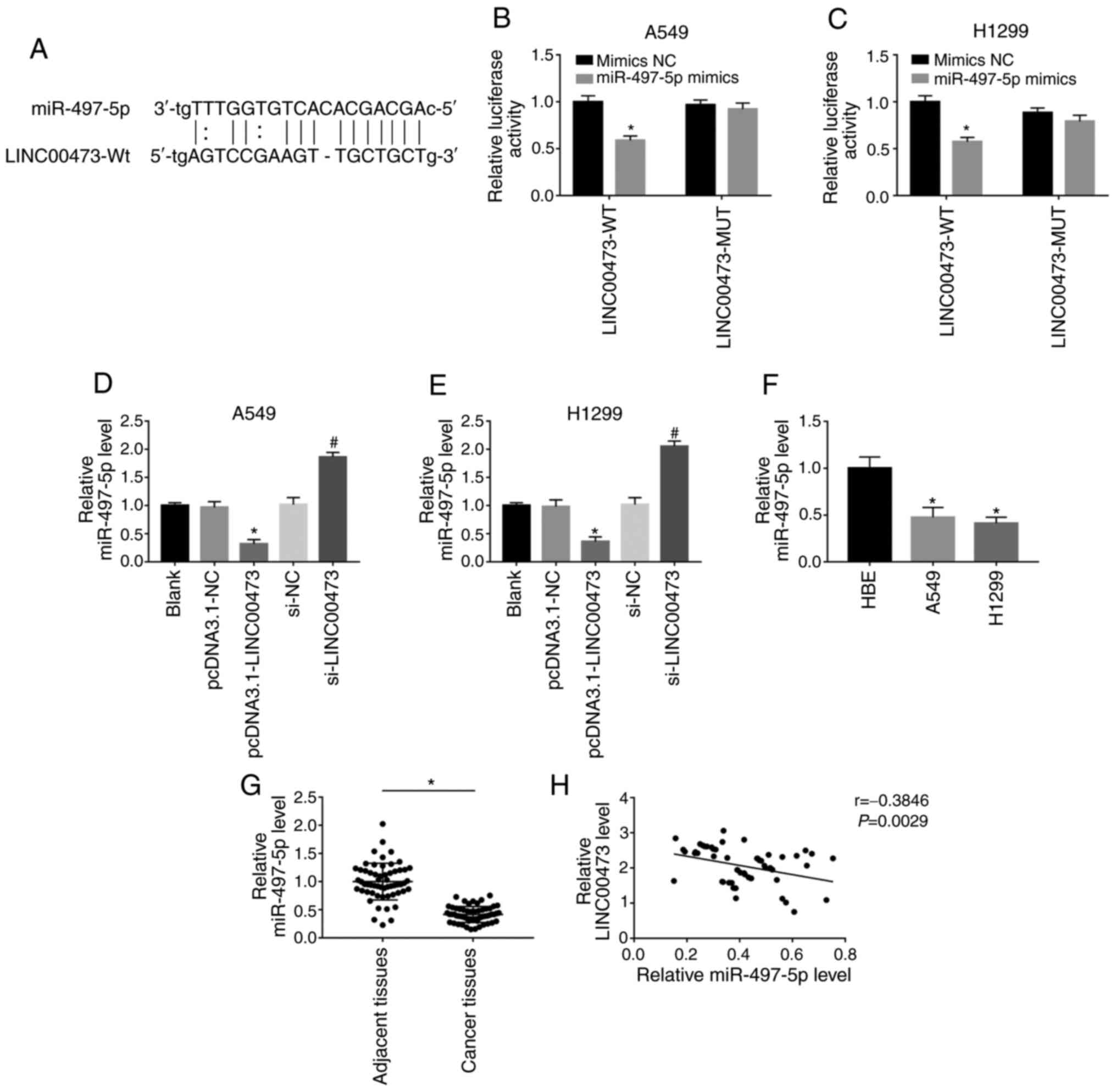
As predicted by DIANA tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/),
miR-497-5p was a possible target miRNA of LINC00473. To confirm
this, two types of luciferase reporter gene vectors (miR-497-5p-WT
and miR-497-5p-MUT) were used to investigate whether LINC00473 acts
as a sponge to miR-497-5p through direct binding (Fig. 4A). As shown in Fig. 4B and C, after transfection with WT
sequences, the miR-497-5p mimics showed significantly inhibited
luciferase activity (both P<0.05). However, when the predicted
binding site was mutated, overexpression of miR-497-5p showed no
change in luciferase activity. Moreover, the expression of
miR-497-5p was significantly reduced after overexpression of
LINC00473 in both cell lines, but significantly increased after
LINC00473-knockdown in A549 and H1299 cells (all P<0.05;
Fig. 4D and E). As shown in Fig. 4F, the expression of miR-497-5p in
A549 and H1299 cells was lower compared with that in HBE (both
P<0.05). Likewise, the expression of miR-497-5p in 58 pairs of
NSCLC and adjacent tissues was compared and it was reported that
miR-497-5p was significantly downregulated in the former
(P<0.05; Fig. 4G). The expression
of miR-497-5p was found to be negatively correlated with LINC00473
(P<0.05; Fig. 4H). The results
indicated that LINC00473 could competitively bind to miR-497-5p and
negatively regulate its expression.
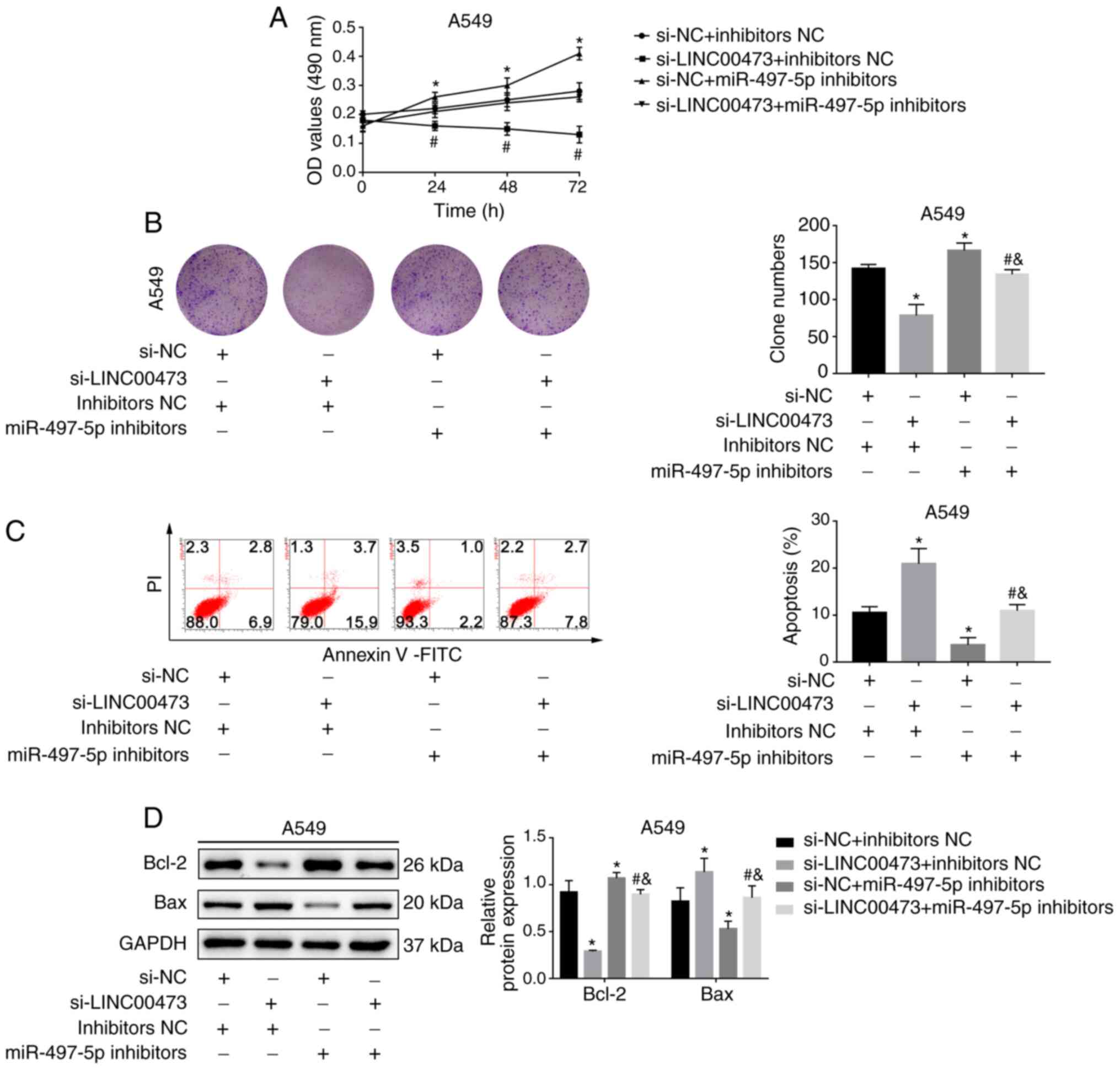
miR-497-5p inhibition attenuates the
effect of LINC00473 silencing
As depicted in Fig.
5A, the cell proliferation rates in the si-LINC00473 group were
lower compared with the NC group. However, cell viability was
enhanced after treatment with miR-497-5p inhibitors (P<0.05).
The clone number of A549 cells decreased after LINC00473-knockdown
but increased after the addition of miR-497-5p inhibitors
(P<0.05; Fig. 5B). Moreover,
miR-497-5p inhibition stabilised the increase in apoptosis and the
expression of apoptosis-related proteins induced by
LINC00473-knockdown (all P<0.05; Fig.
5C and D). Transwell migration assays indicated that both cell
migration and invasion abilities were reduced after silencing
LINC00473 but were reversed after co-transfection with miR-497
inhibitors (both P<0.05; Fig.
6A). Similarly, the expression of MMP-2 and MMP-9 was inhibited
by silencing LINC00473 but was reversed by miR-497 inhibitors (all
P<0.05; Fig. 6B). The current
results suggested that LINC00473 served its role by regulating
miR-497-5p expression.
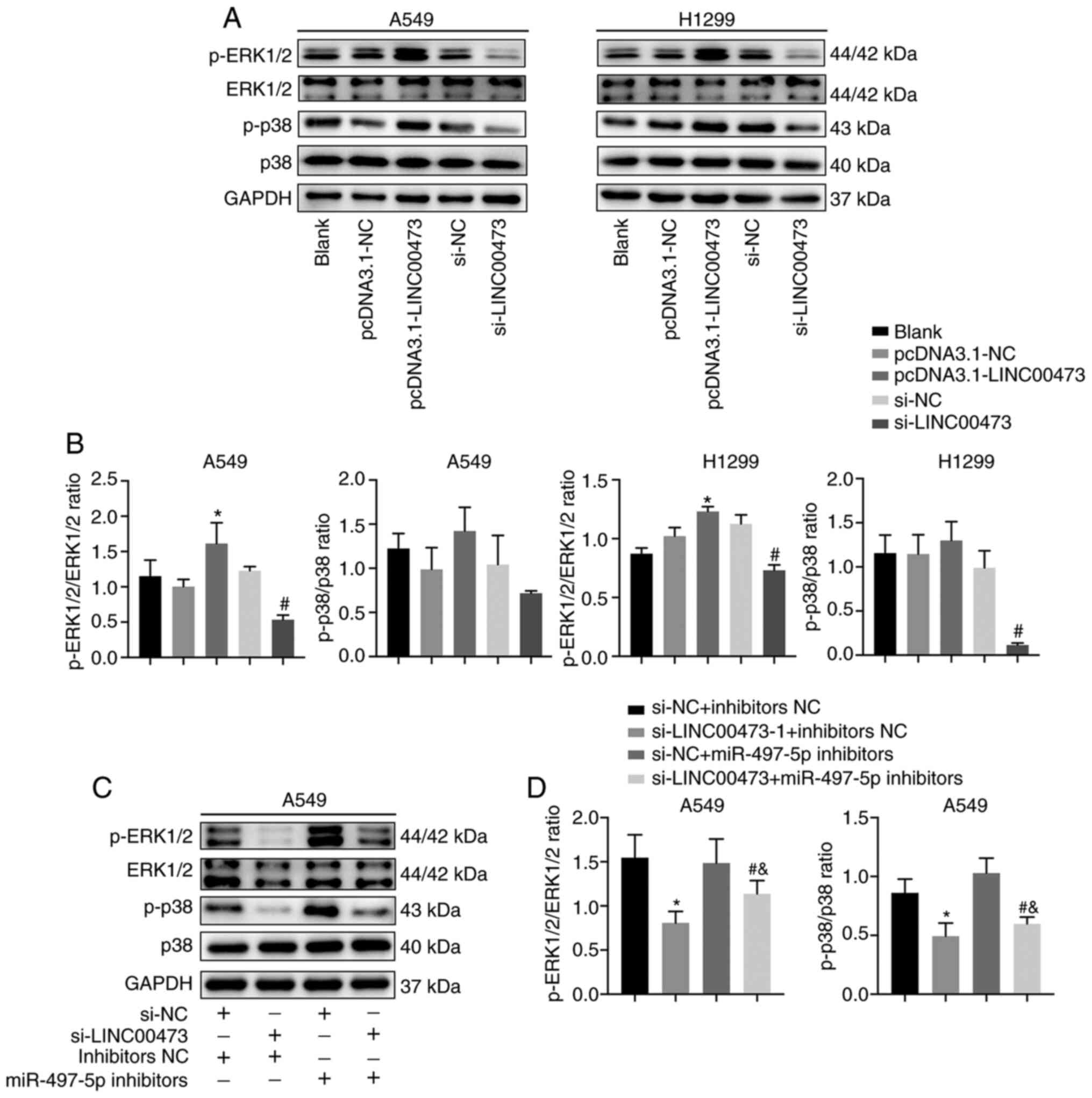
LINC00473 activates the MAPK
signalling pathway
To study the relationship between LINC00473 and the
MAPK signalling pathway, the expression of proteins associated with
the pathway was analysed. The data indicated that p-ERK1/2 and
p-p38 expression increased when LINC00473 was overexpressed and
decreased when LINC00473 was knocked down (Fig. 7A). Moreover, the p-ERK1/2/ERK1/2
ratio (P<0.05; Fig. 7B) and
p-p38/p38 ratio showed the similar trends. In A459 cells, the
expression levels of p-ERK1/2 and p-p38 were significantly
decreased by LINC00473-knockdown and increased after treatment with
miR-497-5p inhibitors (and both p-ERK1/2/ERK1/2 ratio and p-p38/p38
ratio showed the same trends (P<0.05; Fig. 7C and D). These results indicated that
the regulatory mechanism of LINC00473 may be through the MAPK
signalling pathway.
Discussion
Multiple studies have suggested that lncRNAs play
crucial roles in tumour progression, making them promising
therapeutic targets for treatment of diseases such as NSCLC
(16–18). Some studies have reported that the
lncRNA LINC00473 promotes cell migration and invasion in numerous
types of cancer, including pancreatic, mucoepidermoid and breast
cancer (11,19,20). He
(21) reported that LINC00473
regulates the progression of oesophageal squamous cell carcinoma by
affecting 5′-AMP-activated protein kinase catalytic subunit α-1
expression. Furthermore, Mo et al (22) revealed that LINC00473 promotes the
progression of hepatocellular carcinoma by sponging miRNA-195 and
increasing high mobility group protein HMGI-C expression. However,
whether miR-146-5p is involved in the regulation of lung cancer
remains unclear. The present study demonstrated that LINC00473 was
significantly upregulated in both patient lung cancer tissues and
NSCLC cell lines. Additionally, the inhibitory effects of silencing
LINC00473 and the promotional effect of its overexpression on the
proliferation, invasion and migration of NSCLC cells was
investigated.
Several studies have suggested that lncRNAs act as
natural sponges to interact with miRNAs by eliminating the
inhibitory activity of these miRNAs (23–25).
Chen et al (24) showed that
LINC00473 weakens the effect of radiotherapy by targeting
miR-374a-5p. Wang et al (25)
also revealed that in colorectal cancer LINC00473 can promote Taxol
resistance by inhibiting miR-15a. In the present study, to explore
the underlying mechanism of LINC00473 in NSCLC development, target
miRNAs of LINC00473 were investigated. Using bioinformatics
methods, it was demonstrated that miR-497-5p contained a binding
site for LINC00473. Luciferase reporter assays confirmed that
LINC00473 and miR-497-5p bind with each other. Furthermore, the
expression of miR-497-5p was decreased by LINC00473-overexpression
and enhanced by LINC00473 silencing. Moreover, the inhibition of
miR-497-5p attenuated the effect of LINC00473 silencing on cell
proliferation, apoptosis, migration and invasion. These data
suggested that LINC00473 promotes NSCLC progression in an
miR-497-5p-dependent manner.
It has been reported that the MEK/ERK signalling
pathway is involved in cell proliferation, apoptosis and migration
(26). Several lncRNAs, such as
lncRNA-01126 and lncRNA HOTTIP, have been shown to exert their
functions by regulating this pathway (27,28).
This pathway is involved in the migration and invasion of NSCLC
cells induced by the lncRNA SDPR-AS (29). In addition, the lncRNA MALAT1 has
been shown to play a tumour-enhancing role by regulating the
development of NSCLC via the MAPK/ERK pathway (30). The present results indicated that
LINC00473-knockdown decreased the p-ERK and p-p38 levels, which
were reversed by miR-497-5p inhibition. These data indicated that
LINC00473 regulates the activity of the MAPK/ERK signalling pathway
to influence the proliferation, migration and invasion of NSCLC
cells.
Overall, the present results indicated that
LINC00473 is highly expressed in NSCLC tissues, and its
downregulation suppresses the malignant activities of NSCLC cells.
Structurally, LINC00473 competitively binds with miR-497-5p and
regulates the MEK/ERK signalling pathway.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MJS conceived, interpreted the data and designed the
study. MJS and SHX confirmed the authenticity of all the raw data.
SHX, YHB, HCM and HNZ performed the experiments and analysed the
data. SHX wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of The Second Hospital of Shandong University (Ji'nan,
China) and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Inamura K: Lung cancer: Understanding its
molecular pathology and the 2015 WHO classification. Front Oncol.
7:1932017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Laskin JJ and Sandler AB: State of the art
in therapy for non-small cell lung cancer. Cancer Invest.
23:427–442. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Choi SW, Kim HW and Nam JW: The small
peptide world in long noncoding RNAs. Brief Bioinform.
20:1853–1864. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chekulaeva M and Rajewsky N: Roles of long
noncoding RNAs and circular RNAs in translation. Cold Spring Harb
Perspect Biol. 11:a0326802019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li M, Chen Y, Zhu J, Gao Z, Wang T and
Zhou P: Long noncoding RNA CASC15 predicts unfavorable prognosis
and exerts oncogenic functions in non-small cell lung cancer. Am J
Transl Res. 11:4303–4314. 2019.PubMed/NCBI
|
|
8
|
Sun Z, He C, Xiao M, Wei B, Zhu Y, Zhang
G, Zhou H, Yuan J, Hu X and Yi Y: LncRNA FOXC2 antisense transcript
accelerates non-small-cell lung cancer tumorigenesis via silencing
p15. Am J Transl Res. 11:4552–4560. 2019.PubMed/NCBI
|
|
9
|
Yao G, Chen K, Qin Y, Niu Y, Zhang X, Xu
S, Zhang C, Feng M and Wang K: Long non-coding RNA JHDM1D-AS1
interacts with DHX15 protein to enhance non-small-cell lung cancer
growth and metastasis. Mol Ther Nucleic Acids. 18:831–840. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO
and Yang XH: Long noncoding RNA LINC00473 drives the progression of
pancreatic cancer via upregulating programmed death-ligand 1 by
sponging microRNA-195-5p. J Cell Physiol. 234:23176–23189. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Niu L, Zhou Y, Zhang W and Ren Y: Long
noncoding RNA LINC00473 functions as a competing endogenous RNA to
regulate MAPK1 expression by sponging miR-198 in breast cancer.
Pathol Res Pract. 215:1524702019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stavast CJ and Erkeland SJ: The
non-canonical aspects of MicroRNAs: Many roads to gene regulation.
Cells. 8:E14652019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA-LncRNA interactions. Methods Mol Biol.
1402:271–286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu T, Wang Y, Chen D, Liu J and Jiao W:
Potential clinical application of lncRNAs in non-small cell lung
cancer. OncoTargets Ther. 11:8045–8052. 2018. View Article : Google Scholar
|
|
17
|
Zhou Y, Shi H, Du Y, Zhao G, Wang X, Li Q,
Liu J, Ye L, Shen Z, Guo Y, et al: lncRNA DLEU2 modulates cell
proliferation and invasion of non-small cell lung cancer by
regulating miR-30c-5p/SOX9 axis. Aging (Albany NY). 11:7386–7401.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi J, Li J, Yang S, Hu X, Chen J, Feng J,
Shi T, He Y, Mei Z, He W, et al: LncRNA SNHG3 is activated by E2F1
and promotes proliferation and migration of non-small-cell lung
cancer cells through activating TGF-β pathway and IL-6/JAK2/STAT3
pathway. J Cell Physiol. 235:2891–2900. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Z, Lin S, Li J-L, Ni W, Guo R, Lu J,
Kaye FJ and Wu L: CRTC1-MAML2 fusion-induced lncRNA LINC00473
expression maintains the growth and survival of human
mucoepidermoid carcinoma cells. Oncogene. 37:1885–1895. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shi X and Wang X: LINC00473 mediates
cyclin D1 expression through a balance between activation and
repression signals in breast cancer cells. FEBS Lett. 593:751–759.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He Z: LINC00473/miR-497-5p regulates
esophageal squamous cell carcinoma progression through targeting
PRKAA1. Cancer Biother Radiopharm. 34:650–659. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mo J, Li B, Zhou Y, Xu Y, Jiang H, Cheng
X, Wu X and Zhang Y: LINC00473 promotes hepatocellular carcinoma
progression via acting as a ceRNA for microRNA-195 and increasing
HMGA2 expression. Biomed Pharmacother. 120:1094032019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li D, Li H, Yang Y and Kang L: Long
Noncoding RNA urothelial carcinoma-associated 1 promotes the
proliferation and metastasis of human lung tumor cells by
regulating MicroRNA-144. Oncol Res. 26:537–546. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen W, Zhang Y, Wang H, Pan T, Zhang Y
and Li C: LINC00473/miR-374a-5p regulates esophageal squamous cell
carcinoma via targeting SPIN1 to weaken the effect of radiotherapy.
J Cell Biochem. 120:14562–14572. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Zhang X, Sheng L, Qiu C and Luo R:
LINC00473 promotes the Taxol resistance via miR-15a in colorectal
cancer. Biosci Rep. Sep 20–2018.(Epub ahead of print). doi:
10.1042/BSR20180790.
|
|
26
|
Sun Y, Liu W-Z, Liu T, Feng X, Yang N and
Zhou H-F: Signaling pathway of MAPK/ERK in cell proliferation,
differentiation, migration, senescence and apoptosis. J Recept
Signal Transduct Res. 35:600–604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu Y, Ai R, Ding Z, He Q, Zhang X, Dong Y
and He Y: LncRNA-01126 inhibits the migration of human periodontal
ligament cells through MEK/ERK signaling pathway. J Periodontal
Res. 55:631–641. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu J, Hu HB, Liu YM, Li FX, Zhang LP and
Liao ZM: LncRNA HOTTIP promotes the proliferation and invasion of
ovarian cancer cells by activating the MEK/ERK pathway. Mol Med
Rep. 22:3667–3676. 2020.PubMed/NCBI
|
|
29
|
Zhu J, Wang L and Liao R: Long non-coding
RNA SDPR-AS affects the development of non-small cell lung cancer
by regulating SDPR through p38 MAPK/ERK signals. Artif Cells
Nanomed Biotechnol. 47:3172–3179. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu C, Li H, Jia J, Ruan X, Liu Y and
Zhang X: High metastasis-associated lung adenocarcinoma transcript
1 (MALAT1) expression promotes proliferation, migration, and
invasion of non-small cell lung cancer via ERK/mitogen-activated
protein kinase (MAPK) signaling pathway. Med Sci Monit.
25:5143–5149. 2019. View Article : Google Scholar : PubMed/NCBI
|