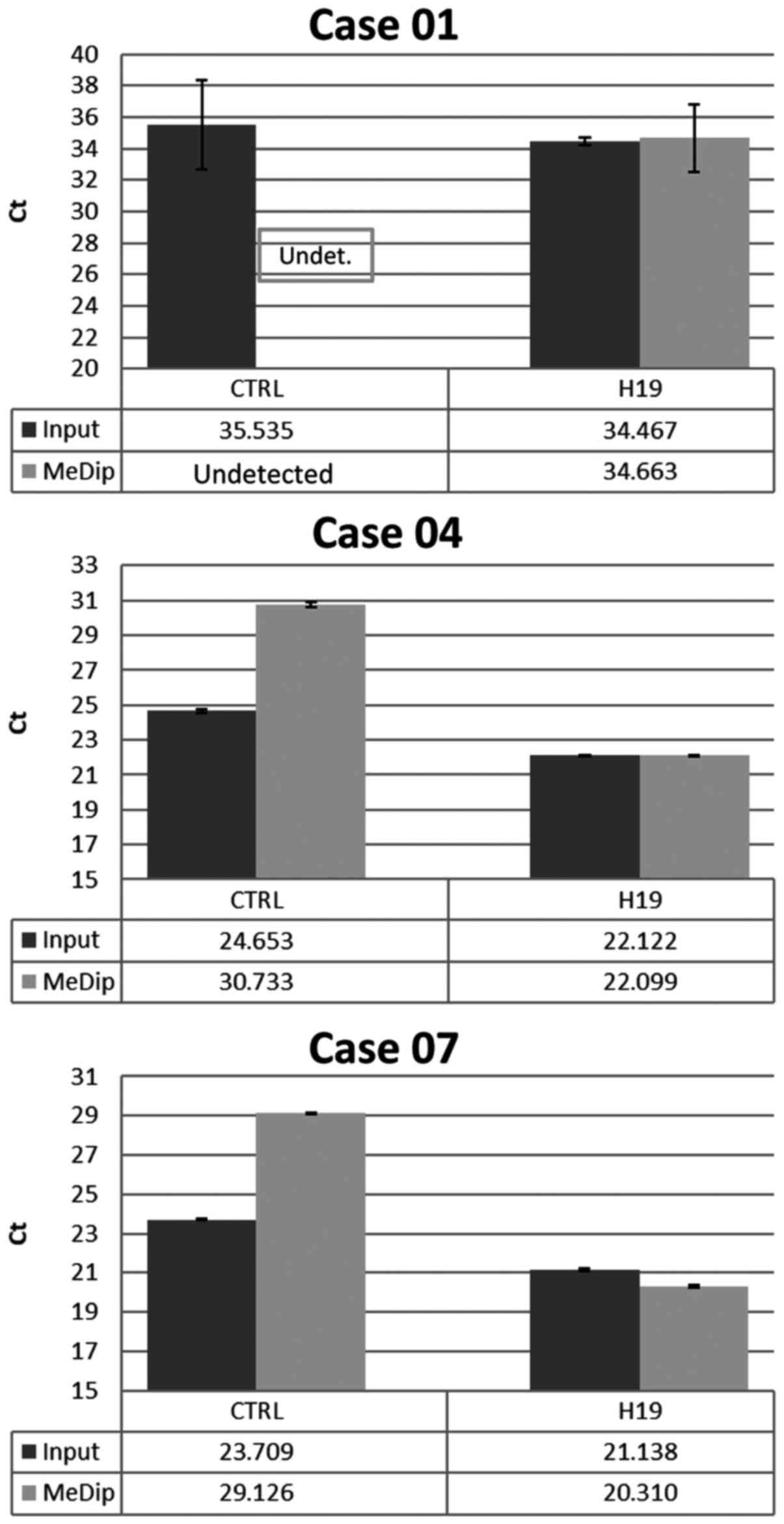
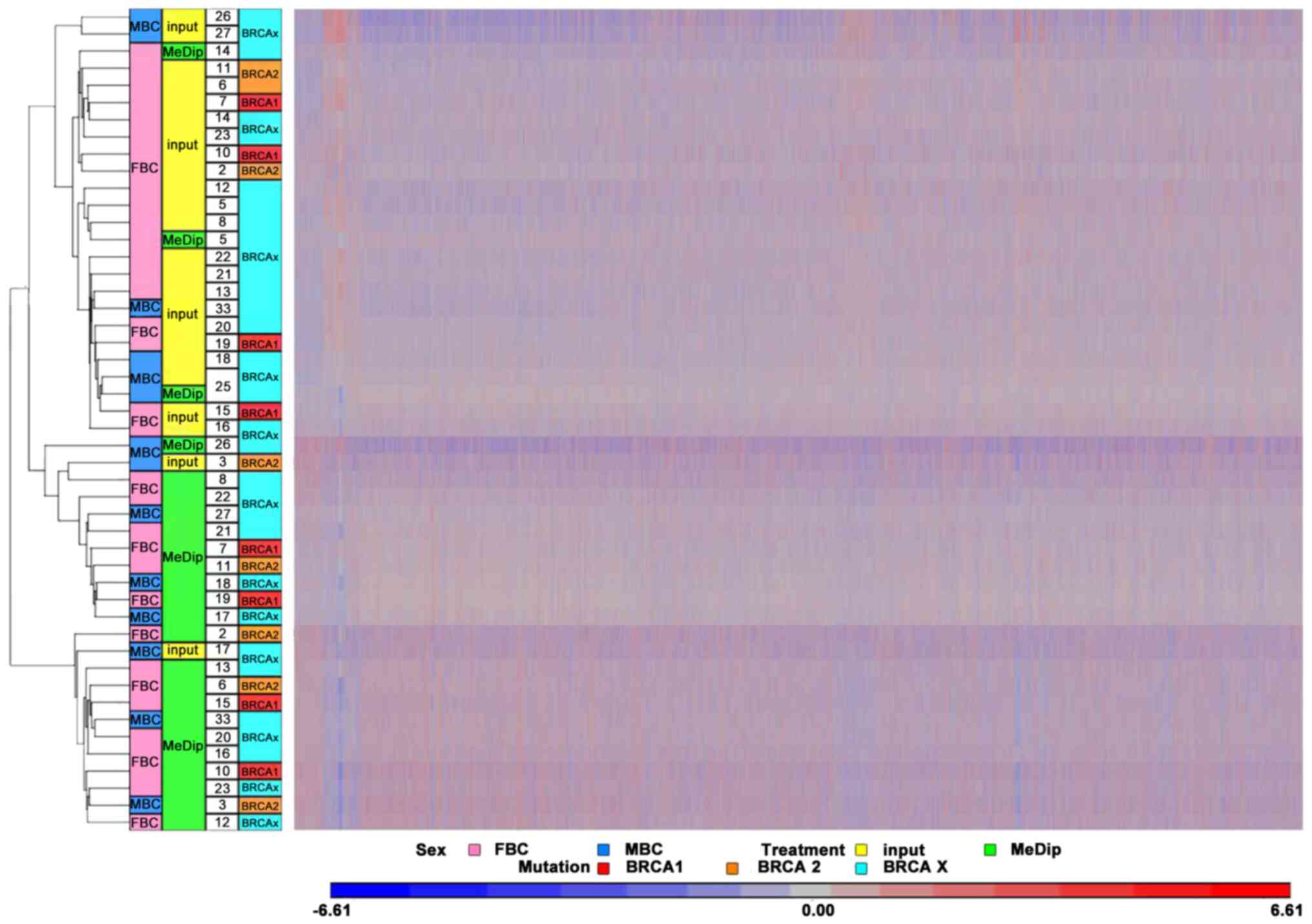
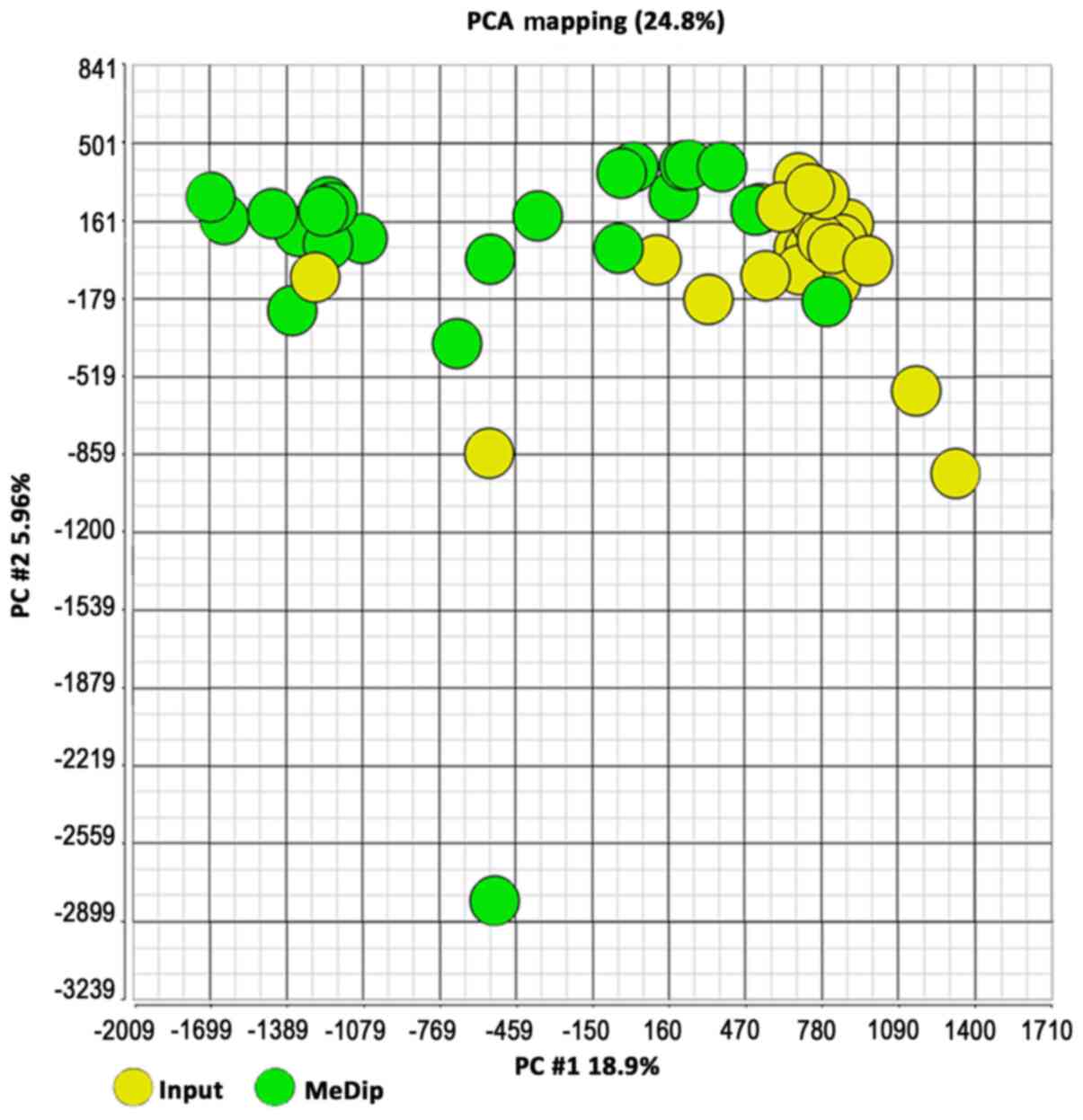
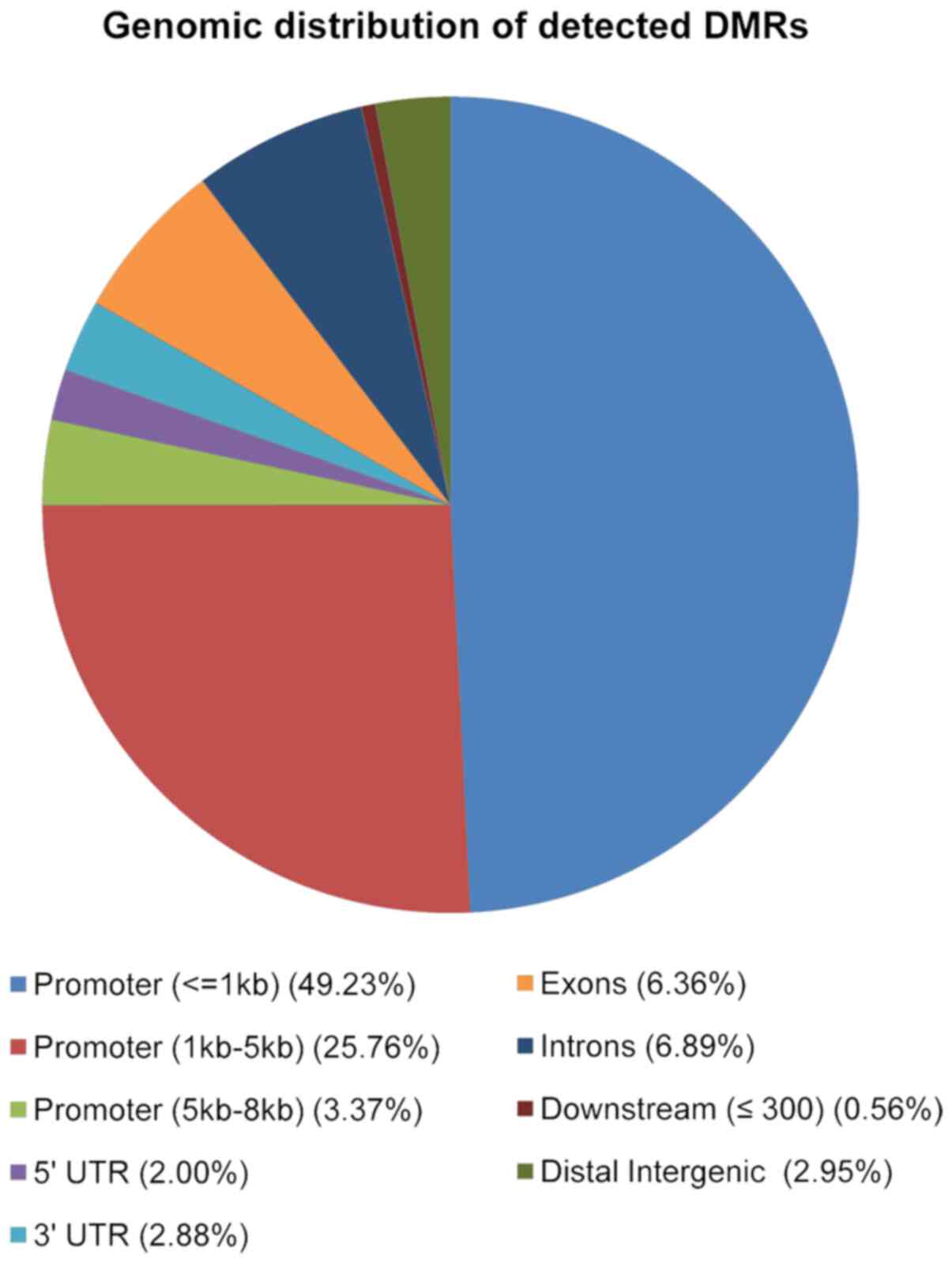
|
1
|
Harbeck N, Penault-Llorca F, Cortes J,
Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J and Cardoso F:
Breast cancer. Nat Rev Dis Prim. 5:662019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kobayashi H, Ohno S, Sasaki Y and Matsuura
M: Hereditary breast and ovarian cancer susceptibility genes
(Review). Oncol Rep. 30:1019–1029. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marchina E, Fontana MG, Speziani M, Salvi
A, Ricca G, Di Lorenzo D, Gervasi M, Caimi L and Barlati S: BRCA1
and BRCA2 genetic test in high risk patients and families:
Counselling and management. Oncol Rep. 24:1661–1667. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang YA, Jian JW, Hung CF, Peng HP, Yang
CF, Cheng HCS and Yang AS: Germline breast cancer susceptibility
gene mutations and breast cancer outcomes. BMC Cancer. 18:3152018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Melchor L and Benítez J: The complex
genetic landscape of familial breast cancer. Hum Genet.
132:845–863. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pinto R, Summa S, Pilato B and Tommasi S:
DNA methylation and miRNAs regulation in hereditary breast cancer:
Epigenetic changes, players in transcriptional and
post-transcriptional regulation in hereditary breast cancer. Curr
Mol Med. 14:45–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Flanagan J, Kugler S, Waddell N, Johnstone
CN, Marsh A, Henderson S, Simpson P, da Silva L; kConFab
Investigators, ; Khanna K, et al: DNA methylome of familial breast
cancer identifies distinct profiles defined by mutation status.
Breast Cancer Res. 86:420–33. 2010.
|
|
9
|
Gucalp A, Traina TA, Eisner JR, Parker JS,
Selitsky SR, Park BH, Elias AD, Baskin-Bey ES and Cardoso F: Male
breast cancer: A disease distinct from female breast cancer. Breast
Cancer Res Treat. 173:37–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kornegoor R, van Diest PJ, Buerger H and
Korsching E: Tracing differences between male and female breast
cancer: Both diseases own a different biology. Histopathology.
67:888–897. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Moncini S, Salvi A, Zuccotti P, Viero G,
Quattrone A, Barlati S, De Petro G, Venturin M and Riva P: The role
of miR-103 and miR-107 in regulation of CDK5R1 expression and in
cellular migration. PLoS One. 6:e200382011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Johansson I, Ringnér M and Hedenfalk I:
The landscape of candidate driver genes differs between male and
female breast cancer. PLoS One. 8:e782992013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rizzolo P, Silvestri V, Tommasi S, Pinto
R, Danza K, Falchetti M, Gulino M, Frati P and Ottini L: Male
breast cancer: Genetics, epigenetics, and ethical aspects. Ann
Oncol. 24 (Suppl 8):viii75–viii82. 2013. View Article : Google Scholar
|
|
14
|
André S, Nunes SP, Silva F, Henrique R,
Félix A and Jerónimo C: Analysis of epigenetic alterations in
homologous recombination dna repair genes in male breast cancer.
Int J Mol Sci. 21:27152020. View Article : Google Scholar
|
|
15
|
Kornegoor R, Moelans CB, Verschuur-Maes
AH, Hogenes MCH, de Bruin PC, Oudejans JJ and van Diest PJ:
Promoter hypermethylation in male breast cancer: Analysis by
multiplex ligation-dependent probe amplification. Breast Cancer
Res. 14:R1012012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vermeulen MA, van Deurzen CHM, Doebar SC,
de Leng WWJ, Martens JWM, van Diest PJ and Moelans CB: Promoter
hypermethylation in ductal carcinoma in situ of the male breast.
Endocr Relat Cancer. 26:575–584. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pinto R, Pilato B, Ottini L, Lambo R,
Simone G, Paradiso A and Tommasi S: Different methylation and
MicroRNA expression pattern in male and female familial breast
cancer. J Cell Physiol. 228:1264–1269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deb S, Gorringe KL, Pang JM, Byrne DJ,
Takano EA; kConFab Investigators, ; Dobrovic A and Fox SB: BRCA2
carriers with male breast cancer show elevated tumour methylation.
BMC Cancer. 17:6412017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rizzolo P, Silvestri V, Valentini V, Zelli
V, Zanna I, Masala G, Bianchi S, Palli D and Ottini L:
Gene-specific methylation profiles in BRCA-mutation positive and
BRCA-mutation negative male breast cancers. Oncotarget.
9:19783–19792. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abeni E, Salvi A, Marchina E, Traversa M,
Arici B and De Petro G: Sorafenib induces variations of the DNA
methylome in HA22T/VGH human hepatocellular carcinoma-derived
cells. Int J Oncol. 51:128–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Omura N, Li CP, Li A, Hong SM, Walter K,
Jimeno A, Hidalgo M and Goggins M: Genome-wide profiling of
methylated promoters in pancreatic adenocarcinoma. Cancer Biol
Ther. 7:1146–1156. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ling G and Waxman DJ: DNase I digestion of
isolated nulcei for genome-wide mapping of DNase hypersensitivity
sites in chromatin. Methods Mol Biol. 977:21–33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johnson WE, Li W, Meyer CA, Gottardo R,
Carroll JS, Brown M and Liu XS: Model-based analysis of
tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA.
103:12457–12462. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Karantza V: Keratins in health and cancer:
More than mere epithelial cell markers. Oncogene. 30:127–138. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shao MM, Chan SK, Yu AM, Lam CC, Tsang JY,
Lui PC, Law BK, Tan PH and Tse GM: Keratin expression in breast
cancers. Virchows Arch. 461:313–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wertheimer E, Gutierrez-Uzquiza A,
Rosemblit C, Lopez-Haber C, Sosa MS and Kazanietz MG: Rac signaling
in breast cancer: A tale of GEFs and GAPs. Cell Signal. 24:353–362.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Subramani D and Alahari SK:
Integrin-mediated function of Rab GTPases in cancer progression.
Mol Cancer. 9:3122010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ishibashi K, Kanno E, Itoh T and Fukuda M:
Identification and characterization of a novel Tre-2/Bub2/Cdc16
(TBC) protein that possesses Rab3A-GAP activity. Genes Cells.
14:41–52. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Callari M, Cappelletti V, De Cecco L,
Musella V, Miodini P, Veneroni S, Gariboldi M, Pierotti MA and
Daidone MG: Gene expression analysis reveals a different
transcriptomic landscape in female and male breast cancer. Breast
Cancer Res Treat. 127:601–610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu W, Koike A, Takeshita T and Ohta T: The
ubiquitin E3 ligase activity of BRCA1 and its biological functions.
Cell Div. 3:12008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bertucci F, Finetti P and Birnbaum D:
Basal breast cancer: A complex and deadly molecular subtype. Curr
Mol Med. 12:96–110. 2011. View Article : Google Scholar
|
|
35
|
Johansson I, Lauss M, Holm K, Staaf J,
Nilsson C, Fjällskog ML, Ringnér M and Hedenfalk I: Genome
methylation patterns in male breast cancer-Identification of an
epitype with hypermethylation of polycomb target genes. Mol Oncol.
9:1565–1579. 2015. View Article : Google Scholar : PubMed/NCBI
|