Introduction
Secreted protein acidic and rich in cysteine (SPARC)
is a protein encoded by a single gene in human chromosome 5q31.1
(1). Mature SPARC has 286 amino
acids with three distinct functional domains, including an
N-terminal acidic domain, a follistatin-like domain and a
C-terminal domain (2). There are two
calcium binding sites on this protein, which are the N-terminus
acidic domain that binds 5 to 8 Ca2+ with a low affinity
and a EF-hand motifs located in the C-terminus domain that bind a
Ca2+ ion with a high affinity (1). As a secreted glycoprotein, SPARC binds
several types of extracellular components, such as collagen, fibrin
and minerals, and plays essential roles in physiological and
pathological conditions, such as cataract formation, wound and
defective organ healing, as well as tumorigenesis (1–3).
SPARC plays a complex and multifaceted role in
tumours, including tumorigenesis, cellular malignant proliferation,
drug resistance and metastasis (4,5). The
function of SPARC is associated with tumour type, cellular origin
and the unique cancer milieu at both primary and metastatic sites.
Different or even contradictory functions have been reported for
SPARC in different tumour types (4–7).
Moreover, the expression of SPARC has been reported to be
associated with the prognosis of multiple tumours. Indeed, high
expression of SPARC indicates good prognosis in diffuse large
B-cell lymphoma (DLBL) (8) but poor
prognosis in bladder urothelial carcinoma (BLAD), breast invasive
carcinoma (BRAD) and colon adenocarcinoma (COAD) (9–11). Thus,
SPARC appears to be oncogene in some tumours and a tumour
suppressor in others.
Pancreatic adenocarcinoma (PAAD) is the most common
pathological type of pancreatic cancer and one of the malignant
diseases with the worst prognosis worldwide, with a median survival
time of 6–10 months for locally advanced disease and 3–5 months for
metastatic disease (12). SPARC has
demonstrated prognostic and therapeutic potential, as it is
expressed by peritumoral fibroblasts, but not PAAD cells, and is
associated with poor prognosis for patients with PAAD (13). Another study demonstrated that
increased SPARC expression in primary PAAD cells is also associated
with poorer overall and disease-free survival (14). However, SPARC has also been reported
to inhibit proliferation, promote apoptosis and enhance the
chemosensitivity of pancreatic cancer cells to gemcitabine
(15–17). In general, SPARC presents as an
oncogene in clinical studies and is associated with poor prognosis
in patients with PAAD (13,14). However, in biological experimental
studies, SPARC is generally presented as a tumour suppressor of
PAAD (15–17).
Therefore, several questions remain to be addressed.
First, a systematic pan-cancer analysis of SPARC is needed to
determine whether it is an oncogene, a tumour suppressor, or both.
Secondly, the function of SPARC in PAAD needs further
clarification, and the contradicting results of clinical and
biological research need to be unified. Finally, while
stroma-derived SPARC is associated with poor prognosis in patients
with PAAD, cancer cell-derived SPARC inhibits the proliferation and
chemoresistance of PAAD cells (13,15,17).
Whether SPARC produced by tumour stromal cells vs. PAAD cells has
the same effects on tumour growth and progression must be
investigated. In the present study, a pan-cancer analysis of SPARC
expression using The Cancer Genome Atlas (TCGA) data was carried
out. This analysis confirmed that SPARC was an oncogene in some
cancer types, but a tumour suppressor in others. Consistent with
clinical data, this study also confirmed SPARC to be an oncogene in
PAAD. This has important clinical implications for patients with
PAAD. Moreover, SPARC was found to promote PAAD cell proliferation
and migration only when secreted.
Materials and methods
Datasets
Mutation, RNA Sequencing (RNASeq) and clinical data
of 10,182 patients were collected from TCGA (https://portal.gdc.cancer.gov/) with 33 cancer types:
Adrenocortical carcinoma (ACC), bladder urothelial carcinoma
(BLCA), BRCA, cervical squamous cell carcinoma (CESC),
cholangiocarcinoma (CHOL), COAD, DLBL, oesophageal carcinoma
(ESCA), glioblastoma multiforme (GBM), head and neck squamous
carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear
cell carcinoma (KIRC), kidney renal papillary cell carcinoma
(KIRP), bone marrow acute myeloid leukaemia (LAML), brain low grade
glioma (LGG), liver hepatocellular carcinoma (LIHC), lung
adenocarcinoma (LUAD), lung squamous carcinoma (LUSC), pleura
mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), PAAD,
adrenal phechromocytoma and paraganglioma (PCPG), prostate
adenocarcinoma (PRAD), rectum adenocarcinoma (READ), soft tissue
sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach
adenocarcinoma (STAD), testicular germ cell tumours (TGCT), thyroid
carcinoma (THCA), thymoma (THYM), uterine corpus endometrial
carcinoma (UCEC), uterine carcinosarcoma (UCS) and uveal melanoma
(UVM). For mutation data, the dataset produced by the Multi-Center
Mutation Calling in Multiple Cancers (MC3) project was downloaded
instead of the respective maf files of 33 types of cancer (18) (https://gdc.cancer.gov/about-data/publications/mc3-2017).
Patients and samples
All surgically removed PAAD and para-cancer
specimens were obtained from the Department of Hepatobiliary
Surgery of The First Affiliated Hospital, Wenzhou Medical
University, between September 2016 and July 2019. Verbal informed
consent was obtained from the patients. A total of 17 pairs of
pathologically diagnosed PAAD and matched, para-cancer specimens
were collected for RNA and protein extraction in accordance with
institutional protocols. Para-cancer specimens were resected
0.3–0.5 cm from the tumour tissue. The patients age ranged between
52 and 77 years, with a mean age of 62.9 years and a median age of
64 years. The patients included 10 men and 7 women. The ethics of
the study were reviewed and approved by the board of Wenzhou
Medical University.
Cell culture and transfection
The PANC-1 and SW1990 human pancreatic cancer cell
lines, the 293T human embryonic kidney cell line and the AR42J rat
immortalized pancreatic acinar cell line were purchased from the
Institute of Biochemistry and Cell Biology, The Chinese Academy of
Science. PANC-1 and 293T cells were cultured in high-glucose DMEM
medium (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS
(Sigma-Aldrich; Merck KGaA), 100 U/ml penicillin and 100 µg/ml
streptomycin. SW1990 and AR42J were cultured in 1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) with the same supplements.
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and polyethyleneimine (Sigma-Aldrich; Merck KGaA)
were used for the transfection of the PAAD cells and 293T cells,
respectively, according to the manufacturers' protocols.
Plasmid construction
The pLenti-CMV-EGFP-3Flag vector was treated with
EcoRI and BamHI at 37°C for linearization.
Full-length SPARC cDNA (NCBI, accession no. NM_003118.4; http://www.ncbi.nlm.nih.gov/nuccore/NM_003118.4)
was amplified from the cDNA of PANC-1 cells and cloned into the
linearized vector using a seamless clone kit (cat. no. D7010S;
Beyotime Institute of Biotechnology) according to the
manufacturer's protocol.
For pLenti-CMV-SPARC∆sig peptide-Flag,
the N-terminal 17 amino acids are the signal peptide of the SPARC
protein and are essential for autocrine secretion (19). Using the full-length cDNA of SPARC as
a template, we obtained cDNA with deleted codons for signal peptide
via polymerase chain reaction (PCR) amplification while retaining
the start codon. This SPARCΔsig peptide cDNA was cloned
into the linearized pLenti-CMV-EGFP-3Flag using the aforementioned
seamless clone kit according to the manufacturer's protocol.
Silencing of SPARC by small
interfering (si)RNA
SPARC was silenced using two siRNA molecules
(Shanghai Biosun Sci&Tech Co., Ltd.). The sequences of the
SPARC siRNAs and the scrambled negative control siRNA are listed in
Table I. The powder of SPARC and
scrambled siRNAs was dissolved in RNase-free water to a
concentration of 20 µM. Subsequently, 5 µl of the siRNA solution
was mixed with 150 µl FBS-free medium at room temperature for 5
min. At the same time, for each group of siRNA, 5 µl Lipofectamine
3000 (Thermo Fisher Scientific, Inc.) was mixed with 150 µl
FBS-free medium at room temperature for 5 min. The siRNA solution
was mixed with the Lipofectamine 3000 solution at room temperature
for 20 min, and the transfection mixture was added to a 6-cm dish
containing PANC-1 and SW1990 cells at 30–40% confluency and 1.7 ml
FBS-free medium. The final siRNA concentration was 50 pM. The
medium for transfection was replaced by medium containing FBS and
antibiotics 5 h later, and cells were lysed for RNA or protein
extraction 48 h later. For Cell Counting Kit-8 (CCK-8) assay, cells
were re-cultured in a 96-well plate 24 h after transfection.
 | Table I.Primers and siRNA molecules. |
Table I.
Primers and siRNA molecules.
| A, Primers |
|---|
|
|---|
| Primer name | Sequence,
5′-3′ |
|---|
| SPARC-mRNA-F |
CGAAGAGGAGGTGGTGGCGGAAA |
| SPARC-mRNA-R |
GGTTGTTGTCCTCATCCCTCTCATAC |
| GAPDH-F |
CTCTCTGCTCCTCCTGTTCGACAG |
| GAPDH-R |
AGGGGTCTTACTCCTTGGAGGCCA |
|
| B,
siRNA |
|
| siRNA
name | Sequence,
5′-3′ |
|
| siRNA-SPARC-1 |
UUAUCUAAUGUAUUCCUCCUG |
| siRNA-SPARC-2 |
UAGUUCUUCUCGAAGUCCCGG |
| Negative
control |
UUCUCCGAACGUGUCACGUTT |
Overexpression (OE) of SPARC
pLenti-CMV-SPARC-Flag and pLenti-CMV-SPARCΔsig
peptide-Flag vector were constructed as aforementioned. A
pLenti-CMV-EGFP-3Flag vector with deleted EGFP cDNA was used as
control. For transfection of PAAD cells, 3 µg of control or OE
vector was mixed with 150 µl FBS-free medium at room temperature
for 5 min. At the same time, for each group of vectors, 5 µl
Lipofectamine 3000 was mixed with 150 µl FBS-free medium at room
temperature for 5 min. Subsequently, the medium containing vector
was mixed with the medium containing Lipofectamine 3000 at room
temperature for 20 min, and the transfection mixture was added to a
6-cm dish containing PANC-1 and SW1990 cells at 30–40% confluency
and 1.7 ml FBS-free medium. The medium for transfection was
replaced by medium containing FBS and antibiotics 5 h later, and
cells were lysed for RNA or protein extraction 48 h later. For
CCK-8 assay, cells were re-cultured in a 96-well plate 24 h after
transfection.
Silencing of SPARC by short hairpin
(sh)RNA
The sequence of siRNA1 was used for the construction
of SPARC shRNA using a pLKO.1-puro plasmid (Sigma-Aldrich; Merck
KGaA). The pLKO.1-puro plasmid without shRNA insert was used as a
negative control. For lentivirus production, 4 µg of the shRNA
vector, 3 µg of psPAX and 2 µg of pMD2.G were mixed with 150 µl
FBS-free medium at room temperature for 5 min. A total of 30 µl of
polyethylenimine (Sigma-Aldrich; Merck KGaA) was mixed with 150 µl
FBS-free medium at room temperature for 5 min. Subsequently, the
medium containing vector was mixed with the medium containing
polyethylenimine at room temperature for 20 min. The transfection
mixture was added to the 10-cm dishes with 293T cells at 50–60%
confluency. 293T cells were incubated at 37°C, and the transfection
medium was replaced 6 h later. Virus-containing medium was
collected 48 h after transfection and was centrifugated at 3,000 ×
g and 4°C for 5 min to remove cell debris. For lentivirus
infection, PAAD cells were cultured in 6-cm dishes at 40–50%
confluency, and the virus-containing medium was supplemented with 5
µg/ml polybrene to infect PAAD cells at 37°C for 24 h. After 48 h
of infection, the infected cells were positively selected with 2.5
µg/ml puromycin to eliminate uninfected cells to generate stable
cell lines.
RNA isolation and reverse
transcription quantitative-PCR (RT-qPCR)
The total RNA from either PAAD or para-cancer
tissues was extracted using TRI Reagent (Sigma-Aldrich; Merck KGaA)
and chloroform according to the manufacturer's protocol. The
RevertAid First Strand cDNA Synthesis kit (Thermo Fisher
Scientific, Inc.) was used for first-strand cDNA synthesis.
Briefly, 1 µg total RNA was mixed with oligo-dT primer and
nuclease-free water to a total volume of 12 µl, and the mixture was
denatured at 65°C for 5 min. Subsequently, reaction buffer, RNase
inhibitor, dNTP, and reverse transcriptase were added to a total
final volume of 20 µl. The mixture was incubated at 25°C for 5 min,
cDNA was synthesized at 42°C for 60 min, and finally reverse
transcriptase was denatured at 70°C for 5 min.
qPCR was performed using the Quantstudio™ DX
Real-Time PCR Instrument (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the Fast SYBR Green Master Mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.). cDNA was mixed with
SYBR Green and primers to a final volume of 20 µl. The mixture was
incubated at 95°C for 10 min for pre-denaturation, followed by 40
cycles at 95°C for 15 sec and 62°C for 30 sec. The relative mRNA
expression levels were normalized to the expression level of GAPDH
using the 2−ΔΔCq method (20). The primers used for qPCR are listed
in Table I.
CCK-8 and clone formation assay
The proliferation of PAAD cells was determined using
a CCK-8 assay. PANC-1 and SW1990 cells were seeded in a 96-well
plate at a density of 2×103 cells/well in triplicate. After a 12-h
incubation, the cells adhered to the bottom of the plate, and this
time was defined as day 0. At days 0–3 and 4, 100 µl FBS-free
medium with 10% CCK-8 reagent (Shanghai Yeasen Biotechnology Co.,
Ltd.) were added to the wells. After incubation at 37°C for 2 h,
the absorbance was measured with a microplate reader (BioTek
Instruments, Inc.; Agilent Technologies, Inc.). A well with 100 µl
FBS-free medium and 10% CCK-8 reagent without cells was used to
determine the background absorbance. Cell viability was determined
by subtracting the background absorbance from the experimental
wells.
For the clone formation assay, 5×102
cells/well of PANC-1 and SW1990 cells were seeded into 6-well
plates and cultured at 37°C for 14 days. The medium was changed
every three days to avoid bias due to different evaporation rates.
The cells were then fixed with methanol for 15 min at room
temperature and stained with crystal violet for 5 min at room
temperature (Beyotime Institute of Biotechnology).
Transwell assay
The migration of the PANC-1 and SW1990 cells was
assessed using 24-well BioCoat cell culture inserts (BD
Biosciences) with an 8-µm porosity polyethylene terephthalate
membrane. The lower compartment contained RPMI-1640 (for SW1990) or
DMEM (for PANC-1) with 10% FBS as a chemoattractant. A total of
2×104 cells in 0.2 ml FBS-free medium were plated in the upper
compartment and incubated at 37°C for 48 h. After incubation, the
cells were fixed with methanol for 15 min at room temperature and
stained with crystal violet for 5 min at room temperature. The
images of Transwell assay were acquired using the Leica DMIL-LED
inverted light laboratory microscope (Leica Microsystems GmbH;
magnification, ×400).
Western blot analysis
Cells were lysed using a radioimmunoprecipitation
assay lysis buffer with 10% phosphatase inhibitor (Shanghai Yeasen
Biotechnology Co., Ltd.) and 1% phenylmethylsulfonyl fluoride
(Sigma-Aldrich; Merck KGaA) for 30 min at 4°C. Total protein lysate
was then collected, and the concentration was determined with a BCA
Protein Assay kit (Beyotime Institute of Biotechnology). After
denaturation at 100°C for 5 min, 20 µg of protein samples from each
group were resolved by SDS-PAGE using 10% gels, then
electrophoretically transferred to a PVDF. The PVDF membrane was
blocked with 5% skimmed milk (Shanghai Yeasen Biotechnology Co.,
Ltd.) for 1.5 h at room temperature. The membrane was incubated
overnight with primary antibodies at 4°C and for 1 h with
HRP-conjugated secondary antibodies (1:10,000) at 37°C (Table II). Amersham ECL Prime (Cytiva) was
used to detect bands. The densities of the specific protein bands
were visualized and captured using ImageQuant™ 400 (GE Healthcare
Life Sciences) and analysed using Image Lab 3.0 software (Bio-Rad
Laboratories).
 | Table II.Antibodies. |
Table II.
Antibodies.
| Antibody name | Supplier | Cat. no. | Dilution |
|---|
| SPARC | Abcam | ab207743 | 1:2,000 |
| β-actin | Sigma-Aldrich,
Merck KGaA | A1978 | 1:5,000 |
| Flag | Sigma-Aldrich,
Merck KGaA | SAB4200071 | 1:5,000 |
| AKT | Cell Signaling
Technology, Inc. | 4691 | 1:1,000 |
| p-AKT | Cell Signaling
Technology, Inc. | 4060 | 1:1,000 |
| β-catenin | Abcam | ab32572 | 1:5,000 |
| EGFR | Abcam | ab32077 | 1:1,000 |
| p-EGFR | Cell Signaling
Technology, Inc. | 3777 | 1:1,000 |
| ERK | Abcam | ab184699 | 1:2,000 |
| p-ERK | Abcam | ab214036 | 1:2,000 |
| Stat3 | Bioworld | AP0365 | 1:1,000 |
| p-stat3 | Abcam | ab76315 | 1:2,000 |
| p-NFκB | Abcam | ab76302 | 1:1,000 |
| PI3K | Abcam | ab32089 | 1:1,000 |
| MMP2 | Abcam | Ab92536 | 1:1,000 |
| MMP7 | Santa Cruz
Biotechnology, Inc. | sc-515703 | 1:100 |
| MMP9 | Santa Cruz
Biotechnology, Inc. | sc-393859 | 1:200 |
| E-Cadherin | Santa Cruz
Biotechnology, Inc. | sc-21791 | 1:200 |
| N-Cadherin | Abcam | ab98952 | 1:2,000 |
| Vimentin | Santa Cruz
Biotechnology, Inc. | sc-6260 | 1:200 |
| Goat anti rabbit
secondary antibody, HRP-conjugated | Sigma-Aldrich,
Merck KGaA | SAB3700885 | 1:10,000 |
| Goat anti mouse
secondary antibody, HRP-conjugated | Sigma-Aldrich,
Merck KGaA | SAB3700885 | 1:10,000 |
Collection of cellular supernatant and
preparation for western blot analysis
The 293T cells (5×106) transfected with a SPARC
siRNA or OE vector, were seeded in two 10-cm dishes. A volume of 8
ml medium containing 10% FBS was added to each dish. The
supernatant was collected 24 h later, centrifuged at 12,000 × g and
4°C for 5 min, then added to 96-well plates containing the PANC-1
and SW1990 cells.
Western blot analysis was used to determine the
difference of SPARC concentration in the supernatant. The
supernatant was collected and centrifuged at 12,000 × g and 4°C for
5 min; then, 5X SDS-PAGE loading buffer was added. After
denaturation at 100°C for 5 min, the samples were used for the
western blot assay. To determine the concentration of the
SPARC-Flag and SPARCΔsig peptide-Flag fusion protein in
the supernatant, the same number of PANC-1 or SW1990 cells
(2×106), transfected with the same amount of the
control, SPARC-Flag, or SPARCΔsig peptide-Flag
expression vector (2 µg), were seeded in 6 cm-dishes. The
supernatant was collected 24 h later for western blot analysis,
which was carried out as aforementioned.
Statistical analysis
The data are presented as the mean ± SD. A paired or
unpaired two-tailed t-test was used for comparisons between two
groups. One-way ANOVA was used for comparisons of the multiple
groups, and Tukey's method was used for the multiple comparison
test. The Kaplan-Meier/log-rank method was used for the comparison
of survival time between two groups if the survival curves did not
cross. Otherwise, a Cramér-von Mises test was used instead. For
survival analysis of each type of cancer, patients were split into
two cohorts (high and low expression groups) where the P-value was
minimal. The cut-off value was achieved using the R packages
‘survival’ and ‘survminer’, with a loop function. To avoid the bias
caused by the difference in the number of people in the two groups,
each group comprising ≥25% of the total population. R-3.4
(https://www.r-project.org/) was used for
the statistical analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
SPARC drives tumorigenesis through
abnormal changes in expression but not gene mutations
First, the mutations in the SPARC gene from
>10,000 cancer specimens across 33 cancer types were analysed.
The mutation frequency of SPARC was very low, and only 101 single
nucleotide variations were identified in the coding region of
SPARC, including 21 synonymous, 71 missense, 8 nonsense and 1
splice site mutations. The 80 non-synonymous mutations were
uniformly distributed across 64 coding base sites, with 1–3
mutations at each site, and no prominent recurrent mutations were
found (Fig. 1A). It is likely that
these mutations were only passengers and mutations of SPARC had no
significant relationship with tumorigenesis. Interestingly, no
insertion-deletion of the SPARC gene was found in the MC3 dataset,
whether in-frame or frame-shift. The low mutation frequency and
high conservation suggested that the normal function of SPARC was
essential in tumour development. Thus, SPARC might be a potential
negative selection gene during tumorigenesis (21).
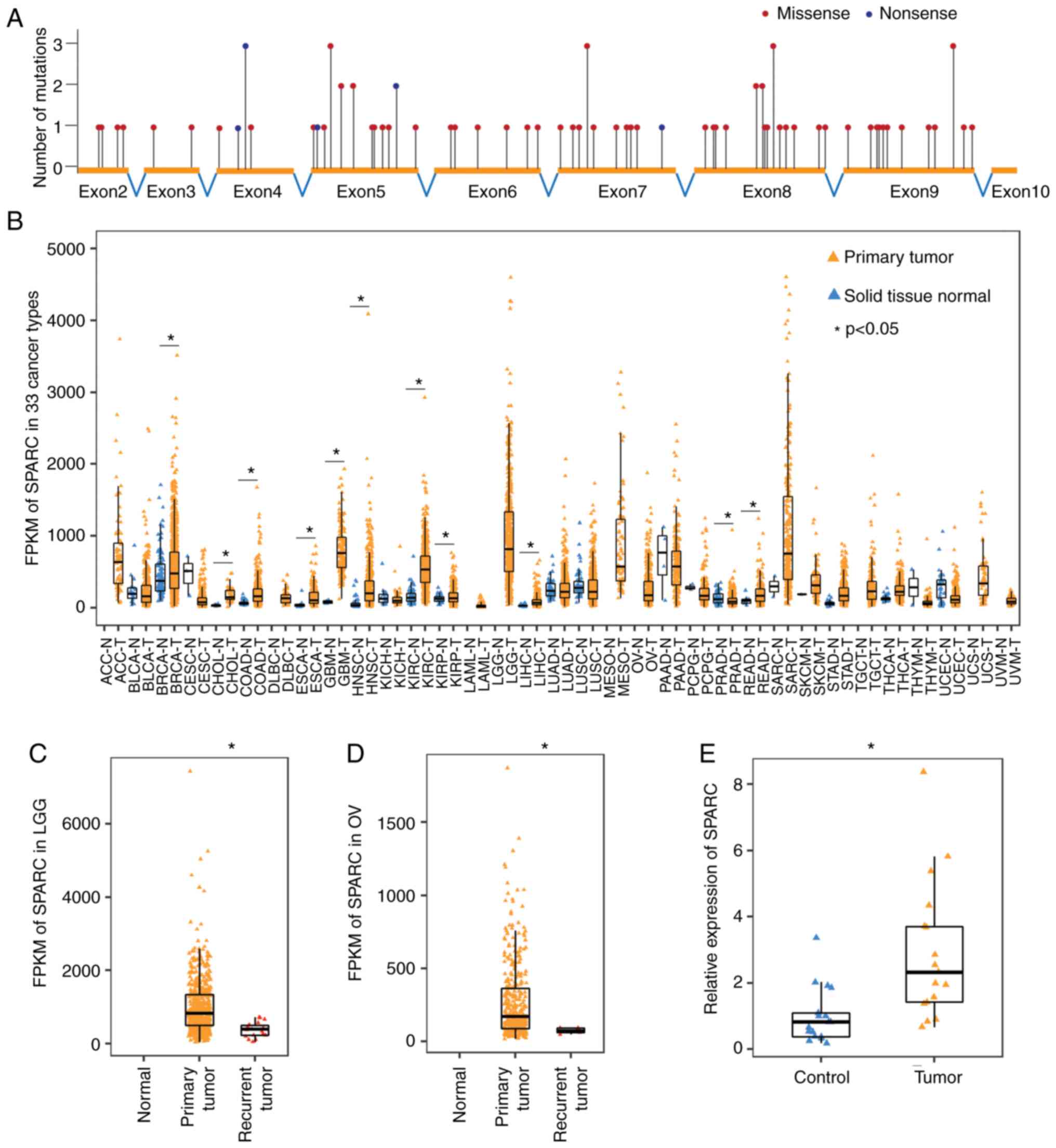 | Figure 1.SPARC drives tumorigenesis through
abnormal changes in expression but not gene mutations. (A)
Single-nucleotide variations are uniformly distributed across the
coding region of the SPARC gene (TCGA data). (B) Expression pattern
of SPARC in 33 cancer types. SPARC expression is generally
increased in various cancer types. Unpaired, two-tailed t-test
(TCGA data). (C) Expression of SPARC decreased significantly in
recurrent LGG, compared with the primary tumour. Unpaired,
two-tailed t-test (TCGA data). (D) Expression of SPARC decreased
significantly in recurrent OV, compared with the primary tumour.
Unpaired, two-tailed t-test (TCGA data). (E) Reverse
transcription-quantitative PCR demonstrated that SPARC expression
increased significantly in PAAD, compared with the paired
para-cancer tissue. n=17, paired, two-tailed t-test. *P<0.05.
LGG, brain low-grade glioma; OV, ovarian serous cystadenocarcinoma;
PAAD, pancreatic adenocarcinoma; TCGA, The Cancer Genome Atlas;
FPKM, fragments per kilobase million; SPARC, secreted protein
acidic and rich in cysteine. |
Since mutations in the SPARC gene had no significant
relationship to tumorigenesis, the expression of SPARC mRNA was
then analysed in tumour and normal tissues. Among the 33 tumour
types included in TCGA database, 24 tumour types contained the
RNASeq data of both cancer and normal tissues. The expression of
SPARC was significantly increased in 10 tumour types (BRCA, CHOL,
COAD, ESCA, GBM, HNSC, KIRC, KIRP, LIHC and READ) and significantly
decreased only in PRAD (Fig. 1B).
These results suggested that SPARC may mainly function as an
oncogene in most cancer types. TCGA database also collected the
RNASeq data of some recurrent and metastatic tumours. SPARC
expression was significantly reduced in tissue from recurrent LGG
and OV tumours compared with primary tumour tissues (Fig. 1C and D), but its clinical implication
is unclear.
In TCGA dataset, the RNASeq data of PAAD contained
only 4 normal specimens, and no significant change in SPARC
expression was found. Thus, 17 pairs of RNA samples from PAAD and
adjacent tissues were obtained and used to determine SPARC
expression by RT-qPCR. The results confirmed that SPARC expression
was significantly increased in PAAD tissues, compared with normal
tissue (Fig. 1E).
SPARC is associated with the prognosis
of various cancers
The expression level of SPARC was associated with
the prognosis of various tumour types. Of the 33 cancer types in
TCGA dataset, high expression of SPARC was associated with
significantly worse prognosis in 9 cancer types, ACC, GBM, KIRP,
LUSC, MESO, PAAD, SARC, STAD and UVM (Fig. 2A-I). However, SPARC may be a tumour
suppressor gene in a few cancer types. Indeed, high SPARC
expression was associated with improved prognosis in DLBL, KIRC,
LGG and SKCM (Fig. 2J-M).
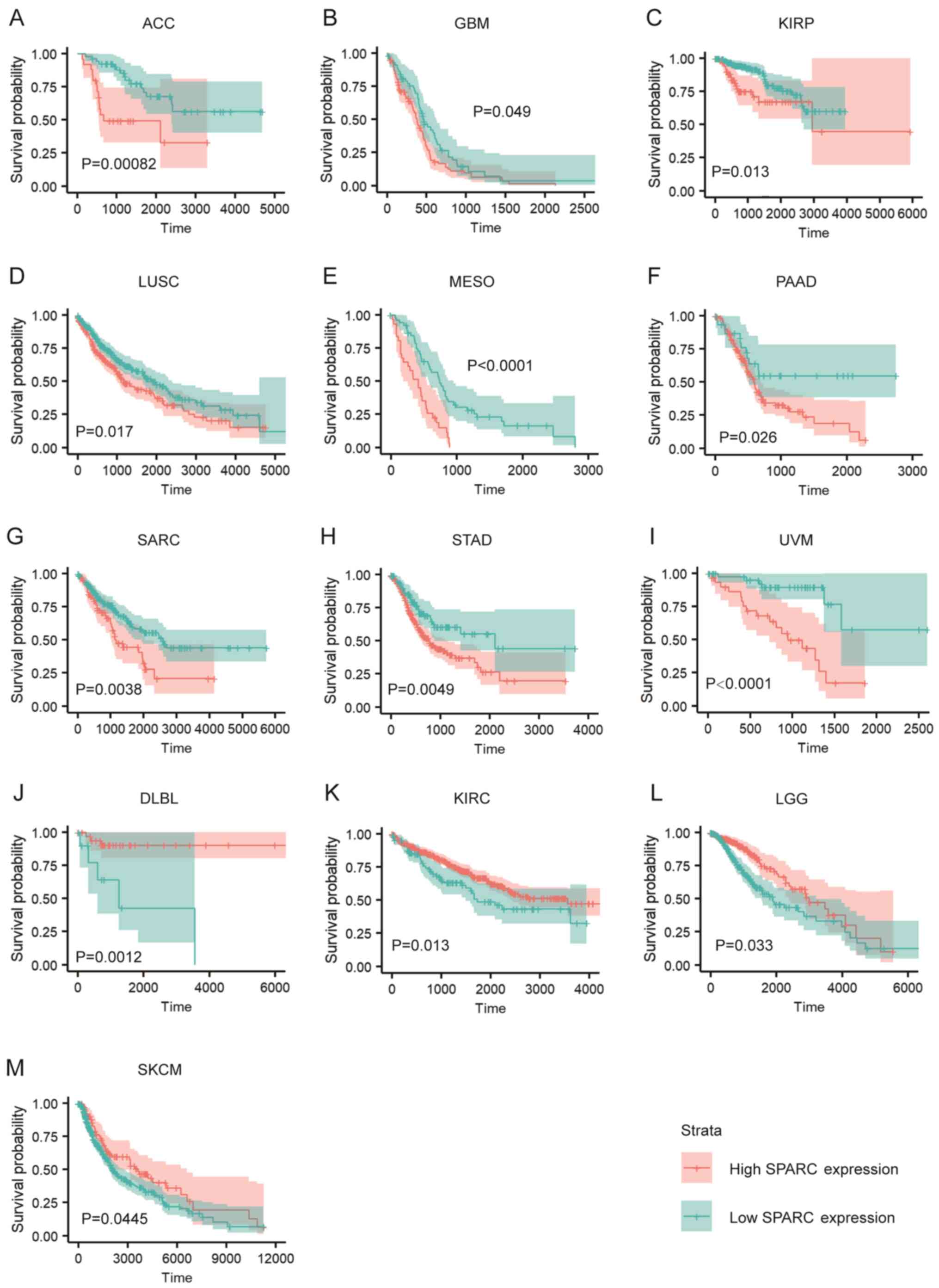 | Figure 2.SPARC is associated with prognosis of
various cancers. (A-I) High SPARC expression is associated with
worse prognosis in ACC, GBM, KIRP, LUSC, MESO, PAAD, SARC, STAD and
UVM (TGCA data). (J-M) High SPARC expression is associated with
improved prognosis in DLBL, KIRC, LGG and SKCM. (TCGA data). The
Kaplan-Meier method with log-rank tests (ACC, GBM, LUSC, MESO,
PAAD, SARC, STAD, UVM, DLBL, KIRC) or Cramér-von Mises tests (KIRP,
LGG, SKCM) were used for comparison of survival time between two
groups. SPARC, secreted protein acidic and rich in cysteine; ACC,
adrenocortical carcinoma; GBM, glioblastoma multiforme; KIRP,
kidney renal papillary cell carcinoma; LUSC, lung squamous
carcinoma; MESO, pleura mesothelioma; SARC, soft tissue sarcoma;
STAD, stomach adenocarcinoma; UVM, uveal melanoma; DLBL, diffuse
large B-cell lymphoma; KIRC, kidney renal clear cell carcinoma;
LGG, brain low-grade glioma; SKCM, skin cutaneous melanoma. |
High expression of SPARC was associated with the
worse prognosis of PAAD (Fig. 2F).
Further analysis revealed that patients with high or low SPARC
expression levels displayed similar survival times in the short
term after diagnosis (<500 days; Fig.
2F). Thus, it was hypothesized that the high early mortality
rate caused by the high aggressiveness of PAAD meant that SPARC had
little effect on early survival. However, when the follow-up time
was extended, patients with different SPARC expression levels
presented with completely different prognoses (Fig. 2F). This suggests that SPARC is
associated with long-term prognosis of patients with PAAD.
SPARC is not associated with the
pathological stage of PAAD
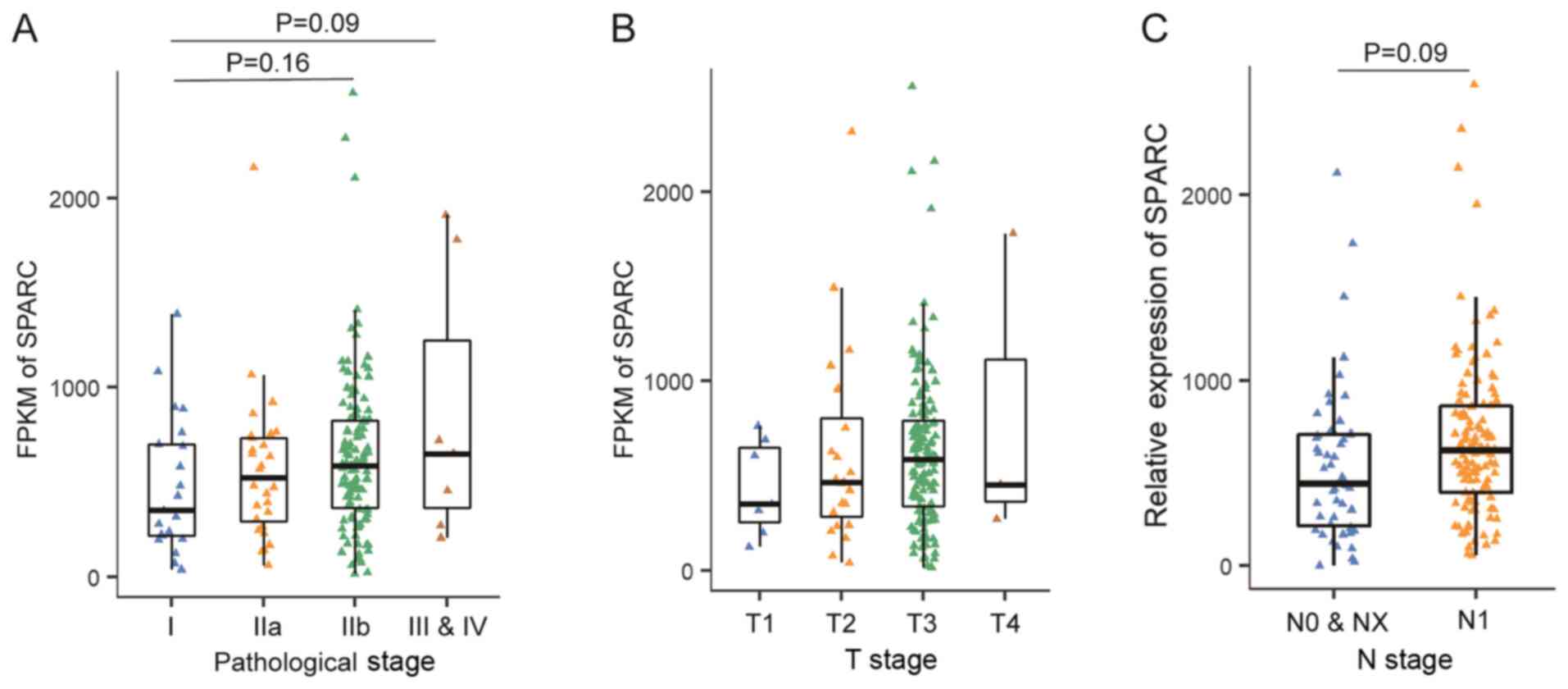
In general, SPARC expression increased with the PAAD
pathological stage, but this was not statistically significant
(Fig. 3A). The difference in SPARC
expression between stage I and stage IIB or stage I and III–IV was
not significant (P=0.16 and 0.09, respectively). The tumour size (T
stage) and lymph node metastasis (N stage) were then analysed.
SPARC protein expression also appeared to be associated with T
stage, but this also did not reach significance (Fig. 3B). Patients with lymph node
metastasis also showed an upward trend in SPARC expression,
although this was not statistically significant (P=0.09; Fig. 3C).
SPARC promotes the proliferation of
PAAD cells
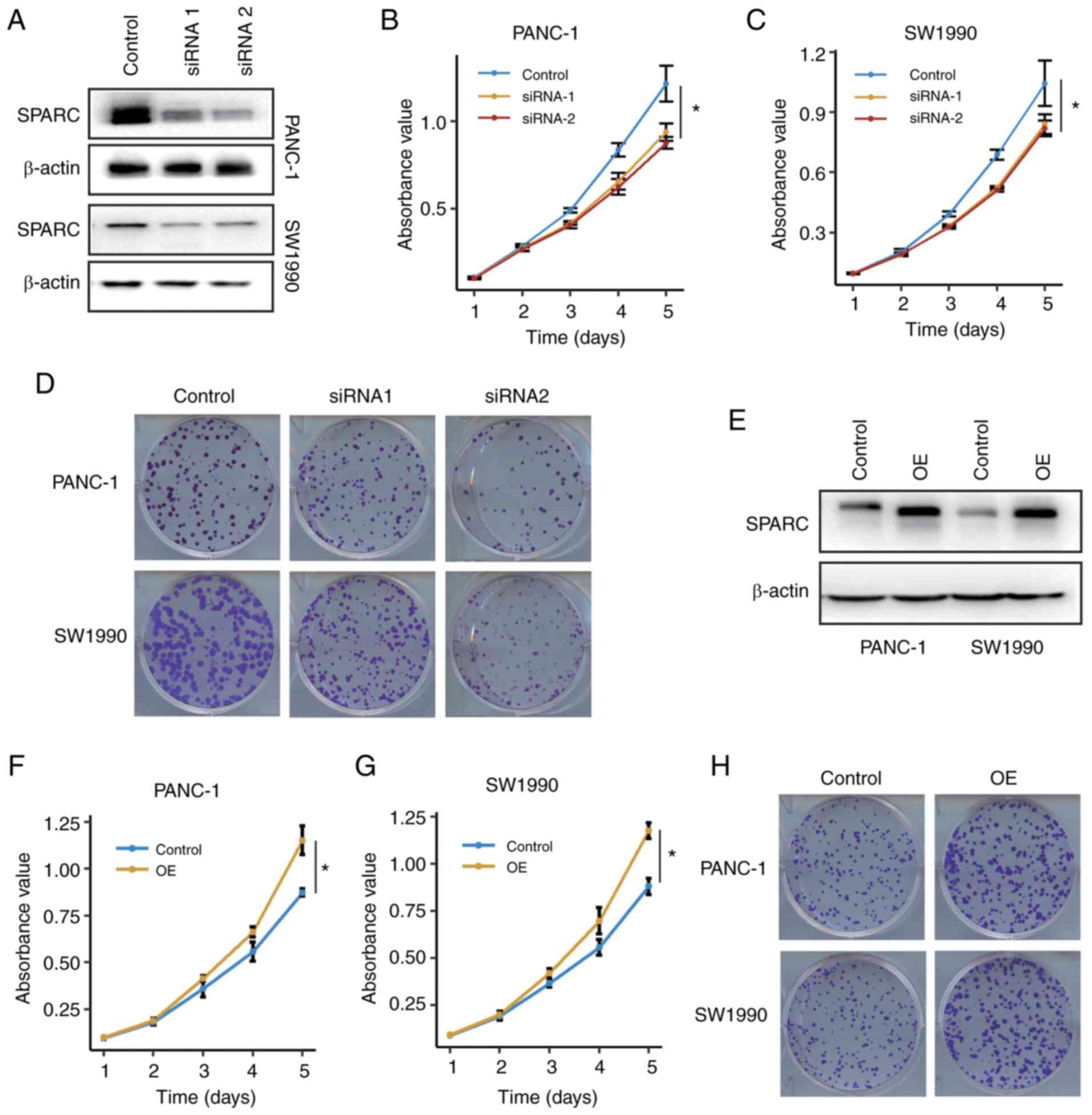
To identify whether SPARC affects the biological
behaviour of PAAD cells in vitro, two siRNAs were designed
to silence the expression of SPARC. Both siRNAs effectively
silenced SPARC expression (Fig. 4A).
SPARC silencing significantly impaired the proliferation of PANC-1
and SW1990 cells (Fig. 4B and C).
Moreover, SPARC silencing also impaired clone formation in PAAD
cells (Fig. 4D), suggesting that
SPARC was associated with proliferation in PAAD cells.
To further provide evidence that SPARC is implicated
in PAAD cell proliferation, a rescue assay was carried out. A SPARC
shRNA expression vector was constructed with the pLKO.1 vector and
siRNA1 sequence. A SPARC OE vector was constructed and transfected
into PANC-1 and SW1990 cells that were stably transfected with the
SPARC shRNA vector. The ectopic expression of SPARC was confirmed
with using a western blot assay (Fig.
4E). SPARC OE restored the proliferative ability of the PAAD
cells (P<0.05; Fig. 4F-H). Thus,
these results indicated that SPARC was associated with the
proliferation of PAAD cells.
SPARC promotes the migration of PAAD
cells
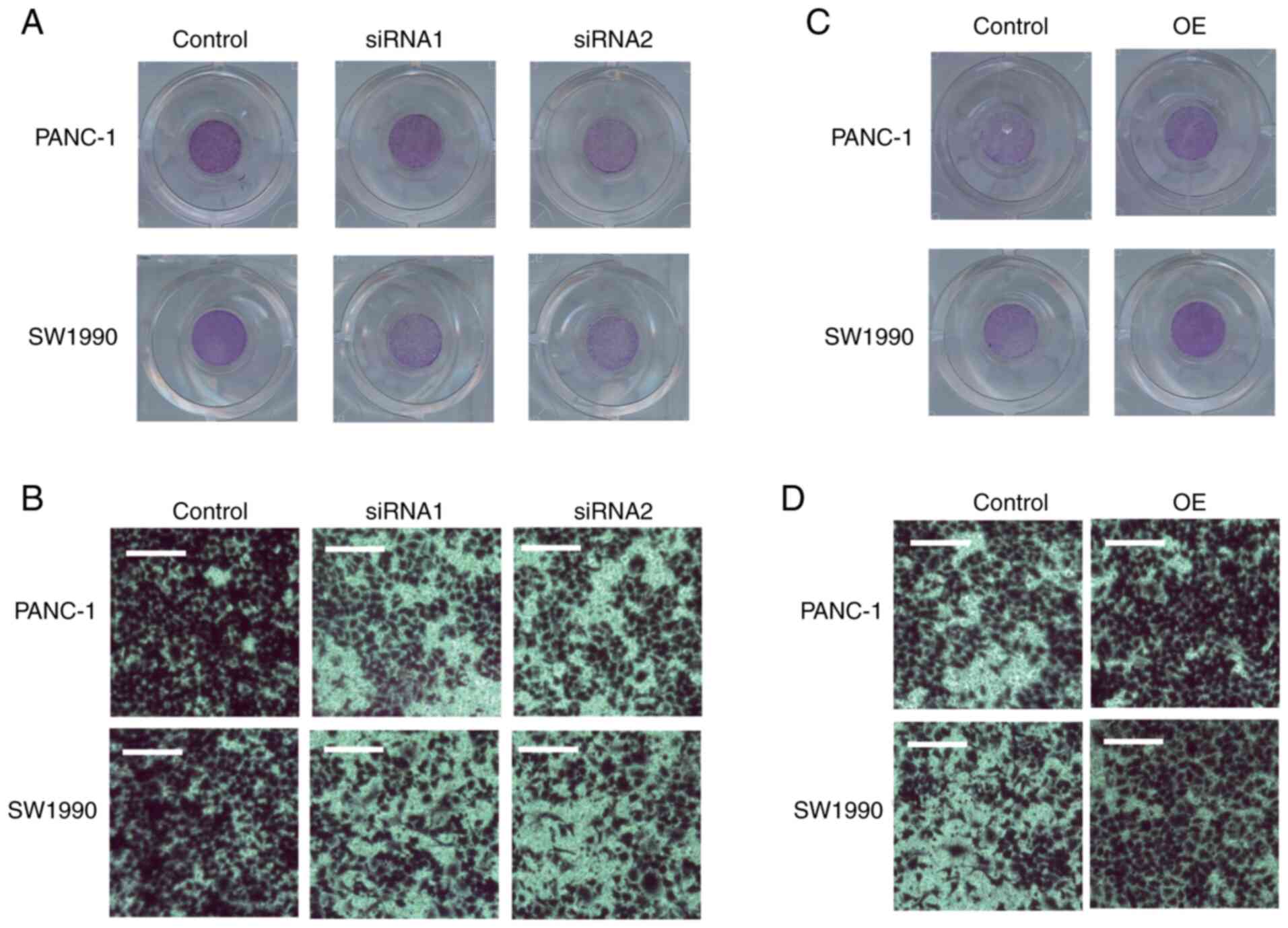
A Transwell assay was performed to determine whether
SPARC could affect the migration of PAAD cells. The ability of the
PAAD cells to cross the Transwell membrane was significantly
impaired after SPARC was silenced (Fig.
5A and B). The changes in the migration ability of the PAAD
cells following SPARC OE were also assessed. The migration of PAAD
cells increased following SPARC OE (Fig.
5C and D).
SPARC regulates pancreatic cancer cell
proliferation through autocrine secretion into the extracellular
milieu
Considering that SPARC is an exocrine protein,
western blot analysis was carried out in the culture supernatant of
PAAD cells. Clear SPARC bands were detected in the supernatant of
PAAD cells, immortalized normal acinar AR42J cells and 293T cells,
confirming that SPARC was secreted into the extracellular milieu
(Fig. 6A).
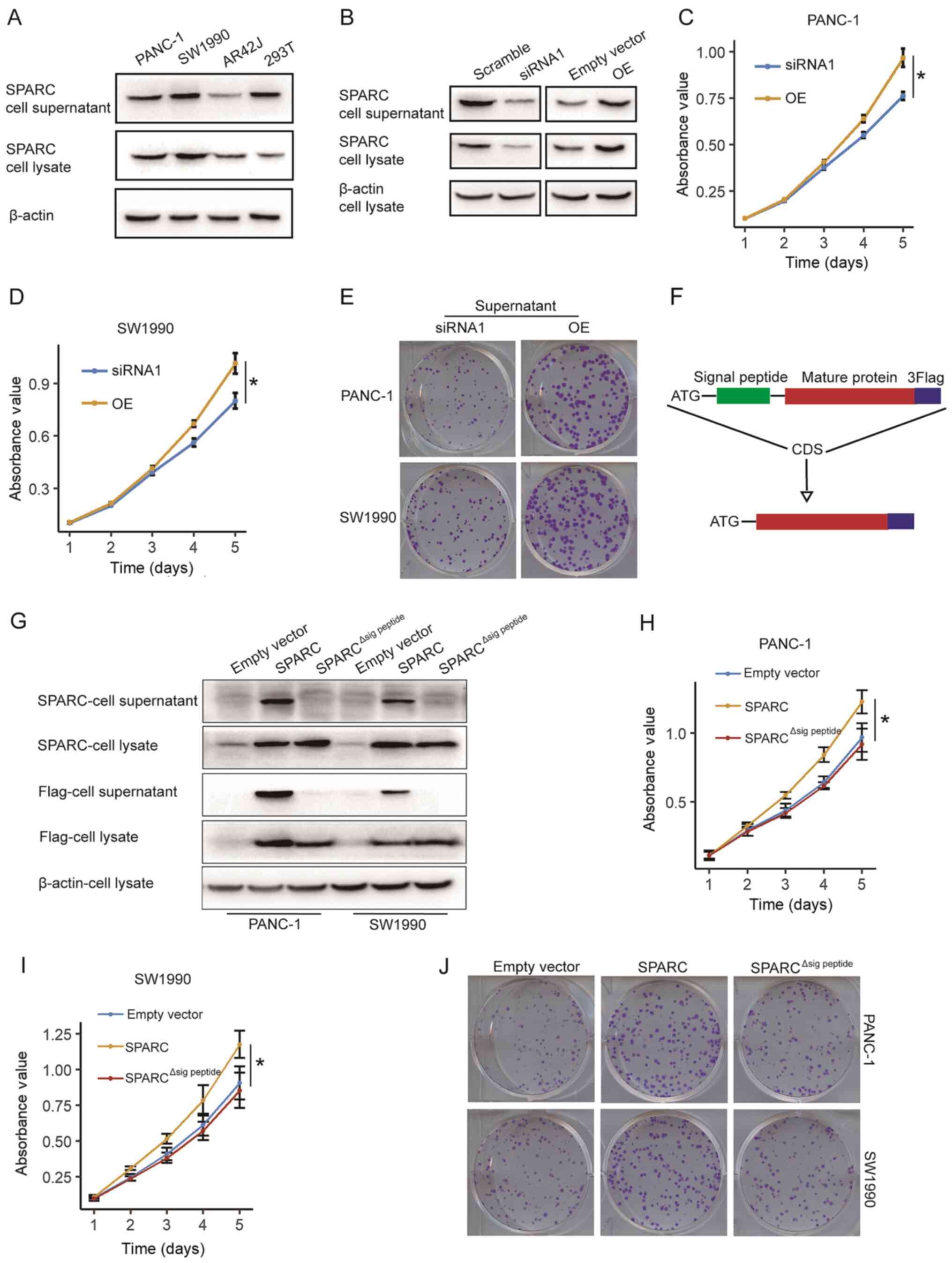 | Figure 6.SPARC regulates pancreatic cancer
cell proliferation through autocrine secretion into the
extracellular milieu. (A) SPARC is detected in cell lysate and
supernatant of PAAD cells, immortalized normal acinar AR42Jccells,
and 293T cells. (B) Supernatant of 293T cells transfected with the
SPARC siRNA has significant decreased concentration of SPARC
protein, compared with cells transfected with scramble siRNA. The
supernatant of 293T cells transfected with SPARC OE vector has
increased concentration of SPARC protein, compared with the empty
vector group. (C) Proliferation of PANC-1 cells transfected with
SPARC OE supernatant increased significantly, compared with the
cells treated with the SPARC-silenced supernatant (n=3).
*P<0.05, unpaired two-tailed t-test. (D) Proliferation of SW1990
cells treated with the SPARC OE supernatant increased
significantly, compared with the cells treated with the
SPARC-silenced supernatant (n=3). *P<0.05, unpaired two-tailed
t-test. (E) Treatment with SPARC OE supernatant restores clone
formation in PANC-1 and SW1990 cells. (F) Schematic plot of the
construction of SPARC∆sig peptide-Flag expression
vector. (G) SPARC∆sig peptide-Flag is expressed but not
effectively secreted to the extracellular milieu. (H and I)
Wild-type SPARC, but not SPARC∆sig peptide, promotes the
proliferation of (H) PANC-1 cells and (I) SW1990 cells (n=3).
*P<0.05, one-way ANOVA with Tukey's post-hoc test. (J) Colony
formation in PAAD cells is restored by wild-type SPARC, but not
SPARC∆sig peptide. Quantitative data from three
independent experiments was presented as mean ± SD (error bars).
OE, overexpression; SPARC, secreted protein acidic and rich in
cysteine; siRNA, small interfering RNA. |
It was hypothesised that the SPARC protein could
affect the proliferation and migration of PAAD cells when produced
either by cancer cells or stromal cells and secreted to the stroma.
This would explain why the prognosis of patients with PAAD is
associated with SPARC expression levels of both tumour cells and
stroma cells (13,14). Therefore, the role of the SPARC
protein in the extracellular milieu was examined. It was confirmed
that the SPARC concentration in the supernatant of 293T cells was
significantly decreased following siRNA silencing and increased by
the SPARC OE vector, compared with scramble RNA or control vector,
respectively (Fig. 6B). Before being
treated with the supernatant from 293T cells treated with siRNA and
OE vector, the PANC-1 and SW1990 cells were stably transfected with
SPARC shRNA. The supernatant from the SPARC OE cells significantly
promoted the proliferation of the PAAD cells, compared with the
control group treated with a supernatant of SPARC-silenced 293T
cells (Fig. 6C and D). PAAD cells
treated with exogenous SPARC also showed increased clone formation
(Fig. 6E). These results confirmed
that SPARC from the extracellular milieu can promote the
proliferation of PAAD cells.
Furthermore, it was hypothesised that this
autocrine- extracellular milieu-cancer cell pathway may be
necessary for SPARC function in cancer cells. As SPARC has a
well-defined signal peptide, which is its N-terminal 17 amino acid
residues, an expression vector that expresses the SPARC protein
with deletion of signal peptide, referred to as
pLenti-CMV-SPARC∆sig peptide, was constructed (Fig. 6F). In both PANC-1 and SW1990 cells,
endogenous SPARC was almost undetectable compared with the
SPARC-Flag or SPARC∆sig peptide-Flag fusion protein.
Using an anti-Flag antibody, it was demonstrated that the
SPARC-Flag fusion protein, but not the SPARC∆sig
peptide-Flag fusion protein, can be effectively secreted to
the extracellular milieu (Fig. 6G).
The endogenous SPARC expression of the PANC-1 and SW1990 cells was
silenced by stably transfecting with SPARC shRNA. Then, the control
vector, SPARC OE vector and SPARC∆sig peptide vector
were transfected into the PAAD cells with the silencing of
endogenous SPARC, and the proliferation of the PAAD cells was
examined. Only the wildtype SPARC, but not SPARC∆sig
peptide, effectively promoted the proliferation of PAAD cells
(Fig. 6H and I). A clone-formation
assay further confirmed this result (Fig. 6J). Thus, SPARC promotes the
proliferation of PAAD cells through autocrine secretion into the
extracellular milieu.
Exploration of the mechanism of the
SPARC protein
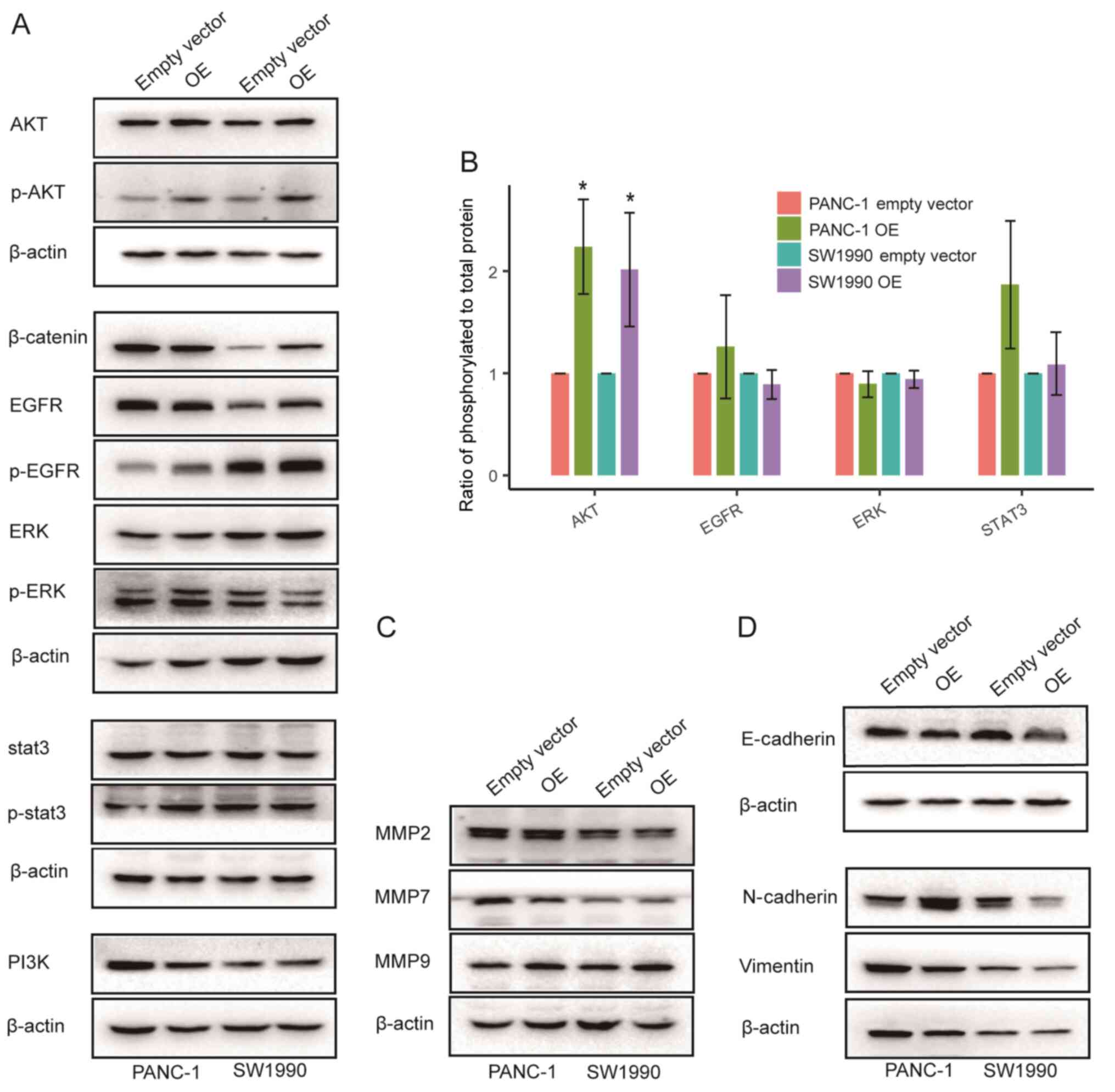
To elucidate how SPARC affects the proliferation and
migration of PAAD cells, associated signalling pathways and
molecules were examined. In PANC-1 and SW1990 cells, SPARC OE had
no significant effect on the phosphorylation of STAT3, EGFR and ERK
(Fig. 7A and B). Moreover, the
expression of β-catenin, a critical molecule in the WNT pathway,
had no significant and repeatable effect (Fig. 7A). However, the phosphorylation of
AKT increased significantly following SPARC OE (Fig. 7A and B). As the abnormal activation
of AKT plays an important role in the malignant proliferation of
PAAD cells (22,23), SPARC may regulate the proliferation
of PAAD cells through the AKT pathway.
Furthermore, the effect of SPARC on matrix
metalloproteinase (MMP) expression and epithelial-to-mesenchymal
transition (EMT) in PAAD cells was also evaluated (Fig. 7C and D). However, SPARC had no
observable effect on the expression of MMP2, MMP7, MMP9, and
EMT-related genes. Thus, the way in which SPARC regulates the
migration of PAAD cells still needs further investigation.
Discussion
One of the major reasons that the SPARC gene has
attracted the attention of scholars is that it plays multifaceted
or even controversial roles in cancer formation and progression. In
terms of cancer treatment, whether SPARC is a friend or a foe is
still unclear. SPARC is associated with poor prognosis in breast
cancer when expressed by cancer cells but improved prognosis when
expressed by tumour stromal cells (10,24).
Moreover, in non-small cell lung cancer, SPARC from either cancer
cells or stromal cells is associated with poor prognosis (25,26). In
addition, SPARC predicts improved prognosis in DLBL but poor
prognosis in melanoma (8,27). To construct a comprehensive picture
of SPARC's function in cancer, a pan-cancer analysis of SPARC was
carried out using a public dataset from TCGA covering 33 cancer
types. By analysing the association between SPARC and the prognosis
of different tumours, it was found that the function of SPARC
seemed to mainly depend on the pathological cancer type rather than
the organic or cellular origin. Moreover, SPARC predicted the
distinct prognosis of LGG and GBM, which both stem from glial cells
and occur in the brain or spine cord (28,29), and
KIRC and KIRP, which both stem from renal tubular epithelial cells
(30). Meanwhile, SPARC seems
associated with poor prognosis of adenocarcinoma (PAAD and STAD),
whereas for COAD, LUAD and READ, the results did not reach
significance. In addition, consistent with a previous report
(8), SPARC was found to predict
improved prognosis of lymphatic and hematopoietic cancer types,
such as DLBL and LAML. Due to the influence of the mRNA turnover
rate, ribosome-bound mRNA abundance and the protein turnover rate,
mRNA abundance does not always accurately reflect protein content
(31). Nevertheless, according to
existing reports, the mRNA and protein expression of the SPARC gene
in tumour tissues show a consistent trend (32,33). To
our knowledge, this study is the first to clarify the function of
SPARC at the pan-cancer level, which could therefore help explain
the inconsistencies seen in previous studies.
It was demonstrated that SPARC acts as an oncogene
in PAAD. Moreover, SPARC is associated with poor prognosis of PAAD,
and in the present in vitro research, we found that it
promotes the proliferation and migration of PAAD cells. To a
certain extent, the in vitro results of our work contradict
some previous reports, in which SPARC was proposed to inhibit the
proliferation and increase the chemosensitivity of PAAD cells
(15–17). Thus, it may be proposed that cell
type and experimental technique differences may be the direct cause
of this inconsistency. However, the root cause may also be the
versatility of SPARC. In PAAD, SPARC may have different functions
in patients with different genomic alterations and molecular
profiles. The possibility that SPARC may act as a tumor suppressor
in some patients and some PAAD cell lines cannot be excluded, since
subtypes of PAAD with significant molecular and clinical difference
have been previously found (34).
Another question regarding SPARC and PAAD is whether
the SPARC proteins that affect tumour prognosis come from tumour
cells or tumour stromal cells. Our work confirmed that SPARC
regulated PAAD proliferation only when secreted extracellularly.
Since the SPARC that affects the proliferation of PAAD cells comes
from the extracellular milieu, it may be inferred that the SPARC
from both stroma cells and tumor cells can promote PAAD cell
proliferation. In TCGA data used on this study, SPARC expression
was determined via RNASeq; however, this method could not
distinguish the cellular origin of SPARC. The optimal method to
elucidate the source of SPARC may be immunohistochemistry (IHC),
which requires paraffin-embedded clinical PAAD specimen or
xenograft tissue. Unfortunately, there were not enough PAAD
specimens to carry out IHC in the study. In addition, according to
our past experience, the subcutaneous injection of PAAD cells in
node mice does not result in massive fibroblasts or other types of
stromal cell infiltration. The subcutaneous xenografts are mainly
composed of cancer cells. Thus, an in vivo experiment may be
useful in determining the effect of SPARC on in vivo growth
of PAAD, but it cannot be used to distinguish the cellular origin
of SPARC. The lack of IHC and in vivo experiments are a
limitation of the present study, and the above questions should be
explored in further studies.
The mechanisms underlying SPARC functions are
complex. SPARC has been reported to facilitate the proliferation
and metastasis of hepatocellular carcinoma via the ERK signalling
pathway, promote cervical cancer cell proliferation by modulating
the bax/bcl-2 ratio, and induce the migration of non-small cell
lung cancer via WNK1/Snail signalling (35–37). In
acute myeloid leukaemia, SPARC interacts with integrin-linked
kinase (ILK) and promotes ILK signalling (38). Since AKT is an important downstream
effector of ILK (39), it was
hypothesised that SPARC may also promote AKT activation by
interacting with ILK in PAAD cells.
In conclusion, the present study partly answered the
aforementioned controversial roles of SPARC. Overall, SPARC
exhibits regulatory potential and may play a role in PAAD
progression, and its significance in cancer therapy merits further
study.
Acknowledgements
The authors would like to thank Dr Yi Zhou (The
First Affiliated Hospital of Wenzhou Medical University, Wenzhou,
China) for her help in finishing supplementary western blot assay
and for providing guidance for survival analysis.
Funding
The present study was supported by The Foundation of
Wenzhou Science & Technology Bureau (grant no. 2020Y0255).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The datasets generated and/or analyzed during the current
study are available in TCGA repository (https://portal.gdc.cancer.gov/).
Authors' contributions
KP, XH and XJ performed the biological experiments,
data analysis, statistical work and manuscript preparation. KP and
XJ collected TCGA data and performed pan-cancer analysis of SPARC.
XJ supervised the experimental work and finished manuscript
proofreading. XJ designed the study and guarantees its integrity.
KP, XH and XJ confirm the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was reviewed and approved by The
Board of Wenzhou Medical University. Verbal informed consent was
obtained from the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SPARC
|
secreted protein acidic and rich in
cysteine
|
|
PAAD
|
pancreatic adenocarcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
DLBL
|
diffuse large B-cell lymphoma
|
|
BRAD
|
breast invasive carcinoma
|
|
COAD
|
colon adenocarcinoma
|
|
CCK-8
|
Cell Counting Kit-8
|
|
MC3
|
Multi-Center Mutation-Calling in
Multiple Cancers
|
|
MMP
|
matrix metalloproteinase
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
IHC
|
immunohistochemistry
|
References
|
1
|
Termine JD, Kleinman HK, Whitson SW, Conn
KM, McGarvey ML and Martin GR: Osteonectin, a bone-specific protein
linking mineral to collagen. Cell. 26:99–105. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mayer U, Aumailley M, Mann K, Timpl R and
Engel J: Calcium-dependent binding of basement membrane protein
BM-40 (osteonectin, SPARC) to basement membrane collagen type IV.
Eur J Biochem. 198:141–150. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gaudet P, Livstone MS, Lewis SE and Thomas
PD: Phylogenetic-based propagation of functional annotations within
the Gene Ontology consortium. Brief Bioinform. 12:449–462. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nagaraju GP, Dontula R, El-Rayes BF and
Lakka SS: Molecular mechanisms underlying the divergent roles of
SPARC in human carcinogenesis. Carcinogenesis. 35:967–973. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Said N: Roles of SPARC in urothelial
carcinogenesis, progression and metastasis. Oncotarget.
7:67574–67585. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Said N, Frierson HF, Sanchez-Carbayo M,
Brekken RA and Theodorescu D: Loss of SPARC in bladder cancer
enhances carcinogenesis and progression. J Clin Invest.
123:751–766. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chin D, Boyle GM, Williams RM, Ferguson K,
Pandeya N, Pedley J, Campbell CM, Theile DR, Parsons PG and Coman
WB: Novel markers for poor prognosis in head and neck cancer. Int J
Cancer. 113:789–797. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meyer PN, Fu K, Greiner T, Smith L,
Delabie J, Gascoyne R, Ott G, Rosenwald A, Braziel R, Campo E, et
al: The stromal cell marker SPARC predicts for survival in patients
with diffuse large B-cell lymphoma treated with rituximab. Am J
Clin Pathol. 135:54–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yamanaka M, Kanda K, Li NC, Fukumori T,
Oka N, Kanayama HO and Kagawa S: Analysis of the gene expression of
SPARC and its prognostic value for bladder cancer. J Urol.
166:2495–2499. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones C, Mackay A, Grigoriadis A, Cossu A,
Reis-Filho JS, Fulford L, Dexter T, Davies S, Bulmer K, Ford E, et
al: Expression profiling of purified normal human luminal and
myoepithelial breast cells: Identification of novel prognostic
markers for breast cancer. Cancer Res. 64:3037–3045. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang JF, Wang HK, Xiao H, Li N, Cheng CX,
Zhao YZ, Ma YB, Gao JZ, Bai RB and Zheng HX: Relationship and
prognostic significance of SPARC and VEGF protein expression in
colon cancer. J Exp Clin Cancer Res. 29:712010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hariharan D, Saied A and Kocher HM:
Analysis of mortality rates for pancreatic cancer across the world.
HPB (Oxford). 10:58–62. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Infante JR, Matsubayashi H, Sato N,
Tonascia J, Klein AP, Riall TA, Yeo C, Iacobuzio-Donahue C and
Goggins M: Peritumoral fibroblast SPARC expression and patient
outcome with resectable pancreatic adenocarcinoma. J Clin Oncol.
25:319–325. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu XZ, Guo ZY, Di Y, Yang F, Ouyang Q, Fu
DL and Jin C: The relationship between SPARC expression in primary
tumor and metastatic lymph node of resected pancreatic cancer
patients and patients' survival. Hepatobiliary Pancreat Dis Int.
16:104–109. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan X, Mao Z, Ma X, Cui L, Qu J, Lv L,
Dang S, Wang X and Zhang J: Secreted protein acidic and rich in
cysteine enhances the chemosensitivity of pancreatic cancer cells
to gemcitabine. Tumour Biol. 37:2267–2273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao Y, Zhang H, Ma Q, Huang R, Lu J,
Liang X, Liu X, Zhang Z, Yu L, Pang J, et al: YAP1-mediated
pancreatic stellate cell activation inhibits pancreatic cancer cell
proliferation. Cancer Lett. 462:51–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mao Z, Ma X, Fan X, Cui L, Zhu T, Qu J,
Zhang J and Wang X: Secreted protein acidic and rich in cysteine
inhibits the growth of human pancreatic cancer cells with G1 arrest
induction. Tumour Biol. 35:10185–10193. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ellrott K, Bailey MH, Saksena G, Covington
KR, Kandoth C, Stewart C, Hess J, Ma S, Chiotti KE, McLellan M, et
al: Scalable open science approach for mutation calling of tumor
exomes using multiple genomic pipelines. Cell Syst. 6:271–281.e7.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Almagro Armenteros JJ, Tsirigos KD,
Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G and
Nielsen H: SignalP 5.0 improves signal peptide predictions using
deep neural networks. Nat Biotechnol. 37:420–423. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martincorena I, Raine KM, Gerstung M,
Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR and Campbell
PJ: Universal patterns of selection in cancer and somatic tissues.
Cell. 171:1029–1041.e21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chio IIC, Jafarnejad SM, Ponz-Sarvise M,
Park Y, Rivera K, Palm W, Wilson J, Sangar V, Hao Y, Öhlund D, et
al: NRF2 promotes tumor maintenance by modulating mRNA translation
in pancreatic cancer. Cell. 166:963–976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Pan YZ, Cheung M, Cao M, Yu C,
Chen L, Zhan L, He ZW and Sun CY: LAMB3 mediates apoptotic,
proliferative, invasive, and metastatic behaviors in pancreatic
cancer by regulating the PI3K/Akt signaling pathway. Cell Death
Dis. 10:2302019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bergamaschi A, Tagliabue E, Sørlie T,
Naume B, Triulzi T, Orlandi R, Russnes HG, Nesland JM, Tammi R,
Auvinen P, et al: Extracellular matrix signature identifies breast
cancer subgroups with different clinical outcome. J Pathol.
214:357–367. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang Y, Zhang J, Zhao YY, Jiang W, Xue C,
Xu F, Zhao HY, Zhang Y, Zhao LP, Hu ZH, et al: SPARC expression and
prognostic value in non-small cell lung cancer. Chin J Cancer.
31:541–548. 2012.PubMed/NCBI
|
|
26
|
Koukourakis MI, Giatromanolaki A, Brekken
RA, Sivridis E, Gatter KC, Harris AL and Sage EH: Enhanced
expression of SPARC/osteonectin in the tumor-associated stroma of
non-small cell lung cancer is correlated with markers of
hypoxia/acidity and with poor prognosis of patients. Cancer Res.
63:5376–5380. 2003.PubMed/NCBI
|
|
27
|
Massi D, Franchi A, Borgognoni L, Reali UM
and Santucci M: Osteonectin expression correlates with clinical
outcome in thin cutaneous malignant melanomas. Hum Pathol.
30:339–344. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cancer Genome Atlas Research Network, .
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Perry A and Wesseling P: Histologic
classification of gliomas. Handb Clin Neurol. 134:71–95. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ricketts CJ, De Cubas AA, Fan H, Smith CC,
Lang M, Reznik E, Bowlby R, Gibb EA, Akbani R, Beroukhim R, et al:
The cancer genome atlas comprehensive molecular characterization of
renal cell carcinoma. Cell Rep. 23:313–326.e5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maier T, Güell M and Serrano L:
Correlation of mRNA and protein in complex biological samples. FEBS
Lett. 583:3966–3973. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang S, Jin J, Tian X and Wu L:
hsa-miR-29c-3p regulates biological function of colorectal cancer
by targeting SPARC. Oncotarget. 8:104508–104524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu X, Gao D, Ren Q, Jiang X, Bai J and
Sheng L: miR-211 inhibits proliferation, invasion and migration of
cervical cancer via targeting SPARC. Oncol Lett. 16:853–860.
2018.PubMed/NCBI
|
|
34
|
Cancer Genome Atlas Research Network.
Electronic address, . simpleandrew_aguirre@dfci.harvard.edu;
Cancer Genome Atlas Research Network: Integrated genomic
characterization of pancreatic ductal adenocarcinoma. Cancer Cell.
32:185–203.e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Feng Y, Wang X, Yang X, Hu Y, Li Y,
Zhang Q, Huang Y, Shi K, Ran C, et al: SPARC negatively correlates
with prognosis after transarterial chemoembolization and
facilitates proliferation and metastasis of hepatocellular
carcinoma via ERK/MMP signaling pathways. Front Oncol. 10:8132020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen J, Shi D, Liu X, Fang S, Zhang J and
Zhao Y: Targeting SPARC by lentivirus-mediated RNA interference
inhibits cervical cancer cell growth and metastasis. BMC Cancer.
12:4642012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hung JY, Yen MC, Jian SF, Wu CY, Chang WA,
Liu KT, Hsu YL, Chong IW and Kuo PL: Secreted protein acidic and
rich in cysteine (SPARC) induces cell migration and epithelial
mesenchymal transition through WNK1/snail in non-small cell lung
cancer. Oncotarget. 8:63691–63702. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Alachkar H, Santhanam R, Maharry K,
Metzeler KH, Huang X, Kohlschmidt J, Mendler JH, Benito JM, Hickey
C, Neviani P, et al: SPARC promotes leukemic cell growth and
predicts acute myeloid leukemia outcome. J Clin Invest.
124:1512–1524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lynch DK, Ellis CA, Edwards PA and Hiles
ID: Integrin-linked kinase regulates phosphorylation of serine 473
of protein kinase B by an indirect mechanism. Oncogene.
18:8024–8032. 1999. View Article : Google Scholar : PubMed/NCBI
|