Introduction
Melanoma is one of the most common malignant tumors
and its incidence increases by 3–5% percent annually worldwide
(1). The common pathological types
of melanoma are superficial spreading type, nodular type, malignant
freckles and acral freckles (2).
Superficial spreading type is the most common in the white
ethnicity, and extremity freckle-like melanoma is more common in
Asians and Pacific Islanders (3).
The 5-year survival rates of patients with melanoma at stage I, II,
III and IV in China are 94, 44, 38 and 4.6%, respectively (4). Moreover, the survival rate is also
closely related to primary ulcers (5).
Excessive ultraviolet radiation is one of the known
causes of skin melanoma, and the effects of other factors such as
endocrine, chemical and physical factors on the occurrence of
melanoma are poorly understood (6).
At present, several treatment strategies for melanoma exist
(7). Chemotherapy alone has limited
efficacy and is hindered by drug resistance (8). Molecular targeting drugs and
immunotherapy drugs have made great progress in the treatment of
tumors due to their specificity, effectiveness, good patient
tolerance and low adverse reactions compared to cytotoxic drugs
(9). Multivariate analysis of the
relationship between genetic variation and survival revealed that
KIT and BRAF gene mutations are independent prognostic factors for
melanoma, with risk factors of 1.989 (95% CI, 1.263–3.131) and
1.536 (95% CI, 1.110–2.214), respectively (10). However, distant metastases can occur
early in malignant melanoma, which is a key factor affecting
clinical treatment and leading to poor prognosis (11).
The mechanism of melanoma progression has not been
fully elucidated. In addition, research on the mechanism underlying
chemotherapy resistance is also of particular interest. It is also
worth mentioning that a large part of current research on melanoma
is carried out at the genomic level, and less attention has been
given to gene expression analysis (12). Studying long non-coding RNA (lncRNA)
molecules related to the pathogenesis and progression of melanoma
may provide insight into the mechanism of melanoma onset, which is
of great significance for the treatment and new drug development.
So far, several lncRNA candidates have been explored. Schmidt et
al (13) demonstrated that the
lncRNA SRA-like non-coding RNA1 could increase the expression of
matrix metalloproteinase-9, accelerate invasion of melanoma and
predict poor survival. Furthermore, homeobox D-antisense1 was
confirmed to be upregulated in patients with melanoma and was also
closely associated with survival time (14). Although DUXAP8 has been shown to
regulate hepatocellular carcinoma and lung cancer development
(15,16), its role in melanoma has not been
reported. Hence, the present study aimed to investigate whether
DUXAP8 is involved in melanoma development and the mechanism of
lncRNA DUXAP8 on the invasion and metastasis of melanoma was
studied, with the aim of identifying a potential molecular-level
treatment against melanoma.
Materials and methods
Patients and tissue samples
A total of 43 patients with skin melanoma (mean age,
52 years; age range, 39–61 years; 27 male patients and 16 female
patients) were enrolled from January 2014 to December 2018 at The
Affiliated Changzhou No. 2 People's Hospital with Nanjing Medical
University (Changzhou, China) or Weihai Central Hospital (Weihai,
China). All patients were diagnosed pathologically by at least 3
experienced and independent pathologists from Weihai Central
Hospital or the Affiliated Changzhou No. 2 People's Hospital with
Nanjing Medical University. Patients who had radiotherapy or
chemotherapy before surgery were excluded. Patients that had not
received radiotherapy or chemotherapy prior to sample collection
were included. The cases included 19 patients at stage I+II and 24
patients at stage III+IV. Tissue samples excised during the
operation were immediately frozen in liquid nitrogen until RNA
extraction. Adjacent normal tissue was used as a control. The
distance between the resected tumor tissue and the healthy tissues
was 2 cm. This study was approved by the ethics committee of The
Affiliated Changzhou No. 2 People's Hospital with Nanjing Medical
University. The patients signed informed consent forms.
Cell lines and transfection
The human epidermal melanocyte HEMa-LP cell lines
and the malignant melanoma A375, A2058 and SK-MEL-2 cell lines were
purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. Cells were cultured in DMEM medium
containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific
Inc.) and 1% penicillin-streptomycin. The cells were incubated at
37°C in an incubator with 5% CO2.
The eukaryotic expression plasmid vector
pSilencerTM3.1-H1neo containing DUXAP8-shRNA
(5′-GGAACTTCCCAAACCTCCATGATTT-3′) and control shRNA
(5′-TTCTCCGAACGTGTCACGTTT-3′) were constructed by Shanghai
GenePharma Co., Ltd. The miR-3182 mimic (5′-GCUUCUGUAGUGUAGUC3′-),
NC mimic (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-3182 inhibitor
(5′-GACUACACUACAGAAGC-3′), NC inhibitor
(5′-CAGUACUUUUGUGUAGUACAA-3′) and NUPR1 overexpression vector
(oeNUPR1; pcDNA3-NUPR1) were designed and synthesized by Shanghai
GenePharma Co., Ltd. Cells were seeded in 6-well plates. pcDNA3
(Invitrogen; Thermo Fisher Scientific Inc.) empty vector was used
as the negative control for oeNUPR1. Transfection was performed
using Lipofectamine® 2000 reagent (Invitrogen, Thermo
Fisher Scientific, Inc.) at 37°C for 48 h with a 100 nM of the
aforementioned plasmids, negative controls, mimics and inhibitors,
according to the manufacturer's protocol. Transfection efficiency
was measured by RT-qPCR and after 48 h of transfection, subsequent
experimentation was performed.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
Total RNA (500 ng) was reverse transcribed into cDNA using
SuperScript III (Invitrogen; Thermo Fisher Scientific, Inc.) at
37°C for 15 min. Subsequently, qPCR was performed using an ABI
PRISM 7500 Sequence Detection system (Thermo Fisher Scientific,
Inc.) and SYBR Select Master mix (Thermo Fisher Scientific, Inc.).
U6 or GAPDH were used as the internal reference genes. The
following thermocycling conditions were used for qPCR: i) Initial
denaturation at 95°C for 5 min; ii) 40 cycles at 95°C for 20 sec,
58°C for 30 sec and 74°C for 30 sec; and iii) final extension step
at 72°C for 5 min. The primer sequences were as follows: i) DUXAP8
forward, 5′-ACCCAAACACTAATTGTAGACT-3′ and reverse,
5′-TGTCTGGGAGACTGCTTACA-3′; miR-3182 forward,
5′-CACTCAGCTGGCTTCTGTAGTG-3′ and reverse, 5′-CTGGTGTCGTGGAGTCG-3′;
NUPR1 forward, 5′-AGGACTTATTCCCGCTGACTGA-3′ and reverse,
5′-TGCCGTGCGTGTCTATTTATTG-3′; GAPDH forward,
5′-GTCGATGGCTAGTCGTAGCATCGAT-3′ and reverse,
5′-TGCTAGCTGGCATGCCCGATCGATC-3′; and U6 forward,
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′. miRNA and mRNA expression levels were
quantified using the 2−ΔΔCq method (17).
Cell-Counting Kit-8 (CCK-8) assay
After cell transfection for 48 h, A375 and A2058
cells were seeded in a 96-well cell culture plate at a density of
2×103 cells/well. After incubation for 24, 48 or 72 h,
10 µl of CCK-8 reagent (Beyotime Institute of Biotechnology) was
added to each well and incubated for 4 h. The absorbance at 450 nm
was measured using a microplate reader (Bio-Rad Laboratories,
Inc.). The average value of each well was taken to evaluate the
cell proliferation.
Transwell assay
Cell migration and invasion were assessed by a
Transwell assay. For the migration assay, 2×104 A375 or
A2058 cells were diluted in FBS-free medium and seeded into the
upper chamber (BD Biosciences). For the invasion assay, cells were
added onto a Matrigel®-pre-coated (Sigma-Aldrich; Merck
KGaA) upper chamber. Subsequently, 500 µl of culture medium
containing 10% FBS was inoculated into the lower chamber. After 24
h, non-migratory/-invasive cells were detached while the
migrated/invaded cells in the lower chamber were fixed in 4%
paraformaldehyde for 30 min at room temperature and stained using
0.5% crystal violet for 30 min at room temperature. The numbers of
migratory/invasive cells were counted in five randomly selected
fields using an inverted light microscope (Olympus Corporation;
magnification, ×200).
RNA pull-down
Biotin-labeled wild-type (WT) or mutant miR-3182
(Shanghai GenePharma Co., Ltd.) were synthesized. A375 and A2058
cells were lysed using specific lysis buffer (Sigma-Aldrich; Merck
KGaA) containing RNase inhibitor (Beyotime Institute of
Biotechnology). The lysate was subjected to centrifugation at
12,000 × g for 12 min at 4°C and the supernatant (500 ml per
reaction) were incubated with biotin-labeled wild-type or mutant
miR-3182, followed by incubation with M-280 streptavidin beads (100
µl; Sigma-Aldrich; Merck KGaA) at 4°C overnight. The beads were
washed twice with cold lysis buffer and precipitated RNAs were
eluted using elution buffer (Sigma-Aldrich; Merck KGaA) according
to the manufacturer's protocol, followed by RT-qPCR analysis as
described above. Transfection conditions were the same as described
above.
Luciferase reporter assay
The bioinformatics online tools miRDB (http://mirdb.org/miRDB/index.html) and TargetScan
7 (http://www.targetscan.org/vert_71/) predicted that
miR-3182 had targeted binding sites with lncRNA DUXAP8 and NUPR1.
The 3′UTR of DUXAP8 and NUPR1 containing binding site sequence was
amplified and constructed into psiCHECK2 luciferase reporter vector
(Promega Corporation), referred to as DUXAP8-WT and NUPR1-WT
reporter plasmids, respectively. The mutant (MUT) sequence was
constructed on the reporter gene vector and recorded as DUXAP8-MUT
and NUPR1-MUT reporter plasmids. A375 and A2058 cells were
transfected with indicated luciferase reporter and miR-3182 mimic
(5′-GCUUCUGUAGUGUAGUC3′-) or NC mimic (5′-UUCUCCGAACGUGUCACGUTT-3′)
(Shanghai GenePharma Co., Ltd.) using Lipofectamine®
2000 reagent (Invitrogen, Thermo Fisher Scientific, Inc.) for 48 h
and then luciferase reporter assay was performed using a Dual
Luciferase Reporter Assay kit (Promega Corporation) according to
according to the manufacturer's protocol. Dual luciferase activity
was measured separately, and the relative luciferase activity of
each cell was calculated based on the fluorescence intensity and
normalized to Renilla luciferase activity.
Statistical analysis
Statistical analysis was carried out using SPSS 22.0
software (IBM Corp.). Continuous data ware expressed as the mean ±
standard deviation. Differences between groups were compared using
an unpaired Student's t-test or one-way ANOVA followed by Tukey's
post hoc test. The prognosis of patients was analyzed using the
Kaplan-Meier method and log-rank tests. The relationship between
DUXAP8 expression and miR-3182 or NUPR1 levels was analyzed using
Pearson's correlation coefficient. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of DUXAP8 and survival
analysis
The expression levels of lncRNA DUXAP8 were examined
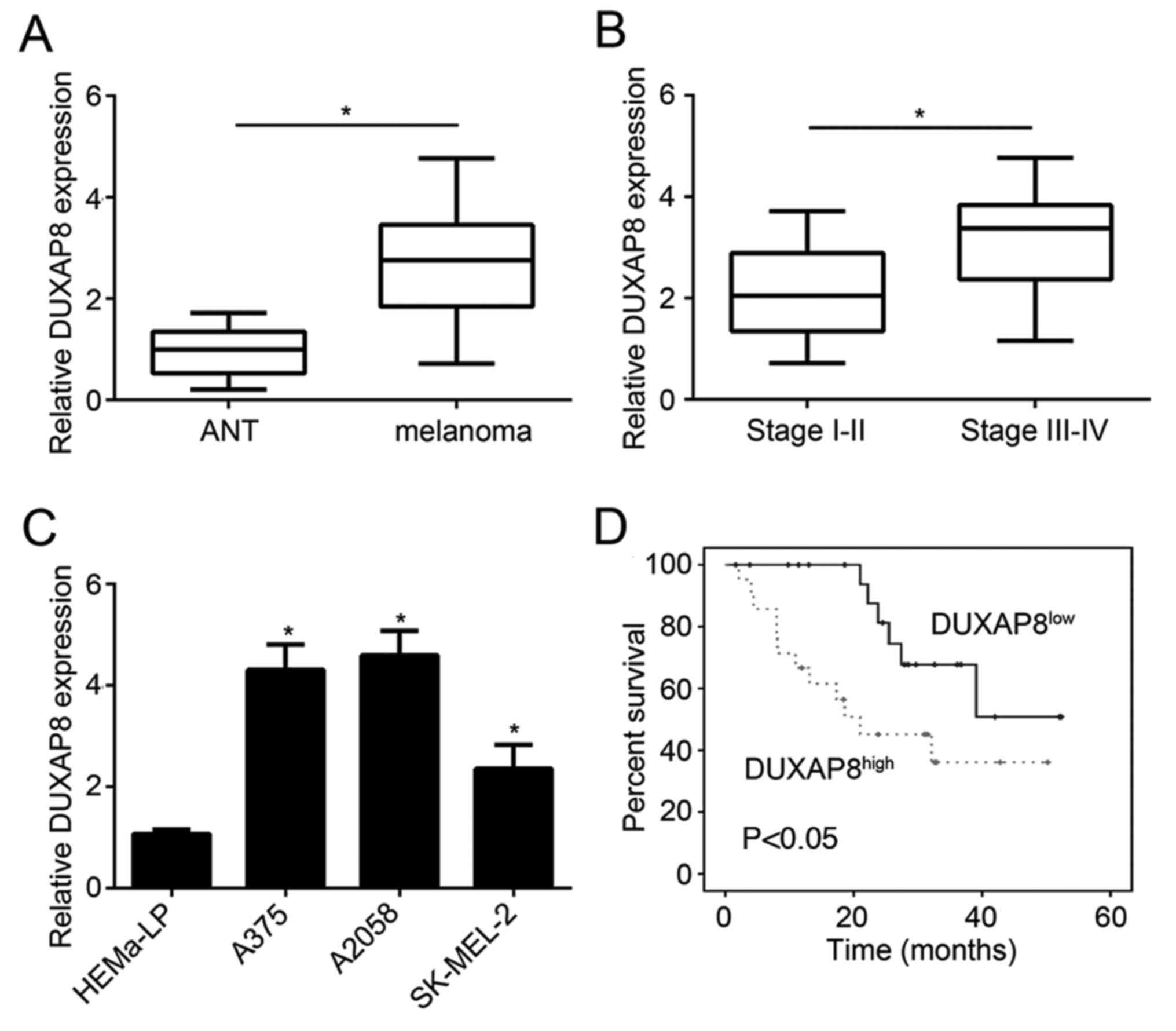
in melanoma tissue samples and cell lines. DUXAP8 was upregulated
in melanoma tissue compared with adjacent normal tissue (Fig. 1A). Moreover, patients with stage
III+IV melanoma displayed higher levels of DUXAP8 than patients in
stage I–II (Fig. 1B). Similar
results were also observed in cell lines. The expression of DUXAP8
in malignant melanoma cell lines (A375, A2058 and SK-MEL-2) was
higher than that in HEMa-LP cells (Fig.
1C). A375 and A2058 cell lines displayed the highest levels of
DUXAP8. Therefore, these two cell lines were used in subsequent
experiments. In addition, the patients were divided into two groups
based on the median value of DUXAP8 expression, and patients with
higher expression of DUXAP8 experienced shorter survival time
(Fig. 1D). High DUXAP8 levels were
associated with poor prognosis.
DUXAP8 knockdown inhibits the
migration and invasion of melanoma cells
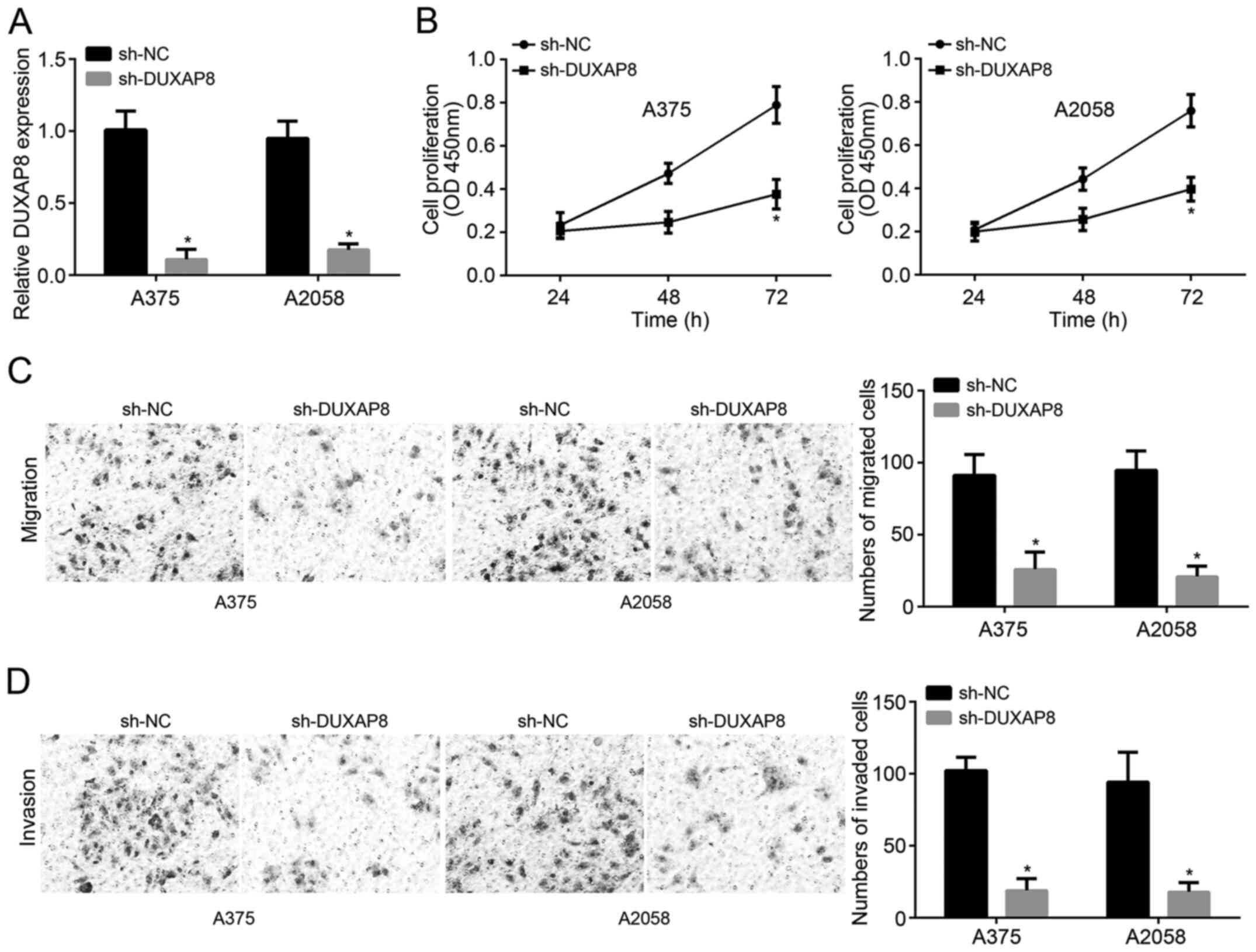
The expression of DUXAP8 was significantly reduced
following transfection with sh-DUXAP8, which confirmed the
transfection was successful (Fig.
2A). In order to examine the effect of sh-DUXAP8 on
proliferation, CCK-8 assays were carried out. After transfection
for 48 h and further culturing for 72 h, the proliferation of A375
and A2058 cells significantly decreased in the sh-DUXAP8 group
compared with that of the sh-NC group (Fig. 2B). In addition, DUXAP8 knockdown
significantly decreased migration and invasion of melanoma cells
(Fig. 2C and D). Therefore, DUXAP8
knockdown inhibited the proliferation, migration and invasion of
melanoma cells.
lncRNA DUXAP8 targets miR-3182,
whereas miR-3182 targetsNUPR1
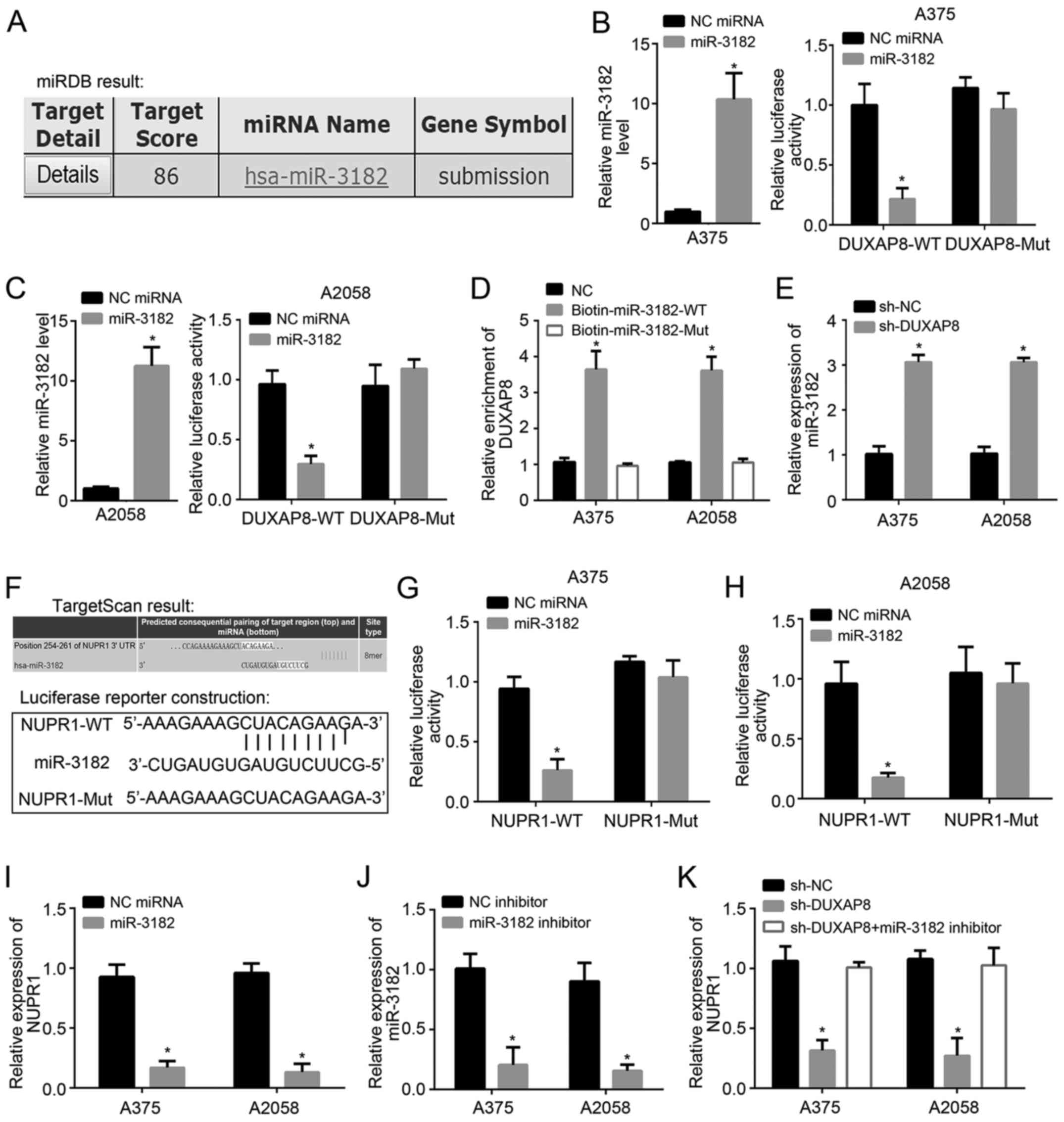
To identify the potential targets of DUXAP8,
bioinformatics analysis was carried out using miRDB. miR-3182 was
identified as the most likely target according to the prediction
results (Fig. 3A). To validate the
interaction between DUXAP8 and miR-3182, a dual luciferase reporter
assay was carried out. Both in A375 and A2058 cell lines,
transfection of miR-3182 reduced the luciferase activity in the
DUXAP8-WT group. However, NC miRNA did not significantly reduce the
luciferase activity of the vector. In the DUXAP8-MUT group,
luciferase activity remained unchanged. These results indicated
that miR-3182 significantly downregulated luciferase activity
(Fig. 3B and C). Pull-down assays
also demonstrated that miR-3182-WT precipitated DUXAP8, proving
their binding (Fig. 3D). Following
DUXAP8 knockdown, the expression of miR-3182 significantly
increased (Fig. 3E). To identify the
targets of miR-3182, bioinformatics analysis was performed using
TargetScan7. The results indicated that NUPR1 was the most likely
candidate. The predicted binding sites of miR-3182 and NUPR1 are
shown in Fig. 3F. Moreover,
luciferase reporter assays confirmed this interaction (Fig. 3G and H). Transfection with miR-3182
downregulated the expression of NUPR1 in both A375 and A2058 cells
(Fig. 3I). More importantly,
sh-DUXAP8 decreased the expression of NUPR1, while the miR-3182
inhibitor reversed this effect (Fig. 3J
and K). Thus, DUXAP8 promotes NUPR1 expression by inhibiting
miR-3182.
Rescue assay and correlation
analysis
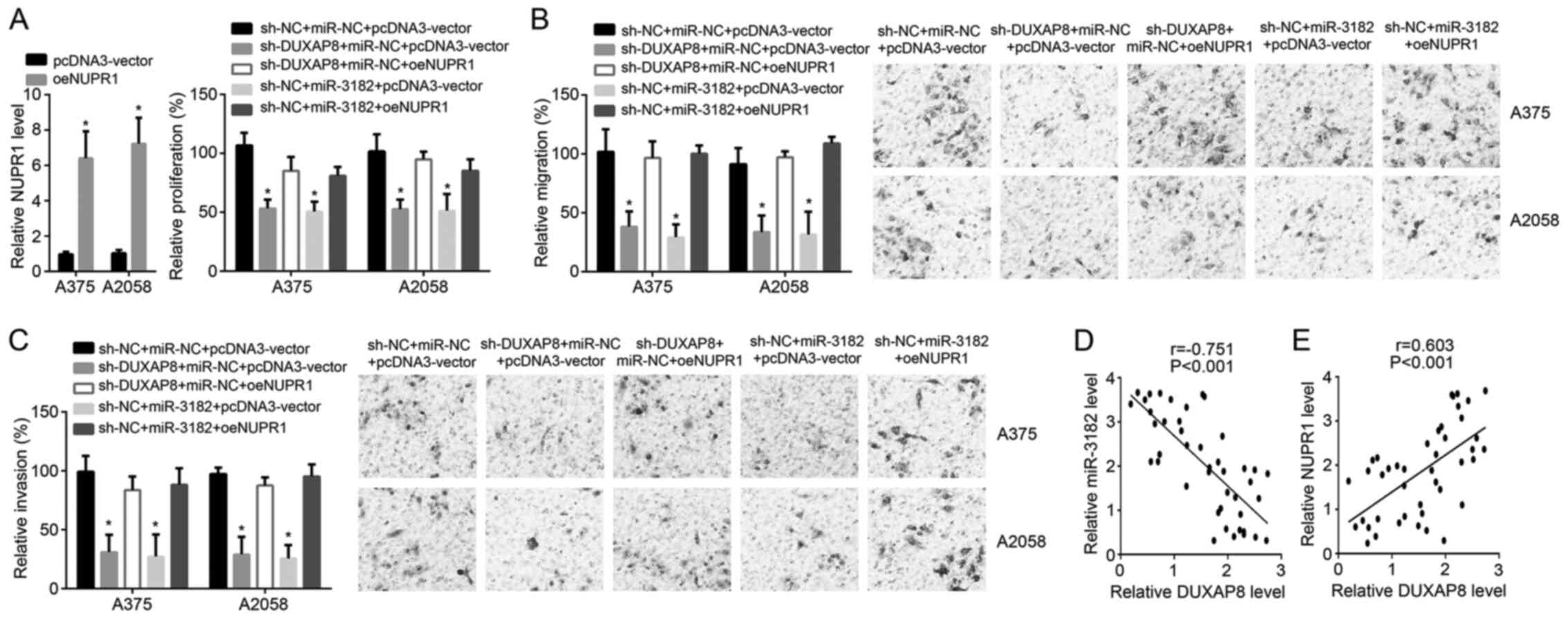
For rescue assays, NUPR1 was overexpressed in A375
cells and A2058 cells (Fig. 1A).
CCK8 and transwell assays demonstrated that the cell proliferation,
migration and invasion were inhibited in the
sh-DUXAP8+miR-NC+pcDNA3-vector group compared with the
sh-NC+miR-NC+pcDNA3-vector group (Fig.
4A-C). However, proliferation, migration and invasion were
rescued by NUPR1 overexpression in sh-DUXAP8+miR-NC+oeNUPR1 group
compared with that in the sh-DUXAP8+miR-NC+pcDNA3-vector group
(Fig. 4A-C). Proliferation,
migration and invasion were suppressed by miR-3182 in the
sh-NC+miR-3182+pcDNA3-vector group compared with the
sh-NC+miR-NC+pcDNA3-vector group (Fig.
4A-C). Proliferation, migration and invasion were also rescued
by NUPR1 overexpression in sh-NC+miR-3182+oeNUPR1 group compared
with the sh-NC+miR-3182+pcDNA3-vector group (Fig. 4A-C). In melanoma tissue samples,
DUXAP8 expression negatively correlated with that of miR-3182, but
positively correlated with that of NUPR1 (Fig. 4D and E). Altogether, these results
confirmed the regulatory relationship between DUXAP8, miR-3182 and
NUPR1.
Discussion
In order to examine the role of DUXAP8 in melanoma,
DUXAP8-related miRNA and genes were explored. In the present study,
DUXAP8 was upregulated in melanoma tissue samples and cell lines.
DUXAP8 levels were negatively associated with the survival time of
patients with melanoma. DUXAP8 knockdown inhibited the
proliferation, migration and invasion of melanoma cells.
Previous studies have shown that lncRNA derived from
pseudogenes could also regulate gene expression through various
mechanisms, such as binding to RNA-binding proteins and regulating
target genes (18,19). Xu et al (20) carried out a genome-wide analysis and
identified the lncRNA DUXAP8 as a significant biomarker of
metastatic renal carcinoma. Ma et al (19) confirmed that DUXAP8 downregulated
pleckstrin homology domain containing O1 level and promoted the
progression of gastric cancer. Moreover, DUXAP8 could also inhibit
Ras homolog family member B (RHOB) and early growth response 1
(EGR1) expression and accelerate development of lung cancer in
vitro (18). Although the role
of DUXAP8 in melanoma has not been reported so far, the effects of
DUXAP8-related genes have been confirmed. Indeed, as reported in a
previous study, the RHO family is important for the activation of
the FAK signaling pathway, which increases aggregation of melanoma
cells (21). Importantly, the
expression of RHOB is closely related to drug resistance in
melanoma cells (22). Furthermore,
RHOB also regulates the RAS/PI3K/AKT pathway, which increases
invasion and migration of melanoma cells and patient survival
(23). Besides, the transcription
factor EGR1 was differentially expressed in saliva samples of mice
with melanoma compared with normal tissues (24). Based on the aforementioned evidence,
it may be hypothesized that DUXAP8 plays an important role in the
development of melanoma.
Bioinformatics prediction and in vitro
experiments confirmed that DUXAP8 targeted miR-3182 in this study.
miR-3182 is involved in different diseases, such as breast cancer
(25), osteosarcoma (26) and lung cancer (27). Zhou et al (28) proposed that miR-3182 was important
for metastasis of nasopharyngeal carcinoma and prognosis of this
disease. In breast cancer, miR-3182 can inhibit mTOR signaling
(25). It is well known that the
mTOR signaling pathway is closely associated with autophagy,
invasion and proliferation of melanoma cell (29). The effect of miR-3182 in melanoma
cells could not be ignored. In the present study, DUXAP8 promoted
melanoma development by downregulating the expression of
miR-3182.
Furthermore, miR-3182 was found to target NUPR1 in
this study. In previous studies, NUPR1 has been confirmed to be
associated with lung cancer, pancreatic cancer and liver cancer
(30–32). In non-small cell lung cancer,
knockdown of NUPR1 inhibited proliferation in vitro and
slowed tumor growth in vivo (30). Emma et al (32) reported that NUPR1 regulated RUNX2
expression and further effect activation of hepatocellular
carcinoma cells. Interestingly, RUNX2 was upregulated in melanoma
tumor cells compared with controls and was also involved in
migration of cells (33). In
malignant tumors, NUPR1 was confirmed to modulate the caspase 3 and
PTEN levels and also regarded as a novel target for treatment
(32). In addition, knockdown of
PTEN is critical for the progression of melanoma development
(34). In the present study, the
direct effect of NUPR1 on melanoma cell lines was examined.
Overexpression of NUPR1 reversed the effect of DUXAP8 knockdown or
miR-3182 mimic transfection on melanoma cells. In melanoma tissue,
DUXAP8 expression positively correlated with that of NUPR1.
Therefore, NUPR1 is critical factor in the function of DUXAP8 in
proliferation, invasion and migration of melanoma.
In conclusion, the lncRNA DUXAP8 may accelerate the
development of melanoma by regulating miR-3182/NUPR1 expression.
This demonstrates the molecular function of the
DUXAP8/miR-3182/NUPR1 axis in melanoma progression and suggests
these molecules may represent potential targets for melanoma
treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XC and NL designed the study. XC, JG and NL
contributed toward performing the experiments, data analysis,
drafting and critically revising the paper, gave final approval of
the version to be published, and confirm the authenticity of all
the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by The Ethics Committee of
The Affiliated Changzhou No. 2 People's Hospital with Nanjing
Medical University (Changzhou, China). All patients signed informed
consent forms.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fodstad O, Aass N and Pihl A: An inverse
relationship between the growth rate of human melanoma xenografts
and their response to some cytostatic drugs. Br J Cancer.
41:829–831. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Farahmand AM, Ehsani AH, Mirzaei M,
Mohsenian M and Ghanadan A: Patients' characteristics,
histopathological findings, and tumor stage in different types of
malignant melanoma: A retrospective multicenter study. Acta Med
Iran. 55:316–323. 2017.PubMed/NCBI
|
|
3
|
Hu DN, Yu GP and McCormick SA:
Population-based incidence of vulvar and vaginal melanoma in
various races and ethnic groups with comparisons to other
site-specific melanomas. Melanoma Res. 20:153–158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Del Vecchio M: AACR update on 5-year
survival rates, effcacy and long-term safety in previously treated
advanced/metastatic melanoma patients receiving mono-immunotherapy
with nivolumab. Recenti Prog Med. 107:414–417. 2016.(In Italian).
PubMed/NCBI
|
|
5
|
Morton DL, Davtyan DG, Wanek LA, Foshag LJ
and Cochran AJ: Multivariate analysis of the relationship between
survival and the microstage of primary melanoma by Clark level and
Breslow thickness. Cancer. 71:3737–3743. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pfahlberg A, Kölmel KF and Gefeller O;
Febim Study Group, : Timing of excessive ultraviolet radiation and
melanoma: Epidemiology does not support the existence of a critical
period of high susceptibility to solar ultraviolet
radiation-induced melanoma. Br J Dermatol. 144:471–475. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thoelke A, Willrodt S, Hauschild A and
Schadendorf D: Primary extracutaneous malignant melanoma: A
comprehensive review with emphasis on treatment. Onkologie.
27:492–499. 2004.PubMed/NCBI
|
|
8
|
Gasparini G: Tamoxifen and chemotherapy in
the treatment of metastatic melanoma: Are there other possible
mechanisms explaining their potentiation? J Clin Oncol.
12:1994–1996. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Moschos SJ and Becker D:
Functional analysis and molecular targeting of aurora kinases a and
B in advanced melanoma. Genes Cancer. 1:952–963. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chi Z, Li S, Sheng X, Si L and Guo J:
Clinical presentation, histology, and prognoses of malignant
melanoma in ethnic Chinese: A study of 522 consecutive cases. BMC
Cancer. 11:852011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Barth A, Wanek L and Morton DL: Prognostic
factors in 1,521 melanoma patients with distant metastases. J Am
Coll Surg. 181:193–201. 1995.PubMed/NCBI
|
|
12
|
Ribero S, Glass D and Bataille V: Genetic
epidemiology of melanoma. Eur J Dermatol. 26:335–339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schmidt K, Joyce CE, Buquicchio F, Brown
A, Ritz J, Distel RJ, Yoon CH and Novina CD: The lncRNA SLNCR1
mediates melanoma invasion through a conserved SRA1-like region.
Cell Rep. 15:2025–2037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gong J, Zhang H, He L, Wang L and Wang J:
Increased expression of long non-coding RNA BCAR4 is predictive of
poor prognosis in patients with non-small cell lung cancer. Tohoku
J Exp Med. 241:29–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei F, Yang L, Jiang D, Pan M, Tang G,
Huang M and Zhang J: Long noncoding RNA DUXAP8 contributes to the
progression of hepatocellular carcinoma via regulating
miR-422a/PDK2 axis. Cancer Med. 9:2480–2490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji X, Tao R, Sun LY, Xu XL and Ling W:
Down-regulation of long non-coding RNA DUXAP8 suppresses
proliferation, metastasis and EMT by modulating miR-498 through
TRIM44-mediated AKT/mTOR pathway in non-small-cell lung cancer. Eur
Rev Med Pharmacol Sci. 24:3152–3165. 2020.PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei
C, Li W, He X and Lu KH: The pseudogene DUXAP8 promotes
non-small-cell lung cancer cell proliferation and invasion by
epigenetically silencing EGR1 and RHOB. Mol Ther. 25:739–751. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma HW, Xie M, Sun M, Chen TY, Jin RR, Ma
TS, Chen QN, Zhang EB, He XZ, De W and Zhang ZH: The pseudogene
derived long noncoding RNA DUXAP8 promotes gastric cancer cell
proliferation and migration via epigenetically silencing PLEKHO1
expression. Oncotarget. 8:52211–52224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu X, Xu Y, Shi C, Wang B, Yu X, Zou Y and
Hu T: A genome-wide comprehensively analyses of long noncoding RNA
profiling and metastasis associated lncRNAs in renal cell
carcinoma. Oncotarget. 8:87773–87781. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goundiam O, Nagel MD and Vayssade M: Akt
and RhoA inhibition promotes anoikis of aggregated B16F10 melanoma
cells. Cell Biol Int. 36:311–319. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Delmas A, Cherier J, Medale-Giamarchi C,
Pradines A and Favre G: Abstract 3401: Vemurafenib-induced RhoB
expression leads to melanoma cell resistance. Cancer Res. 73 (Suppl
8):S34012013.
|
|
23
|
Eide A: The role of the sub-commission on
promotion and protection of human rights and its working groups in
the prevention of conflicts. Int J Minor Group Rights. 8:25–29.
2001. View Article : Google Scholar
|
|
24
|
Gao K, Zhou H, Zhang L, Lee JW, Zhou Q, Hu
S, Farrell J, Eibl G and Wong D: Abstract #314: Induction of
systemic disease-specific salivary biomarker profiles in mouse
models of melanoma and non-small cell lung cancer. Cancer Res. 69
(Suppl 9):S3142009.
|
|
25
|
Razaviyan J, Hadavi R, Tavakoli R, Kamani
F, Paknejad M and Mohammadi-Yeganeh S: Expression of miRNAs
targeting mTOR and S6K1 genes of mTOR signaling pathway including
miR-96, miR-557, and miR-3182 in triple-negative breast cancer.
Appl Biochem Biotechnol. 186:1074–1089. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu KP, Ma XL and Zhang CL: LncRNA ODRUL
contributes to osteosarcoma progression through the miR-3182/MMP2
axis. Mol Ther. 25:2383–2393. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xue M, Shi D, Xu G and Wang W: The long
noncoding RNA linc00858 promotes progress of lung cancer through
miR-3182/MMP2 axis. Artif Cells Nanomed Biotechnol. 47:2091–2097.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou MY, Lan GP, Si YF and Mo XG:
Expression patterns and significances of distant metastasis and
prognosis of MiR-3182 innasopharygeal carcinoma. Chin J New Clin
Med. 5:369–372. 2016.
|
|
29
|
Zhao G, Han X, Zheng S, Li Z, Sha Y, Ni J,
Sun Z, Qiao S and Song Z: Curcumin induces autophagy, inhibits
proliferation and invasion by downregulating AKT/mTOR signaling
pathway in human melanoma cells. Oncol Rep. 35:1065–1074. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo X, Wang W, Hu J, Feng K, Pan Y, Zhang
L and Feng Y: Lentivirus-mediated RNAi knockdown of NUPR1 inhibits
human nonsmall cell lung cancer growth in vitro and in vivo. Anat
Rec (Hoboken). 295:2114–2121. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cano CE, Hamidi T, Garcia MN, Grasso D,
Loncle C, Garcia S, Calvo E, Lomberk G, Dusetti N, Bartholin L, et
al: Genetic inactivation of Nupr1 acts as a dominant suppressor
event in a two-hit model of pancreatic carcinogenesis. Gut.
63:984–995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Emma MR, Iovanna JL, Bachvarov D, Puleio
R, Loria GR, Augello G, Candido S, Libra M, Gulino A, Cancila V, et
al: NUPR1, a new target in liver cancer: Implication in controlling
cell growth, migration, invasion and sorafenib resistance. Cell
Death Dis. 7:e22692016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Boregowda RK, Olabisi OO, Abushahba W,
Jeong BS, Haenssen KK, Chen W, Chekmareva M, Lasfar A, Foran DJ,
Goydos JS and Cohen-Solal KA: RUNX2 is overexpressed in melanoma
cells and mediates their migration and invasion. Cancer Lett.
348:61–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu H, Goel V and Haluska FG: PTEN
signaling pathways in melanoma. Oncogene. 22:3113–3122. 2003.
View Article : Google Scholar : PubMed/NCBI
|