Introduction
Lung cancer remains the leading cause of
cancer-associated morbidity (~787,000 cases) and mortality
(~622,000 deaths) in China in 2015 (1). The mortality rate of male patients is
particularly high in the Yunnan province, where tobacco is a
mainstay of the local economy (2).
Overall, ~85% of patients with lung cancer are diagnosed with
non-small cell carcinoma (NSCLC), which comprises large cell
carcinoma, lung adenocarcinoma and squamous cell carcinoma subtypes
(3,4). More than 80% of patients are dead by 5
years (5–7). Due to the poor prognosis and
ineffective screening methods available for lung adenocarcinoma,
the clinical cure rate for lung adenocarcinoma remains low.
Transfer RNA (tRNA) is involved in protein synthesis
by recognizing and transporting specific amino acids (8). Previous research has focused on
investigating the additional functions of tRNA. For example,
several studies have identified tRNA-derived fragments (tRFs) and
tRNA halves (tiRNAs), which are classes of non-coding small RNA
fragments of <40 nucleotides in length derived from tRNA
transcripts (9,10). In the context of stress, mature tRNA
or precursor tRNA can be precisely cleaved by specific nucleases
(such as Dicer and angiogenin) into specific-sized fragments with
regulatory functions, instead of undergoing random tRNA degradation
(11–13). Stress-induced tRFs and tiRNAs have
been discovered to serve a role in cancer, metabolic diseases and
nervous system disorders (14). tRFs
and tiRNAs are enriched in biological body fluids, and their
composition and quantity are highly dependent on the cell type and
disease state (15). Consequently,
tRFs and tiRNAs may represent novel non-invasive biomarkers for
disease diagnosis (16).
Previous studies have categorized tRFs into the
following types: tRF-1, tRF-2, tRF-3 and tRF-5 (9,17–19). In
addition, there are currently two known types of tiRNAs, namely
5′-tiRNAs and 3′-tiRNAs (20). tRFs
and tiRNAs are similar to microRNA (miRNA) in their structure and
function (21,22); however, they are more stable and
abundant than miRNA. tRFs and tiRNAs have been reported to suppress
mRNA translation of ribosomal proteins (23) and to decrease mRNA stability by
binding to Y-box-binding protein 1 (24). In addition, tRFs and tiRNAs have been
demonstrated to regulate translation initiation and elongation
(25,26). A previous study has revealed that
tiRNAs can prevent apoptosis by binding to cytochrome c
(27). These findings provided
insight into the role of tRFs and tiRNAs on proliferation,
apoptosis, invasion and metastasis in tumor cells (28). Thus, it is of great significance to
study the molecular mechanisms of tRFs and tiRNAs with regards to
their role in the development of lung adenocarcinoma to identify
reliable biomarkers and novel drug targets. However, the regulatory
effects and molecular mechanisms of tRFs and tiRNAs remain poorly
understood.
High-throughput sequencing and analysis of small RNA
fragments can be used to identify tRFs and their corresponding
known tRNA sequences. The present study aimed to use RNA sequencing
(RNA-seq) technologies to analyze the expression levels of tRFs and
tiRNAs in lung adenocarcinoma and adjacent tissues, some of which
were reported for the first time. In addition, the biological
functions of tRFs and tiRNAs were evaluated using bioinformatics
analysis. The current findings may provide a novel perspective into
the molecular mechanisms underlying lung adenocarcinoma
development.
Materials and methods
Patient studies
A total of three pairs of lung adenocarcinoma and
adjacent tissues were collected from patients in August 2018 at the
Department of Pathology in The First People's Hospital of Yunnan
Province (Kunming, China). Two patients were female and one was
male. All patients were between 51–70 years of age, with a mean age
of 58.3 years. The tissues were preserved in RNAlater (Invitrogen;
Thermo Fisher Scientific, Inc.) at −80°C. Both the intraoperative
frozen and paraffin-embedded tissues (~3-µm-thick) were diagnosed
as infiltrating lung adenocarcinoma. The protocol of the present
study was approved by the Ethics Committee of The First People's
Hospital of Yunnan Province. All patients provided written informed
consent prior to participation.
RNA extraction and quality
control
Total RNA was extracted from tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The integrity and
quantity of each RNA sample were subsequently analyzed using 1%
agarose gel electrophoresis and a NanoDrop® ND-1000
spectrophotometer (Thermo Fisher Scientific, Inc.).
Pretreatment of tRF and tiRNA
tRFs and tiRNAs have heavy RNA modifications that
interfere with small RNA-seq library construction. Therefore, total
RNA was pretreated to remove the RNA modifications, including
3′-aminoacyl deacetylation, 2′,3′-cyclic phosphate, 5′-OH (hydroxyl
group) phosphorylation and m1A and m3C demethylation, using a
rtStar™ tRF and tiRNA Pretreatment kit (cat. no. AS-FS-005;
Arraystar Inc.). Preprocessed total RNA samples were subsequently
used for tRF- and tiRNA-seq library preparation.
Library preparation
Sequencing libraries were size-selected for the RNA
biotypes to be sequenced using an automated gel cutter. The
sequencing library was quantified on an Agilent 2100 Bioanalyzer
(Agilent Technologies, Inc.) using an Agilent DNA 1000 chip kit
(Agilent Technologies, Inc.). Libraries were mixed in equal amounts
and used for sequencing.
Sequencing
The libraries were qualified and absolutely
quantified using an Agilent BioAnalyzer 2100 (Agilent Technologies,
Inc.). Mixed DNA fragments in the libraries were denatured with 0.1
M NaOH to generate single-stranded molecules, and loaded onto the
reagent cartridge at a concentration of 1.8 pM. The sequencing was
performed on an Illumina NextSeq 500 system (Illumina, Inc.) using
a NextSeq 500/550 V2 kit (cat. no. FC-404-2005; Illumina, Inc.)
according to the manufacturer's protocol. The sequencing type was
50-bp single-read.
Data analysis
The tRFs and tiRNAs were identified by mapping to
tRFdb (http://genome.bioch.virginia.edu/trfdb/). Image
analysis and base calling were performed using Solexa Pipeline v1.8
software (Off-Line Base Caller software; Illumina, Inc.).
Sequencing quality was first examined using FastQC v0.11.7
(http://www.bioinformatics.babraham.ac.uk/projects/fastqc/),
and trimmed reads were aligned to allow for only one mismatch to
the mature tRNA sequences. Reads that did not map were aligned to
allow for only one mismatch to precursor tRNA sequences using
bowtie v1.2.2 software (29). The
remaining reads were aligned to allow for only one mismatch to
miRNA reference sequences with miRDeep2 v2.0.0.8 (30). The abundance of tRF, tiRNA and miRNA
was evaluated using their sequencing counts, which were normalized
as counts per million (CPM) of total aligned reads. Differentially
expressed tRFs and tiRNAs were screened based on the count value
using the R package edgeR (31).
Principal component analysis (PCA), Pearson correlation analysis,
pie plots, Venn plots, hierarchical clustering, scatter plots and
volcano plots were generated in R (https://www.r-project.org/) or Perl (https://www.perl.org/) for statistical computing and
visualization of the differentially expressed tRFs and tiRNAs. The
cut-off values for differentially expressed tRFs and tiRNAs were
log2 fold-change (FC) ≥1.4 and P≤0.05. The differentially expressed
tRFs and tiRNAs were selected for further analysis.
Reverse transcription-quantitative PCR
(RT-qPCR)
Preprocessed total RNA samples of the three lung
adenocarcinoma and adjacent tissues were reverse transcribed into
cDNA using a rtStar™ First-Strand cDNA Synthesis kit (cat. no.
AS-FS-003; Arraystar, Inc.) according to the manufacturer's
protocol, and 2X SYBR Green qPCR master mix (cat. no. AS-MR-005;
Arraystar, Inc.) was used for qPCR analysis. Primers were designed
using Primer v5.0 (Premier Biosoft International) and synthesized
by Aksomics, Inc. Primer sequences are presented in Table I. qPCR was performed on a
QuantStudio™ 5 Real-time PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) using the following thermocycling
conditions: Initial denaturation at 95°C for 10 min, followed by 40
cycles at 95°C for 10 sec and 60°C for 60 sec. Following
amplification, the system was slowly heated from 60 to 99°C at a
ramp rate of 0.05°C/sec to establish the melting curve of the PCR
products. Data for relative quantification of tRFs and tiRNAs was
obtained by normalization to U6 expression. The expression levels
were determined using the 2−ΔΔCq method for relative
quantification of gene expression (32).
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) | Product length,
bp |
|---|
| U6 | F:
GCTTCGGCAGCACATATACTAAAAT | 89 |
|
| R:
CGCTTCACGAATTTGCGTGTCAT |
|
|
tiRNA-Lys-CTT-002 | F:
ATCGCCCGGCTAGCTCAGT | 45 |
|
| R:
TCCGATCTGAGTCTCATGCTCTAC |
|
|
tRF-Val-CAC-011 | F:
CTTCTGTAGTGTAGTGGTTATCACG | 46 |
|
| R:
GTGCTCTTCCGATCTGGC |
|
|
tRF-Val-CAC-010 | F:
CTTCTGTAGTGTAGTGGTTATCACG | 45 |
|
| R:
GTGCTCTTCCGATCTGCG |
|
|
tRF-Val-CAC-007 | F:
CGATCGCTTCTGTAGTGTAGTGG | 46 |
|
| R:
GCTCTTCCGATCTAACGTGATAA |
|
|
tRF-His-GTG-008 | F:
CGATCGCCGTGATCGTATAGT | 44 |
|
| R:
TCCGATCTCGCAGAGTACTAACC |
|
|
tRF-Ser-AGA-006 | F:
AGTCCGACGATCGTAGTCGTG | 42 |
|
| R:
CGATCTCCTTAACCACTCGGC |
|
|
tRF-Glu-TTC-027 | F:
CGACGATCTGACTGGACCTTT | 45 |
|
| R:
GACGTGTGCTCTTCCGATCTAA |
|
|
tRF-Lys-CTT-001 | F:
GATCGCCCAGCTAGCTCAGT | 47 |
|
| R:
GTGCTCTTCCGATCTTATGCTCT |
|
|
tRF-Ser-TGA-005 | F:
GATCGAAGCGGGTGCTCT | 41 |
|
| R:
GACGTGTGCTCTTCCGATCTAT |
|
|
tRF-His-GTG-012 | F:
AGTCCGACGATCTCGAATCC | 40 |
|
| R:
CGATCTTGGTGCCGTGACTC |
|
Bioinformatics analysis
The structures of tRNA were downloaded from the
Leipzig tRNA database (http://trna.bioinf.uni-leipzig.de/DataOutput/). As
tRFs and tiRNA have been reported to be similar to miRNA in
function (21,22), miRanda v3.3a (33) and TargetScan v7.1 software (34) were used to predict the target mRNAs
of validated tRFs and tiRNAs (35–37).
Gene Ontology (GO) functional term enrichment analysis (http://www.geneontology.org/) was used to determine
functional terms of the target genes. Signaling pathway enrichment
analysis was used to investigate the significant pathways of the
target genes using the Kyoto Encyclopedia of Genes and Genomes
(KEGG) database (http://www.genome.jp/kegg/).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 (GraphPad Software, Inc.). Results are represented as the
mean ± SD of 3 parallel samples. Statistical differences between
two groups were determined using a two-tailed paired Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
tRF and tiRNA expression profiling in
patients with lung adenocarcinoma
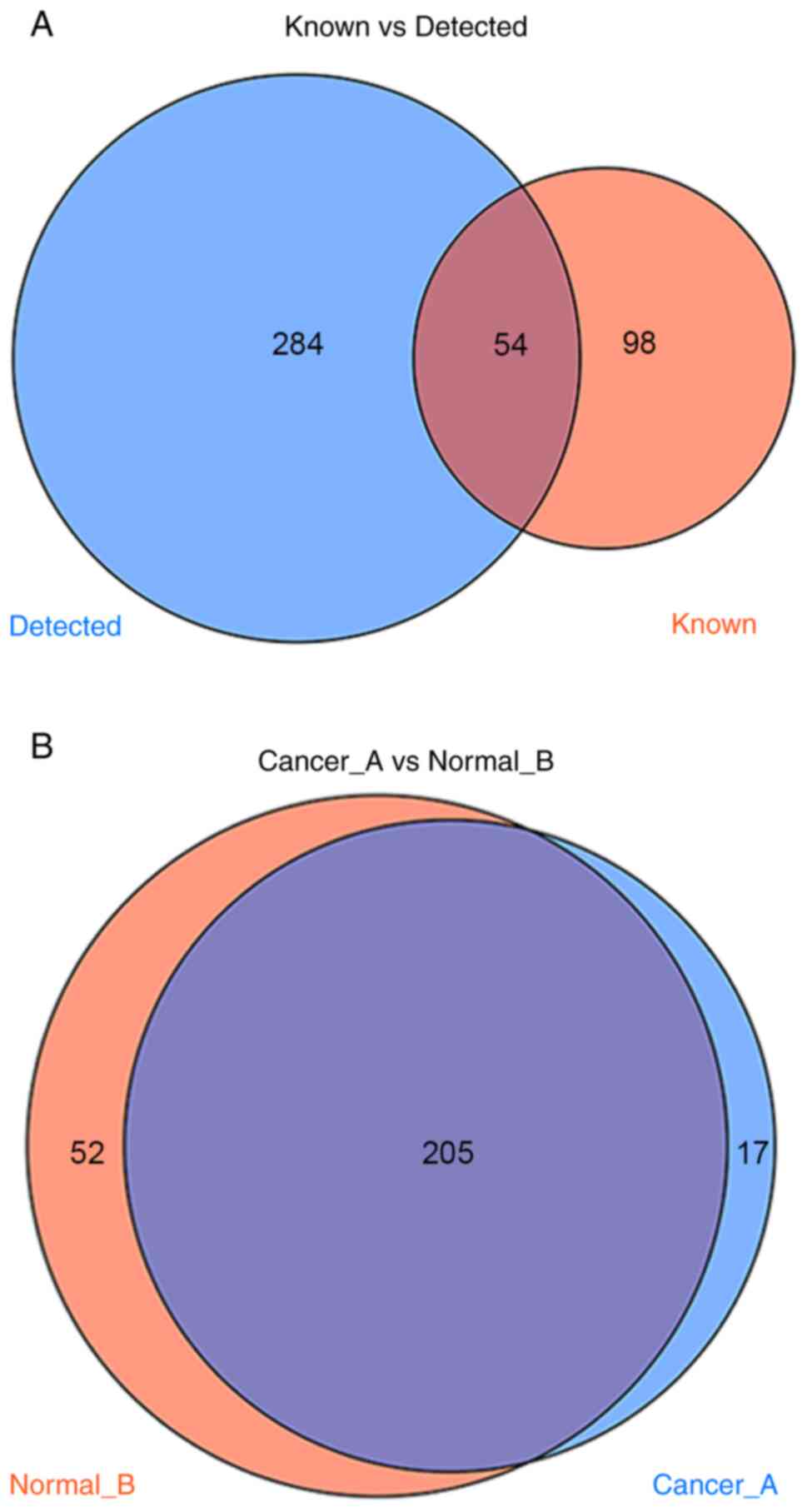
There are currently 152 known types of
tumor-associated tRFs and tiRNAs that have been reported in the tRF
database. A total of 338 types were detected in the sequencing
performed in the present study, of which 54 overlapped with the
database and the other 284 were novel, to the best of our knowledge
(Fig. 1A). Compared with adjacent
tissues, 17 differentially expressed tRFs and tiRNAs were
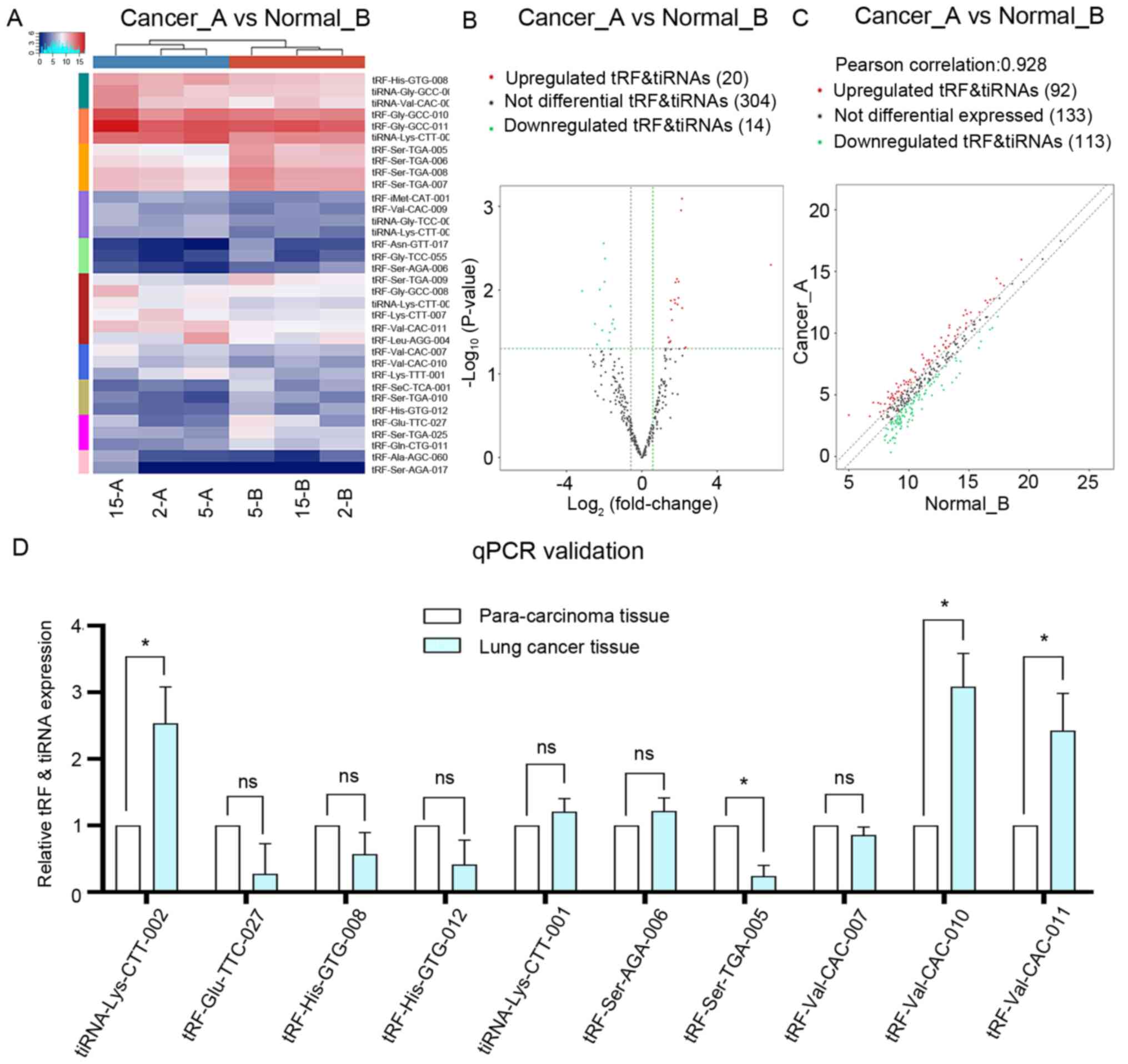
identified in all three cancer tissues during sequencing (Fig. 1B). Clustering results revealed that
there were 34 differently expressed subtypes of tRF and tiRNA
between the two groups (Fig. 2A),
among which 20 were upregulated and 14 were downregulated (Fig. 2B). The correlation in identified tRFs
and tiRNAs between lung adenocarcinoma and adjacent tissues was
examined using a scatter plot (Fig.
2C). In both the lung adenocarcinoma and adjacent normal
tissues, the three samples were clustered together. The
differentially expressed tRFs and tiRNAs are listed in Table II. To validate these changes, ten
types of tRF and tiRNA were selected and their expression levels
were analyzed using RT-qPCR. As shown in Fig. 2D, four genes out of the 10 measured
transcripts demonstrated differential expression between the three
lung adenocarcinoma and adjacent tissues, namely tiRNA-Lys-CTT-002,
tRF-Ser-TGA-005, tRF-Val-CAC-010 and tRF-Val-CAC-011. In the lung
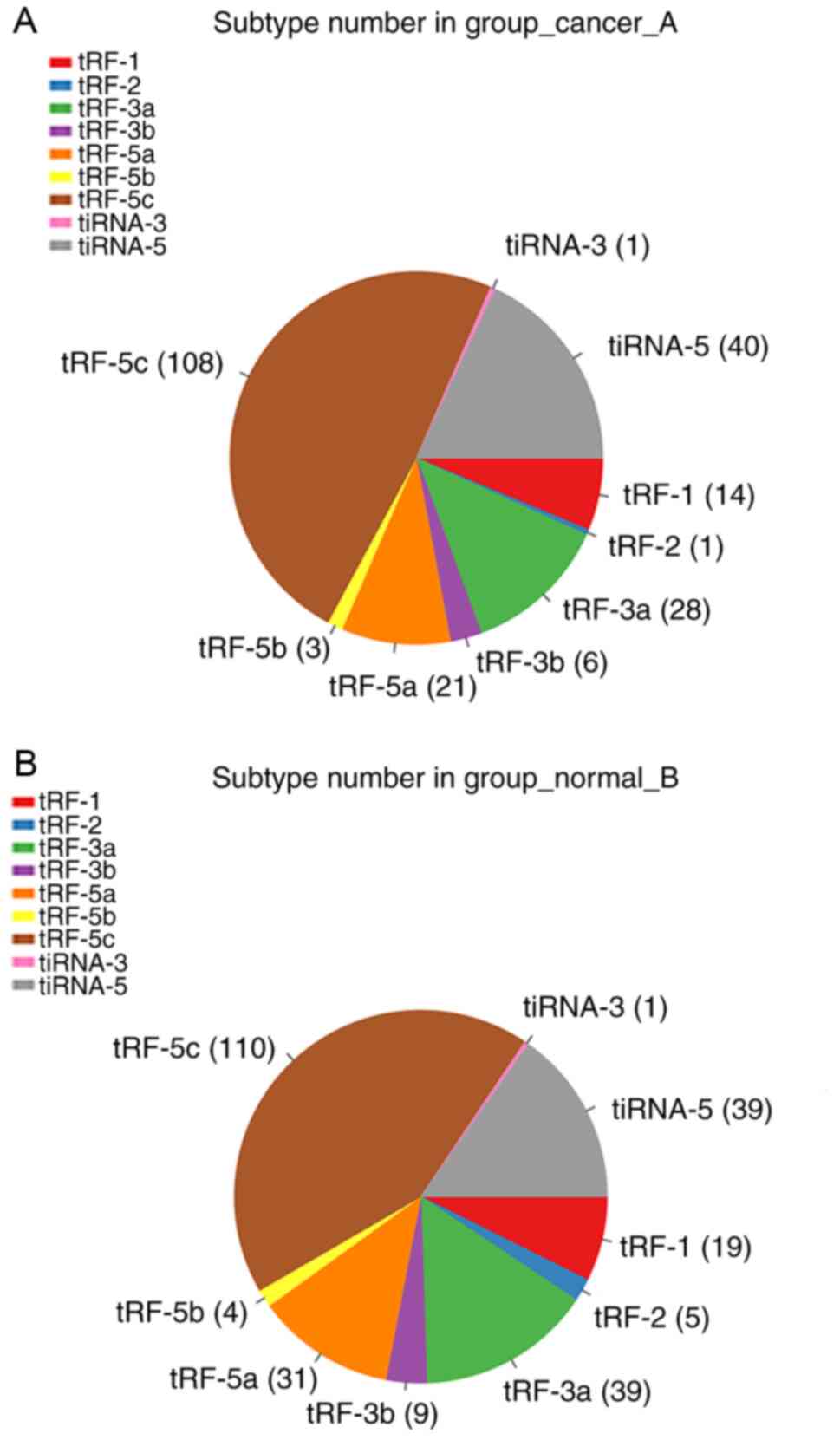
adenocarcinoma group, 14 tRF-1, 1 tRF-2, 28 tRF-3a, 6 tRF-3b, 21
tRF-5a, 3 tRF-5b, 108 tRF-5c, 1 tiRNA-3 and 40 tiRNA-5 were
identified (Fig. 3A). In the
adjacent tissues, 19 tRF-1, 5 tRF-2, 39 tRF-3a, 9 tRF-3b, 31
tRF-5a, 4 tRF-5b, 110 tRF-5c, 1 tiRNA-3 and 39 tiRNA-5 were
identified (Fig. 3B).
 | Table II.Differentially expressed tRFs and
tiRNAs in lung adenocarcinoma. |
Table II.
Differentially expressed tRFs and
tiRNAs in lung adenocarcinoma.
| tRF_ID | Length, nt |
log2FC | FC | P-value | Q-value | Cancer_CPM | Normal_CPM | Regulation |
|---|
|
tRF-Ser-AGA-017 | 32 | 6.84 | 114.92 | 0.00500 | 0.30239 | 3.35 | −3.49 | Up |
|
tRF-Ala-AGC-060 | 18 | 2.33 | 5.02 | 0.04836 | 0.42981 | 4.37 | 2.04 | Up |
|
tRF-Leu-AAG-004 | 16 | 2.27 | 4.83 | 0.04968 | 0.42981 | 10.64 | 8.37 | Up |
|
tRF-Lys-TTT-001 | 15 | 2.14 | 4.41 | 0.01641 | 0.30239 | 8.25 | 6.11 | Up |
|
tiRNA-Lys-CTT-002 | 34 | 2.13 | 4.39 | 0.00081 | 0.18848 | 14.43 | 12.30 | Up |
|
tRF-Val-CAC-011 | 32 | 2.08 | 4.23 | 0.00112 | 0.18848 | 10.44 | 8.36 | Up |
|
tiRNA-Lys-CTT-005 | 34 | 1.95 | 3.86 | 0.00794 | 0.30239 | 5.91 | 3.96 | Up |
|
tRF-Lys-CTT-007 | 31 | 1.94 | 3.85 | 0.01241 | 0.30239 | 9.63 | 7.69 | Up |
|
tiRNA-Val-CAC-001 | 34 | 1.90 | 3.72 | 0.01487 | 0.30239 | 11.44 | 9.54 | Up |
|
tRF-Val-CAC-010 | 31 | 1.86 | 3.64 | 0.00737 | 0.30239 | 7.07 | 5.20 | Up |
|
tiRNA-Lys-CTT-001 | 34 | 1.78 | 3.44 | 0.00809 | 0.30239 | 8.93 | 7.14 | Up |
|
tRF-Val-CAC-007 | 28 | 1.77 | 3.40 | 0.01435 | 0.30239 | 7.98 | 6.21 | Up |
|
tiRNA-Gly-GCC-002 | 33 | 1.74 | 3.34 | 0.01310 | 0.30239 | 11.55 | 9.82 | Up |
|
tRF-Gly-GCC-011 | 32 | 1.60 | 3.03 | 0.02293 | 0.36900 | 15.95 | 14.35 | Up |
|
tiRNA-Gly-TCC-001 | 34 | 1.55 | 2.93 | 0.01700 | 0.30239 | 6.14 | 4.59 | Up |
|
tRF-His-GTG-008 | 31 | 1.54 | 2.91 | 0.01337 | 0.30239 | 11.68 | 10.14 | Up |
|
tRF-Gly-GCC-008 | 29 | 1.52 | 2.87 | 0.04077 | 0.42981 | 9.80 | 8.28 | Up |
|
tRF-Val-CAC-009 | 30 | 1.48 | 2.80 | 0.04184 | 0.42981 | 5.60 | 4.12 | Up |
|
tRF-iMet-CAT-001 | 30 | 1.44 | 2.71 | 0.04220 | 0.42981 | 5.77 | 4.33 | Up |
|
tRF-Gly-GCC-010 | 31 | 1.41 | 2.65 | 0.03684 | 0.42981 | 14.02 | 12.61 | Up |
|
tRF-Asn-GTT-017 | 15 | −3.17 | 0.11 | 0.01026 | 0.30239 | 0.34 | 3.51 | Down |
|
tRF-Glu-TTC-027 | 17 | −2.49 | 0.18 | 0.02539 | 0.37306 | 5.40 | 7.89 | Down |
|
tRF-Gly-TCC-055 | 14 | −2.38 | 0.19 | 0.04466 | 0.42981 | 1.49 | 3.86 | Down |
|
tRF-SeC-TCA-001 | 15 | −2.24 | 0.21 | 0.00987 | 0.30239 | 3.84 | 6.08 | Down |
|
tRF-Ser-AGA-006 | 24 | −2.10 | 0.23 | 0.03021 | 0.40839 | 1.86 | 3.96 | Down |
|
tRF-Ser-TGA-005 | 17 | −2.02 | 0.25 | 0.00277 | 0.30239 | 8.80 | 10.82 | Down |
|
tRF-Ser-TGA-025 | 15 | −1.98 | 0.25 | 0.00797 | 0.30239 | 5.75 | 7.74 | Down |
|
tRF-Gln-CTG-011 | 14 | −1.96 | 0.26 | 0.00421 | 0.30239 | 4.95 | 6.91 | Down |
|
tRF-Ser-TGA-010 | 22 | −1.74 | 0.30 | 0.04032 | 0.42981 | 3.52 | 5.26 | Down |
|
tRF-His-GTG-012 | 22 | −1.70 | 0.31 | 0.03203 | 0.41640 | 3.45 | 5.15 | Down |
|
tRF-Ser-TGA-006 | 18 | −1.66 | 0.32 | 0.01559 | 0.30239 | 9.33 | 10.99 | Down |
|
tRF-Ser-TGA-009 | 21 | −1.58 | 0.33 | 0.02526 | 0.37306 | 7.71 | 9.29 | Down |
|
tRF-Ser-TGA-008 | 20 | −1.52 | 0.35 | 0.02253 | 0.36900 | 10.42 | 11.94 | Down |
|
tRF-Ser-TGA-007 | 19 | −1.44 | 0.37 | 0.02905 | 0.40839 | 10.29 | 11.73 | Down |
Functional enrichment analysis reveals
a significant association between tRFs, tiRNAs and
carcinogenesis
Several previous studies have demonstrated that tRFs
and tiRNAs are involved in translational regulation and gene
silencing (18,38). tRFs and tiRNAs have a miRNA-like
structure and function, and can therefore inhibit protein
translation (21,22). Based on these characteristics, target
genes of tRFs and tiRNAs were identified using miRanda and
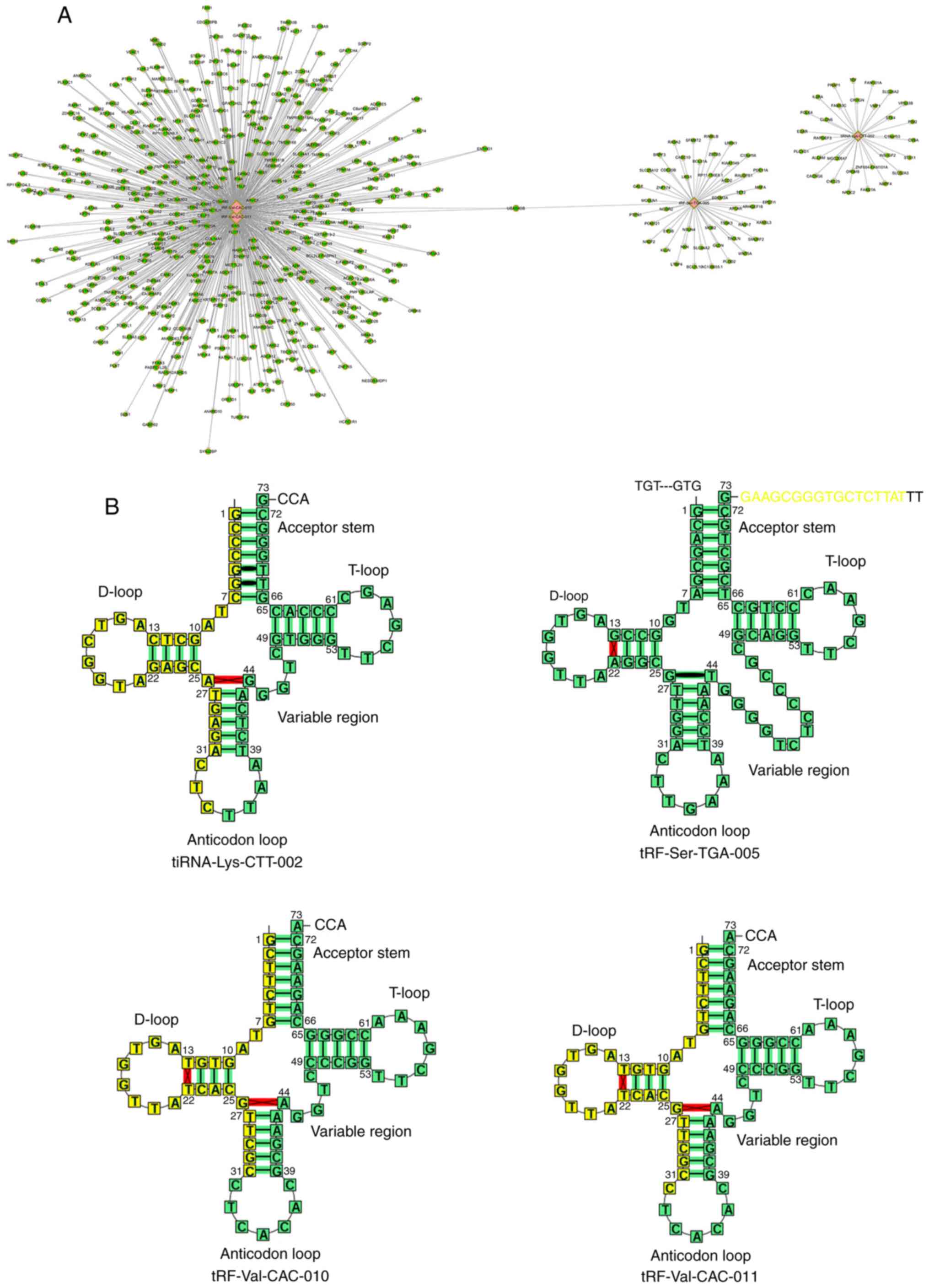
TargetScan software. Fig. 4A
presents the association between tiRNA-Lys-CTT-002,
tRF-Ser-TGA-005, tRF-Val-CAC-010 and tRF-Val-CAC-011 and their
potential targets, such as cyclin D1 (CCND1), C-X-C motif chemokine
ligand (CXCL)1, CXCL2 and glioma-associated oncogene homolog 2
(Gli2). The cleavage site of tiRNA-Lys-CTT-002, tRF-Ser-TGA-005,
tRF-Val-CAC-010 and tRF-Val-CAC-011 was further investigated
(Fig. 4B). tiRNA-Lys-CTT-002,
tRF-Val-CAC-010 and tRF-Val-CAC-011 were generated by the cleavage
in or near the tRNA anticodon loop.
To further investigate the possible mechanisms and
potential functions of the significant heterogeneity in the
expression levels of tRFs and tiRNAs, GO functional term and KEGG
signaling pathway analyses based on the predicted target genes were
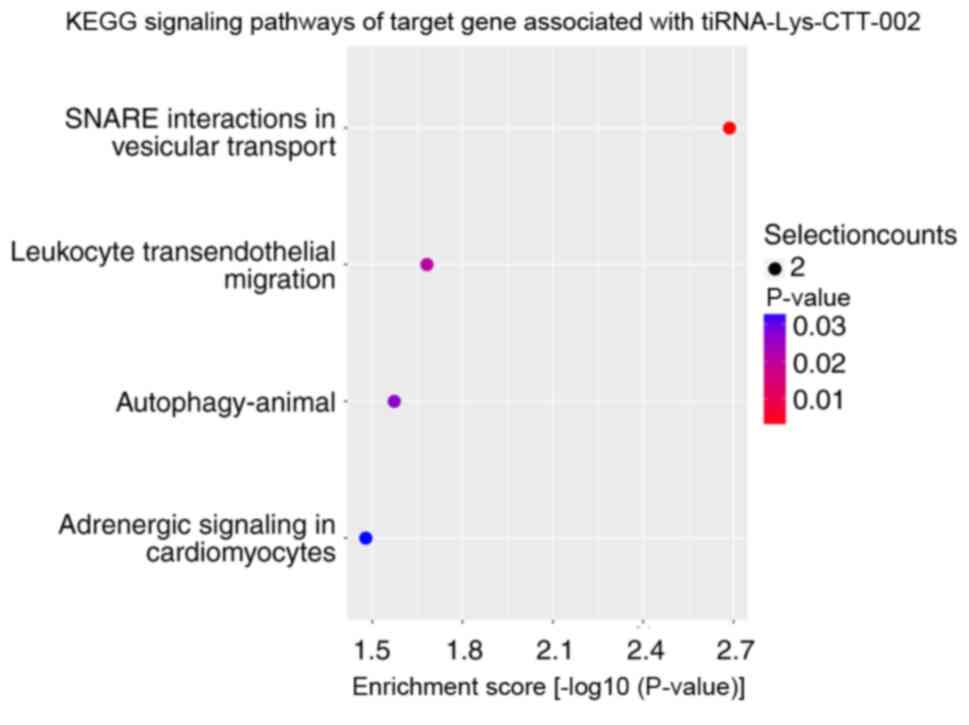
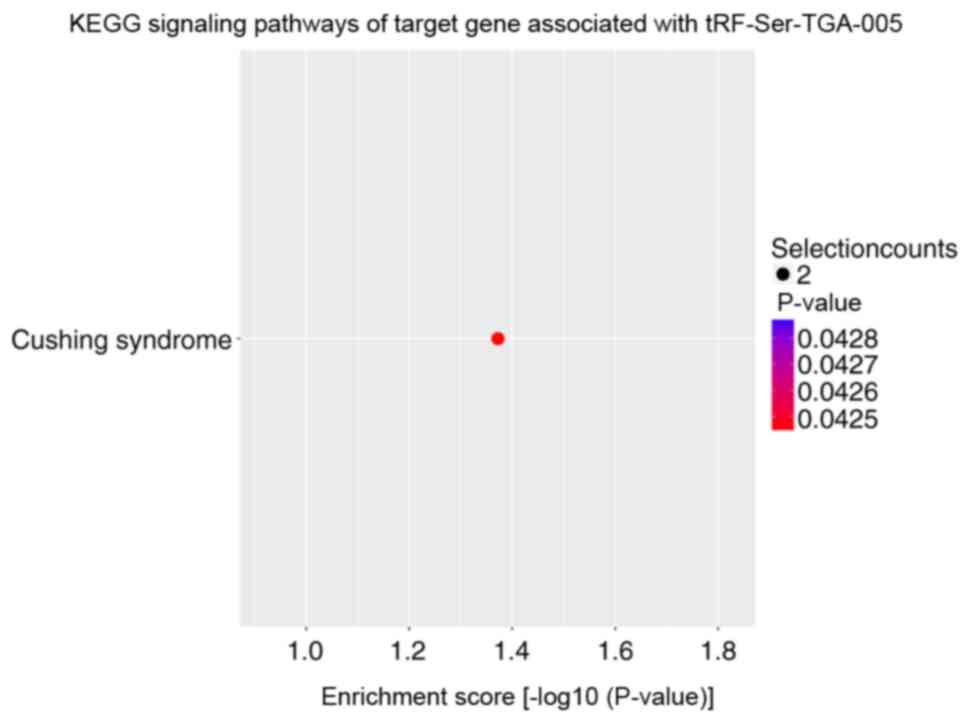
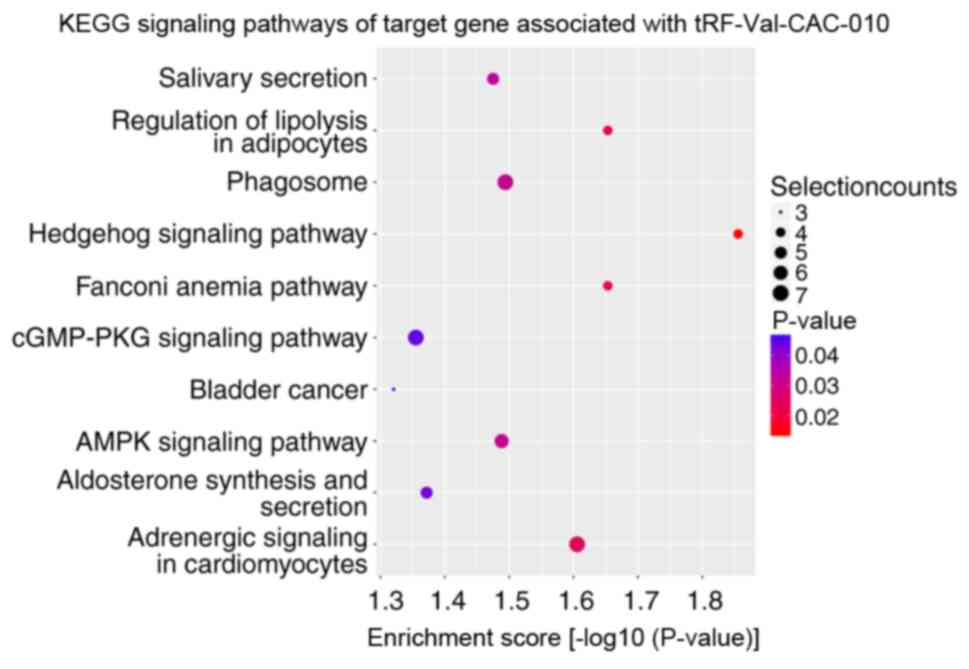
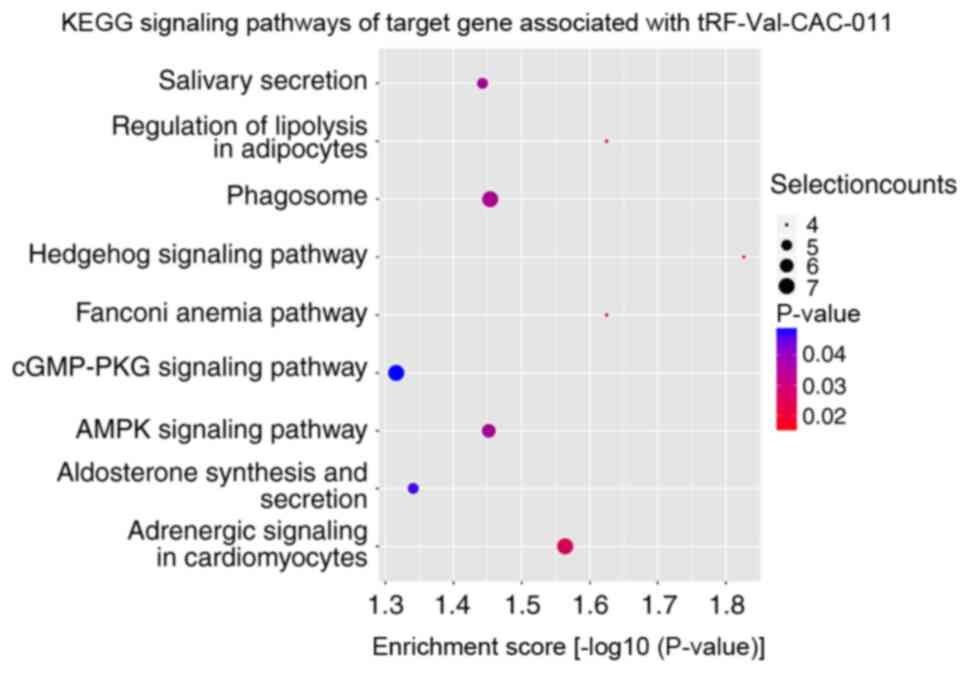
performed. KEGG signaling pathway enrichment analysis revealed that
the altered target genes of differentially expressed tRFs and
tiRNAs were mostly enriched in ‘SNARE interactions in vesicular
transport’ with tiRNA-Lys-CTT-002 (Fig.
5), ‘cushing syndrome’ with tRF-Ser-TGA-005 (Fig. 6) and ‘Hedgehog signaling pathway’
with tRF-Val-CAC-010 and tRF-Val-CAC-011 (Figs. 7 and 8). The significantly enriched GO terms of
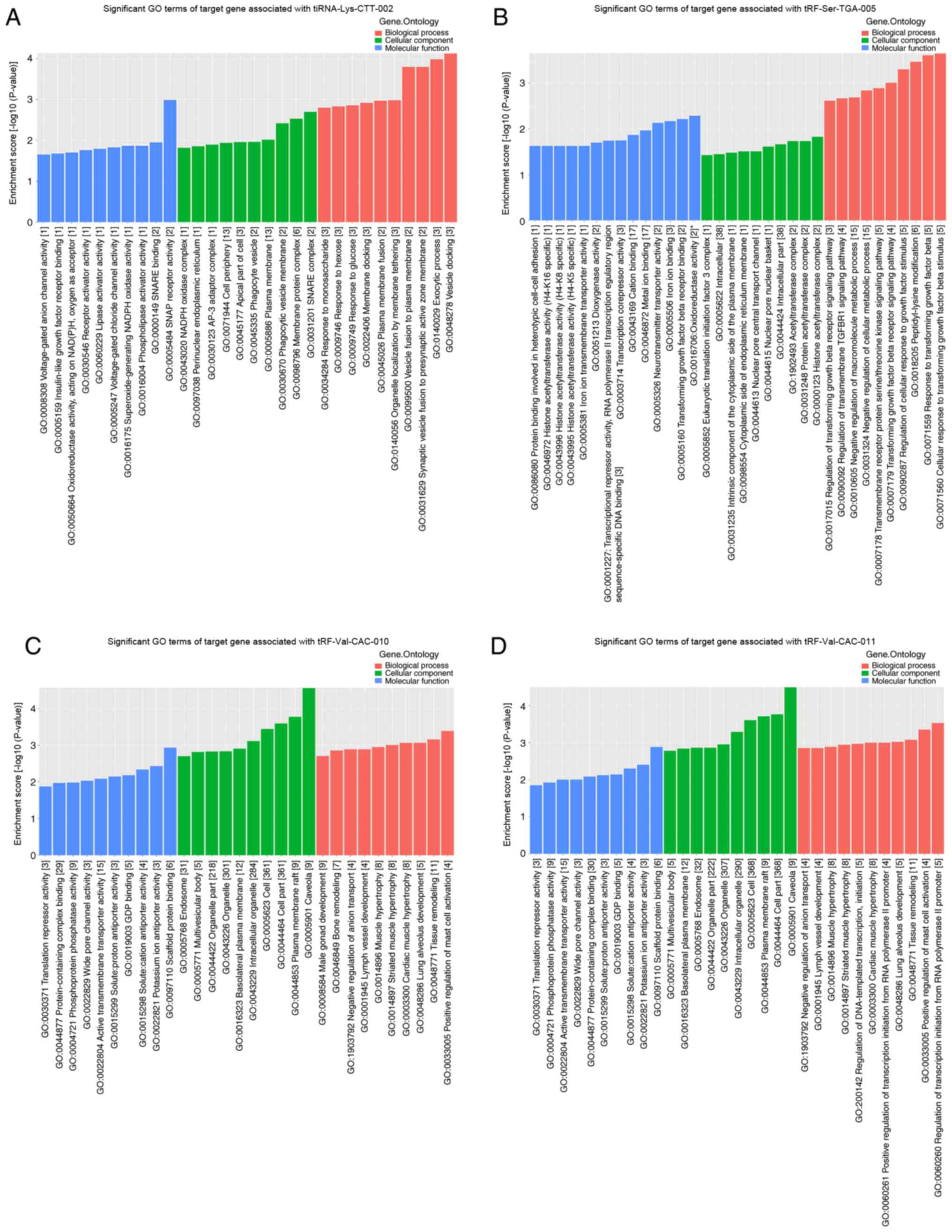
tiRNA-Lys-CTT-002 included ‘vesicle docking’ (GO:0048278),
‘exocytic process’ (GO:0140029) and ‘synaptic vesicle fusion to
presynaptic active zone membrane’ (GO:0031629) (Fig. 9A). The target genes of
tiRNA-Lys-CTT-002 were also enriched in ‘response to glucose’
(GO:0009749), ‘response to hexose’ (GO:0009746) and ‘response to
monosaccharide’ (GO:0034284) (Fig.
9A). Notably, several proliferation-associated terms of
tRF-Ser-TGA-005 were detected, including ‘cellular response to
transforming growth factor β stimulus’ (GO:0071560), ‘response to
transforming growth factor β’ (GO:0071559), ‘regulation of cellular
response to growth factor stimulus’ (GO:0090287), ‘transforming
growth factor β receptor signaling pathway’ (GO:0007179) and
‘regulation of transforming growth factor β receptor signaling
pathway’ (GO:0017015) (Fig. 9B).
Several cellular metabolism-associated terms of tRF-Ser-TGA-005
were enriched, including ‘negative regulation of cellular metabolic
process’ (GO:0031324) and ‘negative regulation of macromolecule
metabolic process’ (GO:0010605) (Fig.
9B). The most significantly enriched GO terms of
tRF-Val-CAC-010 were ‘caveola’ (GO:0005901), ‘plasma membrane raft’
(GO:0044853) and ‘cell part’ (GO:0044464) (Fig. 9C), which were similar to those of
tRF-Val-CAC-011 (Fig. 9D). Moreover,
the terms of tRF-Val-CAC-010 and tRF-Val-CAC-011 included
‘translation repressor activity’ (GO:0030371) and ‘lung alveolus
development’ (GO:0048286) (Fig. 9C and
D).
Discussion
The presence of tRFs and tiRNAs has been detected in
several types of human cells or tissues, including breast cancer
cells (24), ovarian cancer cells
(39) and peripheral blood
mononuclear cells (40). Due to the
lack of oxygen and nutrients in the microenvironment, tumor cells
adapt to the microenvironment through different regulatory
mechanisms to ensure their survival and proliferation (41). The production of tRFs and tiRNAs from
tRNAs under stress is one of the most important pathways of tRF and
tiRNA production. tRFs and tiRNAs have been reported to serve a
role in lung cancer (42,43). Lung adenocarcinoma represents ~40% of
all types of lung cancer and usually evolves from the mucosal
glands (44). Despite recent
improvements in the early diagnosis and clinical treatment
strategies, the prognosis for patients with lung adenocarcinoma
remains poor (44). The development
of lung adenocarcinoma is influenced by a number of factors, such
as post-translational protein modifications, activation of
oncogenes and silencing of tumor suppressor genes (45). Current research has increasingly
focused on the biological function of small non-coding RNAs in
numerous types of disease (42). The
results of the present study demonstrated that the expression
levels of tiRNA-Lys-CTT-002, tRF-Ser-TGA-005, tRF-Val-CAC-010 and
tRF-Val-CAC-011 were significantly different in lung adenocarcinoma
tissues compared with in adjacent tissues. The current findings
suggested that the altered expression levels of tRFs and tiRNAs may
be associated with the tumorigenesis of lung adenocarcinoma.
However, despite breakthroughs in determining the roles of tRFs and
tiRNAs in previous studies (10,43), the
current understanding of the regulatory mechanisms of the majority
of tRFs and tiRNAs remains limited.
Previous studies have reported that tRFs and tiRNAs
are associated with the occurrence of lung cancer (42,43). To
the best of our knowledge, the present study was the first to
report that the expression levels of tiRNA-Lys-CTT-002,
tRF-Val-CAC-010 and tRF-Val-CAC-011 were significantly upregulated,
while those of tRF-Ser-TGA-005 were downregulated in lung
adenocarcinoma tissues. Pekarsky et al (10) found that the expression levels of
ts-4521 (derived from tRNA-Ser) and ts-3676 (derived from tRNA-Thr)
were significantly downregulated in lung cancer tissue samples
compared with matched normal lung tissue samples. Furthermore,
Balatti et al (43)
demonstrated that the low ts-4521 expression was associated with
cell proliferation and apoptosis signaling pathways. The findings
of the aforementioned studies indicated that tRFs and tiRNAs may
participate in lung carcinogenesis.
To determine the potential role of the altered
expression levels of tRFs and tiRNAs, the present study performed
GO functional term and KEGG signaling pathway enrichment analyses
on the target genes of altered tRFs and tiRNAs. Several
proliferation-associated terms were discovered to be associated
with tRF-Ser-TGA-005, including ‘cellular response to transforming
growth factor β stimulus’ (GO:0071560), ‘response to transforming
growth factor β’ (GO:0071559), ‘transforming growth factor β
receptor signaling pathway’ (GO:0007179) and ‘regulation of
transforming growth factor β receptor signaling pathway’
(GO:0017015). During the multistep development of tumors, an
important hallmark is the ability to modify, or reprogram, cellular
metabolism to support cell proliferation (46). The findings of the present analysis
revealed that several cellular metabolism-associated terms were
enriched. For example, the target genes of tRF-Ser-TGA-005 were
enriched in ‘negative regulation of cellular metabolic process’
(GO:0031324) and ‘negative regulation of macromolecule metabolic
process’ (GO:0010605). Several GO terms of tiRNA-Lys-CTT-002 were
also associated with metabolism, such as ‘response to glucose’
(GO:0009749), ‘response to hexose’ (GO:0009746) and ‘response to
monosaccharide’ (GO:0034284). Notably, the target genes of
tRF-Val-CAC-010 and tRF-Val-CAC-011 were also enriched in ‘lung
alveolus development’ (GO:0048286).
It has been established that tRFs and tiRNAs exert
similar functions to miRNAs, which directly bind to target mRNAs to
regulate their stability (21,22). For
instance, 3-tRFs derived from tRNA-Leu-CAG were found to inhibit
protein translation or cleave complementary targets in NSCLC cells
(42). The present study constructed
a network of mRNAs and differentially expressed tRFs and tiRNAs,
and identified the tRF-mRNA pairs involved in the regulation of
lung adenocarcinoma progression, such as CCND1, CXCL1, CXCL2 and
Gli2 associated with tRF-Val-CAC-010 and tRF-Val-CAC-011. CCND1,
alongside cyclin-dependent kinases, drives G1 to S phase
progression through retinoblastoma phosphorylation (47). CXCL1 has been closely associated with
the formation of tumor blood vessels, and it modulates
angiogenesis, tumorigenesis and leukocyte migration (48). The Gli2 protein has been reported to
be involved in cancer progression through canonical regulation of
the Hedgehog signaling pathway (49). Thus, these genes may also be involved
in the development of cancer. The hallmarks of cancer comprise
sustained proliferative signaling, induction of angiogenesis and
activation of invasion and metastasis, amongst others (46). Further investigations into the
underlying mechanisms of differentially expressed tRFs and tiRNAs
and their respective targets in cancer development are
required.
According to a previous study, tiRNAs may decrease
the global translation speed by ~10% (50). Several other studies have reported
that tiRNAs assemble into a G-quadruplex structure that
competitively binds with eukaryotic translation initiation factor
(EIF) 4γ1/EIF4A1 in the translation initiation complex, which in
turn inhibits cap-dependent mRNA translation (25,26). In
the present study, GO functional term enrichment analysis revealed
that target genes of tRF-Val-CAC-010 and tRF-Val-CAC-011 were
enriched in ‘translation repressor activity’ (GO:0030371), which
involves EIF4E binding protein 2 (EIF4EBP2). In a previous study,
EIF4EBP2 was discovered to regulate EIF4E activity by preventing
its assembly into the EIF4A2 complex (51). Therefore, future studies should aim
to determine how tRF-Val-CAC-010 and tRF-Val-CAC-011 may regulate
the translation process.
In conclusion, the present study investigated tRF
and tiRNA profiles in lung adenocarcinoma and adjacent tissues, and
identified several dysregulated tRFs and tiRNAs, whose expression
may be closely associated with the pathogenesis and development of
lung adenocarcinoma. However, the development of lung
adenocarcinoma is a complicated multistep process. Therefore,
further research is required to elucidate the detailed molecular
mechanisms of these tRFs and tiRNAs in lung adenocarcinoma.
Additionally, the present study included a small sample size; thus,
a larger sample size is required to validate the current
findings.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Yunnan Health
Training Project of High Level Talents (grant no. H-2019001), the
National Natural Science Foundation of China (grant no. 81660238),
National Natural Science Foundation of China (grant no. 81660302)
and Yunnan Province Clinical Center for Hematologic Disease (grant
no. 2020LCZXKF-XY13).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository (http://www.ncbi.nlm.nih.gov/geo/; project no.
GSE168270).
Authors' contributions
JHZ and LHL wrote the paper. JHZ, LHL, LLL, XTY and
RL contributed to data interpretation. LLL, XTY and RL contributed
to critical revision of the article. JJZ contributed to the
acquisition of reagents and materials. YXX collected lung
adenocarcinoma fresh tissues. JJZ, YXX, JHZ and LHL performed the
experiments. WPW and SYL designed the study and reviewed the
manuscript. WPW approved the study. JHZ and WPW confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
The First People's Hospital of Yunnan Province (Kunming, China) and
all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Xia C, Zheng R, Zhou M, Lin C,
Zeng H, Zhang S, Wang L, Yang Z, Sun K, et al: Disparities by
province, age, and sex in site-specific cancer burden attributable
to 23 potentially modifiable risk factors in China: A comparative
risk assessment. Lancet Glob Health. 7:e257–e269. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Travis DW, Brambilla E, Burke AP, Marx A
and Nicholson AG: WHO classification of tumours of the lung,
pleura, thymus and heart. 4th edition. Lyon, France: IARC Press;
2015
|
|
4
|
Rolfo C, Castiglia M, Perez A, Reclusa P,
Pauwels P, Sober L, Passiglia F and Russo A: Liquid biopsy in
non-small cell lung cancer (NSCLC). In Liquid Biopsy in Cancer
Patients. 3rd edition. Giordano A, Russo A and Rolfo C: Springer
International Publishing AG; Cham, Switzerland: pp. 103–115.
2017
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Howlader N, Noone AM, Krapcho M, Miller D,
Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, et
al: SEER cancer statistics review, 1975–2013, based on November
2015 SEER data submission, posted to the SEER web site, April 2016,
Bethesda, MD, National Cancer Institute. 2016.
|
|
8
|
Banerjee R, Chen S, Dare K, Gilreath M,
Praetorius-Ibba M, Raina M, Reynolds NM, Rogers T, Roy H, Yadavalli
SS and Ibba M: tRNAs: Cellular barcodes for amino acids. FEBS Lett.
584:387–395. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee YS, Shibata Y, Malhotra A and Dutta A:
A novel class of small RNAs: tRNA-derived RNA fragments (tRFs).
Genes Dev. 23:2639–2649. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pekarsky Y, Balatti V, Palamarchuk A,
Rizzotto L, Veneziano D, Nigita G, Rassenti LZ, Pass HI, Kipps TJ,
Liu CG and Croce CM: Dysregulation of a family of short noncoding
RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci USA.
113:5071–5076. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anderson P and Ivanov P: tRNA fragments in
human health and disease. FEBS Lett. 588:4297–4304. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pliatsika V, Loher P, Telonis AG and
Rigoutsos I: MINTbase: A framework for the interactive exploration
of mitochondrial and nuclear tRNA fragments. Bioinformatics.
32:2481–2489. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zheng LL, Xu WL, Liu S, Sun WJ, Li JH, Wu
J, Yang JH and Qu LH: tRF2Cancer: A web server to detect
tRNA-derived small RNA fragments (tRFs) and their expression in
multiple cancers. Nucleic Acids Res. 44((W1)): W185–W193. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blanco S, Dietmann S, Flores JV, Hussain
S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et
al: Aberrant methylation of tRNAs links cellular stress to
neuro-developmental disorders. EMBO J. 33:2020–2039. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maute RL, Schneider C, Sumazin P, Holmes
A, Califano A, Basso K and Dalla-Favera R: tRNA-derived microRNA
modulates proliferation and the DNA damage response and is
down-regulated in B cell lymphoma. Proc Natl Acad Sci USA.
110:1404–1409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Torres AG: Enjoy the silence: Nearly half
of human tRNA genes are silent. Bioinform Biol Insights.
13:11779322198684542019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu L, Ge J, Li T, Shen Y and Guo J:
tRNA-derived fragments and tRNA halves: The new players in cancers.
Cancer Lett. 452:31–37. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li S, Xu Z and Sheng J: tRNA-derived small
RNA: A novel regulatory small non-coding RNA. Genes (Basel).
9:2462018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumar P, Mudunuri SB, Anaya J and Dutta A:
tRFdb: A database for transfer RNA fragments. Nucleic Acids Res.
43:D141–D145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li S and Hu GF: Emerging role of
angiogenin in stress response and cell survival under adverse
conditions. J Cell Physiol. 227:2822–2826. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karaiskos S, Naqvi AS, Swanson KE and
Grigoriev A: Age-driven modulation of tRNA-derived fragments in
Drosophila and their potential targets. Biol Direct.
10:512015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kumar P, Anaya J, Mudunuri SB and Dutta A:
Meta-analysis of tRNA derived RNA fragments reveals that they are
evolutionarily conserved and associate with AGO proteins to
recognize specific RNA targets. BMC Biol. 12:782014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luo S, He F, Luo J, Dou S, Wang Y, Guo A
and Lu J: Drosophila tsRNAs preferentially suppress general
translation machinery via antisense pairing and participate in
cellular starvation response. Nucleic Acids Res. 46:5250–5268.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goodarzi H, Liu X, Nguyen HC, Zhang S,
Fish L and Tavazoie SF: Endogenous tRNA-derived fragments suppress
breast cancer progression via YBX1 displacement. Cell. 161:790–802.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ivanov P, Emara MM, Villen J, Gygi SP and
Anderson P: Angiogenin-induced tRNA fragments inhibit translation
initiation. Mol Cell. 43:613–623. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ivanov P, O'Day E, Emara MM, Wagner G,
Lieberman J and Anderson P: G-quadruplex structures contribute to
the neuroprotective effects of angiogenin-induced tRNA fragments.
Proc Natl Acad Sci USA. 111:18201–18206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saikia M, Jobava R, Parisien M, Putnam A,
Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, et al:
Angiogenin-cleaved tRNA halves interact with cytochrome c,
protecting cells from apoptosis during osmotic stress. Mol Cell
Biol. 34:2450–2463. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi
J, Feng GH, Peng H, Zhang X, Zhang Y, et al: Sperm tsRNAs
contribute to intergenerational inheritance of an acquired
metabolic disorder. Science. 351:397–400. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Langmead B, Trapnell C, Pop M and Salzberg
SL: Ultrafast and memory-efficient alignment of short DNA sequences
to the human genome. Genome Biol. 10:R252009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Friedländer MR, Mackowiak SD, Li N, Chen W
and Rajewsky N: miRDeep2 accurately identifies known and hundreds
of novel microRNA genes in seven animal clades. Nucleic Acids Res.
40:37–52. 2012. View Article : Google Scholar
|
|
31
|
Robinson MD, McCarthy DJ and Smyth GK:
EdgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fu Y, Lee I, Lee YS and Bao X: Small
non-coding transfer RNA-Derived RNA fragments (tRFs): Their
biogenesis, function and implication in human diseases. Genomics
Inform. 13:94–101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou K, Diebel KW, Holy J, Skildum A,
Odean E, Hicks DA, Schotl B, Abrahante JE, Spillman MA and Bemis
LT: A tRNA fragment, tRF5-Glu, regulates BCAR3 expression and
proliferation in ovarian cancer cells. Oncotarget. 8:95377–95391.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu H, Chen W, Zheng F, Tang D, Dai W,
Huang S, Zhang C, Zeng J, Wang G and Dai Y: The potential role of
tRNAs and small RNAs derived from tRNAs in the occurrence and
development of systemic lupus erythematosus. Biochem Biophys Res
Commun. 527:561–567. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Keith B and Simon MC: Hypoxia-inducible
factors, stem cells, and cancer. Cell. 129:465–472. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shao Y, Sun Q, Liu X, Wang P, Wu R and Ma
Z: tRF-Leu-CAG promotes cell proliferation and cell cycle in
non-small cell lung cancer. Chem Biol Drug Des. 90:730–738. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Balatti V, Nigita G, Veneziano D, Drusco
A, Stein GS, Messier TL, Farina NH, Lian JB, Tomasello L, Liu CG,
et al: tsRNA signatures in cancer. Proc Natl Acad Sci USA.
114:8071–8076. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Myers DJ and Wallen JM: Lung
adenocarcinoma. National Institutes of Health 2020.
|
|
45
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kato J, Matsushime H, Hiebert SW, Ewen ME
and Sherr CJ: Direct binding of cyclin D to the retinoblastoma gene
product (pRb) and pRb phosphorylation by the cyclin D-dependent
kinase CDK4. Genes Dev. 7:331–342. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Amiri KI and Richmond A: Fine tuning the
transcriptional regulation of the CXCL1 chemokine. Prog Nucleic
Acid Res Mol Biol. 74:1–36. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hui CC and Angers S: Gli proteins in
development and disease. Annu Rev Cell Dev Biol. 27:513–537. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yamasaki S, Ivanov P, Hu GF and Anderson
P: Angiogenin cleaves tRNA and promotes stress-induced
translational repression. J Cell Biol. 185:35–42. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Martineau Y, Azar R, Bousquet C and
Pyronnet S: Anti-oncogenic potential of the eIF4E-binding proteins.
Oncogene. 32:671–677. 2013. View Article : Google Scholar : PubMed/NCBI
|