Breast cancer is a heterogeneous disease with
varying biological and clinical characteristics. It is the most
common cancer among women worldwide, accounting for 25% of all
cancer cases (1). According to
GLOBOCAN 2020, the incidence and mortality of breast cancer
reported worldwide were 34,65,951 new cases and 11,21,413 deaths,
respectively; in India, 1,204,532 new cases and 436,417 deaths were
recorded in 2020 (2).
Immunohistochemical analysis of breast tumors is the
gold-standard method used in clinics to classify them based on the
hormone receptor expression for improved therapeutic decisions.
Based on this, breast cancer can be broadly grouped into five
types, namely: i) Progesterone receptor (PR)-positive, estrogen
receptor (ER)-positive and human epidermal growth factor 2
(Her2)-negative (luminal A); ii) ER-positive, PR-positive/negative
and Her2-positive (luminal B); iii) Her2-overexpressing, ER- and
PR-negative; iv) ER-, PR- and Her2-negative (basal-like or
triple-negative), and v) normal-like (expression status similar to
luminal A and resemble normal breast profile) (3–5).
Additionally, molecular breast cancer analysis identified a
distinctive phenotype with low claudin expression, immune receptor,
and EMT markers expression (6).
Cancer types with the claudinlow phenotype are highly
metastatic and associated with poor prognosis (7). Her2-overexpressing cancer also displays
high metastasis and poor prognosis (8). Among ER-positive subtypes, luminal B is
associated with a significantly worse prognosis than luminal A
(9,10). Patients with basal subtypes of cancer
with BRCA1 mutations have a poor prognosis (9).
Based on specific gene expression patterns, breast
cancers are categorized into five intrinsic or molecular subtypes.
Among the intrinsic subtypes, basal-like triple-negative breast
cancer (TNBC) accounts for 12–20% of breast cancers (11). TNBC has drawn specific attention due
to the lack of expression of all three receptors (ER, PR, and
Her2). Thus, it cannot be treated using anti-estrogen hormonal
therapies or trastuzumab (12).
Morphologically, TNBC is characterized by hyperdense masses without
calcification, usually occurring in women <50 years of age.
Histological features include significant lymphocyte infiltration,
central necrosis, pushing tumor borders, and fibrosis (13). Cytokeratins, fascin, epidermal growth
factor receptor (EGFR), caveolin, and vimentin are usually
expressed in basal-like TNBC (14,15).
TNBC is challenging to treat, as it is quite complex due to poor
cell differentiation, molecular heterogeneity, and rapid
metastasis, often leading to chemoresistance and recurrence of the
disease (16). Fast relapse and
invasions are common features of TNBC tumors and show poor
prognosis (17). Recent advances in
omics technologies have provided insight into the molecular
mechanisms underlying TNBC (18).
The present review focuses on the different subtypes of TNBC and
therapeutic approaches currently employed in the treatment of
TNBC.
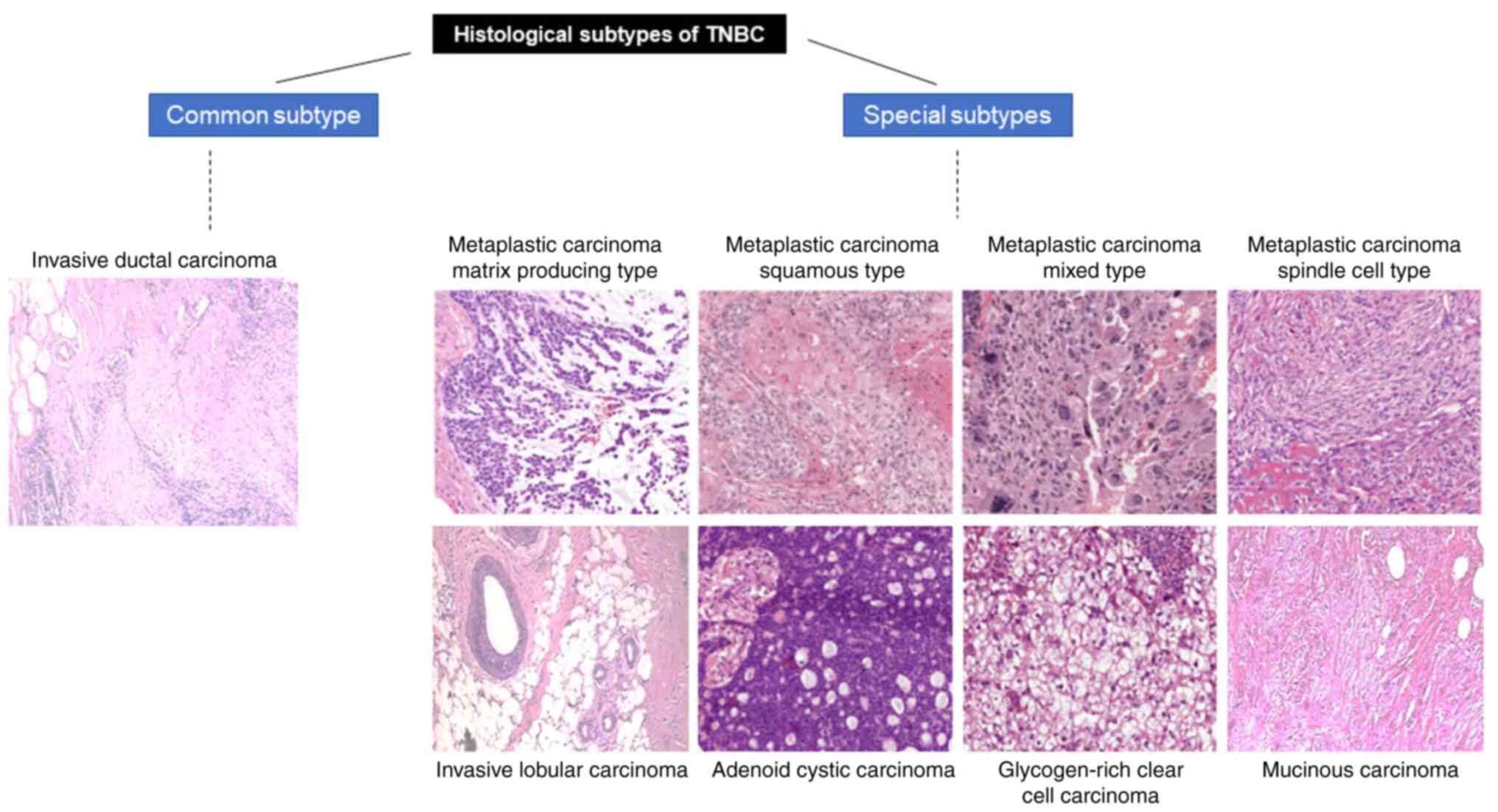
Among the histological subtypes, adenoid cystic
carcinoma has a median recurrence of only 2 months, and metaplastic
carcinoma has ~9.9 months (33),
compared to IDC-NST and matrix-producing metaplastic carcinoma,
which are less aggressive, with 34 and 31.4 months of median time
to recurrence, respectively (20).
Although the histological assessments were pointing
to the presence of WBCs in and around the TNBC subtypes, focus on
the presence of WBCs has led to the identification of TILS and
TAMs, which are the parameters defining prognosis and therapy of
TNBC. The TNBCs might be immunogenic due to mutations that lead to
aberrant protein expression on the cell membrane (34). Tumor-infiltrating lymphocytes (TILs)
are white blood cells that migrate towards the tumor from the
bloodstream via the newly formed blood vessels (angiogenesis),
which cancer cells use for their nutritional and oxygen
requirements (35). They consist of
a mixture of B cells, macrophages, natural killer cells and are
dominated by T cells (35). TILs are
present in ~20% of TNBC tumors and carry a pivotal prognostic and
predictive value (36). The presence
of TILs indicates a good prognosis (37). High number of TILs indicate that
there is an equilibrium between the immune status and cancer
(38). The ratio of cancer cells:
TILs is tilted towards TILs after surgical removal of a tumor,
resulting in an improved prognosis in TNBC (38). A high mutation load and clonal
heterogeneity are associated with a low number of TILs, which may
provide an escape route to tumor cells from immune surveillance
(39). However, in addition to TILs,
the tumor microenvironment components also influence the outcome of
patients with TNBC (39). Relapsing
patients with TNBC have been shown to have low levels of TILs and a
high number of CD163+ tumor-associated macrophages
(TAMs) compared with that of patients without relapse (39). High levels of CD8+ T cells
may reflect improved sensitivity to chemotherapy, whereas high
levels of TAMs correlate with poor patient outcomes (36). Nevertheless, a previous study in TNBC
has reported paradoxical findings, with high levels of
CD8+ T cells in the tumor stroma leading to the low
infiltration of the tumor epithelium, thereby indicating a poor
outcome (40). Therefore,
immunohistological assessment for TILS or TAMS will help develop
immunotherapies detailed in section 7.
Profiling based on gene expression has led to
improved insight into tumor heterogeneity at the molecular level
and has generated an impartial classification (Fig. 1). The PAM50 microarray set of 50
genes is used to identify breast cancer intrinsic subtypes
(41). A set of 374 TNBC samples
taken from 14 microarray datasets was analyzed to characterize TNBC
subtypes using PAM50. The results from this analysis categorized
most of the TNBC as basal-like (80.6%). The rest of the tumours
were classified as Her2-positive(0.2%), normal-like (14.6%),
luminal B (3.5%) and luminal A (1.1%) (Table I) (41).
Genomic/transcriptomic data from a set of 997
primary tumors were extracted, and an integrated analysis was
performed by Curtis et al (47). A set of 995 tumors from the Molecular
Taxonomy of Breast Cancer International Consortium (METABRIC)
cohort was used as a validation set that divided TNBC into ten
groups, named Integrated Clusters (IntClust) 1–10 (47). Basal-like breast cancer mostly fell
in IntClust 4 and 10 (~80%). IntClust 4 is known to have greater
TIL counts, while IntClust 10 subtype can display genomic
instability and chromosomal aberrations (Table I) (47–49).
Through whole-exome and whole-genome data, it is
evident that most of the genetic alterations in TNBC are copy
number alterations and somatic mutations (40). The BRCA1 and BRCA2 tumor suppressor
genes are required for the maintenance of genomic stability. These
genes play a role in DNA repair and replication error control
(50,51). A total of 10% of patients with TNBC
are known to harbor germline mutations in BRCA1 or BRCA2 (12,26,27). The
lifetime risk of breast cancer becomes 60–70% in the presence of
such mutations (52). Gene
alterations leading to homologous recombination (HR) defects other
than germline BRCA mutations are termed ‘BRCAness’ (53). Moreover, ~35% of TNBC tumors show
abnormalities in the HR pathway, making them sensitive to poly
(ADP-ribose) polymerase (PARP) inhibitors and DNA-damaging agents
(54).
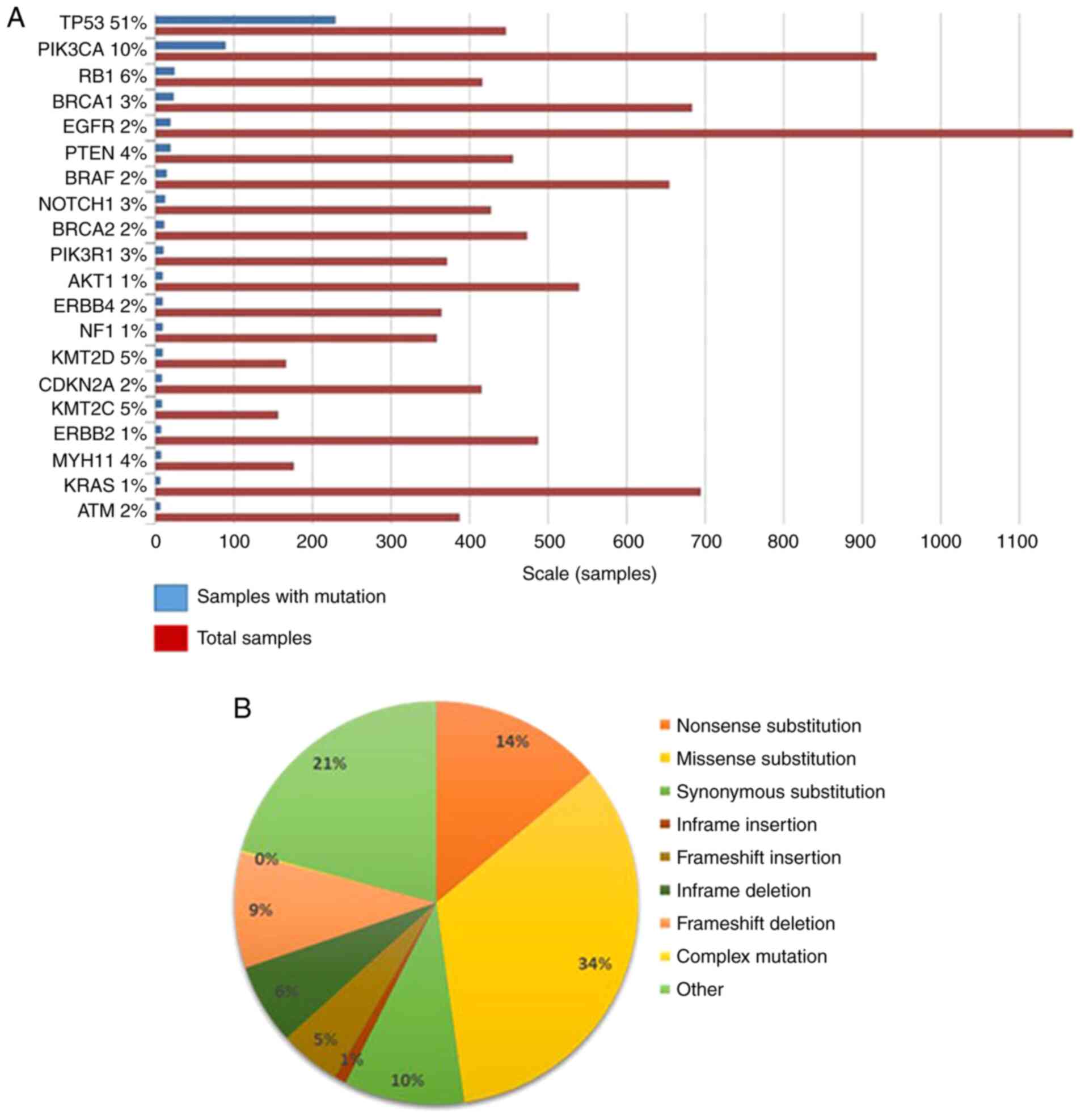
Other common mutations observed in TNBC patients
include those in TP53 (50–60%) and PIK3CA (~10%) (18,42). An
analysis from the Catalogue of Somatic Mutations in Cancer (COSMIC)
database revealed that the top genes mutated in TNBC, apart from
BRCA1/2, TP53, and PIK3CA, were RB1, PTEN, NOTCH1 and BRAF
(Fig. 2A). Among the point mutations
observed, 34% of them were nonsense substitutions (where a base
change leads to a stop codon in the DNA sequence), 21% were
synonymous mutations (where a change in a base in the exon of a
coding gene does not change the structure of the protein) (Fig. 2B). The rest of the mutations were
missense mutations, frameshift insertion/deletions, and in-frame
insertions/deletions. In the metastatic disease setting, genes from
HR repair showed a larger frequency of biallelic loss-of-function
mutations than in early TNBC (55).
Recently, much focus has been put on bringing liquid
biopsies, such as circulating tumor cells (CTC) and circulating
tumor DNA (ctDNA), into the clinical setting for diagnostic and
prognostic use (59). CTCs are
nucleated cancer cells present in the bloodstream that can be
detected using techniques, such as reverse
transcription-quantitative PCR, flow cytometry, and
immunohistochemistry (60). Tumor
cells that undergo necrosis or apoptosis release DNA fragments into
the plasma are referred to as ctDNA (61). In breast cancer, ctDNA and CTCs have
been studied as potential biomarkers for prognosis (60). Stover et al (62) performed studies in metastatic breast
cancer patients receiving chemotherapy and identified an
association between CTCs and ctDNA and tumor burden, indicating
that these could be used to measure early-treatment response in
patients. A retrospective study in 164 patients with metastatic
TNBC revealed that >10% of patients with ctDNA had worse
disease-free survival (62). A study
by Bidard et al (63), with
metastatic breast cancer, revealed that patients with CTC levels
>5 per 7.5 ml were associated with lower progression-free
survival (PFS) and OS compared with patients who had CTC levels
<5 per 7.5 ml. Cristofanilli et al (64) reported that CTC counts could be
utilized to classify metastatic patients into two groups. Patients
with CTCs levels >5 per 7.5 ml were categorized as aggressive
stage IV and those <5 per 7.5 ml as indolent stage IV (64). ctDNA has been associated with
chemotherapy in studies by Riva et al (65), in which ctDNA-positive patients
before and after chemotherapy experienced poor OS and disease-free
survival (DFS). Additionally, Radovich et al (66) reported that patients with early-stage
TNBC and positive ctDNA after chemotherapy had a higher risk of
disease relapse. Therefore, liquid biopsies are being developed as
a non-invasive method to study recurrence, treatment response, and
survival in the clinical setting.
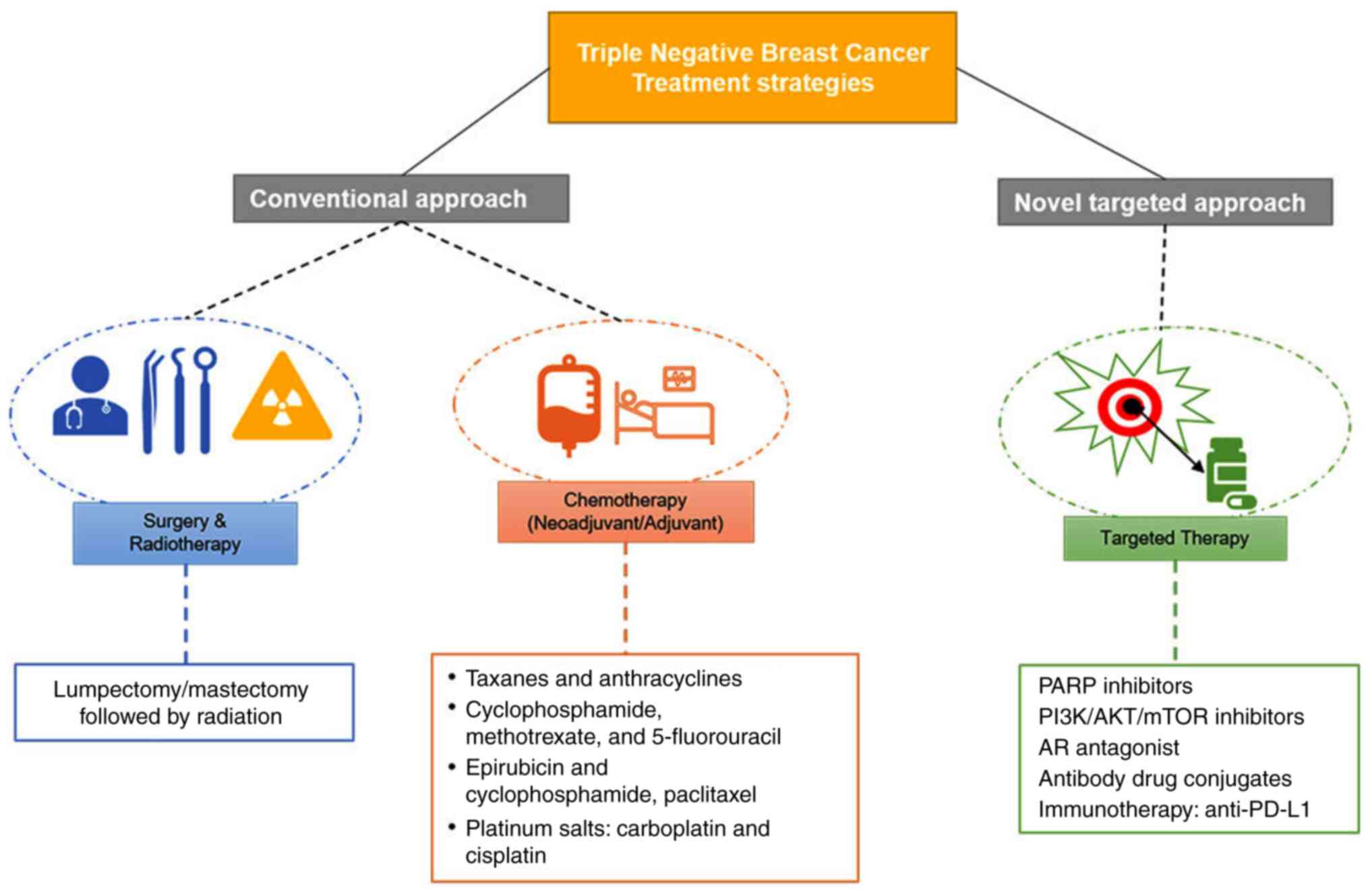
TNBC treatment involves a combination of surgery,
radiotherapy, and chemotherapy. New methods, such as targeted
therapy and immunotherapy, have been developed to improve patient
survival and prognosis. Lumpectomy and mastectomy are the surgical
procedures performed for TNBC patients and are usually followed by
radiotherapy and chemotherapy (67).
Neoadjuvant therapy is given before the surgery, which may help
shrink the tumor size and avoid mastectomy (Fig. 3) (61). Taxanes and anthracyclines form the
current standard of care for TNBC in both the neoadjuvant and the
adjuvant settings. Epirubicin and doxorubicin are the most common
anthracyclines (anticancer antibiotics known to disrupt DNA
replication and mitochondrial functions to activate apoptosis)
(68,69). Taxanes block angiogenesis by
inhibiting epidermal growth factor receptor signaling (70).
Paclitaxel and docetaxel are familiar examples of
taxanes used in the first line of therapy (71). TNBC shows a 40% pathological complete
response (pCR) for taxane and anthracycline-based therapy in the
neoadjuvant setting (72–74). Adjuvant therapy guidelines are
usually identical for all the subtypes of breast cancer and TNBC.
Chemotherapy in the adjuvant setting is recommended for tumors
>0.5 cm in size, as they exhibit increased aggressiveness, with
a faster growth rate and metastasis (75). Anthracycline chemotherapy
(cyclophosphamide and 5-fluorouracil) in patients with metastatic
TNBC exhibited a response to survival within 22 months (69). However, acute toxicity is a major
concern with anthracycline-based chemotherapy (76). Metastatic patients who develop
resistance to anthracycline have shown sensitivity to capecitabine,
gemcitabine and vinorelbine (77–79). The
combination of docetaxel with capecitabine has improved the OS of
patients with metastatic TNBC (78).
Carboplatin and cisplatin are platinum salts that
are used in the treatment of TNBC. These generate DNA lesions, and
apoptosis occurs in cells unable to repair these breaks (80). For TNBC, carboplatin as a neoadjuvant
addition increases the response rate from 37 to 52.1% (81). A phase-II study of 86 patients
evaluating the efficacy of platinum monotherapy demonstrated a 32%
overall response rate (ORR) for cisplatin and 19% for carboplatin
in early TNBC. Patients with BRCA1/2 mutations showed an improved
response compared with patients without BRCA1/2 mutations (82). Moreover, phase-II trials showed an
improved ORR of 72% in metastatic patients with BRCA mutation with
neoadjuvant cisplatin monotherapy (83,84).
Recently, the PEARLY trial (NCT02441933) has explored combination
therapy of taxanes and carboplatin in the neoadjuvant setting
(85). Carboplatin with docetaxel or
paclitaxel combination has demonstrated promising efficacy in
patients with TNBC and brain metastasis (86). Although TNBC is sensitive to
chemotherapy, early relapse is a major concern (75). Therefore, optimizing a tailored
standard regime to address chemotherapy issues, such as toxicity,
and relapse has led to customizing personalized therapy based on
tumor type.
Therapies targeted to TNBC are being developed based
on the expression of specific pathways and genes. Targeted therapy
focuses on customizing cancer therapy to an individual patient's
tumor (87,88). TNBC being heterogenous, targeting
alterations specific to the tumor would be the most effective
treatment option. A study using genomics and transcriptomics has
led to identifying molecular markers that could be effectively
targeted in TNBC (89). PARP
inhibitors, PI3K/AKT inhibitors, and anti-androgen therapy are
under clinical investigation (Fig.
3) (58).
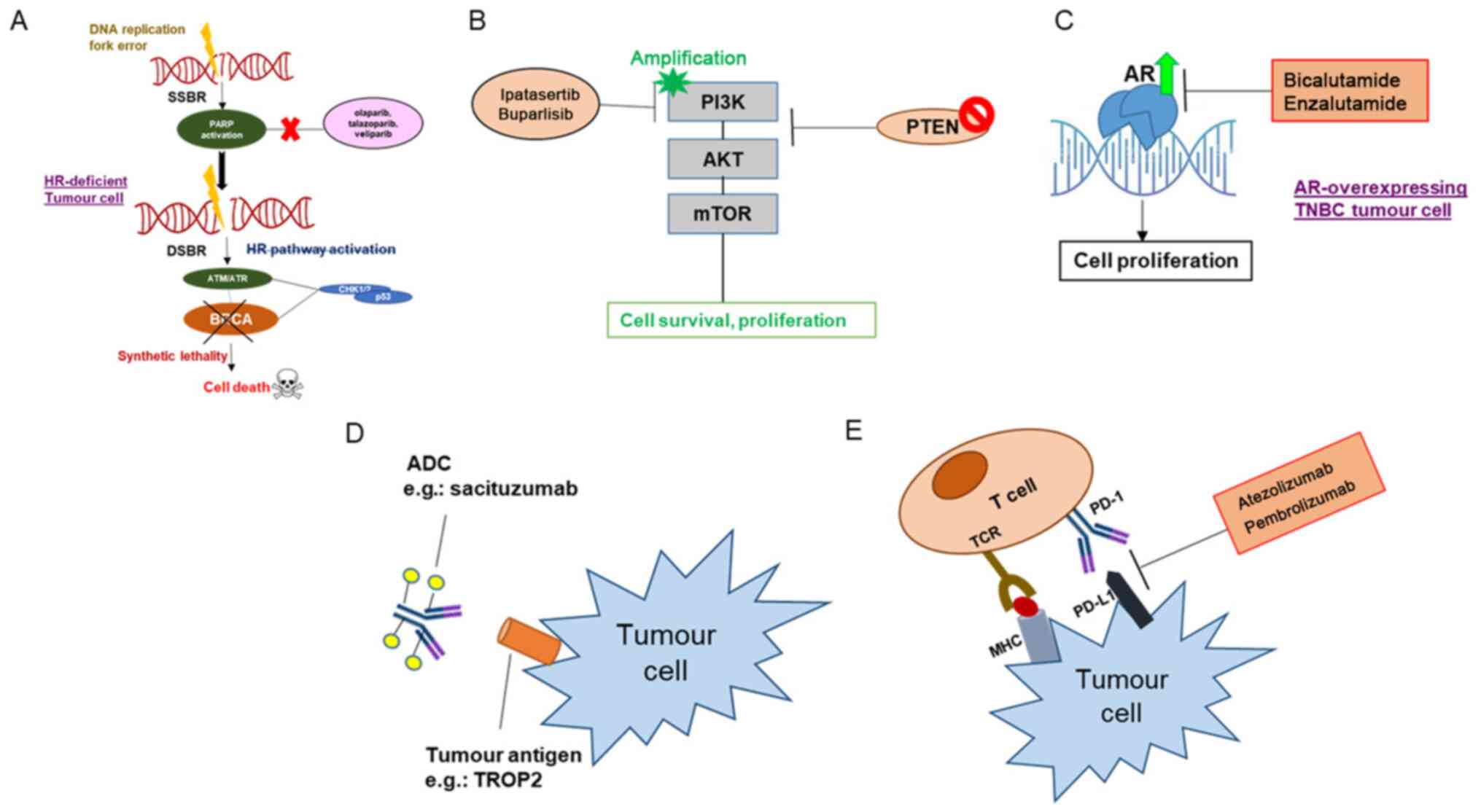
PARP is expressed in ample amounts as a nuclear
enzyme that plays a critical role in DNA repair, cell
proliferation, and signaling. It transfers ADP-ribose to target
proteins from NAD+ and ribosylates them (90). In response to DNA damage, PARP is
known to activate the DNA repair process through poly
(ADP)-ribosylation of multiple nuclear proteins that play a role in
chromatin architecture and DNA metabolism (91). Therefore, PARP inhibition leads to
the accumulation of double-strand breaks (DSBs) in cells undergoing
replication. The presence of wild-type BRCA1/2 in cells results in
a homologous recombination mode of repair of DSBs. However, in the
cells deficient of BRCA1/2, homologous recombination is disrupted,
and PARP repairs the breaks (92–94).
Therefore, in these BRCA1/2-deficient cases, inhibiting PARP will
result in severe, selective toxicity called ‘synthetic lethality’
(95). Using PARP inhibitors in
treatment sensitizes the tumor cells to chemotherapy and
radiotherapy, causing synthetic lethality in patients with
hereditary BRCA1/2 mutations identified in several TNBC subtypes
(Fig. 4A) (96).
Olaparib and talazoparib are two of the PARP
inhibitors approved by the United States Food and Drug
Administration (FDA) for use in patients with deficient BRCA1/2 in
metastatic Her2-negative breast cancer as a single agent, based on
the phase-III OlympiAD and EMBRACA clinical trials (86–88).
Patients with a germline BRCA1/2 mutation (gBRCA1/2+)
with metastatic breast cancer were grouped into 2:1 to olaparib vs.
chemotherapy (capecitabine, eribulin, or vinorelbine) of
physician's choice in OlympiAD trial (NCT02000622) (97,98). The
ORR was 59.9% in the TNBC patient subgroup for olaparib (n=102) and
29.9% in the case of patients who underwent chemotherapy (n=48).
Olaparib showed less toxicity in tumorgrade3 and 4 patients than
the chemotherapy arm (98,99). In the EMBARCA trial (NCT01945775),
gBRCA1/2+ metastatic patients were given 2:1 Talazoparib
1 mg daily vs. chemotherapy of physician's choice. The ORR was
62.6% in patients given with Talazoparib (n=219) and 27.2% in
patients treated with chemotherapy (n=144) (99). Several other PARP inhibitors are
currently under phase-II/III clinical trials, including veliparib
(NCT02163694) and niraparib (NCT01905592) (100–103).
PARP inhibitors are being investigated in
combination with chemotherapy and immunotherapy. BrighTNess trial
(NCT02032277) is a phase-III trial for stage-II and -III TNBC
evaluating the combination of carboplatin with the PARP inhibitor
veliparib followed by doxorubicin (104). The ongoing phase-I/II trial
(MEDIOLA trial) involves a combination of olaparib and anti-PDL1
checkpoint inhibitor durvalumab (105). The phase-III OlympiA trial
(NCT02032823) for early TNBC is currently assessing patients with
BRCA1/2 mutation treated with olaparib as monotherapy following
neoadjuvant chemotherapy (106).
PARTNER (NCT03150576) (107) is a
phase-II/III trial that is currently ongoing checking the efficacy
of olaparib and carboplatin combination in a neoadjuvant setting
(107). Table II summarizes the clinical trials
taken from clinicaltrials.gov.
PI3K/AKT pathway is involved in cell growth and
glucose metabolism. Under normal conditions, growth factors, such
as insulin-activated receptor tyrosine kinases (RTKs) result in
PI3K activation (108). This is
followed by phosphorylation of
phosphatidylinositol-4,5-trisphosphate (PIP2) by PI3K and
conversion to phosphatidylinositol-3,4,5-trisphosphate (PIP3)
(109). AKT binds to membrane-bound
PIP3, bringing AKT close to phosphoinositide-dependent kinase 1
(PDK1) (110). PDK1 phosphorylates
AKT resulting in the activation of multiple downstream pathways
like cell growth, cell cycle, and metabolic pathways. This pathway
is negatively regulated by the PTEN phosphatase (108,109,111).
In TNBC, this pathway is active in 9.6% of patients due to the loss
of PTEN activity (110) (Fig. 4B). Therefore, studies using PI3K
inhibitors have been conducted in patients with TNBC (Table II), such as the LOTUS trial
(NCT02162719), which is a phase-II trial evaluating ipatasertib in
124 patients (ORR in the PTENlow group was 48% compared
with patients with PTEN expression) (112). The oral pan-PI3K inhibitor
buparlisib has also been used in combination with paclitaxel in a
phase-II trial (NCT01572727) involving metastatic Her2-negative
patients; the ORR was 22.6% compared with placebo and paclitaxel
(113).
Capivasertib and AZD5363 are AKT inhibitors that are
currently being investigated for PFS in patients with metastatic
TNBC in the CAPItello-290 (NCT03997123) and PAKT (NCT02423603)
trials, respectively (114,115). In the Phase-II trial under
neoadjuvant setting, mTOR inhibitor and chemotherapy combined did
not show any effect in early TNBC treatment (116). The mTOR inhibitors temsirolimus or
everolimus in combination with doxorubicin and bevacizumab
displayed an objective response rate of 21% in mesenchymal subtype
of TNBC (117).
AR belongs to the nuclear steroid hormone family of
receptors, is highly expressed in the LAR subtype of TNBC (118). AR antagonists have shown an effect
in vitro and in vivo in the LAR type (Fig. 4C). Gucalp et al (119) used the AR inhibitor bicalutamide in
a phase-II trial involving 424 AR-positive patients, which showed a
clinical benefit rate of 19% and a median progression-free survival
of 12 weeks (119). Among the
ongoing clinical trials, Bicalutamide treatment response is being
compared to standard chemotherapy in patients with metastatic TNBC
in an ongoing phase-III as the first line of therapy (NCT03055312).
Enzalutamide is another AR antagonist with which a phase-II trial
(NCT01889238) was conducted in AR-positive patients with advanced
TNBC, in which a clinical benefit of 25% was observed (120). Androgen-driven gene expression
signature (Dx-signature) stratified patients into a Dx-positive and
a Dx-negative group. Dx-positive patients had an improved response
to enzalutamide compared with Dx-negative patients (120,121).
In AR-positive patients with early-stage TNBC, enzalutamide is
currently under investigation both as a monotherapy (NCT02750358)
and in combination with paclitaxel (NCT02689427). Around 40% of
AR-positive TNBC patients show activation of the PI3K-AKT pathway
(122). Therefore, the combined
effect of enzalutamide and the PI3K inhibitor taselisib was
evaluated in the TBRC032 trial(NCT02457910) where CBR was 35.7%
(123). Further details are
provided in Table II.
Antibody-drug conjugates (ADC) are made up of a
linker, an inhibitor, and an antibody. The antibody is selected to
be specific to cell surface molecules of cancer cells and not
normal cells. The payload of cytotoxic agents must be potent to
kill the cancer cell. Usually, a stable molecule is used as a
linker that will bind strongly to the inhibitor (124,125)
(Fig. 4D). Elevated expression of
tumor-associated calcium-linked signal-transducer two cell surface
glycoprotein (Trop-2) has been reported in TNBC and often
correlated with poor prognosis (126). Sacituzumab Govitecan (IMMU-132) is
an ADC used to target Trop-2 that delivers a topoisomerase-I
inhibiting payload resulting in DSBs. Bardia et al (127) conducted a phase-I/II study
involving patients with advanced-stage TNBC who had previously
received two lines of treatment, and the ORR was 33.33%. A
phase-III study (NCT02574455) of sacituzumab govitecan in relapsed
patients with TNBC is ongoing. SKB264 is another anti-Trop2
currently under investigation in the NCT04152499 phase-I trial with
metastatic TNBC patients (128).
Another ADC, ladiratuzumab vedotin, an immunoglobulin G1 antibody
with a microtubule inhibitor (MMAE), has shown an ORR of 25% of
patients with TNBC (129).
In addition to PARP and PI3K inhibitors, inhibitors
of other molecular targets are being investigated in TNBC. HDAC
inhibitors are currently being investigated as monotherapy
(NCT02623751) and in combination with cisplatin (NCT02393794).
Various Ataxia Telangiectasia and Rad3-Related Protein (ATR) and
Wee inhibitors are also in clinical trials for TNBC (1). MEK inhibitors and inhibitors of cell
cycle-regulating agents, such as Aurora kinase, showed antitumor
effects in animal xenografts (130,131).
Palbociclib, a cyclin-dependent kinase 4/6 inhibitor, was used in a
phase-I study along with paclitaxel in patients with metastatic
TNBC (n=9). Clinical benefit was experienced in one-third of the
patients (132). BCL2 inhibitors in
TNBC cell lines have shown to decrease cell proliferation (133). In TNBC cells, BCL2 expression is
high (134). Therefore, BCL2
inhibitors should be further investigated for their impact as
monotherapy and in combination.
In the last decade, substantial evidence has been
generated describing the immune system's role in guiding the
disease progression of TNBC (135).
It is one of the rapidly progressing areas of breast cancer
research. The T cell receptor (TCR) recognizes antigen presented on
major histocompatibility complex molecules by cancer cells
(136). It is followed by signaling
from co-stimulatory factors such as CD28, modulated by
immune-checkpoint (co-inhibitory) molecules (137). In TNBC, programmed death-ligand 1
(PD-L1) functions as a critical mediator of the balance and escape
stages of cancer immunoediting (138–140).
Around 20% of TNBC tumors express PD-L1, which is associated with
poor prognostic features, such as higher grade, HER2-positive
status, ER-negative status and large tumor size (141). Quantification of PD-L1 can be
carried out on immune cells or tumor cells using
immunohistochemistry (141–143). Studies have suggested that PD-L1
expression varies depending on the stage of TNBC and cell type
(141–143). Expression of PD-L1 in TNBC has been
associated with improved pCR (50% vs. 21%) (39,144).
Along with PD-L1, TILs are also high in number in TNBC (144,145).
TILs are considered to be a good prognostic marker in TNBC
(146). Inhibitors of PD-1/PD-L1
block the interaction between PD-1 and PD-L1, thereby initiating a
positive immune response that results in tumor killing (123). Over the last few years, immune
checkpoint inhibitors (CPIs) have been in the limelight due to
improved efficacy shown during clinical trials (Fig. 4E). Pembrolizumab (NCT04191135 and
NCT01042379), nivolumab (NCT03818685 and NCT03414684), atezolizumab
(NCT03281954 and NCT03498716) and durvalumab (NCT03167619 and
NCT03616886) are some of the CPIs currently used in ongoing
clinical trials for TNBC (147).
The IMpassion130 trial (NCT02425891) evaluated the use of
atezolizumab with paclitaxel as the first line of therapy for
patients with metastatic TNBC (n=901), showing PD-L1 positivity.
Atezolizumab is a PD-LA inhibitor that blocks the interaction
between PD-L1 and PD-1, thereby promoting T cell activity. It is
now an FDA-approved drug for PD-L1-positive patients with TNBC
(148). The KEYNOTE-119 phase-III
clinical trial (149) evaluated
pembrolizumab's effect as monotherapy in patients with metastatic
TNBC vs. physician's choice chemotherapy (capecitabine,
vinorelbine, gemcitabine, or eribulin). The OS of this study was
not encouraging (149). In the
recent trial KEYNOTE-355 (NCT02819518), PD-L1-positive patients
with metastatic TNBC showed improved PFS when pembrolizumab was
given in combination with chemotherapy, in comparison with patients
given chemotherapy alone (150).
Currently, two trials, IMpassion131 (NCT03125902) and IMpassion132
(NCT03371017), are being carried out: The former is investigating
the outcomes for paclitaxel and atezolizumab in untreated
metastatic patients who are PD-L1 positive, while the latter is for
atezolizumab along with chemotherapy (gemcitabine, capecitabine and
carboplatin) in early relapsing recurrent patients with TNBC (PD-L1
positive). For early-stage breast cancer, the KEYNOTE-173 phase-Ib
trial evaluated pembrolizumab along with taxane and anthracycline
neoadjuvant therapy, which resulted in an ORR of 100% (151). The ISPY-2 trial was a phase-III
trial evaluating pembrolizumab in combination with chemotherapy
(vs. placebo) in patients with stage-II/III TNBC, which
demonstrated an ORR of 60 and 20%, respectively (152). The SWOG S1418 (NCT02954874) trial
is investigating anti-PD-1/-PD-L1 in the adjuvant setting for a
year in order to determine whether there is an improvement in DFS.
The NSABP B-59 (NCT03281954) and IMpassion030 (NCT03498716) trials
are addressing whether the combination of neoadjuvant/adjuvant
chemotherapy and atezolizumab might improve DFS compared with
chemotherapy alone (153).
Sequencing of all the RNA species in a given cell
using RNA-seq identified several RNA species, including mRNA. The
two major classes of non-coding RNA studied in TNBC development and
treatment are miRNA and Long non-coding RNA.
MicroRNA (miRNA/miR) is a small non-coding RNA,
usually 20–22 nucleotides in length, regulating gene expression.
miRNA is known to bind to the 3′untranslated region of mRNA. This
binding either degrades mRNA or represses translation (154). miRNA is a key player in
tumorigenesis, stemness, and drug resistance in TNBC (155–157).
For instance, tumour suppressor miRNAs, involved in tumour
development, miR-190a, miR-136-5p, miR-126-5p, miR-135b-5p and
miR-182-5p are downregulated in TNBC (158). miR-22 is downregulation in TNBC, is
associated with migration and metastasis. miR-22 exerts its effect
through eukaryotic elongation factor 2 kinase (eEF2K) expression,
which activates PI3K signaling pathway (159). Also, oncosuppressor, miR-200b,
activate target genes like SRY-box transcription factor 2 (SOX2),
CD133, and zinc finger E-box binding homeobox 1 (ZEB1), aiding in
migration and invasion and stemness (157,160).
High expression of miR-95 in TNBC indicates radiotherapy resistance
that occurs by targeting sphingosine-1-phosphate signaling
(161). Downregulated miR-449
upregulates CDK2, CCNE2 causing doxorubicin resistancein TNBC
(162,163) (Table
III). Multiple studies also show that miRNAs are expressed in
different stages of TNBC Multiple studies also show that miRNAs are
expressed in different stages of TNBC (164–166).
These studies give hope for miRNA-based therapies, as the use of
miRNA mimics or inhibitor oligonucleotides could serve as a
therapeutic approach for TNBC (167). A study conducted by Shu et
al (168) used miR-21 combined
with aptamer targeting EGFR, blocking tumor growth in murine
models. Yin et al (169)
designed an RNA aptamer bound to CD133 with a sequence
complementary to miR-21 carried by a three-way junction motif
scaffold that reduced cell migration in TNBC cells (169). Non-coding RNA is being pursued as
one of the TNBC therapy.
lncRNA (long non-coding RNA), ~200 nucleotides in
length, regulates gene expression at the epigenetic, transcription,
post-transcription levels, and post-translation modification
(16). The long intergenic
non-coding RNA for kinase activation activates HIF-1α by
phosphorylating it via leucine-rich repeat kinase 2to promote
glycolysis and tumorigenesis in TNBC (170). Yang et al (171) demonstrated the involvement of POU
domain class 3 transcription factor 3 (POU3F3) in inhibiting
apoptosis and promoting proliferation in TNBC (171). Nuclear paraspeckle assembly
transcript 1 (NEAT1) plays a role in TNBC metastasis (172–174).
Some lncRNAs (HOTAIR, LncRNA-ATB, LincRNA-ROR) are known to be
co-expressed with transcription factors involved in EMT and
proliferation (175). Vaidya et
al (176) demonstrated that
nanoparticle-mediated transfer of RNA interference molecules
targeting differentiation antagonizing non-protein coding RNA, a
lncRNA that is enriched in TNBC, showed some efficacy in a murine
xenograft model of TNBC (Table
III). These studies have shed light on the use of antisense
oligonucleotides against oncogenic lncRNA as a potential approach
to TNBC therapy.
TNBC is associated with poor prognosis compared to
other breast cancer subtypes, and its treatment remains
challenging. New technology and tools have provided insight into
the molecular mechanism of the disease. This knowledge has led to
the identification of druggable targets and the development of
biomarker-driven therapy. The FDA-approved drugs for TNBC to date
include PARP inhibitors for patients with BRCA1/2 mutations and
atezolizumab for PD-L1+ tumors. Emerging targeted
therapies have given hope for the treatment of TNBC. The inclusion
of immunotherapy has shown promising results. Additionally,
attempts to identify combinations that work effectively against
TNBC are ongoing. A combination of the molecular profiles,
including non-coding RNA and histology, has improved the prognosis
and guided the treatment for TNBC.
The authors would like to thank Dr Raksha Rao K.
(Institute of Bioinformatics and Applied Biotechnology, Bengaluru,
Karnataka, India) for the critical reading of the manuscript and
suggestions.
Financial support was provided by The Department of
Science and Technology Fund for Improvement of S&T
Infrastructure in Higher Educational Institutions (grant no.
SR/FST/LSI-5361/2012), The Department of Biotechnology, India, Glue
grant (BTIPR23078/MED/29/1253/2017), and The Departments
Information Technology, Biotechnology and Science and Technology,
Government of Karnataka, India. MM is supported by The Senior
Research Fellowship from Department of Science and
Technology-Innovation in Science Pursuit for Inspired Research,
India (DST/INSPIRE Fellowship/2016/IF160535).
Not applicable.
MM and BC conceived the article, performed the
literature search and data analysis, drafted and critically revised
the work, and confirm the authenticity of the raw data. All authors
read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Hwang SY, Park S and Kwon Y: Recent
therapeutic trends and promising targets in triple negative breast
cancer. Pharmacol Ther. 199:30–57. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. Feb 4–2021.(Epub ahead
of print). doi: 10.3322/caac.21660. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Perue CM, Sorlie T, Elsen MB, van de Rijn
M, Jeffrey S and Rees C: Molecular portraits of human breast
tumors. Nature. 406:747–52. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Penault-Llorca F and Viale G: Pathological
and molecular diagnosis of triple-negative breast cancer: A
clinical perspective. Ann Oncol. 23:vi19–vi22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yeh IT and Mies C: Application of
immunohistochemistry to breast lesions. Arch Pathol Lab Med.
132:349–358. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fedele M, Cerchia L and Chiappetta G: The
epithelial-to-mesenchymal transition in breast cancer: Focus on
basal-like carcinomas. Cancers. 9:1342017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dias K, Dvorkin-Gheva A, Hallett RM, Wu Y,
Hassell J, Pond GR, Levine M, Whelan T and Bane AL: Claudin-low
breast cancer; clinical & pathological characteristics. PLoS
One. 12:e01686692017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spigel DR and Burstein HJ: HER2
overexpressing metastatic breast cancer. Curr Treat Options Oncol.
3:163–174. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sorlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu Z, Fan C, Oh DS, Marron JS, He X,
Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al: The
molecular portraits of breast tumors are conserved across
microarray platforms. BMC Genomics. 7:962006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Q, Xu M, Sun Y, Chen J, Chen C, Qian
C, Chen Y, Cao L, Xu Q, Du X and Yang W: Gene expression profiling
for diagnosis of triple-negative breast cancer: A multicenter,
retrospective cohort study. Front Oncol. 9:3542019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Slamon D, Eiermann W, Robert N, Pienkowski
T, Martin M, Press M, Mackey J, Glaspy J, Chan A, Pawlicki M, et
al: Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J
Med. 365:1273–1283. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marotti JD, de Abreu FB, Wells WA and
Tsongalis GJ: Triple-negative breast cancer: Next-generation
sequencing for target identification. Am J Pathol. 187:2133–2138.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reis-Filho JS and Tutt ANJ: Triple
negative tumours: A critical review. Histopathology. 52:108–118.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kaplan HG, Malmgren JA and Atwood M: T1N0
triple negative breast cancer: Risk of recurrence and adjuvant
chemotherapy. Breast J. 15:454–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang-Qing Y, Jie L, Shi-Qi Z, Kun Z,
Zi-Qian G, Ran X, Hui-Meng L, Ren-Bin Z, Gang Z, Da-Chuan Y and
Chen-Yan Z: Recent treatment progress of triple negative breast
cancer. Prog Biophys Mol Biol. 151:40–53. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weigelt B and Reis-Filho JS: Histological
and molecular types of breast cancer: Is there a unifying taxonomy?
Nat Rev Clin Oncol. 6:7182009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shah SP, Roth A, Goya R, Oloumi A, Ha G,
Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al: The clonal
and mutational evolution spectrum of primary triple-negative breast
cancers. Nature. 486:395–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Malhotra GK, Zhao X, Band H and Band V:
Histological, molecular and functional subtypes of breast cancers.
Cancer Biol Ther. 10:955–960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Balkenhol MC, Vreuls W, Wauters CA, Mol
SJ, van der Laak JA and Bult P: Histological subtypes in triple
negative breast cancer are associated with specific information on
survival. Ann Diagn Pathol. 46:1514902020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Romero P, Benhamo V, Deniziaut G, Fuhrmann
L, Berger F, Manié E, Bhalshankar J, Vacher S, Laurent C, Marangoni
E, et al: Medullary breast carcinoma, a triple-negative breast
cancer associated with BCLG overexpression. Am J Pathol.
188:2378–2391. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huober J, Gelber S, Thurlimann B,
Goldhirsch A, Coates AS, Viale G, Öhlschlegel C, Price KN, Gelber
RD, Regan MM and Thürlimann B: Prognosis of medullary breast
cancer: Analyses of 13 International Breast Cancer Study Group
(IBCSG) trials. Ann Oncol. 23:2843–2851. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Geyer FC, Weigelt B, Natrajan R, Lambros
MB, de Biase D, Vatcheva R, Savage K, Mackay A, Ashworth A and
Reis-Filho JS: Molecular analysis reveals a genetic basis for the
phenotypic diversity of metaplastic breast carcinomas. J Pathol.
220:562–573. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hennessy BT, Gonzalez-Angulo AM,
Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J,
Sahin A, Agarwal R, Joy C, et al: Characterization of a naturally
occurring breast cancer subset enriched in
epithelial-to-mesenchymal transition and stem cell characteristics.
Cancer Res. 69:4116–4124. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hayes MJ, Thomas D, Emmons A, Giordano TJ
and Kleer CG: Genetic changes of Wnt pathway genes are common
events in metaplastic carcinomas of the breast. Clin Cancer Res.
14:4038–4044. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Thomas DN, Asarian A and Xiao P: Adenoid
cystic carcinoma of the breast. J Surg Case Rep. 2019:rjy3552019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ichikawa K, Mizukami Y, Takayama T,
Takemura A, Miyati T and Taniya T: A case of adenoid cystic
carcinoma of the breast. J Med Ultrasonics. 34:193–196. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun JY, Wu SG, Chen SY, Li FY, Lin HX,
Chen YX and He ZY: Adjuvant radiation therapy and survival for
adenoid cystic carcinoma of the breast. Breast. 31:214–218. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aktepe F, Sarsenov D and Özmen V:
Secretory carcinoma of the breast. J Breast Health. 12:1742016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Wu N, Li F, Li L, Wei L and Liu J:
Clinicopathologic and molecular characteristics of 44 patients with
pure secretory breast carcinoma. Cancer Biol Med. 16:1392019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pareja F, Geyer FC, Marchiò C, Burke KA,
Weigelt B and Reis-Filho JS: Triple-negative breast cancer: The
importance of molecular and histologic subtyping, and recognition
of low-grade variants. NPJ Breast Cancer. 2:160362016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuroda H, Sakamoto G, Ohnisi K and Itoyama
S: Clinical and pathological features of glycogen-rich clear cell
carcinoma of the breast. Breast Cancer. 12:189–195. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Geyer FC, Pareja F, Weigelt B, Rakha E,
Ellis IO, Schnitt SJ and Reis-Filho JS: The spectrum of
triple-negative breast disease: High-and low-grade lesions. Am J
Pathol. 187:2139–2151. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Degnim AC, Brahmbhatt RD, Radisky DC,
Hoskin TL, Stallings-Mann M, Laudenschlager M, Mansfield A, Frost
MH, Murphy L, Knutson K and Visscher DW: Immune cell quantitation
in normal breast tissue lobules with and without lobulitis. Breast
Cancer Res Treat. 144:539–549. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Aaltomaa S, Lipponen P, Eskelinen M, Kosma
VM, Marin S, Alhava E and Syrjänen K: Lymphocyte infiltrates as a
prognostic variable in female breast cancer. Eur J Cancer.
28:859–864. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matsumoto H, Koo S, Dent R, Tan PH and
Iqbal J: Role of inflammatory infiltrates in triple negative breast
cancer. J Clin Pathol. 68:506–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fridman WH, Galon J, Pagès F, Tartour E,
Sautès-Fridman C and Kroemer G: Prognostic and predictive impact of
intra-and peritumoral immune infiltrates. Cancer Res. 71:5601–5605.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Karn T, Jiang T, Hatzis C, Sänger N,
El-Balat A, Holtrich U, Becker S, Bianchini G and Pusztai L:
Abstract S1-07: Immune sculpting of the triple negative breast
cancer genome. Cancer Res. 772017.doi:
10.1158/1538-7445.SABCS16-S1-07.
|
|
39
|
Bottai G, Raschioni C, Losurdo A, Di
Tommaso L, Tinterri C, Torrisi R, Reis-Filho JS, Roncalli M,
Sotiriou C, Santoro A, et al: An immune stratification reveals a
subset of PD-1/LAG-3 double-positive triple-negative breast
cancers. Breast Cancer Res. 18:1212016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gruosso T, Gigoux M, Bertos N, Manem VSK,
Guiot MC, Buisseret L, Salgado R, Van den Eyden G, Haibe-Kains B
and Park M: Distinct immune microenvironments stratify
triple-negative breast cancer and predict outcome. Ann Oncol.
28:i162017. View Article : Google Scholar
|
|
41
|
Abramson VG, Lehmann BD, Ballinger TJ and
Pietenpol JA: Subtyping of triple-negative breast cancer:
Implications for therapy. Cancer. 121:8–16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lehmann BD and Pietenpol JA:
Identification and use of biomarkers in treatment strategies for
triple-negative breast cancer subtypes. J Pathol. 232:142–150.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Burstein MD, Tsimelzon A, Poage GM,
Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK,
Hilsenbeck SG, Chang JC, et al: Comprehensive genomic analysis
identifies novel subtypes and targets of triple-negative breast
cancer. Clin Cancer Res. 21:1688–1698. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ahn SG, Kim SJ, Kim C and Jeong J:
Molecular classification of triple-negative breast cancer. J Breast
Cancer. 19:223–230. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu YR, Jiang YZ, Xu XE, Yu KD, Jin X, Hu
X, Zuo WJ, Hao S, Wu J, Liu GY, et al: Comprehensive transcriptome
analysis identifies novel molecular subtypes and subtype-specific
RNAs of triple-negative breast cancer. Breast Cancer Res.
18:332016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yin L, Duan JJ, Bian XW and Yu S:
Triple-negative breast cancer molecular subtyping and treatment
progress. Breast Cancer Res. 22:612020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Venkitaraman AR: Linking the cellular
functions of BRCA genes to cancer pathogenesis and treatment. Ann
Rev Pathol. 4:461–487. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Atchley DP, Albarracin CT, Lopez A, Valero
V, Amos CI, Gonzalez-Angulo AM, Hortobagyi GN and Arun BK: Clinical
and pathologic characteristics of patients with BRCA-positive and
BRCA-negative breast cancer. J Clin Oncol. 26:4282–4288. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Foulkes WD, Stefansson IM, Chappuis PO,
Bégin LR, Goffin JR, Wong N, Trudel M and Akslen LA: Germline BRCA1
mutations and a basal epithelial phenotype in breast cancer. J Natl
Cancer Inst. 95:1482–1485. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Antoniou A, Pharoah PD, Narod S, Risch HA,
Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et
al: Average risks of breast and ovarian cancer associated with
BRCA1 or BRCA2 mutations detected in case series unselected for
family history: A combined analysis of 22 studies. Am J Hum Genet.
72:1117–1130. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Turner N, Tutt A and Ashworth A: Hallmarks
of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 4:814–819. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lord CJ and Ashworth A: BRCAness
revisited. Nat Rev Cancer. 16:110–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bertucci F, Ng CK, Patsouris A, Droin N,
Piscuoglio S, Carbuccia N, Soria JC, Dien AT, Adnani Y, Kamal M, et
al: Genomic characterization of metastatic breast cancers. Nature.
569:560–564. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X,
Xiao Y, Yu KD, Liu YR, Yu Y, et al: Genomic and transcriptomic
landscape of triple-negative breast cancers: Subtypes and treatment
strategies. Cancer Cell. 35:428–440.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nik-Zainal S, Davies H, Staaf J,
Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB,
Martin S, Wedge DC, et al: Landscape of somatic mutations in 560
breast cancer whole-genome sequences. Nature. 534:47–54. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bianchini G, Balko JM, Mayer IA, Sanders
ME and Gianni L: Triple-negative breast cancer: Challenges and
opportunities of a heterogeneous disease. Nat Rev Clin Oncol.
13:674–690. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhao Y, Sheng M, Zheng L, Xiong D, Yang K
and Luo Y: Application of circulating tumor DNA in breast cancer.
Breast J. 26:1797–1800. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lustberg MB, Stover DG and Chalmers JJ:
Implementing liquid biopsies in clinical trials: State of affairs,
opportunities and challenges. Cancer J. 24:61–64. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Thompson AM and Moulder-Thompson SL:
Neoadjuvant treatment of breast cancer. Ann Oncol. 23 (Suppl
10):x231–x236. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Stover DG, Parsons HA, Ha G, Freeman SS,
Barry WT, Guo H, Choudhury AD, Gydush G, Reed SC, Rhoades J, et al:
Association of cell-free DNA tumor fraction and somatic copy number
alterations with survival in metastatic triple-negative breast
cancer. J Clin Oncol. 36:543–553. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bidard FC, Peeters DJ, Fehm T, Nolé F,
Gisbert-Criado R, Mavroudis D, Grisanti S, Generali D, Garcia-Saenz
JA, Stebbing J, et al: Clinical validity of circulating tumour
cells in patients with metastatic breast cancer: A pooled analysis
of individual patient data. Lancet Oncol. 15:406–414. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Cristofanilli M, Pierga JY, Reuben J,
Rademaker A, Davis AA, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado
R, Mavroudis D, et al: The clinical use of circulating tumor cells
(CTCs) enumeration for staging of metastatic breast cancer (MBC):
International expert consensus paper. Crit Rev Oncol Hematol.
134:39–45. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Riva F, Bidard FC, Houy A, Saliou A, Madic
J, Rampanou A, Hego C, Milder M, Cottu P, Sablin MP, et al:
Patient-specific circulating tumor DNA detection during neoadjuvant
chemotherapy in triple-negative breast cancer. Clin Chem.
63:691–699. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Radovich M, Jiang G, Chitambar C, Nanda R,
Falkson C, Lynce FC, Gallagher C, Isaacs C, Blaya M, Paplomata E,
et al: Abstract GS5-02: Detection of circulating tumor DNA (ctDNA)
after neoadjuvant chemotherapy is significantly associated with
disease recurrence in early-stage triple-negative breast cancer
(TNBC): Preplanned correlative results from clinical trial
BRE12-158. Cancer Res. 802020.doi:
10.1158/1538-7445.SABCS19-GS5-02.
|
|
67
|
Becker S: A historic and scientific review
of breast cancer: The next global healthcare challenge. Int J
Gynecol Obstet. 131 (Suppl 1):S36–S39. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Blum JL, Flynn PJ, Yothers G, Asmar L,
Geyer CE Jr, Jacobs SA, Robert NJ, Hopkins JO, O'Shaughnessy JA,
Dang CT, et al: Anthracyclines in early breast cancer: The ABC
Trials-USOR 06-090, NSABP B-46-I/USOR 07132, and NSABP B-49 (NRG
Oncology). J Clin Oncol. 35:2647–2655. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mansel RE, Fodstad O and Jiang WG:
Metastasis of breast cancer. Springer; 2007, View Article : Google Scholar
|
|
70
|
Mosca L, Ilari A, Fazi F, Assaraf YG and
Colotti G: Taxanes in cancer treatment: Activity, chemoresistance
and its overcoming. Drug Resist Updat. 54:1007422021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bachegowda LS, Makower DF and Sparano JA:
Taxanes: Impact on breast cancer therapy. Anticancer Drugs.
25:512–521. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Cortazar P, Zhang L, Untch M, Mehta K,
Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L,
Valagussa P, et al: Pathological complete response and long-term
clinical benefit in breast cancer: The CTNeoBC pooled analysis.
Lancet. 384:164–172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Von Minckwitz G, Untch M, Blohmer JU,
Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich
J, Huober J, et al: Definition and impact of pathologic complete
response on prognosis after neoadjuvant chemotherapy in various
intrinsic breast cancer subtypes. J Clin Oncol. 30:1796–1804. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple-negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Park JH, Ahn JH and Kim SB: How shall we
treat early triple-negative breast cancer (TNBC): From the current
standard to upcoming immuno-molecular strategies. ESMO Open.
3:e0003572018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Greene J and Hennessy B: The role of
anthracyclines in the treatment of early breast cancer. J Oncol
Pharm Pract. 21:201–212. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Park JS, Jeung HC, Rha SY, Ahn JB, Kang B,
Chon HJ, Hong MH, Lim S, Yang WI, Nam CM and Chung HC: Phase II
gemcitabine and capecitabine combination therapy in recurrent or
metastatic breast cancer patients pretreated with anthracycline and
taxane. Cancer Chemother Pharmacol. 74:799–808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Karachaliou N, Ziras N, Syrigos K,
Tryfonidis K, Papadimitraki E, Kontopodis E, Bozionelou V, Kalykaki
A, Georgoulias V and Mavroudis D: A multicenter phase II trial of
docetaxel and capecitabine as salvage treatment in
anthracycline-and taxane-pretreated patients with metastatic breast
cancer. Cancer Chemother Pharmacol. 70:169–176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Anton A, Lluch A, Casado A, Provencio M,
Muñoz M, Lao J, Bermejo B, Paules AB, Gayo J and Martin M: Phase I
study of oral vinorelbine and capecitabine in patients with
metastatic breast cancer. Anticancer Res. 30:2255–2261.
2010.PubMed/NCBI
|
|
80
|
Kennedy RD, Quinn JE, Mullan PB, Johnston
PG and Harkin DP: The role of BRCA1 in the cellular response to
chemotherapy. J Natl Cancer Inst. 96:1659–1668. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Huang L, Liu Q, Chen S and Shao Z:
Cisplatin versus carboplatin in combination with paclitaxel as
neoadjuvant regimen for triple negative breast cancer. Onco Targets
Ther. 10:5739–5744. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Isakoff SJ, Mayer EL, He L, Traina TA,
Carey LA, Krag KJ, Rugo HS, Liu MC, Stearns V, Come SE, et al:
TBCRC009: A multicenter phase II clinical trial of platinum
monotherapy with biomarker assessment in metastatic triple-negative
breast cancer. J Clin Oncol. 33:1902–1909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Byrski T, Dent R, Blecharz P,
Foszczynska-Kloda M, Gronwald J, Huzarski T, Cybulski C, Marczyk E,
Chrzan R, Eisen A, et al: Results of a phase II open-label,
non-randomized trial of cisplatin chemotherapy in patients with
BRCA1-positive metastatic breast cancer. Breast Cancer Res.
14:R1102012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Isakoff SJ: Triple negative breast cancer:
Role of specific chemotherapy agents. Cancer J. 16:53–61. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kim GM, Jeung HC, Jung KH, Kim HJ, Lee KH,
Park KH, Lee JE, Anh MS, Kohn S, Lee SS, et al: PEARLY: A
randomized, multicenter, open-label, phase III trial comparing
anthracyclines followed by taxane versus anthracyclines followed by
taxane plus carboplatin as (neo) adjuvant therapy in patients with
early triple-negative breast cancer. J Clin Oncol. 35
(15_suppl):TPS587. 2017. View Article : Google Scholar
|
|
86
|
Chen XS, Nie XQ, Chen CM, Wu JY, Wu J, Lu
JS, Shao ZM, Shen ZZ and Shen KW: Weekly paclitaxel plus
carboplatin is an effective nonanthracycline-containing regimen as
neoadjuvant chemotherapy for breast cancer. Ann Oncol. 21:961–967.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Sawyers C: Targeted cancer therapy.
Nature. 432:294–297. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dancey JE and Chen HX: Strategies for
optimizing combinations of molecularly targeted anticancer agents.
Nat Rev Drug Dis. 5:649–659. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Jhan JR and Andrechek ER: Triple-negative
breast cancer and the potential for targeted therapy.
Pharmacogenomics. 18:1595–1609. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Audebert M, Salles B and Calsou P:
Involvement of poly (ADP-ribose) polymerase-1 and XRCC1/DNA ligase
III in an alternative route for DNA double-strand breaks rejoining.
J Biol Chem. 279:55117–55126. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Shall S and de Murcia G: Poly(ADP-ribose)
polymerase-1: What have we learned from the deficient mouse model?
Mutat Res. 460:1–15. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Farmer H, McCabe N, Lord CJ, Tutt AN,
Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I,
Knights C, et al: Targeting the DNA repair defect in BRCA mutant
cells as a therapeutic strategy. Nature. 434:917–921. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Turner N, Tutt A and Ashworth A: Targeting
the DNA repair defect of BRCA tumours. Curr Opin Pharmacol.
5:388–393. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Turner NC, Lord CJ, Iorns E, Brough R,
Swift S, Elliott R, Rayter S, Tutt AN and Ashworth A: A synthetic
lethal siRNA screen identifying genes mediating sensitivity to a
PARP inhibitor. EMBO J. 27:1368–1377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Calabrese CR, Almassy R, Barton S, Batey
MA, Calvert AH, Canan-Koch S, Durkacz BW, Hostomsky Z, Kumpf RA,
Kyle S, et al: Anticancer chemosensitization and radiosensitization
by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J
Natl Cancer Inst. 96:56–67. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Robson ME, Im SA, Senkus E, Xu B, Domchek
SM, Masuda N, Delaloge S, Li W, Tung NM, Armstrong A, et al:
OlympiAD: Phase III trial of olaparib monotherapy versus
chemotherapy for patients (pts) with HER2-negative metastatic
breast cancer (mBC) and a germline BRCA mutation (gBRCAm). J Clin
Oncol. 352017.PubMed/NCBI
|
|
98
|
Robson ME, Tung N, Conte P, Im SA, Senkus
E, Xu B, Masuda N, Delaloge S, Li W, Armstrong A, et al: OlympiAD
final overall survival and tolerability results: Olaparib versus
chemotherapy treatment of physician's choice in patients with a
germline BRCA mutation and HER2-negative metastatic breast cancer.
Ann Oncol. 30:558–566. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Litton JK, Rugo HS, Ettl J, Hurvitz SA,
Gonçalves A, Lee KH, Fehrenbacher L, Yerushalmi R, Mina LA, Martin
M, et al: Talazoparib in patients with advanced breast cancer and a
germline BRCA mutation. N Engl J Med. 379:753–763. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Poggio F, Bruzzone M, Ceppi M, Conte B,
Martel S, Maurer C, Tagliamento M, Viglietti G, Del Mastro L, de
Azambuja E and Lambertini M: Single-agent PARP inhibitors for the
treatment of patients with BRCA-mutated HER2-negative metastatic
breast cancer: A systematic review and meta-analysis. ESMO Open.
3:e0003612018. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Miller K, Tong Y, Jones DR, Walsh T, Danso
MA and Ma CX; MCSSSM, : Cisplatin with or without rucaparib after
preoperative chemotherapy in patients with triple negative breast
cancer: Final efficacy results of Hoosier Oncology Group BRE09-146.
J Clin Oncol. 33:10822015. View Article : Google Scholar
|
|
102
|
Isakoff SJ, Puhalla S, Domchek SM,
Friedlander M, Kaufman B, Robson M, Telli ML, Diéras V, Han HS,
Garber JE, et al: A randomized phase II study of veliparib with
temozolomide or carboplatin/paclitaxel versus placebo with
carboplatin/paclitaxel in BRCA1/2 metastatic breast cancer: Design
and rationale. Future Oncol. 13:307–320. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zimmer AS, Gillard M, Lipkowitz S and Lee
JM: Update on PARP inhibitors in breast cancer. Curr Treat Options
Oncol. 19:212018. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Rugo HS, Olopade OI, DeMichele A, Yau C,
van't Veer LJ, Buxton MB, Hogarth M, Hylton NM, Paoloni M,
Perlmutter J, et al: Adaptive randomization of
veliparib-carboplatin treatment in breast cancer. N Engl J Med.
375:23–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Domchek SM, Postel-Vinay S, Im SA, Hee
Park Y, Delord JP, Italiano A, Alexandre J, You B, Bastian S, Krebs
MG, et al: Abstract PD5-04: An open-label, phase II basket study of
olaparib and durvalumab (MEDIOLA): Updated results in patients with
germline BRCA-mutated (gBRCAm) metastatic breast cancer (MBC).
Cancer Res. 792019.doi: 10.1158/1538-7445.SABCS18-PD5-04.
|
|
106
|
Tutt A, Kaufman B, Gelber RD, McFadden E,
Goessl C, Viale G, Geyer G, Zardavas D, Arahmani A, Fumagalli D, et
al: OlympiA: A randomized phase III trial of olaparib as adjuvant
therapy in patients with high-risk HER2-negative breast cancer (BC)
and a germline BRCA1/2 mutation (gBRCAm). Ann Oncol. 28:V672017.
View Article : Google Scholar
|
|
107
|
Earl HM, Vallier AL, Qian W, Grybowicz L,
Thomas S, Mahmud S, Harvey C, McAdam K, Hughes-Davies L, Roylance
R, et al: PARTNER: Randomised, phase II/III trial to evaluate the
safety and efficacy of the addition of olaparib to platinum-based
neoadjuvant chemotherapy in triple negative and/or germline BRCA
mutated breast cancer patients. J Clin Oncol. 35:TPS5912017.
View Article : Google Scholar
|
|
108
|
Cantley LC: The phosphoinositide 3-kinase
pathway. Science. 296:1655–1657. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Engelman JA: Targeting PI3K signalling in
cancer: Opportunities, challenges and limitations. Nat Rev Cancer.
9:550–562. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Delaloge S and DeForceville L: Targeting
PI3K/AKT pathway in triple-negative breast cancer. Lancet Oncol.
18:1293–1294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Katso R, Okkenhaug K, Ahmadi K, White S,
Timms J and Waterfield MD: Cellular function of phosphoinositide
3-kinases: Implications for development, immunity, homeostasis, and
cancer. Annu Rev Cell Dev Biol. 17:615–675. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Kim SB, Dent R, Im SA, Espié M, Blau S,
Tan AR, Isakoff SJ, Oliveira M, Saura C, Wongchenko MJ, et al:
Ipatasertib plus paclitaxel versus placebo plus paclitaxel as
first-line therapy for metastatic triple-negative breast cancer
(LOTUS): A multicentre, randomised, double-blind,
placebo-controlled, phase 2 trial. Lancet Oncol. 18:1360–1372.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Martín M, Chan A, Dirix L, O'Shaughnessy
J, Hegg R, Manikhas A, Shtivelband M, Krivorotko P, Batista López
N, Campone M, et al: A randomized adaptive phase II/III study of
buparlisib, a pan-class I PI3K inhibitor, combined with paclitaxel
for the treatment of HER2-advanced breast cancer (BELLE-4). Ann
Oncol. 28:313–320. 2017. View Article : Google Scholar
|
|
114
|
Schmid P, Cortes J, Robson ME, Iwata H,
Hegg R, Nechaeva M, Xu B, Verma S, Haddad V, Imedio R, et al: A
phase III trial of capivasertib and paclitaxel in first-line
treatment of patients with metastatic triple-negative breast cancer
(CAPItello290). J Clin Oncol. 38:TPS11092020. View Article : Google Scholar
|
|
115
|
Schmid P, Abraham J, Chan S, Wheatley D,
Brunt M, Nemsadze G, Baird R, Park YH, Hall P, Perren T, et al:
AZD5363 plus paclitaxel versus placebo plus paclitaxel as
first-line therapy for metastatic triple-negative breast cancer
(PAKT): A randomised, double-blind, placebo-controlled, phase II
trial. J Clin Oncol. 36:10072018. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Gonzalez-Angulo AM, Green MC, Murray JL,
Palla SL, Koenig KH, Valero Brewster NK; SLJKDJ, ; et al: Open
label, randomized clinical trial of standard neoadjuvant
chemotherapy with paclitaxel followed by FEC (T-FEC) versus the
combination of paclitaxel and RAD001 followed by FEC (TR-FEC) in
women with triple receptor-negative breast cancer (TNBC). J Clin
Oncol. 29:10162011. View Article : Google Scholar
|
|
117
|
Basho RK, Gilcrease M, Murthy RK, Helgason
T, Karp DD, Meric-Bernstam F, Hess KR, Herbrich SM, Valero V,
Albarracin C, et al: Targeting the PI3K/AKT/mTOR pathway for the
treatment of mesenchymal triple-negative breast cancer: Evidence
from a phase 1 trial of mTOR inhibition in combination with
liposomal doxorubicin and bevacizumab. JAMA Oncol. 3:509–515. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Kim Y, Jae E and Yoon M: Influence of
androgen receptor expression on the survival outcomes in breast
cancer: A meta-analysis. J Breast Cancer. 18:134–142. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Gucalp A, Tolaney S, Isakoff SJ, Ingle JN,
Liu MC, Carey LA, Blackwell K, Rugo H, Nabell L, Forero A, et al:
Phase II trial of bicalutamide in patients with androgen
receptor-positive, estrogen receptor-negative metastatic breast
cancer. Clin Cancer Res. 19:5505–5512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Traina TA, Miller K, Yardley DA, Eakle J,
Schwartzberg LS, O'Shaughnessy J, Gradishar W, Schmid P, Winer E,
Kelly C, et al: Enzalutamide for the treatment of androgen
receptor-expressing triple-negative breast cancer. J Clin Oncol.
36:884–890. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Traina TA, Miller K, Yardley DA,
O'Shaughnessy J, Cortes J, Kelly AACM, Trudeau ME, Schmid P, Gianni
L, García-Estevez A, et al: Results from a phase 2 study of
enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in
advanced AR+ triple-negative breast cancer (TNBC). J Clin Oncol.
33:10032015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lehmann BD, Bauer JA, Schafer JM,
Pendleton CS, Tang L, Johnson KC, Chen X, Balko JM, Gómez H,
Arteaga CL, et al: PIK3CA mutations in androgen receptor-positive
triple negative breast cancer confer sensitivity to the combination
of PI3K and androgen receptor inhibitors. Breast Cancer Res.
16:4062014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Lehmann BD, Abramson VG, Sanders ME, Mayer
EL, Haddad TC, Nanda R, Van Poznak C, Storniolo AM, Nangia JR,
Gonzalez-Ericsson PI, et al: TBCRC 032 IB/II multicenter study:
Molecular insights to AR antagonist and PI3K inhibitor efficacy in
patients with AR+ metastatic triple-negative breast cancer. Clin
Cancer Res. 26:2111–2123. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Panowski S, Bhakta S, Raab H, Polakis P
and Junutula JR: Site-specific antibody drug conjugates for cancer
therapy. MAbs. 6:34–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Nejadmoghaddam MR, Minai-Tehrani A,
Ghahremanzadeh R, Mahmoudi M, Dinarvand R and Zarnani AH:
Antibody-drug conjugates: Possibilities and challenges. Avicenna J
Med Biotechnol. 11:3–23. 2019.PubMed/NCBI
|
|
126
|
Goldenberg DM, Stein R and Sharkey RM: The
emergence of trophoblast cell-surface antigen 2 (TROP-2) as a novel
cancer target. Oncotarget. 9:28989–29006. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Bardia A, Mayer IA, Vahdat LT, Tolaney SM,
Isakoff SJ, Diamond JR, O'Shaughnessy J, Moroose RL, Santin AD,
Abramson VG, et al: Sacituzumab govitecan-hziy in refractory
metastatic triple-negative breast cancer. N Engl J Med.
380:741–751. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Liu Y, Lian W, Zhao X, Diao Y, Xu J, Xiao
L, Qing Y, Xue T and Wang J: SKB264 ADC: A first-in-human study of
SKB264 in patients with locally advanced unresectable/metastatic
solid tumors who are refractory to available standard therapies. J
Clin Oncol. 38((15_suppl)): TPS36592020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Lyons TG: Targeted therapies for
triple-negative breast cancer. Curr Treat Options Oncol. 20:822019.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Giltnane JM and Balko JM: Rationale for
targeting the Ras/MAPK pathway in triple-negative breast cancer.
Dis Med. 17:275–283. 2014.PubMed/NCBI
|
|
131
|
Romanelli A, Clark A, Assayag F,
Chateau-Joubert S, Poupon MF, Servely JL, Fontaine JJ, Liu X,
Spooner E, Goodstal S, et al: Inhibiting aurora kinases reduces
tumor growth and suppresses tumor recurrence after chemotherapy in
patient-derived triple-negative breast cancer xenografts. Mol
Cancer Ther. 11:2693–2703. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Finn RS: Östrogenrezeptor-positiver
Brustkrebs: Erfolgreiche Palbociblib-Letrozol-Kombination. Breast
Cancer. 375:1925–1936. 2016.PubMed/NCBI
|
|
133
|
Lucantoni F, Lindner AU, O'Donovan N,
Düssmann H and Prehn JH: Systems modeling accurately predicts
responses to genotoxic agents and their synergism with BCL-2
inhibitors in triple negative breast cancer cells. Cell Death Dis.
9:422018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Inao T, Iida Y, Moritani T, Okimoto T,
Tanino R, Kotani H and Harada M: Bcl-2 inhibition sensitizes
triple-negative human breast cancer cells to doxorubicin.
Oncotarget. 9:25545–25556. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Marra A, Viale G and Curigliano G: Recent
advances in triple negative breast cancer: The immunotherapy era.
BMC Med. 17:902019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Weber S, Traunecker A, Oliveri F, Gerhard
W and Karjalainen K: Specific low-affinity recognition of major
histocompatibility complex plus peptide by soluble T-cell receptor.
Nature. 356:793–796. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Peggs KS, Quezada SA and Allison JP:
Cancer immunotherapy: Co-stimulatory agonists and co-inhibitory
antagonists. Clin Exp Immunol. 157:9–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Planes-Laine G, Rochigneux P, Bertucci F,
Chrétien A-S, Viens P, Sabatier R and Gonçalves A: PD-1/PD-L1
targeting in breast cancer: The first clinical evidences are
emerging. A literature review. Cancers (Basel). 11:10332019.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Mahoney KM, Rennert PD and Freeman GJ:
Combination cancer immunotherapy and new immunomodulatory targets.
Nat Rev Drug Dis. 14:561–584. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Alsaab HO, Sau S, Alzhrani R, Tatiparti K,
Bhise K, Kashaw SK and Iyer AK: PD-1 and PD-L1 checkpoint signaling
inhibition for cancer immunotherapy: Mechanism, combinations, and
clinical outcome. Front Pharmacol. 8:5612017. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Sabatier R, Finetti P, Mamessier E,
Adelaide J, Chaffanet M, Ali HR, Viens P, Caldas C, Birnbaum D and
Bertucci F: Prognostic and predictive value of PDL1 expression in
breast cancer. Oncotarget. 6:5449–5464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Mittendorf EA, Philips AV, Meric-Bernstam
F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM,
Akcakanat A, et al: PD-L1 expression in triple-negative breast
cancer. Cancer Immunol Res. 2:361–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Patel SP and Kurzrock R: PD-L1 expression
as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther.
14:847–856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Schmidt M, Böhm D, Von Törne C, Steiner E,
Puhl A, Pilch H, Lehr HA, Hengstler JG, Kölbl H and Gehrmann M: The
humoral immune system has a key prognostic impact in node-negative
breast cancer. Cancer Res. 68:5405–5413. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Aguiar PN Jr, Santoro IL, Tadokoro H, de
Lima Lopes G, Filardi BA, Oliveira P, Mountzios G and de Mello RA:
The role of PD-L1 expression as a predictive biomarker in advanced
non-small-cell lung cancer: A network meta-analysis. Immunotherapy.
8:479–488. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Beckers RK, Selinger CI, Vilain R, Madore
J, Wilmott JS, Harvey K, Holliday A, Cooper CL, Robbins E, Gillett
D, et al: Programmed death ligand 1 expression in triple-negative
breast cancer is associated with tumour-infiltrating lymphocytes
and improved outcome. Histopathology. 69:25–34. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Vagia E, Mahalingam D and Cristofanilli M:
The landscape of targeted therapies in TNBC. Cancers (Basel.
12:9162020. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Schmid P, Adams S, Rugo HS, Schneeweiss A,
Barrios CH, Iwata H, Dieras V, Henschel V, Molinero L, Chui SY, et
al: IMpassion130: Updated overall survival (OS) from a global,
randomized, double-blind, placebo-controlled, Phase III study of
atezolizumab (atezo)+ nab-paclitaxel (nP) in previously untreated
locally advanced or metastatic triple-negative breast cancer
(mTNBC). J Clin Oncol. 37 (Suppl 15):S10032019. View Article : Google Scholar
|
|
149
|
Cortés J, Lipatov O, Im SA, Gonçalves A,
Lee KS, Schmid P, Tamura K, Testa L, Witzel I, Ohtani S, et al:
LBA21 KEYNOTE-119: Phase III study of pembrolizumab (pembro) versus
single-agent chemotherapy (chemo) for metastatic triple negative
breast cancer (mTNBC). Ann Oncol. 30:v859–v860. 2019. View Article : Google Scholar
|
|
150
|
Cortes J, Guo Z, Karantza V and Aktan G:
Abstract CT069: KEYNOTE-355: Randomized, double-blind, phase III
study of pembrolizumab plus chemotherapy vs placebo plus
chemotherapy for previously untreated, locally recurrent,
inoperable or metastatic triple-negative breast cancer (mTNBC).
Cancer Res. 772017.doi: 10.1158/1538-7445.AM2017-CT069.
|
|
151
|
Schmid P, Salgado R, Park YH,
Muñoz-Couselo E, Kim SB, Sohn J, Im SA, Foukakis T, Kuemmel S, Dent
R, et al: Pembrolizumab plus chemotherapy as neoadjuvant treatment
of high-risk, early-stage triple-negative breast cancer: Results
from the phase 1b open-label, multicohort KEYNOTE-173 study. Ann
Oncol. 31:569–581. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Barker AD, Sigman CC, Kelloff GJ, Hylton
NM, Berry DA and Esserman LJ: I-SPY 2: An adaptive breast cancer
trial design in the setting of neoadjuvant chemotherapy. Clin
Pharmacol Ther. 86:97–100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Keenan TE and Tolaney SM: Role of
immunotherapy in Triple-negative breast cancer. J Natl Compr Canc
Netw. 18:479–489. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Qattan A: Novel miRNA targets and
therapies in the triple-negative breast cancer microenvironment: An
emerging Hope for a challenging disease. Int J Mol Sci.
21:89052020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Si W, Shen J, Zheng H and Fan W: The role
and mechanisms of action of microRNAs in cancer drug resistance.
Clin Epigenetics. 11:252019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W,
Yu M, Lin J and Cui Q: MicroRNAs involved in carcinogenesis,
prognosis, therapeutic resistance, and applications in human
triple-negative breast cancer. Cells. 8:14922019. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Lyng MB, Lænkholm AV, Søkilde R, Gravgaard
KH, Litman T and Ditzel HJ: Global microRNA expression profiling of
high-risk ER+ breast cancers from patients receiving adjuvant
tamoxifen mono-therapy: A DBCG study. PLoS One. 7:e361702012.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Gorur A, Bayraktar R, Ivan C, Mokhlis HA,
Bayraktar E, Kahraman N, Karakas D, Karamil S, Kabil NN,
Kanlikilicer P, et al: ncRNA therapy with miRNA-22-3p suppresses
the growth of triple-negative breast cancer. Mol Ther Nucleic
Acids. 23:930–943. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Pang Y, Liu J, Li X, Xiao G, Wang H, Yang
G, Li Y, Tang SC, Qin S, Du N, et al: MYC and DNMT 3A-mediated DNA
methylation represses micro RNA-200b in triple negative breast
cancer. J Cell Mol Med. 22:6262–6274. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Huang X, Taeb S, Jahangiri S, Emmenegger
U, Tran E, Bruce J, Mesci A, Korpela E, Vesprini D, Wong CS, et al:
miRNA-95 mediates radioresistance in tumors by targeting the
sphingolipid phosphatase SGPP1. Cancer Res. 73:6972–6986. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Tormo E, Ballester S, Adam-Artigues A,
Burgués O, Alonso E, Bermejo B, Menéndez S, Zazo S, Madoz-Gúrpide
J, Rovira A, et al: The miRNA-449 family mediates doxorubicin
resistance in triple-negative breast cancer by regulating cell
cycle factors. Sci Rep. 9:53162019. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Naorem LD, Muthaiyan M and Venkatesan A:
Identification of dysregulated miRNAs in triple negative breast
cancer: A meta-analysis approach. J Cell Physiol. 234:11768–11779.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Malla RR, Kumari S, Gavara MM, Badana AK,
Gugalavath S, Kumar DKG and Rokkam P: A perspective on the
diagnostics, prognostics, and therapeutics of microRNAs of
triple-negative breast cancer. Biophys Rev. 11:227–234. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Liu Y, Cai Q, Bao PP, Su Y, Cai H, Wu J,
Ye F, Guo X, Zheng W, Zheng Y and Shu XO: Tumor tissue microRNA
expression in association with triple-negative breast cancer
outcomes. Breast Cancer Res Treat. 152:183–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Kahraman M, Röske A, Laufer T, Fehlmann T,
Backes C, Kern F, Kohlhaas J, Schrörs H, Saiz A, Zabler C, et al:
MicroRNA in diagnosis and therapy monitoring of early-stage
triple-negative breast cancer. Sci Rep. 8:115842018. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Mei J, Hao L, Wang H, Xu R, Liu Y, Zhu Y
and Liu C: Systematic characterization of non-coding RNAs in
triple-negative breast cancer. Cell Prolif. 53:e128012020.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Shu D, Li H, Shu Y, Xiong G, Carson WE
III, Haque F, Xu R and Guo P: Systemic delivery of anti-miRNA for
suppression of triple negative breast cancer utilizing RNA
nanotechnology. ACS Nano. 9:9731–9740. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Yin H, Xiong G, Guo S, Xu C, Xu R, Guo P
and Shu D: Delivery of anti-miRNA for triple-negative breast cancer
therapy using RNA nanoparticles targeting stem cell marker CD133.
Mol Ther. 27:1252–1261. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L,
Wang C, Hawke DH, Wang S, Zhang Y, et al: The LINK-A lncRNA
activates normoxic HIF1α signalling in triple-negative breast
cancer. Nat Cell Biol. 18:213–224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Yang J, Meng X, Yu Y, Pan L, Zheng Q and
Lin W: LncRNA POU3F3 promotes proliferation and inhibits apoptosis
of cancer cells in triple-negative breast cancer by inactivating
caspase 9. Biosci Biotechnol Biochem. 83:1117–1123. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Jiang X, Zhou Y, Sun AJ and Xue JL: NEAT1
contributes to breast cancer progression through modulating miR-448
and ZEB1. J Cell Physiol. 233:8558–8566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q,
Su X, Peng L and Jiao B: NEAT1 is required for survival of breast
cancer cells through FUS and miR-548. Gene Regul Syst Biol. 10
(Suppl 1):S11–S17. 2016.PubMed/NCBI
|
|
174
|
Wang LI, Liu D, Wu X, Zeng Y, Li L, Hou Y,
Li W and Liu Z: Long non-coding RNA (LncRNA) RMST in
triple-negative breast cancer (TNBC): Expression analysis and
biological roles research. J Cell Physiol. 233:6603–6612. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Xu Q, Deng F, Qin Y, Zhao Z, Wu Z, Xing Z,
Ji A and Wang QJ: Long non-coding RNA regulation of
epithelial-mesenchymal transition in cancer metastasis. Cell Death
Dis. 7:e22542016. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Vaidya AM, Sun Z, Ayat N, Schilb A, Liu X,
Jiang H, Sun D, Scheidt J, Qian V, He S, et al: Systemic delivery
of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA
facilitates effective triple-negative breast cancer therapy.
Bioconjug Chem. 30:907–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Sha S, Yuan D, Liu Y, Han B and Zhong N:
Targeting long non-coding RNA DANCR inhibits triple negative breast
cancer progression. Biol Open. 6:1310–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|