Introduction
Esophageal cancer is a deadly type of cancer that
ranks 8th in terms of worldwide cancer mortality (1). The predominant histological subtype in
most parts of the world, especially in East and Central Asia and
parts of East Africa and South Africa, is esophageal squamous cell
carcinoma (ESCC) (2). The
development of squamous cell carcinomas (SCCs) is associated with
various genomic and genetic aberrations that lead to the
deregulation of the squamous cell lineage and terminal
differentiation program; thus, these processes must be tightly
controlled to maintain normal squamous epithelium development
(3).
Zinc finger protein 750 (ZNF750) is a member of the
zinc finger family proteins, which are involved in numerous
biological processes, including keratinocyte and epithelial cell
differentiation, as well as adipogenesis (4). These proteins have also been implicated
in different diseases including diabetes, psoriasis and cancer
(4,5). ZNF750 has been reported to be involved
in the maintenance of epithelial homeostasis and the epidermal
terminal differentiation process in primary and organotypic human
keratinocyte culture through its interaction with transcription
factors and epigenetic modifiers (6,7). In
addition, knockout of ZNF750 in squamous cells silenced the
expression levels of differentiation-related genes Krüppel-like
factor 4, S100 calcium-binding protein A8 and loricrin (8). Frequent somatic mutations within the
exonic regions have been reported in cervical, head and neck, lung
and esophageal SCCs (7–9), implicating the silencing of
ZNF750 in these malignancies.
The whole-exome sequencing analysis in our previous
study demonstrated the clinical relevance of ZNF750 in ESCC
(10). ZNF750 levels are
downregulated in ESCC tumors compared with those in distant
non-tumor tissues, and mutations in this gene are significantly
more frequent in primary ESCC tumors with lymph node (LN)
metastasis compared with those without LN metastasis (10). The levels of differentiation-related
genes, including small proline-rich protein 3 (SPRR3) and
transglutaminase 3 (TGM3), have been reported to be
downregulated in primary tumor samples compared with those in
adjacent non-tumor tissues (11,12). In
addition, SPRR3 exhibits tumor-suppressive functions in ESCC cells
(13), indicating a link between
silenced differentiation-related genes and tumor-suppressive
functions in ESCC. Therefore, we hypothesized that ZNF750 as
a frequently mutated and downregulated differentiation-related gene
may function similarly to other downregulated
differentiation-related genes, such as SPRR3, and act as a tumor
suppressor in ESCC.
The function of ZNF750 in the suppression of SCCs,
including ESCC, has been demonstrated in an in vitro cell
viability and an in vivo tumorigenesis models (9,14),
whereas its functions in keratinocyte differentiation induction
have been reported in keratinocyte cell line and organoid models
(5–7); however, the functional link between
keratinocyte differentiation induction and tumor suppression in
ESCC has not been fully explored. The aim of the present study was
to elucidate this relationship.
Materials and methods
Cell culture
The KYSE-series cell lines (ACC 351, ACC 375, ACC
379, ACC 363, ACC 374 ACC 380, ACC 387; DSMZ) were cultured in
RPMI-1640 medium (cat. no. 23400) supplemented with 10% fetal
bovine serum (FBS; cat. no. 10270) and 1% penicillin-streptomycin
(cat. no. 15070). NE-1 cells were cultured in Defined Keratinocyte
SFM medium with Defined Keratinocyte SFM Growth Supplement (cat.
no. 10744019). 293T, HKESC-2 and SLMT cells were cultured in DMEM
(cat. no. 12100) supplemented with 5% FBS and 5% Newborn Calf Serum
(NCS; cat. no. 16010), and 1% penicillin-streptomycin. 293T cells
served as host cells for lentiviral production. Cells were seeded
and incubated until full confluence and passaged every 2–3 days
using Trypsin/EDTA (cat. no. 25200) for continuous culture at 37°C
with 5% CO2. All reagents used for cell culture were
purchased from Invitrogen; Thermo Fisher Scientific, Inc. The cell
lines were genotyped, authenticated and tested for mycoplasma prior
to the experiments.
Mouse normal esophageal epithelial
organoid (mNEEO) generation and culture
Mouse esophageal tissue isolation, organoid
generation and culture were performed as previously described with
modifications (15). In brief,
8–10-week old BALB/c/nu/nu athymic mice (n=4) were sacrificed
through anesthetic overdose by intraperitoneal injection of
ketamine/xylazine (300 mg/kg ketamine and 30 mg/kg xylazine)
followed by carbon dioxide asphyxiation by placing mice in a carbon
dioxide-filled container. The esophagus was surgically removed, and
the mucosa was pulled away from the submucosa layer using forceps.
The mucosa layer was cut into small pieces with a scalpel,
dissociated in Trypsin-EDTA at 37°C for 30 min and subsequently
neutralized with RPMI-1640 supplemented with 10% FBS. To generate
organoids, the dissociated cells were suspended in
Matrigel® (cat. no. 356234; Corning, Inc.) and placed in
24-well low-bind culture plates (cat. no. 174930; Thermo Fisher
Scientific, Inc.) at 37°C to solidify. The mNEEO was cultured in
growth medium comprising advanced DMEM/F12 (cat. no. 12634), 1X N2
(cat. no. 17502) and 1X B27 Supplements (cat. no. 17504), 1X
Glutamax (cat. no. 35050), 1X HEPES (cat. no. 15630), 1X
penicillin/streptomycin, 1 mM N-acetyl-L-cysteine (cat. no. A9165,
Sigma-Aldrich; Merck KGaA), 100 µm gastrin (cat. no. G9020,
Sigma-Aldrich; Merck KGaA), 10 mM nicotinamide (cat. no. N0636,
Sigma-Aldrich; Merck KGaA), 10 µm SB202190 (cat. no. S7067,
Sigma-Aldrich; Merck KGaA), 50 ng/ml epidermal growth factor (cat.
no. PHG0314), 100 ng/ml Noggin (cat. no. 120-10C; PeproTech, Inc.),
100 ng/ml Wnt3A (cat. no. 315-20; PeproTech, Inc.), 100 ng/ml
R-Spondin 2 (cat. no. 120-43; PeproTech, Inc.) and 500 nM A8301
(cat. no. 2939; Tocris Bioscience) as previously described
(15) and passaged every 7 days for
continuous culture. All reagents were supplied by Thermo Fisher
Scientific, Inc. unless otherwise specified. Ethics approval for
animal studies was obtained from the Animal Ethics Committee of The
University of Hong Kong (approval no. CULATR 3377-14).
Chemically induced manipulation of
mNEEO differentiation
For all-trans retinoic acid (ATRA; cat. no. R2625;
Sigma-Aldrich; Merck KGaA) and Compound E (cat. no. sc-221433;
Santa Cruz Biotechnology, Inc.) treatment of mNEEO, 2,000
dissociated esophageal epithelial cells were embedded in 4
wells/group in a low-bind 96-well culture plate (cat. no. 15237905;
Thermo Fisher Scientific, Inc.). The esophageal epithelial cells
were cultured in growth medium for 24 h prior to ATRA and Compound
E treatment. Growth medium was replaced with growth medium
containing 100 nM ATRA, 10 µM Compound E or DMSO (cat. no. D418;
Sigma-Aldrich; Merck KGaA) vehicle after 24 h, and the cells were
cultured for 9 days to generate organoids.
Molecular cloning of ZNF750
ZNF750 (NM_024702.3) was amplified using Q5
High-Fidelity DNA Polymerase (cat. no. M0491; New England Biolabs,
Inc.) from KYSE30 cell line cDNA by PCR. The PCR procedure was
performed using a Veriti™ 96-Well Thermal Cycler (cat. no. 4375786;
Thermo Fisher Scientific, Inc.) under the following cycling
conditions: Initial denaturation at 98°C for 30 sec, followed by 30
cycles of annealing at 56°C for 30 sec and extension at 72°C for 2
min. The oligonucleotide pairs used for amplification were
ZNF750 forward, 5′-GGAATTCGCCACCATGAGTCTCCTCAAAGAGCGG-3′ and
reverse, 5′-GCTCTAGATTACTTGTCGTCATCGTCTTTGTAGTCGGACACCCGGGCCCTC-3′.
The amplified DNA was subsequently cloned into a pLVX-EF1α-puro
vector. The ZNF750 sequence (pLVX-ZNF750) was validated by
Sanger sequencing (16) at the
Centre for PanorOmic Sciences (CPOS) at the University of Hong Kong
(Hong Kong, SAR, China). pLVX-puro (cat. no. 632164; Clontech
Laboratories, Inc.) was modified by removing the CMV promoter and
replacing it with an EF1α promoter to construct a pLVX-EF1α-puro
vector (17). The pLVX-EF1α-puro
vector was used to insert a GFP gene to create the pLVX-GFP control
plasmid (17).
Lentivirus production and
infection
293T cells were seeded in preparation for viral
production in T25 culture flasks. When 293T cells reached 60%
confluence, the transfection mixture was prepared using 8 µl
X-tremeGENE™ HP DNA Transfection Reagent (cat. no. 6366546001;
Roche Molecular Systems, Inc.), 2 µg plasmid DNA (pLVX-GFP or
pLVX-ZNF750), 1.5 µg viral packaging vector psPAX2 (cat. no. 12260;
Addgene, Inc.) and 0.5 µg viral envelope vector pMD2.G (cat. no.
12259; Addgene, Inc.) in 400 µl serum-free DMEM and incubated for
20 min at room temperature. The transfection mixture was added
dropwise to the culture flask of HEK293T cells and incubated for 72
h at 37°C. Subsequently, the viral supernatant was passed through a
0.45-µm filter and aliquoted. The virus was stored at −80°C until
further experiments. For lentiviral infection, KYSE30 and KYSE180
cells were seeded to achieve a confluency of ~30% at 24 h
post-seeding. A 1 ml aliquot of pre-prepared virus expressing GFP
or ZNF750 with 0.006 µl 5 µg/ml polybrene (cat. no. H9268;
Sigma-Aldrich; Merck KGaA) was added to cells in tissue culture
flasks to facilitate lentiviral infection. The medium was changed
at 24 h post-infection to allow the cells to recover. The cells
were passaged at day 2 post-infection for expansion and prepared
for assays after the second passage.
RNA isolation, reverse transcription
(RT) and quantitative (q)PCR
RNA was extracted from cells and organoids using the
TRI Reagent™ solution (cat. no. AM9738; Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions and
reverse-transcribed to complementary DNA using PrimeScript RT
Master Mix (Perfect Real Time) (cat. no. RR036; Takara Bio, Inc.)
according to the manufacturer's instructions. FastStart Universal
SYBR®-Green Master (Rox) (cat. no. 4913914001; Roche
Life Science) was used to determine the mRNA expression levels of
target genes. Each reaction comprised 1X FastStart Universal
SYBR® Green Master mix, 30 ng/µl cDNA, and 0.4 mM each
of forward and reverse primer. The reactions were performed in the
LightCycler® 480 Instrument II (cat. no. 05015243001;
Roche Life Science) using the default SYBR®-Green
program. mRNA expression levels were quantified using the
2−∆∆Cq method (18).
Human GAPDH forward, 5′-AAGGTGAAGGTCGGAGTC-3′ and reverse,
5′-GAAGATGGTGATGGGATTTC-3′, was used as the internal control. The
primer sequences were as follows: Human: ZNF750 forward,
5′-AAAGCACAGAATGCCTACCTG-3′ and reverse,
5′-GGGTCTCCGTTCACAACATT-3′; keratin 4 (KRT4) forward,
5′-GATGCTCTTCTGGGGGATT-3′ and reverse, 5′-GCTCTGGTTGATGGTGACCT-3′;
SPRR1A forward, 5′-CCGTATACCAGCTTTCTGTCTC-3′ and reverse,
5′-GCTGGCAAGGTTGTTTCAC-3′; SPRR2E forward,
5′-TGGTACTTGAGCACTGATCTGC-3′ and reverse,
5′-TGCACTGCTGCTGTTGATAA-3′; TGM3 forward,
5′-TGGAAGGACTCTGCCACAAT-3′ and reverse,
5′-TGAGCGTACGAGATCTTTATGG-3′; and involucrin (IVL) forward,
5′-GCAGGAGGGACAGCTGAA-3′ and reverse, 5′-ATCTGCTCCTCTGGGACCTC-3′;
mouse: Zfp750 forward, 5′-GAGCCAGCGTGAGAACAGA-3′ and
reverse, 5′-CAGGAGAGTTCCTTCCGTCA-3′; Ivl forward,
5′-CCCTAGTTGCTGCTCAGCTC-3′ and reverse,
5′-TGGAAGCTTCTGTAGACCAAAA-3′; Krt4 forward,
5′-AACAAATTCGCGTCCTTCAT-3′ and reverse, 5′-CTGCTGGAGGAGGTTCCAT-3′;
and Krt14 forward, 5′-ATCGAGGACCTGAAGAGCAA-3′ and reverse,
5′-TCGATCTGCAGGAGGACATT-3′.
Protein extraction and western
blotting
Protein lysates were obtained by lysing KYSE30 and
KYSE180 cells for 10 min at 4°C in RIPA buffer (cat. no. 89900;
Thermo Fisher Scientific, Inc.) supplemented with 1X Halt™ Protease
and Phosphatase Inhibitor Cocktail (cat. no. 78442; Thermo Fisher
Scientific, Inc.), and 1 unit of Pierce Universal Nuclease (cat.
no. 88700; Thermo Fisher Scientific, Inc.). Protein lysates were
collected by centrifugation at 12,000 × g at 4°C for 10 min. The
proteins were denatured at 95°C for 10 min prior to
electrophoresis. A total of 50 µg protein per sample were
electrophoresed using 12% SDS-PAGE at 80V for 30 min and 120V for
1.5 h using the Mini Gel Tank (cat. no. A25977; Thermo Fisher
Scientific, Inc.). Following electrophoresis, the proteins within
the gel were transferred to methanol-activated 0.45-µm PVDF
membranes (cat. no. IPVH00010; MilliporeSigma) using a Mini Blot
system (cat. no. B1000; Thermo Fisher Scientific, Inc.) at 30V for
1 h. Subsequently, the membranes were blocked with 3% bovine serum
albumin (v/v) in Tris-buffered saline with 0.1% Tween (TBST) for 45
min at room temperature. The membranes were hybridized with the
primary antibodies at 4°C overnight with constant shaking. The
membranes were briefly washed with TBST and probed with a
horseradish peroxidase (HRP)-conjugated secondary antibody diluted
1:5,000 for 1 h at room temperature with gentle agitation, followed
by extensive washing with TBST prior to signal detection. Signals
were detected by WesternBright ECL HRP substrate (cat. no. K-12045;
Advansta, Inc.) and Super RX X-ray films (Fujifilm Corporation).
Band intensities were measured using ImageJ software version 1.53a
(National Institutes of Health) (19) and quantified by normalization to the
GFP control. The primary antibodies used were polyclonal rabbit
anti-human ZNF750 (1:1,000; cat. no. 21752-1-AP; Proteintech Group,
Inc.), polyclonal rabbit anti-human SPRR3 (1:1,000; cat. no.
A12041; ABclonal, Inc.), monoclonal mouse anti-human IVL (1:1,000;
cat. no. MA5-11803; Thermo Fisher Scientific, Inc.), monoclonal
mouse anti-human p84 (1:1,000; cat. no. GTX70220; GeneTex, Inc.),
monoclonal rabbit anti-human GAPDH (1:5,000; cat. no. 5174; Cell
Signaling Technology, Inc.) and monoclonal rabbit anti-GFP
(1:1,000; cat. no. 2956; Cell Signaling Technology, Inc.). The
secondary antibodies used were polyclonal anti-mouse IgG (1:5,000;
cat. no. GTX213111-01) and polyclonal anti-rabbit IgG (1:5,000;
cat. no. GTX213110-01) (both from GeneTex, Inc.).
RNA sequencing (RNA-seq) and
transcriptomic analysis of ZNF750-upregulation in ESCC cells
Total RNAs of pLVX-GFP- or pLVX-ZNF750-transduced
KYSE180 cells were extracted with the AllPrep DNA/RNA Micro kit
(cat. no. 80284; Qiagen, Inc.) according to the manufacturer's
instructions for RNA sequencing. Library preparation and Illumina
sequencing (paired-end sequencing of 151 bp) were performed at
CPOS. Bioanalyzer RNA Nano 6000 (Agilent Technologies) was used to
determine the quality of the RNA. Ribosomal RNA (rRNA) depletion
was performed using NEBNext® rRNA Depletion Kit
(Human/Mouse/Rat) (cat. no. E6310; New England BioLabs, Inc.). cDNA
libraries were prepared using the NEBNext® Ultra II
Directional RNA Library Prep Kit (cat. no. E7760; New England
BioLabs, Inc.). A total of 500 ng RNA was used as starting material
for ribosomal RNA depletion. Cytoplasmic and mitochondrial rRNA
were firstly hybridized to ssDNA capture probes and then digested
by Ribonuclease H. Subsequently, the residual ssDNA probes were
digested by DNase I. The rRNA-depleted RNA was purified by
Agencourt® RNAClean™ XP beads (cat. no. A63987; Beckman
Coulter, Inc.). The purified rRNA-depleted RNA was fragmented to
200–500 bp by incubating at 94°C for 10 min in the presence of
magnesium ions (NEBNext® Magnesium RNA Fragmentation
Module; cat no. E6150; New England BioLabs, Inc.). The fragmented
rRNA-depleted RNA was then applied as a template to synthesize the
first-strand cDNA using random hexamer-primer and a reverse
transcriptase. In the second strand cDNA synthesis, the mRNA
template was removed, and a replacement strand was generated to
form the blunt-end double-stranded (ds) cDNA. The ds cDNA underwent
3′adenylation and indexed adaptor ligation, followed by treatment
with User Enzyme. The adaptor-ligated libraries were enriched and
indexed using NEBNext® Multiplex Oligos for
Illumina® (96 Unique Dual Index Primer Pairs) (cat. no.
E6440; New England BioLabs, Inc.) by 8 cycles of PCR according to
the manufacturer's instructions. The concentration was determined
using qPCR following library construction, and the libraries were
denatured and diluted to 1.2 nM. Illumina NovaSeq 6000 Reagent Kit
(cat. no. 20028312; Illumina, Inc.) was used for paired-end 151bp
sequencing. Raw RNA-Seq data were cleaned and aligned to the hg19
reference genome using Tophat (version 2.0.14; bowtie version
2.2.4) (20) with library-type
fr-firststrand parameter. Gene expression profile and
differentially expressed genes were calculated by Cufflinks
(version 2.2.1) with the Ensemble gene annotation file and Cuffdiff
(21), respectively. Differential
gene expression was calculated by log2 fold-change (FC)
expression. Genes with log2 (FC) ≥2 were used for
further analysis.
Determination of enriched pathways in
ZNF750-upregulated ESCC cells
Gene Set Enrichment Analysis (GSEA) (22) and ToppFun (https://toppgene.cchmc.org/enrichment.jsp) (23) were used for the analysis of enriched
pathways in ZNF750-overexrpressing cells. Using GSEA version
4.1.0, transcriptomic data were analyzed to identify the enriched
gene sets in the ZNF750-overexpressing group compared with
the GFP-expressing group to identify the genes driving the
enrichment. In ToppFun, differentially upregulated genes in the
ZNF750-overexpressing group compared with the control group
were used for analysis. The gene sets from the Molecular Signatures
Database (MsigDB; http://www.broad.mit.edu/gsea/msigdb/index.jsp) were
used for both analyses. The consensus gene sets between the two
analyses with a false discovery rate (FDR) ≤0.25 were considered to
be enriched.
Extraction and analysis of the cancer
genome atlas (TCGA) esophageal carcinoma (ESCA) RNA-seq data
TCGA data were downloaded from the National Cancer
Institute Genomic Data Commons (https://gdc.cancer.gov/) (24). Normalized gene expression profiles of
SCC samples with RNA-seq data from the TCGA-ESCA project (dbGaP
study accession no. phs000178) (25)
were extracted using R version 4.0.2 (26) and the TCGAbiolinks R/Bioconductor
package (27) (http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html).
ESCC data were divided into high-ZNF750 expressing tumors
(ZNF750-high; n=40) and low-ZNF750 expressing tumors
(ZNF750-low; n=41) using median normalized expression levels
as the cut-off value. GSEA was used to determine the enriched gene
sets in the ZNF750-high compared with the ZNF750-low
group using the same cut-off values as aforementioned.
MTT cell viability assay
To assess cell viability, 2.5×103 KYSE30
and 5×103 KYSE180 cells per well were seeded in a
96-well plate with 100 µl medium. For visualization, 30 µl/well 5
mg/ml MTT dye (cat. no. M6494; Thermo Fisher Scientific, Inc.) was
added to the cells and incubated at 37°C for 2 h. Subsequently, the
medium was aspirated, and 100 µl DMSO was added to dissolve the
formazan crystals. Absorbance was measured at 540 nm using an Ao
Absorbance Microplate Reader (Azure Biosystems) daily for 3 days
post-seeding.
ATRA treatment of parental ESCC cell
lines
For the ATRA treatment, parental KYSE30 and KYSE180
cells were seeded as aforementioned and cultured for 24 h.
Subsequently, the culture medium in each well was replaced with
medium containing 50 or 100 nM ATRA or DMSO and cultured for 3
days. Concurrently, cells from the same batch were seeded in T25
culture flasks and treated with ATRA. RNA was extracted as
aforementioned from the ATRA-treated cells at 48 h post-treatment
and subjected to RT-qPCR analysis. Experiments were performed on
KYSE30 and KYSE180 cell lines using previously reported ATRA
concentrations as a reference to determine the optimal ATRA
concentration for each cell line (28).
Colony formation assay
For the colony formation assay, 1×103
KYSE30 or KYSE180 cells were seeded into a single well of a 6-well
culture plate to form colonies. The colonies were fixed with 37%
formaldehyde for 30 min at room temperature and stained with 1:20
Giemsa solution in water overnight at room temperature. The
solution was washed away with 1X PBS until the buffer was clear.
Colonies were counted using the Gel Doc XR+ system (cat. no.
170-8195; Bio-Rad Laboratories, Inc.).
Subcutaneous tumor mouse model
ESCC tumorigenicity was evaluated by subcutaneous
inoculation of ESCC cells into the flanks of 8–10-week-old
BALB/c/nu/nu athymic nude mice. A total of 1×106 KYSE30
cells were resuspended in 150 µl of serum-free medium and injected
in both flanks of each mouse (n=4 mice per group). Tumor growth was
monitored weekly for ≥4 weeks and measured by calipers to calculate
the tumor volume as follows: Volume=(length × width × height). At
the experimental endpoint, mice were sacrificed by anesthetic
overdose through intraperitoneal injection of ketamine/xylazine
(300 mg/kg ketamine and 30 mg/kg xylazine) followed by carbon
dioxide asphyxiation by placing mice in a carbon dioxide-filled
container. Cessation of cardiac function was used as the method for
verification of death. All experimental procedures were approved by
the Committee on the Use of Live Animals in Teaching and Research
at The University of Hong Kong (approval no. CULATR 3377-14).
Statistical analysis
Data are presented as the mean ± SEM. Each in
vitro experiment was repeated at least three times in
duplicate, unless otherwise stated. Unpaired Student's t-test was
performed for all statistical analyses using GraphPad Prism version
8.4.3 (GraphPad Software, Inc.). P<0.05 was considered to
indicate a statistically significant difference.
Results
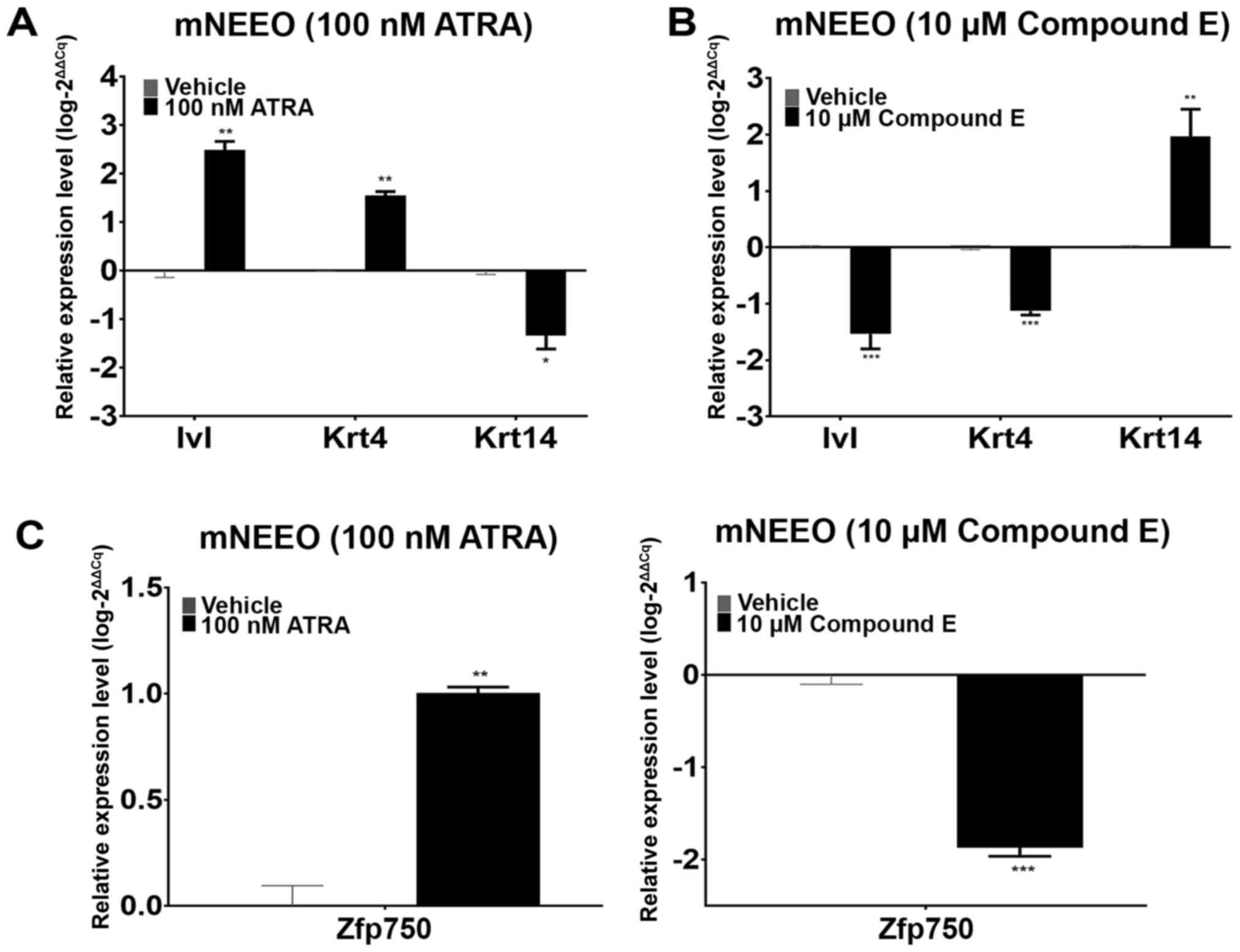
Zfp750 is involved in the esophageal
keratinocyte differentiation program in mouse epithelial cells
mNEEOs were used to determine the role of
Zfp750 in the esophageal keratinocyte differentiation
program. Following ATRA treatment, the organoids demonstrated
increased expression levels of Ivl and Krt4 and decreased
levels of Krt14 compared with those in the vehicle-treated
mNEEOs (Fig. 1A), suggesting an
induction of the keratinocyte differentiation program and
subsequent decrease in the basal cell layers within the organoid.
By contrast, following Compound E treatment, the expression levels
of Ivl and Krt4 were markedly decreased, whereas
those of Krt14 were increased compared with those in the
vehicle-treated group (Fig. 1B),
suggesting the inhibition of the differentiation program and an
increase in the basal cell layers in these organoids. Changes in
Zfp750 expression corresponded with the expression levels of
the markers of differentiation, Ivl and Krt4, after
both treatments (Fig. 1C). These
results suggested that Zfp750 expression may be
proportionally affected by the stimulation and inhibition of the
esophageal keratinocyte differentiation program, and that
Zfp750 may be involved in this cellular process in
esophageal keratinocytes.
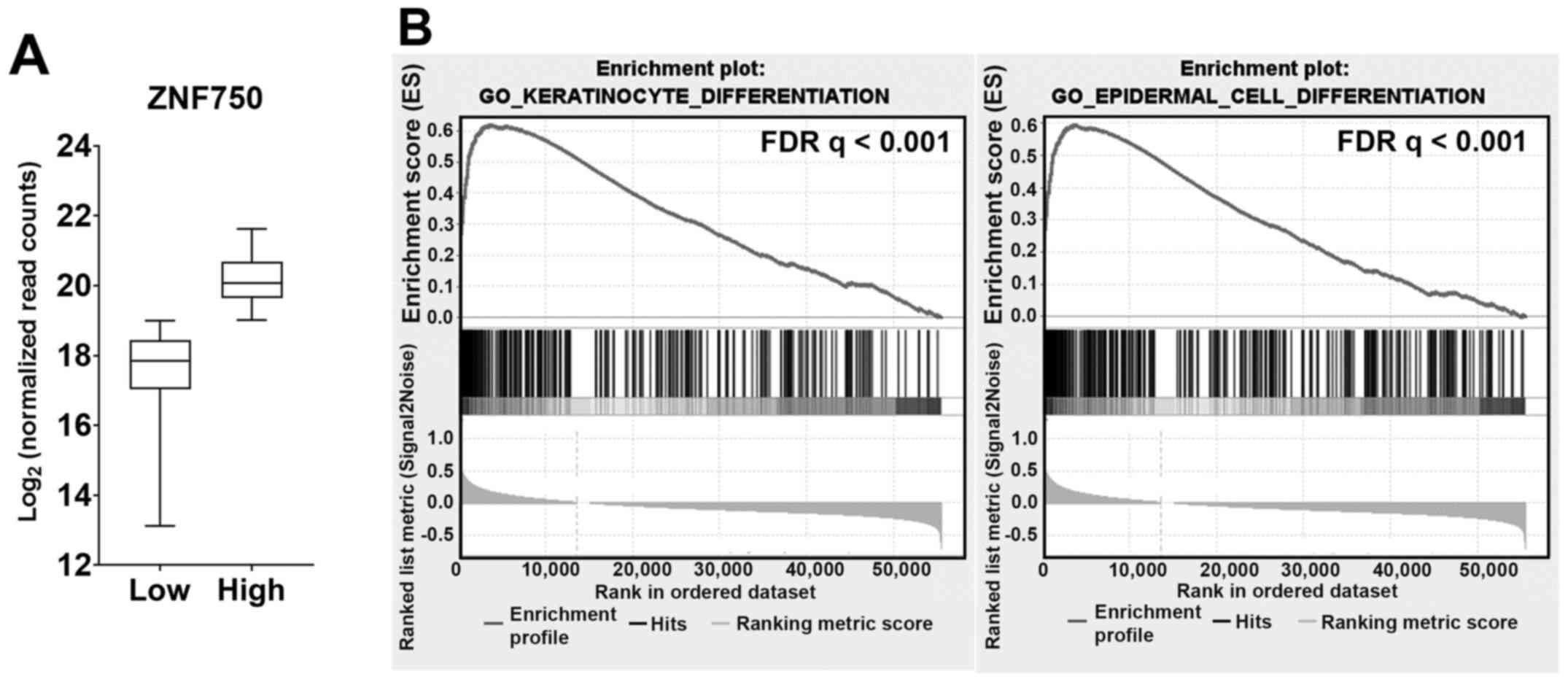
High ZNF750 expression in ESCC tumors
is associated with the enrichment of keratinocyte and epidermal
cell differentiation gene sets
To determine whether ZNF750 expression may
affect the keratinocyte differentiation program in ESCC tumors,
RNA-seq data of ESCC tumor samples (n=81) from the TCGA-ESCA
dataset were analyzed using GSEA. The samples were stratified into
ZNF750-high [median, 20.07; interquartile range (IQR),
19.03–21.11] and ZNF750-low (median, 17.86; IQR,
16.45–19.27) groups (Fig. 2A). The
results of GSEA confirmed that epithelial and epidermal
differentiation-related cellular processes including
‘cornification’, ‘keratinization’, ‘keratinocyte differentiation’
and ‘epidermal cell differentiation’ (Table SI) were enriched in the
ZNF750-high group compared with the ZNF750-low group.
‘Keratinocyte differentiation’ and ‘epidermal cell differentiation’
gene sets were among the most enriched sets (Fig. 2B), whereas SPRR, TGM and
IVL were among the most enriched genes (Table SII). These results supported the
involvement of ZNF750 in keratinocyte differentiation in
ESCC.
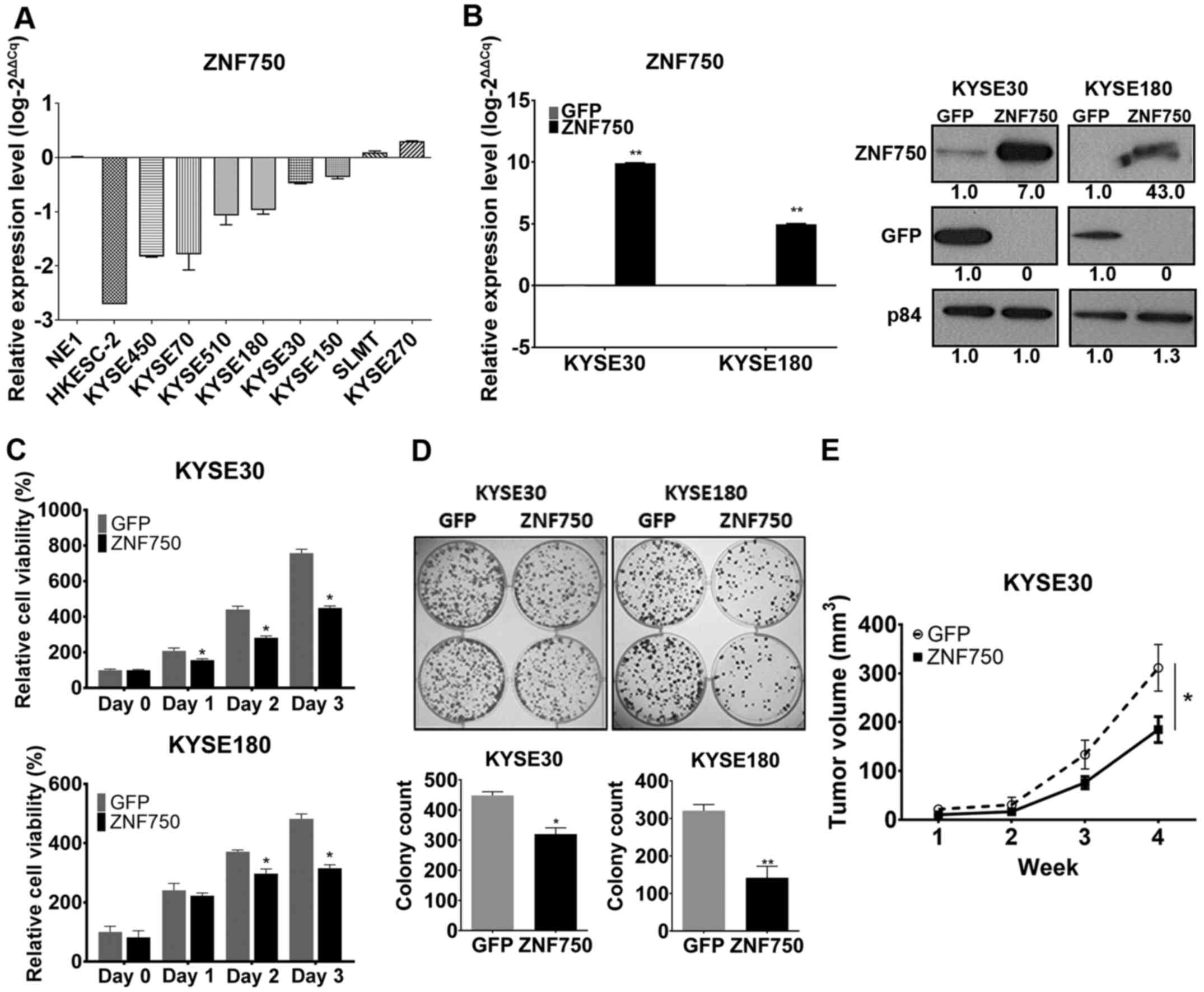
ZNF750 overexpression suppresses ESCC
cell viability and growth in vitro and in vivo
ZNF750 expression levels were downregulated
in the ESCC cell line models compared with those in the
immortalized human esophageal epithelial cell line NE1 (Fig. 3A); therefore, to study the
suppressive function of ZNF750 in ESCC, ZNF750 was overexpressed in
KYSE30 and KYSE180 cells by lentiviral transduction of pLVX-GFP and
pLVX-ZNF750. These cell lines were selected as the models for
further experiments as they exhibited decreased expression of
ZNF750 at the mRNA and protein level (Fig. 3A and B). ZNF750 expression levels
were upregulated in both cell lines in the pLVX-ZNF750 groups
compared with those in the corresponding pLVX-GFP groups, as
demonstrated by RT-qPCR and western blot analyses (Fig. 3B). In vitro MTT cell viability
and colony formation assays and in vivo subcutaneous
tumorigenicity model were used to assess the tumor-suppressive
ability of ZNF750. In vitro, ZNF750 overexpression
suppressed ESCC cell viability compared with that in the control
group (Fig. 3C). Colony formation
ability was also reduced in the ZNF750-overexpressing groups
compared with that in the controls (Fig.
3D). In the in vivo subcutaneous tumor model, mice in
the ZNF750 overexpression group developed smaller tumors (n=8; mean
± SEM, 184±26.5 mm3) in mice compared to the GFP control
(n=8; mean ± SEM, 311.17±47.76 mm3) (P<0.05; Fig. 3E). These results demonstrate the
ability of ZNF750 to suppress cell viability and growth in
vitro and in vivo.
ZNF750 overexpression in ESCC cells
induces the expression of esophageal keratinocyte differentiation
genes
To determine the pathways affected by ZNF750
overexpression in the in vitro model, ZNF750 and GFP
lentivirus-transduced KYSE180 cells were prepared for RNA-seq and
subjected to subsequent bioinformatics analyses. The results of the
analyses using the MSigDB revealed enrichment of gene sets involved
in epithelial differentiation including ‘epithelial cell
differentiation’, ‘epidermal cell differentiation’ and
‘keratinocyte differentiation’ (Table
SIII). Consistent with the GSEA analysis of ZNF750-high
ESCC tumors, these results confirmed the enrichment of keratinocyte
differentiation and epidermal cell differentiation gene sets in
ZNF750-overexpressing ESCC cells compared with the controls
(Fig. 4A). The upregulation of
differentiation-related genes KRT4, SPRR1A, SPRR2E, TGM3,
IVL and SPRR3 were validated by RT-qPCR or western
blotting (Fig. 4B). These results
suggested that ZNF750 overexpression induced the expression
of differentiation-related genes in the ESCC cell line model.
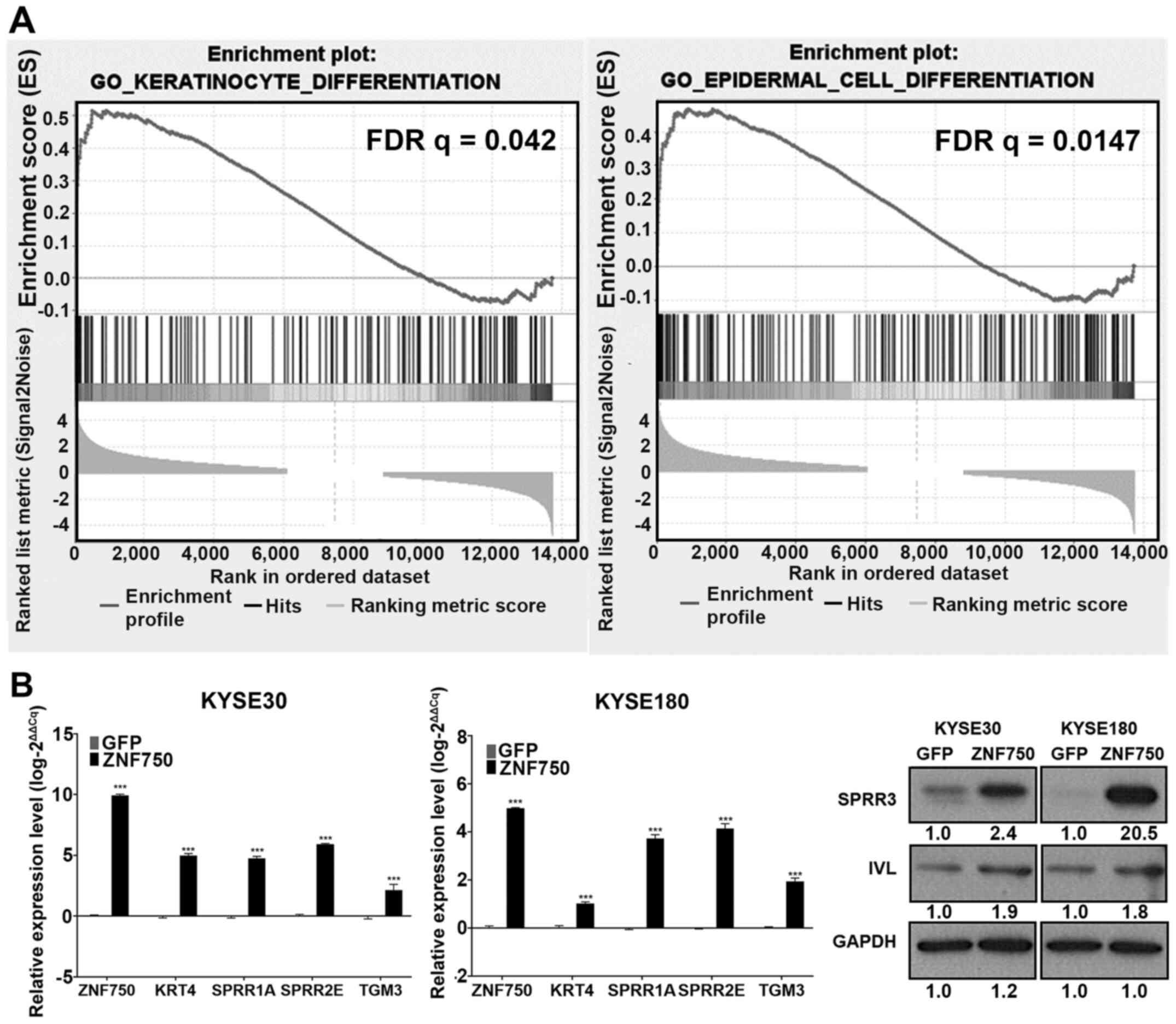 | Figure 4.Overexpression of ZNF750 leads
to enrichment of differentiation-related gene sets in ESCC cells.
(A) Enrichment plots of ‘keratinocyte differentiation’ and
‘epidermal cell differentiation’ gene sets analyzed by Gene Set
Enrichment Analysis and Toppfun with a false discovery rate
<0.25. (B) Reverse transcription-quantitative PCR and western
blot analyses confirmed the upregulation of differentiation-related
gene KRT4, SPRR1A, SPRR2E, SPRR3, TGM3 and IVL
expression levels in ZNF750-overexpressing ESCC cells compared with
those in the control group. Numbers represent band intensities
normalized to the GFP control for the presented blot. Data are
presented as the mean ± SEM. ***P<0.001 vs. GFP. ZNF750, zinc
finger protein 750; GFP, control green fluorescent protein
lentivirus; ESCC, esophageal squamous cell carcinoma; SPRR,
small proline-rich protein; TGM3, transglutaminase 3;
IVL, involucrin. |
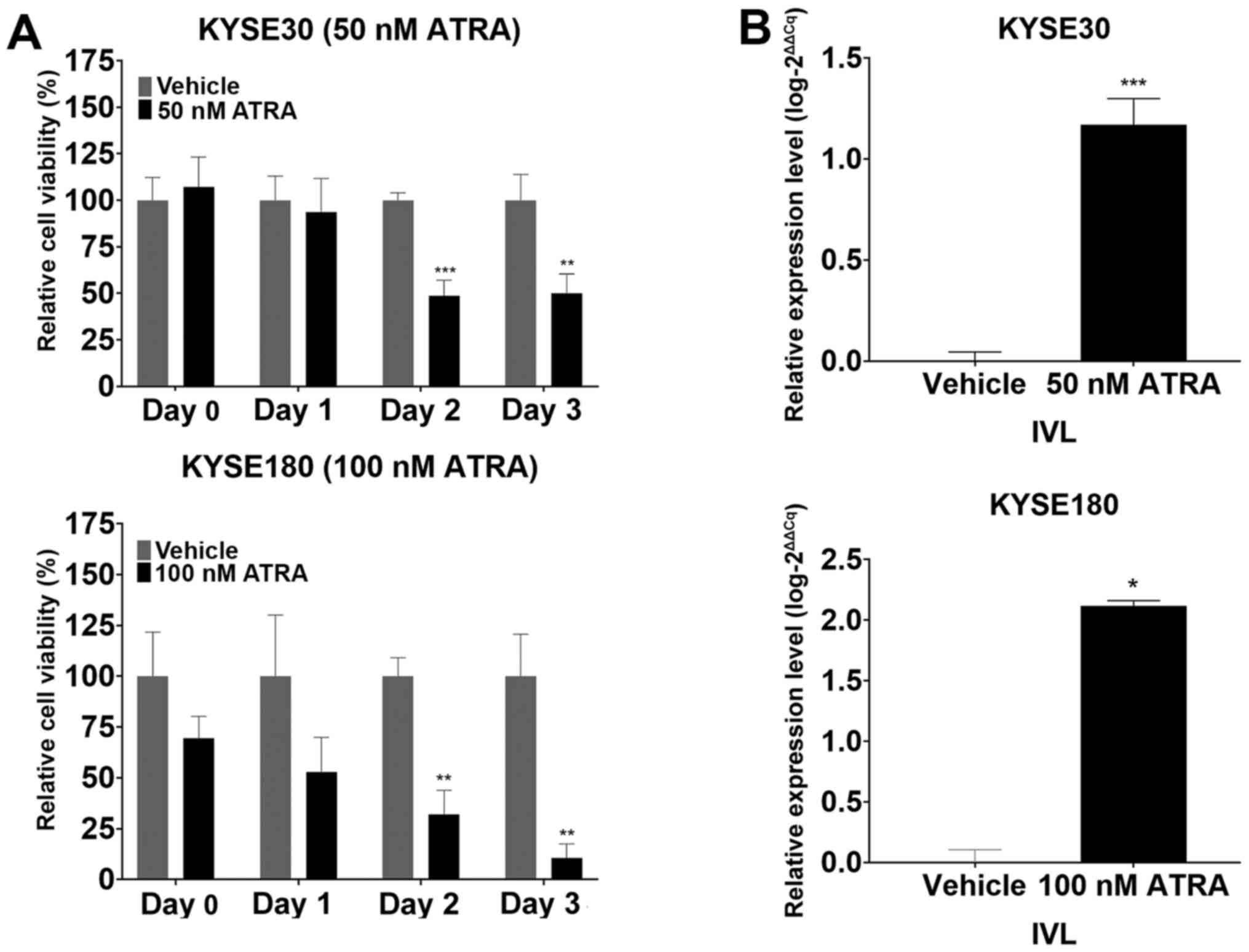
ATRA-induced terminal differentiation
decreases the viability of ESCC cells
To determine the effects of the induction of the
keratinocyte differentiation program on ESCC cell viability,
parental KYSE30 and KYSE180 cells were treated with ATRA and
subsequently tested for changes in cell viability using the MTT
assay. The results demonstrated a significant suppression in the
viability of the ATRA-treated parental ESCC cells compared with
that in the vehicle-treated group at 48 h post-treatment (Fig. 5A). RT-qPCR analysis revealed that
ATRA treatment upregulated the expression levels of IVL
compared with those in the vehicle group (Fig. 5B), indicating the induction of
terminal differentiation. These results suggested that induction of
the keratinocyte differentiation program through to terminal
differentiation may contribute to the suppression of ESCC cell
viability following ATRA-induced stimulation of
differentiation.
Discussion
ESCC is a highly malignant SCC of the esophagus
(29). Although SCCs arise from
various organ systems, they have been reported to share common
genetic, genomic and epigenetic modifications relevant to
malignancy, and the development of SCCs is associated with the
deregulation of squamous cell lineage commitment that may
ultimately lead to carcinogenesis (3). Numerous landscape studies have aimed to
identify the genetic and genomic changes that contribute to the
development of ESCC (9,14,30,31).
These studies identified frequently mutated genes in ESCC including
common cancer-related (TP53, RB1, CDKN2A, PIK3CA, NOTCH1 and
NFE2L2) and previously unreported (ADAM29, FAM135B,
ZNF750 and FAT1) Genes. ZNF750 is a gene that
encodes a transcription factor involved in the terminal
differentiation of SCCs (8,14) and has been reported to be one of the
most frequently mutated genes in ESCC, suggesting that it may
function as a differentiation-related tumor suppressor gene.
Although in vitro and in vivo studies have provided
evidence for ZNF750 functioning as a tumor suppressor and as
an inducer of differentiation in ESCC (9,14), the
link between the functions of ZNF750 in inducing differentiation
and tumor suppression in ESCC is yet to be fully explored.
The results of the present study demonstrated the
involvement of Zfp750 in the esophageal keratinocyte
differentiation program through the upregulation and downregulation
of Zfp750 expression with chemical stimulation and
inhibition, by ATRA and Compound E, respectively, of the
differentiation program in mNEEOs. ATRA is a chemical promoter of
keratinocyte differentiation in esophageal cells (28,29) and
was used in the present study to induce the epithelial
differentiation program. Alternatively, γ-secretase inhibitor
Compound E was used to mimic the inhibition of the keratinocyte
differentiation program through the inhibition of the NOTCH
signaling pathway (30,31). Through the analysis of public data,
the present study also confirmed the ability of ZNF750 to
induce differentiation-related gene expression through the
enrichment of differentiation-related gene sets in
ZNF750-high ESCC tumors. In addition, the results of the
present study demonstrated that ZNF750 suppressed ESCC cell
viability and proliferation in vitro, and its overexpression
resulted in smaller tumors in the mouse model compared with those
in the corresponding controls. RNA-seq study of the ESCC cell line
model further confirmed the involvement of ZNF750 in
keratinocyte differentiation in ESCC cells through the upregulation
of differentiation-related genes SPRR1A, SPRR2E, SPRR3, TGM3,
KRT4 and IVL. Finally, chemically induced stimulation of
keratinocyte differentiation through to terminal differentiation by
ATRA functionally suppressed ESCC through the observed decrease in
cell viability in ATRA-treated cells compared with the controls.
This decrease in cell viability was similar to that of
ZNF750-overexpressing ESCC cells. Collectively, these
results demonstrated the ability of ZNF750 to induce ESCC cell
differentiation through to terminal differentiation, as well as the
link between this process and the suppression of ESCC cell
viability.
The surface of the esophagus is lined with
stratified squamous epithelium, which comprises the basal
proliferative (keratinocyte) layer that gives rise to the layers of
non-dividing terminally differentiated cells that form the
stratified layer (3). The regulation
of this proliferation-differentiation gradient ensures homeostasis
within the epithelium (32). The
squamous cell commitment to differentiate involves the regulation
of the expression levels of genes relevant to this process, and
deregulation of this system has been reported to initiate
malignancy (33). NOTCH is a major
player in epithelial differentiation and one of the most frequently
mutated genes in ESCC (3,9,30),
suggesting that it may be a candidate tumor suppressor.
Differentiation-related genes including SPRR1A, SPRR2E, SPRR3,
TGM3 and IVL have been reported to be downregulated in
ESCC (11,34). SPRR3, also termed esophagin,
is one of the genes upregulated by ZNF750 in ESCC cells in the
current study, and has been implicated as a tumor suppressor in
ESCC (13). Exogenous expression of
SPRR3 attenuates ESCC tumorigenicity by promoting apoptosis
(13) and enhances sensitization to
DNA damage-related apoptosis (35).
In addition, TGM3 has been reported as a potential prognostic
marker in ESCC (12). Proteomics
analysis in tissues from patients with ESCC has revealed that the
presence of TGM3 in ESCC tissues is correlated with a longer
survival time (12). These results
suggest that epithelial differentiation-related genes are
downregulated or silenced in ESCC and may function as putative
tumor suppressors or prognostic markers, providing evidence of the
relevance of this process in ESCC development.
The ability of ZNF750 to induce keratinocyte
terminal differentiation indicated by the expression of IVL
in the present study resembled the chemically induced
differentiation stimulation that caused increased IVL
expression in ESCC cell lines. ESCC viability suppression was
consistently observed in ZNF750-overexpressing and
differentiation-induced cells, suggesting a functional association
between the two groups. This provides a novel functional indication
of the link between the differentiation-related functions of ZNF750
and its tumor-suppressive ability in ESCC.
In conclusion, the results of the present study
demonstrated that ZNF750 may be a differentiation-related
silenced tumor suppressor gene in ESCC. Overexpression of ZNF750 in
ESCC cells led to the upregulation of differentiation-related genes
that may restore epithelial homeostasis by driving differentiation
through to terminal differentiation and, thus, restoring the
proliferation-differentiation balance. In addition, the ability of
ZNF750 to induce the expression of genes such as SPRR3,
which exhibit tumor-suppressive functions in ESCC, may contribute
to its ability to suppress ESCC. Further studies are required to
fully elucidate the link between the differentiation-related
functions of ZNF750 and its growth-suppressive ability in ESCC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Wei Dai
(Department of Clinical Oncology, The University of Hong Kong) for
insights and guidance with bioinformatics analysis. The authors
would also like to thank Professor George Tsao (School of
Biomedical Sciences) and Professor Gopesh Srivastava (Department of
Pathology) of The University of Hong Kong for providing the cell
lines used in the study.
Funding
This work was funded by the Research Grants Council
Collaborative Research Fund (grant no. 106150246), the Theme-based
Research Scheme (grant no. T12-701/17-R) and the Asian Cancer
Research Fund.
Availability of data and materials
The RNA sequencing data generated and/or analyzed
during the current study are available in the National Center for
Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under the
accession no. PRJNA707329.
Authors' contributions
SSAC contributed to the design and execution of the
study and data interpretation, and wrote the manuscript. JMYK and
VZY contributed to the design of the study, data interpretation and
the preparation of the manuscript. SSAC and JMYK confirm the
authenticity of the raw data. LN contributed to the processing of
the raw RNA-sequencing data and its subsequent analysis. MLL
contributed to the study design, preparation of the manuscript and
overall supervision of the study. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval for animal studies was obtained from
the Animal Ethics Committee of The University of Hong Kong
(approval no. CULATR 3377-14). The tumor burden did not exceed the
recommended dimensions set by The University of Hong Kong, and the
animals were anesthetized and sacrificed using acceptable
methods/techniques.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dotto GP and Rustgi AK: Squamous cell
cancers: A unified perspective on biology and genetics. Cancer
Cell. 29:622–637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cassandri M, Smirnov A, Novelli F, Pitolli
C, Agostini M, Malewicz M, Melino G and Raschellà G: Zinc-finger
proteins in health and disease. Cell Death Discov. 3:170712017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jen J and Wang YC: Zinc finger proteins in
cancer progression. J Biomed Sci. 23:532016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sen GL, Boxer LD, Webster DE, Bussat RT,
Qu K, Zarnegar BJ, Johnston D, Siprashvili Z and Khavari PA: ZNF750
is a p63 target gene that induces KLF4 to drive terminal epidermal
differentiation. Dev Cell. 22:669–677. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boxer LD, Barajas B, Tao S, Zhang J and
Khavari PA: ZNF750 interacts with KLF4 and RCOR1, KDM1A, and
CTBP1/2 chromatin regulators to repress epidermal progenitor genes
and induce differentiation genes. Genes Dev. 28:2013–2026. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hazawa M, Lin DC, Handral H, Xu L, Chen Y,
Jiang YY, Mayakonda A, Ding LW, Meng X, Sharma A, et al: ZNF750 is
a lineage-specific tumour suppressor in squamous cell carcinoma.
Oncogene. 36:2243–2254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Zhou Y, Cheng C, Cui H, Cheng L,
Kong P, Wang J, Li Y, Chen W, Song B, et al: Genomic analyses
reveal mutational signatures and frequently altered genes in
esophageal squamous cell carcinoma. Am J Hum Genet. 96:597–611.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dai W, Ko JMY, Choi SSA, Yu Z, Ning L,
Zheng H, Gopalan V, Chan KT, Lee NP, Chan KW, et al: Whole-exome
sequencing reveals critical genes underlying metastasis in
oesophageal squamous cell carcinoma. J Pathol. 242:500–510. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo A, Kong J, Hu G, Liew CC, Xiong M,
Wang X, Ji J, Wang T, Zhi H, Wu M and Liu Z: Discovery of
Ca2+-relevant and differentiation-associated genes
downregulated in esophageal squamous cell carcinoma using cDNA
microarray. Oncogene. 23:1291–1299. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uemura N, Nakanishi Y, Kato H, Saito S,
Nagino M, Hirohashi S and Kondo T: Transglutaminase 3 as a
prognostic biomarker in esophageal cancer revealed by proteomics.
Int J Cancer. 124:2106–2115. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Feng YB, Shen XM, Chen BS, Du XL,
Luo ML, Cai Y, Han YL, Xu X, Zhan QM and Wang MR: Exogenous
expression of Esophagin/SPRR3 attenuates the tumorigenicity of
esophageal squamous cell carcinoma cells via promoting apoptosis.
Int J Cancer. 122:260–266. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L,
Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al: Genomic and
molecular characterization of esophageal squamous cell carcinoma.
Nat Genet. 46:467–473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
DeWard AD, Cramer J and Lagasse E:
Cellular heterogeneity in the mouse esophagus implicates the
presence of a nonquiescent epithelial stem cell population. Cell
Rep. 9:701–711. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sanger F, Nicklen S and Coulson AR: DNA
sequencing with chain-terminating inhibitors. Proc Natl Acad Sci
USA. 74:5463–5467. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shuen WH, Kan R, Yu Z, Lung HL and Lung
ML: Novel lentiviral-inducible transgene expression systems and
versatile single-plasmid reporters for in vitro and in vivo cancer
biology studies. Cancer Gene Ther. 22:207–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen J, Bardes EE, Aronow BJ and Jegga AG:
ToppGene Suite for gene list enrichment analysis and candidate gene
prioritization. Nucleic Acids Res. 37:305–311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grossman RL, Heath AP, Ferretti V, Varmus
HE, Lowy DR, Kibbe WA and Staudt LM: Toward a shared vision for
cancer genomic data. N Engl J Med. 375:1109–1112. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Collins FS and Barker AD: Mapping the
cancer genome. Pinpointing the genes involved in cancer will help
chart a new course across the complex landscape of human
malignancies. Sci Am. 296:50–57. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
R Core Team (2014), . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: http://www.R-project.org/
|
|
27
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koterazawa Y, Koyanagi-Aoi M, Uehara K,
Kakeji Y and Aoi T: Retinoic acid receptor γ activation promotes
differentiation of human induced pluripotent stem cells into
esophageal epithelium. J Gastroenterol. 55:763–774. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ohashi S, Miyamoto S, Kikuchi O, Goto T,
Amanuma Y and Muto M: Recent advances from basic and clinical
studies of esophageal squamous cell carcinoma. Gastroenterology.
149:1700–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao YB, Chen ZL, Li JG, Hu XD, Shi XJ, Sun
ZM, Zhang F, Zhao ZR, Li ZT, Liu ZY, et al: Genetic landscape of
esophageal squamous cell carcinoma. Nat Genet. 46:1097–1102. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Whelan KA, Muir AB and Nakagawa H:
Esophageal 3D culture systems as modeling tools in esophageal
epithelial pathobiology and personalized medicine. Cell Mol
Gastroenterol Hepatol. 5:461–478. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He H, Li S, Hong Y, Zou H, Chen H, Ding F,
Wan Y and Liu Z: Krüppel-like factor 4 promotes esophageal squamous
cell carcinoma differentiation by Up-regulating keratin 13
expression. J Biol Chem. 290:13567–1377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhong X, Huang G, Ma Q, Liao H, Liu C, Pu
W, Xu L, Cai Y and Guo X: Identification of crucial miRNAs and
genes in esophageal squamous cell carcinoma by miRNA-mRNA
integrated analysis. Medicine (Baltimore). 98:e162692019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Luo A, Chen H, Ding F, Zhang Y, Wang M,
Xiao Z and Liu Z: Small proline-rich repeat protein 3 enhances the
sensitivity of esophageal cancer cells in response to DNA
damage-induced apoptosis. Mol Oncol. 7:955–967. 2013. View Article : Google Scholar : PubMed/NCBI
|