Introduction
Glucose-induced degradation-deficient (GID) genes
are required for the glucose-induced degradation of yeast
fructose-1,6-bisphosphatase (1).
Required for meiotic nuclear division 5 homolog A (RMND5A) is the
human ortholog of the yeast gene, Gid2, which contains the Lis
homology (LisH), C-terminal to LisH, CT11-RanBPM and E3 typical
RING domains (1,2). RMND5A functions as an E3 ubiquitin
ligase, which combines with E1 and E2 enzymes to catalyze protein
ubiquitination (1,3). Previous studies have reported that
RMND5A regulated c-Raf expression and that downregulation of RMND5A
inhibited the migration of HeLa cells (4–6).
Abnormal expression levels of RMND5A have been found to be
associated with poor prognosis in breast cancer (7). However, there have been few reports on
the roles of RMND5A in cancer.
MicroRNAs (miRNAs/miRs) are small non-coding RNA
molecules, 21–23 nucleotides in length, which can regulate gene
expression at the post-transcriptional level by degrading targeted
mRNAs or suppressing their translation (8–11).
Numerous studies have demonstrated the important roles of miRNAs
during angiogenesis and tumorigenesis (12–15). To
date, only miR-21 and miR-138 have been reported to target RMND5A
(6,16). The aim of the present study was to
investigate the differential expression of RMND5A and its
prognostic role in multiple types of cancer. The present results
revealed a crucial function of RMND5A in pancreatic adenocarcinoma
(PAAD) and identified a novel miRNA, miR-590-5p, that targets
RMND5A in PAAD cell lines.
Materials and methods
Cell lines and culture conditions
Two human PAAD cell lines, AsPC-1 and PANC-1, were
purchased from the American Type Culture Collection and were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (HyClone; Cytiva) at
37°C in a humidified incubator with 5% CO2.
Identification of potential miRNAs
targeting RMND5A
Four miRNA-target gene databases were used to
predict the miRNAs targeting RMND5A: TargetScan (http://www.targetscan.org/), miRDB (http://www.mirdb.org/), miRcode (http://www.mircode.org/) and TarBase v8 (http://www.microrna.gr/tarbase). In addition, we
further evaluated their possible interactions by using starBase
(http://starbase.sysu.edu.cn/).
Cell transfection
miR-590-5p mimics (5′GAGCUUAUUCAUAAAAGU-GCAG-3′) and
negative control (5′CGGUACGAUCGCGGCGGGAUAUC-3′) were synthesized by
Guangzhou RiboBio Co., Ltd., and transfected into the PAAD cell
lines using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at the concentration of 100 nM. After
transfection for 48 h, reverse transcription-quantitative PCR
(RT-qPCR) analysis was performed to detect the expression levels of
miR-590-5p. The plasmid for overexpressing human RMND5A was
constructed using the pHS-AVC (pLV-hef1a-mScarlet-P2A-neo-WPRE-CMV)
vector (Beijing Syngentech Co., Ltd.), according to the human
RMND5A gene sequence in Genbank (NM_022780.4). HEK293 (CRL-1573,
ATCC) cultured in 6-cm dishes were used to pack viral particles for
RMND5A overexpression. Briefly, 27 µl FuGENE® 6 (Promega
Corporation), 4 µg of recombinant plasmids or empty vector as
control, 3 µg PAX2 (cat. no. 12260; Addgene, Inc.) and 2 µg VSVG
(cat. no. 8454; Addgene Inc.) were mixed in 300 ul OPTI-MEM (Gibco;
Thermo Fisher Scientific, Inc.), followed by being dropped into
6-cm dish. Then, the cells were placed back in the incubator at
37°C. After 6 h, all the medium was replaced by pre-warmed DMEM
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum. After 48 h, ~4 ml of the lentiviral
particles in culture media were collected. For infection, 1 ml of
the lentiviral particles and 10 µg/ml polybrene (Sigma-Aldrich;
Merck KGaA) were used to infect 0.5 million cells at 37°C for 4 h.
After 4 h, more prewarmed medium was added for culturing at 37°C.
After 48 h, RT-qPCR analysis was performed to detect the mRNA
expression levels of RMND5A.
RT-qPCR
Total RNA was extracted either using the mirVana
miRNA kit (Ambion; Thermo Fisher Scientific, Inc.) or the
E.Z.N.A® Total RNA kit (Omega Bio-Tek, Inc.). For mRNA
detection, 1,000 ng RNA was reverse transcribed into cDNA using
qScript cDNA Synthesis kit (cat. no. 95047; Quantabio) according to
the manufacturer's protocol and quantified using the Fast Start
Universal SYBRGreen master mix (cat. no. 4913850001; Merck KGaA).
For miRNA detection, 250 ng RNA was reverse-transcribed into cDNA
and quantified using the Mir-X™ miRNA qRT-PCR TB Green™ kit (cat.
no. 638314; Takara Biotechnology Co., Ltd.). GAPDH and U6 were used
as the internal control for mRNA and miRNA, respectively. The
QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.) was used for qPCR. The thermocycling
conditions used were as follows: Pre-denaturation at 95°C for 1
min; 40 cycles of denaturation at 95°C for 15 sec; annealing at
60°C for 15 sec; and extension at 72°C for 30 sec followed by
melting curve analysis. For quantification, 2−ΔΔCq
method (17) was used and the primer
sequences are listed in Table I.
 | Table I.Primer sequences used for reverse
transcription-quantitative PCR analysis. |
Table I.
Primer sequences used for reverse
transcription-quantitative PCR analysis.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| miR-590-5p |
GAGCTTATTCATAAAAGT |
TCCACGACACGCACTGGATACGAC |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| RMND5A |
AATGAGGTGATGGTGGAGCA |
GCATTTCCCGGTTTGACACT |
| GAPDH |
GGAGCGAGATCCCTCCAAAAT |
GGCTGTTGTCATACTTCTCATGG |
Western blot analysis
Total protein was extracted from the AsPC-1 cells
using the radioimmunoprecipitation assay (RIPA) lysis buffer
(Beyotime Institute of Biotechnology). The Pierce BCA Protein Assay
kit (Thermo Fisher Scientific, Inc.) was used to quantify the total
extracted protein according to the manufacturer's protocol. A total
of 30 µg of proteins were separated using 10% SDS-PAGE and
transferred onto PVDF membranes (EMD Millipore) at room temperature
(RT). The membranes were subsequently incubated with rabbit
polyclonal anti-RMND5A (1:1,000; cat. no. ab254902; Abcam) and
rabbit polyclonal anti-GAPDH antibodies (1:1,000; cat. no.
10494-1-AP; ProteinTech Group, Inc.) overnight at 4°C. Then, the
membranes were incubated with goat polyclonal anti-rabbit IgG
secondary antibody (1:1,000; cat. no. ab6721; Abcam) for 1 h at RT.
The protein expression levels were measured using an enhanced
chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.).
Wound healing assay
The human PAAD cell lines AsPC-1 and PANC-1 were
seeded in a 6-well plate at 1×106 cells/well. When the
cells reached 80–90% confluence, the cell monolayer was scraped
with a 200-µl pipette tip. After washing with PBS, the cells were
cultured for another 24 h. The medium used for this assay was
serum-free DMEM. The migrated cells were observed using a
phase-contrast light microscope at ×10 objective magnification
(Olympus Corporation).
Statistical analysis
Data are presented as mean ± SD and analysed using
GraphPad Prism 8 (GraphPad Software Inc.). A total of 3 biological
replicates and 3 technical repeats were performed. For the
comparison of two groups, Student's t-test was used. One-way ANOVA
followed by Tukey's multiple comparisons test was performed when
comparing more than two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
RMND5A is highly expressed and
associated with overall survival in patients with PAAD
To investigate the role of RMND5A in cancer, the
expression levels of RMND5A in 31 types of cancer were analyzed
using the Gene Expression Profiling Interactive Analysis platform
(http://gepia.cancer-pku.cn). As shown in
Fig. 1A and B, the expression levels
of RMND5A were significantly higher in PAAD, stomach adenocarcinoma
(STAD) and thymoma (THYM) tissues compared with that in normal
tissues. Notably, a lower RMND5A expression was associated with
better overall survival in patients with PAAD (Fig. 1C). However, in patients with STAD and
THYM, RMND5A expression levels were not significantly associated
with overall survival (Fig. 1C).
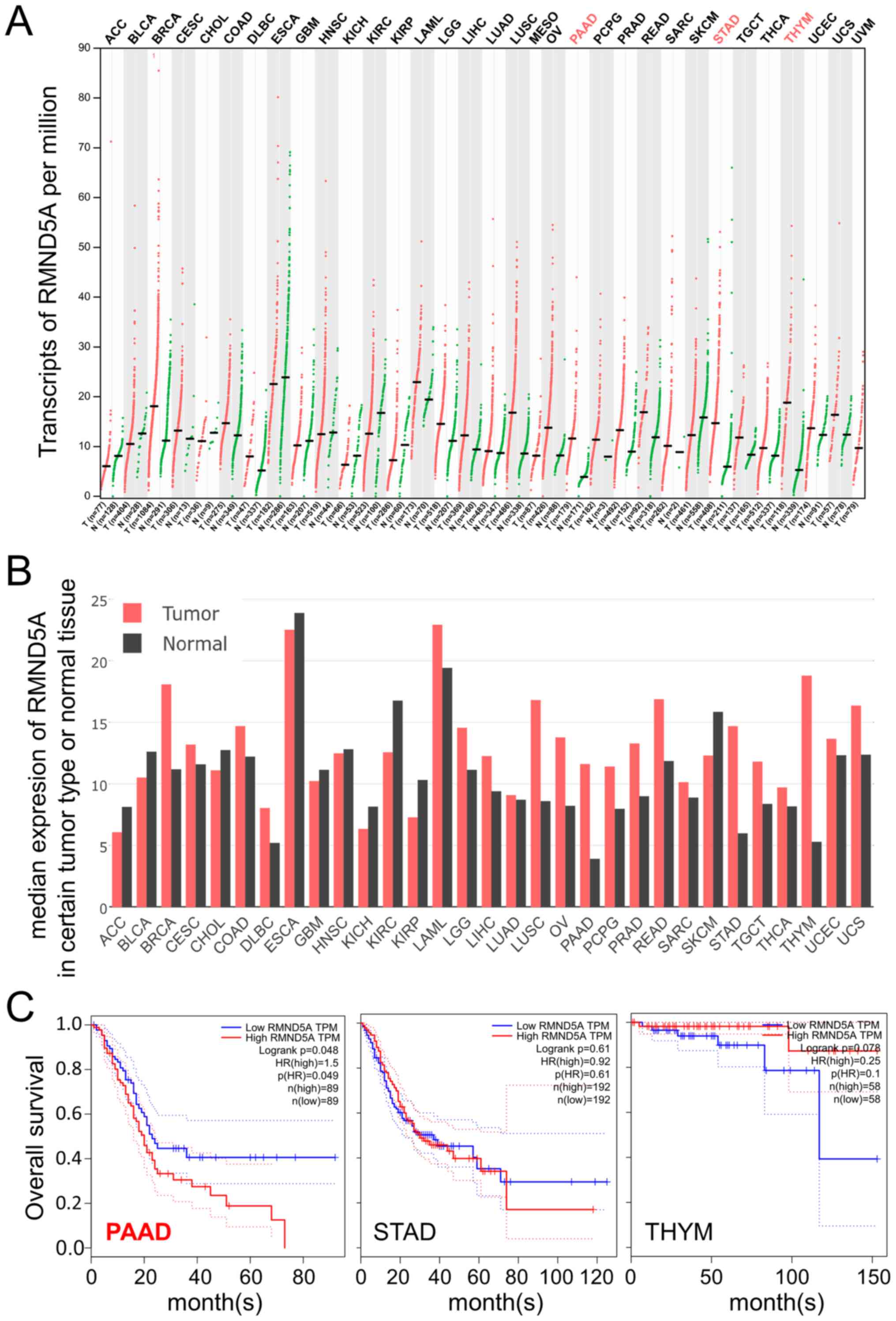 | Figure 1.RMND5A is highly expressed in the
tumor tissues of patients with PAAD and is associated with overall
survival. Data in the Gene Expression Profiling Interactive
Analysis platform (http://gepia.cancer-pku.cn) were analyzed in respect
to RMND5A expression. (A) Transcripts per million and (B) median
expression of RMND5A are plotted in multiple types of cancer. (C)
Association between RMND5A expression levels and overall survival
in patients with PAAD, STAD and THYM. RMND5A, required for meiotic
nuclear division 5 homolog A; ACC, adenoid cystic carcinoma; BLCA,
bladder urothelial carcinoma; BRCA, breast cancer; CESC, cervical
squamous cell carcinoma and endocervical adenocarcinoma; CHOL,
cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large
B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma;
HNSC, head and neck squamous cell carcinoma; KICH, kidney
chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney
renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG,
brain lower grade glioma; LIHC, hepatocellular carcinoma; LUAD,
lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO,
mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD,
pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD,
stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA,
thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial
carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma. |
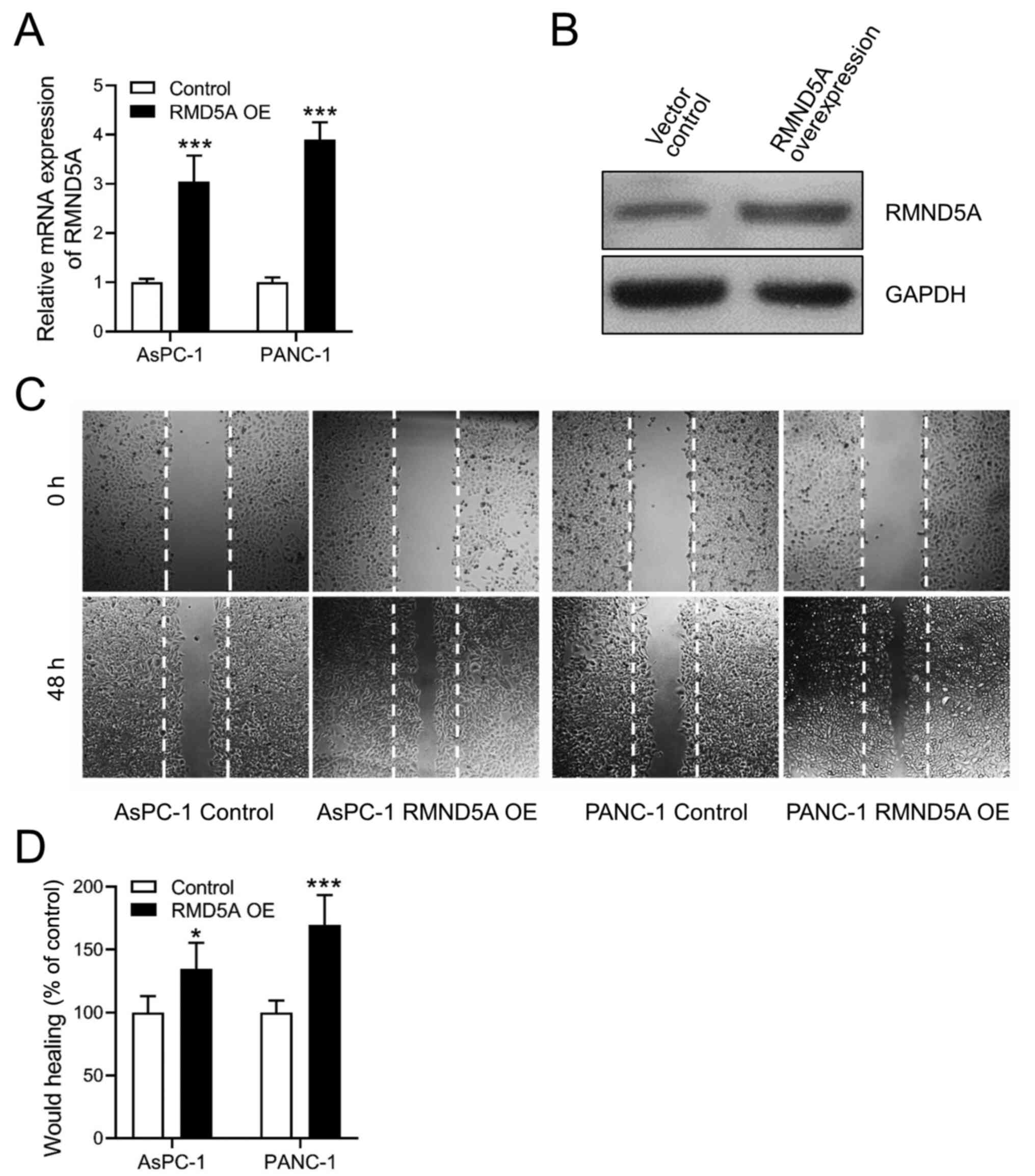
RMND5A promotes migration in PAAD cell
lines
Based on the aforementioned data, the effects of
RMND5A overexpression were investigated in PAAD cell lines to
determine its function. The transfection efficiency of the RMND5A
overexpression plasmid was confirmed using RT-qPCR and western blot
analyses (Fig. 2A and B). Notably,
it was found that overexpression of RMND5A significantly increased
cell migration in both the AsPC-1 and PANC-1 cell lines (Fig. 2C and D).
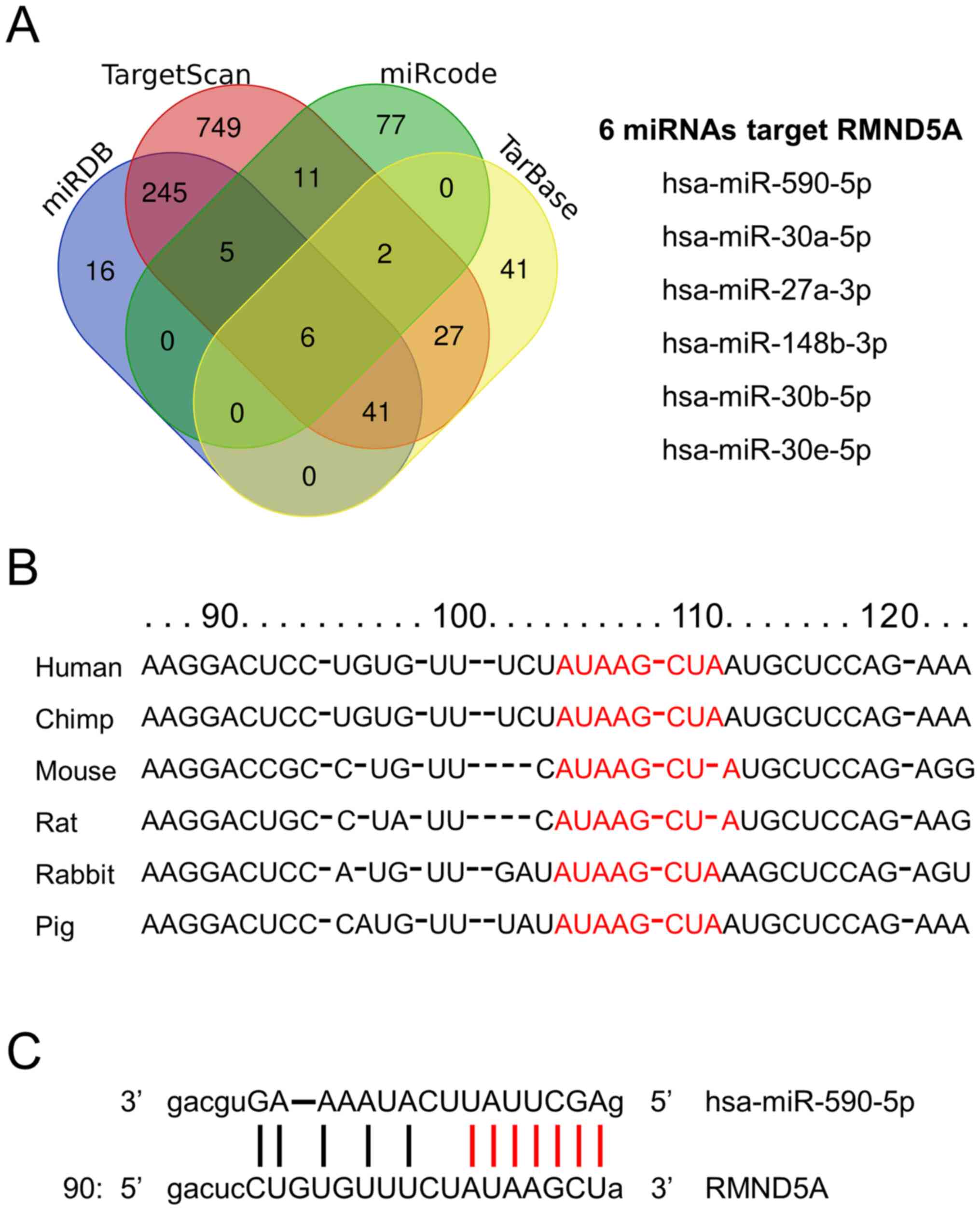
miR-590-5p targets the RMND5A
gene
To predict the potential miRNAs that target RMND5A,
in silico analysis was performed by using the TargetScan,
miRDB, miRcode and TarBase databases. The overlap analysis
identified six potential miRNAs, namely miR-590-5p, miR-30a-5p,
miR-27a-3p, miR-148b-3p, miR-30b-5p, and miR-30e-5p (Fig. 3A). In addition, the results of the
targeting sites, Cross-linking immunoprecipitation (ClipSeq)
PeakCluster and ReadNumber of miRNA-RMND5A interaction in StarBase
were checked, which revealed that miR-590-5p was among the top
hits. The 3′ untranslated region (UTR) of both miR-590-5p and
RMND5A were highly conserved among mammals (Fig. 3B). Finally, the mature miR-590-5p
seeding region exhibited a perfect alignment with the RMND5A 3′UTR
(Fig. 3C).
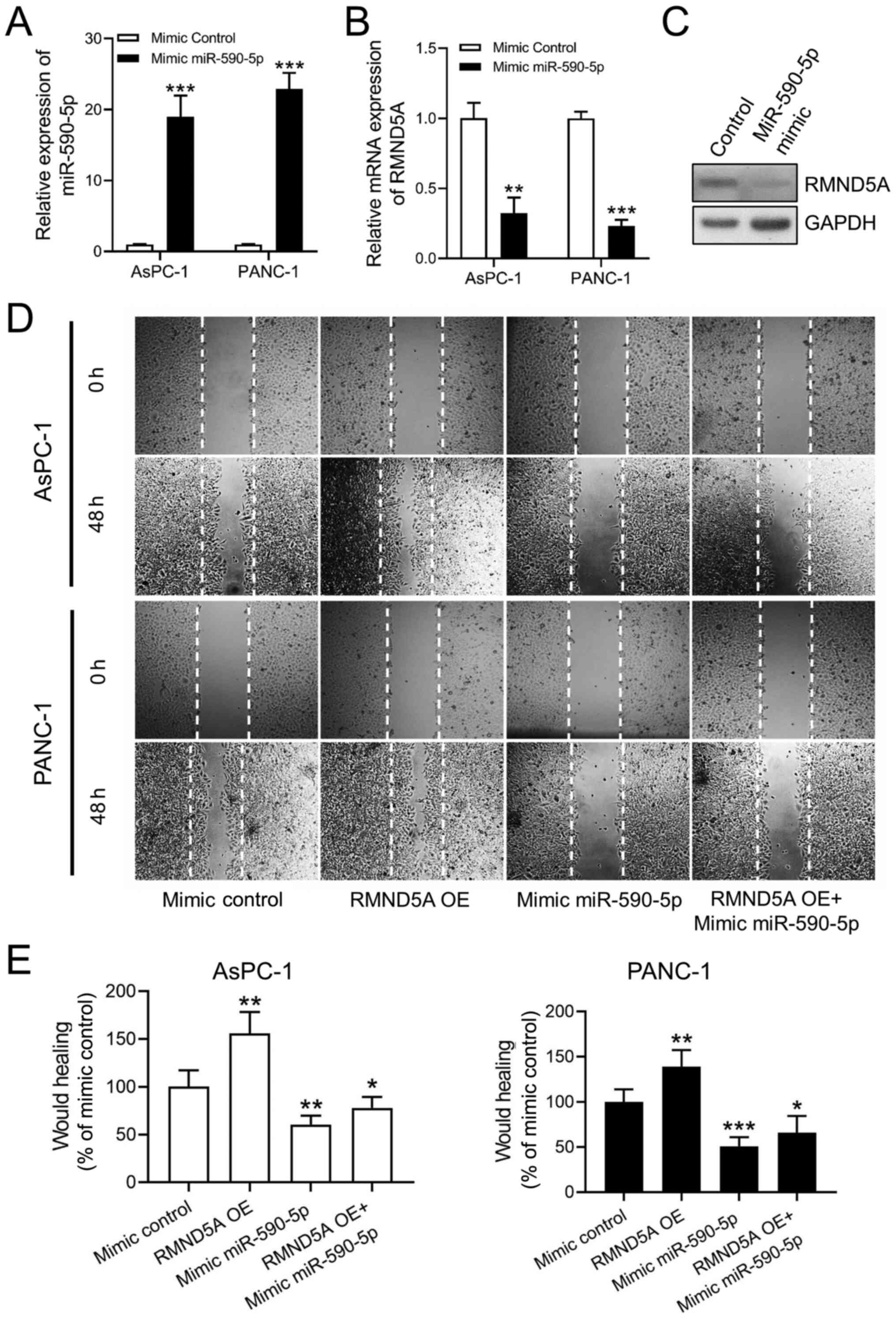
Overexpression of miR-590-5p inhibits
RMND5A expression and cell migration in PAAD cell lines
To confirm the hypothesis that miR-590-5p targets
the RMND5A gene, miR-590-5p mimics were transfected into the PAAD
cell lines. The transfection efficiency of miR-590-5p mimics was
confirmed using RT-qPCR (Fig. 4A).
Following transfection with miR-590-5p mimics, the AsPC-1 and
PANC-1 cell lines exhibited significantly decreased RMND5A
expression levels (Fig. 4B and C).
The wound healing assay demonstrated that miR-590-5p mimics
decreased the migration ability of the AsPC-1 and PANC-1 cells
(Fig. 4D and 4E). Furthermore, miR-590-5p mimics
attenuated the promoting effects of RMND5A overexpression on the
migration of the PAAD cells (Fig. 4D and
E).
Discussion
PAAD is a common gastrointestinal malignancy with
poor prognosis (18,19). Only few effective drugs have been
developed for the treatment of PAAD (19). The 5-year survival rate is <5% and
the majority of patients with PAAD succumb to metastatic disease
(18,20). It is well-known that distant
metastasis is associated with the migration of tumor cells
(19,21). Therefore, it is important to
elucidate the molecular pathogenic processes underlying the
migration of PAAD cells. Previous studies have demonstrated that
RMND5A is important for cell migration and adhesion (5,7). RMND5A
was also found to be involved in the metastasis of primary tumors
(5,22). However, to the best of our knowledge,
the association between RMND5A and PAAD has not been investigated.
In the present study, it was observed that the RMND5A expression
levels were significantly upregulated in PAAD tissues compared with
the control normal tissues. High levels of RMND5A in patients with
PAAD were associated with poor overall survival. Mechanistically,
RMND5A was found to have a key role in the migration of PAAD cells,
with RMND5A overexpression significantly increasing the PAAD cell
migratory ability. The results of the present study indicated that
RMND5A may be a novel prognostic biomarker for patients with
PAAD.
In silico analysis identified miR-590-5p as a
potential miRNA that may target RMND5A. Notably, the effects of
miR-590-5p on tumorigenesis are controversial. Numerous studies
have demonstrated that miR-590-5p functions as a tumor suppressor
in breast cancer, non-small cell lung cancer and tongue squamous
cell carcinoma (23–25). miR-590-5p may also inhibit the
migration or proliferation of breast cancer and hepatocellular
carcinoma cells by targeting the Wnt signaling pathway (26,27). In
addition, miR-590-5p may inhibit the proliferation of malignant
melanoma cells and suppress the chemoresistance of hepatocellular
carcinoma cells by targeting Yes1 associated transcriptional
regulator (28,29). By contrast, it has also been reported
that miR-590-5p may promote cell proliferation and invasion in
cervical cancer, renal cell carcinoma and endometrial cancer
(30–33). In addition, miR-590-5p may promote
malignant behaviors in vulvar squamous cell carcinoma and
hepatocellular carcinoma cells by targeting TGFβ receptor 2
(34,35). To the best of our knowledge, the
present study is the first to report that miR-590-5p inhibited the
migration of PAAD cells by decreasing RMND5A expression.
Furthermore, overexpression of miR-590-5p attenuated the promoting
effects of RMND5A on the migration of PAAD cells. However, as with
majority of studies, the design of the current study is subject to
limitations. There are two major limitations in this study that
could be addressed in future research. Firstly, the specific
interaction between RMND5A and miR-590-5p should be further
confirmed in the future with more experiments, such as
dual-luciferase reporter assay. Secondly, the role of RMND5A in
other biological functions associated with PAAD, such as
proliferation, invasion and metastasis, requires further
investigation.
In summary, to the best of our knowledge, the
present study was the first to report the increased expression
levels of RMND5A in PAAD tissues and its association with poor
overall survival. Functionally, RMND5A was demonstrated to promote
the migration of PAAD cells in vitro. In addition,
miR-590-5p may inhibit the migration of PAAD cells by targeting
RMND5A. These results may shed new light on the mechanisms
underlying PAAD progression and provide novel targets for the
treatment of PAAD.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant nos. 31872315 and 81670290), the
Cooperative Innovation Center of Engineering and New Products for
Developmental Biology of Hunan Province (grant no. 2013-448-6), the
National Key Research and Development Program of China (grant no.
2018YFA0108700), the NSFC Projects of International Cooperation and
Exchanges (grant. no. 81720102004), the Research Team Project of
Natural Science Foundation of Guangdong Province of China (grant
no. 2017A030312007), the Science and Technology Planning Projects
of Guangdong Province of China (grant no. 2019B020230003), the
Special Project of Dengfeng Program of Guangdong Provincial
People's Hospital (grant no. DFJH201802) and the Key Program of
Guangzhou Science Research Plan (grant no. 805212639211).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Author's contributions
SC contributed to the design of the study, data
acquisition and analysis, and drafted the manuscript. YC and YoW
performed data acquisition and analysis. WC, PZ, WY, YL, XF, YaW,
FL and JZ performed data analysis. ZJ, XW and YuW contributed to
the design of the experiments and critically revised the
manuscript. SC and YC have seen and can confirm the authenticity of
the raw data. All authors have read and approved the final
manuscript and agree to be accountable for all aspects of the
study.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing
interests.
References
|
1
|
Pfirrmann T, Villavicencio-Lorini P,
Subudhi AK, Menssen R, Wolf DH and Hollemann T: RMND5 from Xenopus
laevis is an E3 ubiquitin-ligase and functions in early embryonic
forebrain development. PLoS One. 10:e01203422015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maitland MER, Onea G, Chiasson CA, Wang X,
Ma J, Moor SE, Barber KR, Lajoie GA, Shaw GS and Schild-Poulter C:
The mammalian CTLH complex is an E3 ubiquitin ligase that targets
its subunit muskelin for degradation. Sci Rep. 9:98642019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
van Wijk SJ, de Vries SJ, Kemmeren P,
Huang A, Boelens R, Bonvin AM and Timmers HT: A comprehensive
framework of E2-RING E3 interactions of the human
ubiquitin-proteasome system. Mol Syst Biol. 5:2952009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McTavish CJ, Berube-Janzen W, Wang X,
Maitland MER, Salemi LM, Hess DA and Schild-Poulter C: Regulation
of c-Raf Stability through the CTLH Complex. Int J Mol Sci.
20:9342019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huffman N, Palmieri D and Coppola V: The
CTLH complex in cancer cell plasticity. J Oncol. 2019:42167502019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Chen Y, Qin X, Wen J, Ding H, Xia W,
Li S, Su X, Wang W, Li H, et al: miR-138 downregulates miRNA
processing in HeLa cells by targeting RMND5A and decreasing
Exportin-5 stability. Nucleic Acids Res. 42:458–474. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu H and Ye H: Screening of the
prognostic targets for breast cancer based co-expression modules
analysis. Mol Med Rep. 16:4038–4044. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Djuranovic S, Nahvi A and Green R: A
parsimonious model for gene regulation by miRNAs. Science.
331:550–553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kusenda B, Mraz M, Mayer J and Pospisilova
S: MicroRNA biogenesis, functionality and cancer relevance. Biomed
Pap Med Fac Univ Palacky Olomouc Czech Repub. 150:205–215. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang JT, Wang F, Chapin W and Huang RS:
Identification of MicroRNAs as breast cancer prognosis markers
through the cancer genome atlas. PLoS One. 11:e01682842016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu K, Zhang Z, Zhang H, Wang Z and Wang
F: miR-142-3p targeting NUCKS1 inhibits proliferation and invasion
of pancreatic cancer cells. Artif Cells Nanomed Biotechnol.
48:415–424. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamauchi A, Yamamura M, Katase N, Itadani
M, Okada N, Kobiki K, Nakamura M, Yamaguchi Y and Kuribayashi F:
Evaluation of pancreatic cancer cell migration with multiple
parameters in vitro by using an optical real-time cell mobility
assay device. BMC Cancer. 17:2342017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roussos ET, Condeelis JS and Patsialou A:
Chemotaxis in cancer. Nat Rev Cancer. 11:573–587. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lambert AW, Pattabiraman DR and Weinberg
RA: Emerging biological principles of metastasis. Cell.
168:670–691. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu BB, Gu ZF, Ma M, Wang JY and Wang HN:
MicroRNA-590-5p suppresses the proliferation and invasion of
non-small cell lung cancer by regulating GAB1. Eur Rev Med
Pharmacol Sci. 22:5954–5963. 2018.PubMed/NCBI
|
|
24
|
Xu Y, Han T, Zhu Y and Chen Q: miR-590-5P
inhibits the progression of tongue squamous cell carcinoma by
targeting FasL. Int J Clin Exp Pathol. 10:11880–11887.
2017.PubMed/NCBI
|
|
25
|
Yan M, Ye L, Feng X, Shi R, Sun Z, Li Z
and Liu T: MicroRNA-590-3p inhibits invasion and metastasis in
triple-negative breast cancer by targeting Slug. Am J Cancer Res.
10:965–974. 2020.PubMed/NCBI
|
|
26
|
Gao J, Yu SR, Yuan Y, Zhang LL, Lu JW,
Feng JF and Hu SN: MicroRNA-590-5p functions as a tumor suppressor
in breast cancer conferring inhibitory effects on cell migration,
invasion, and epithelial-mesenchymal transition by downregulating
the Wnt-β-catenin signaling pathway. J Cell Physiol. 234:1827–1841.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shan X, Miao Y, Fan R, Qian H, Chen P, Liu
H, Yan X, Li J and Zhou F: miR-590-5P inhibits growth of HepG2
cells via decrease of S100A10 expression and Inhibition of the Wnt
pathway. Int J Mol Sci. 14:8556–8569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen M, Wu L, Tu J, Zhao Z, Fan X, Mao J,
Weng Q, Wu X, Huang L, Xu M and Ji J: miR-590-5p suppresses
hepatocellular carcinoma chemoresistance by targeting YAP1
expression. EBioMedicine. 35:142–154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mou K, Liu B, Ding M, Mu X, Han D, Zhou Y
and Wang LJ: lncRNA-ATB functions as a competing endogenous RNA to
promote YAP1 by sponging miR-590-5p in malignant melanoma. Int J
Oncol. 53:1094–1104. 2018.PubMed/NCBI
|
|
30
|
Chu Y, Ouyang Y, Wang F, Zheng A, Bai L,
Han L, Chen Y and Wang H: MicroRNA-590 promotes cervical cancer
cell growth and invasion by targeting CHL1. J Cell Biochem.
115:847–853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiao X, Tang C, Xiao S, Fu C and Yu P:
Enhancement of proliferation and invasion by MicroRNA-590-5p via
targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res.
20:537–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang L, Wei WQ, Wu ZY and Wang GC:
MicroRNA-590-5p regulates cell viability, apoptosis, migration and
invasion of renal cell carcinoma cell lines through targeting
ARHGAP24. Mol Biosyst. 13:2564–2573. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu M, Zhou X, Zhang J and Zhang Y:
MicroRNA-590-5p promotes cell survival in endometrioid endometrial
cancer by suppressing tumor suppressor PTEN. Int J Clin Exp Pathol.
10:7836–7846. 2017.PubMed/NCBI
|
|
34
|
Jiang X, Xiang G, Wang Y, Zhang L, Yang X,
Cao L, Peng H, Xue P and Chen D: MicroRNA-590-5p regulates
proliferation and invasion in human hepatocellular carcinoma cells
by targeting TGF-β RII. Mol Cells. 33:545–551. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang X and Wu X: miRNA expression profile
of vulvar squamous cell carcinoma and identification of the
oncogenic role of miR-590-5p. Oncol Rep. 35:398–408. 2016.
View Article : Google Scholar : PubMed/NCBI
|