Introduction
Primary liver cancer is estimated to be the third
leading cause of cancer-related deaths worldwide, accounting for
830,000 deaths each year (1).
Hepatocellular carcinoma (HCC) is the most common type of primary
liver cancer comprising 75–85% of cases (1). Despite recent advances in multikinase
inhibitors, such as sorafenib, regorafenib, and lenvatinib, as well
as anti-vascular endothelial growth factor therapies and immune
check point inhibitors, advanced HCC has a dismal prognosis
(2–5). An exploration of the molecular
characteristics of liver cancer is needed to develop more effective
therapeutics.
A well characterized oncoprotein, c-Myc contributes
to the pathogenesis of a broad range of human cancers, including
liver cancer (6). Overexpression of
c-Myc is associated with a poor prognosis (7). The amplification of c-Myc and
alterations of proximal c-Myc network members have been identified
in over 30% and 70% of HCC cases, respectively (8). Such findings highlight c-Myc as an
attractive target for liver cancer therapeutics. However, its
structure, which lacks a druggable hydrophobic pocket, and its
nuclear localization have hampered the development of specific
inhibitors of c-Myc (9). Certainly,
ongoing clinical trials of c-Myc inhibitors are non-existent,
except for a trial involving 90-amino acid peptide as a dominant
negative inhibitor (10).
Specifically, it is critical to seek druggable targets in the c-Myc
pathway to combat c-Myc-driven liver cancer.
Six-transmembrane epithelial antigen of the prostate
1 (STEAP1), which was initially identified in prostate cancer cells
and is expressed at low levels in normal cells, is a cell surface
protein (11) that is over-expressed
in many human cancers (12). STEAP1
has thus emerged as an ideal target in cancer therapeutics. STEAP1
is believed to play a physiological role as an ion channel and
transporter (13). Additionally,
structural analyses using a cryo-electron microscopy revealed that
STEAP1 works as a ferric reductase when binding to the
NADPH-binding domain of STEAP4 (14). In contrast, the pathological
functions of STEAP1 in cancer cells have been largely unexplored.
We recently discovered that high expression of STEAP1 lead to the
suppression of reactive oxygen species (ROS) that escaped from
apoptosis via a NF-E2-related factor 2 (NRF2) pathway in colorectal
cancer cells (15). However, the
roles of STEAP1 in liver cancer pathogenesis remain completely
unknown.
Here, we sought to characterize the biological roles
of STEAP1 in liver cancer. We identified that STEAP1
transcript levels were significantly increased in liver cancer
compared to normal liver cells, and that such high levels were
associated with a poor prognosis. The knockdown of STEAP1
led to cell-growth inhibition accompanied by G1 arrest
by targeting the suppression of c-Myc, which was discovered by
mining publicly available databases. Our findings yield a new
treatment strategy targeting the STEAP1-c-Myc axis in liver
cancer.
Materials and methods
Databases and gene expression data
analysis
Gene expression levels of STEAP1 in non-tumor
and liver cancer tissues were evaluated using gene expression
profiles of GSE14520 and GSE36376 from the Gene Expression Omnibus,
a public and freely available database. The GSE14520 dataset
includes 488 samples of 241 non-cancerous and 247 cancerous hepatic
tissues. These datasets have been widely used and well accepted in
bioinformatics analysis of liver cancer. The GSE36376 dataset
includes 433 samples consisting of 193 non-cancerous hepatic
tissues and 240 cancerous tissues. The correlation between
STEAP1 levels and clinical outcomes of patients with liver
cancer was investigated using GSE14520 and the Cancer Genome Atlas
Program (TCGA) (16). We used a
receiver operating characteristic curve to determine the cutoff
value. In total, 247 patients from the GSE14520 dataset and 360
patients from TCGA, all with liver cancer, were divided into two
groups having high or low levels of STEAP1, respectively.
Kaplan-Meier analyses of survival were performed based on these
groups. Statistical analyses were performed using EZR software
version 1.33 (17).
Gene set enrichment analysis (GSEA) was performed
using the open source software, GSEA 4.0.3. Initially, we set two
groups (STEAP1_high and STEAP1_low) in
GSE14520-GPL3921, which includes 225 liver cancer samples in total.
We conducted GSEA of the two groups using Hallmark gene sets. Gene
sets showing a NOM P-val. (P-value) <0.05 and false discovery
rate (FDR) Q-val. (FDR) <0.25 were considered significant.
Differentially expressed genes (DEGs) between these two groups were
identified using an online tool, GEO2R, with |logFC| >1.5 and an
adjusted P-value <0.05.
Cell lines and culture conditions
HepG2 and Hep3B cell lines were purchased from the
American Type Culture Collection; these were authenticated by short
tandem repeat DNA profiling prior to all experiments. Both cell
lines were cultured in DMEM containing 10% fetal bovine serum
(FBS), 2 µM L-glutamine and 1% penicillin-streptomycin (the medium
and all supplements from Sigma-Aldrich; Merck KGaA).
Inhibition of STEAP1 expression by
small-interfering RNA
Control small-interfering RNA (siRNA; Control;
#4390843; Thermo Fisher Scientific, Inc.) and two independent
siRNAs targeting human STEAP1 (siSTEAP1; D-003713-01:
5′-GGAGAGAAUUUCACUAUAU-3′ and D-003713-02:
5′-UAAAGAAGAUGCCUGGAUU-3′; Dharmacon) were transfected using
Lipofectamine RNAiMAX (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Cells were seeded at a density of
3×105 cells/well into 6-well plates and cultured for 24
h at 37°C. Subsequently, cells were transfected with control siRNA
or siRNA targeting human STEAP1, and incubated for 72 h at
37°C. Final siRNA used per well was 25 pmol. After incubation,
floating cells in media were collected, adhesive cells were washed
and collected, and both were immediately used for experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using TRIzol Reagent (Thermo
Fisher Scientific) according to the manufacturer's protocol.
Subsequently, complementary (c)DNA was synthesized from the RNA
using a SuperScript VILO cDNA synthesis kit (Thermo Fisher
Scientific). qPCR was performed with an Applied Biosystems 7300
Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The analysis of target genes (STEAP1 and
c-Myc) was conducted in quadruplicate using a POWER
SYBR-Green Master Mix (Thermo Fisher Scientific, Inc.) as
previously described (18). The
thermal profile of the qPCR program consisted of 2 min at 50°C, 10
min at 95°C, 40 cycles of 15 sec at 95°C and 1 min at 60°C, and a
dissociation stage at the end of the run from 60°C to 95°C.
Transcript levels were normalized to β-actin expression and
analyzed using the 2−ΔΔCq method. The following PCR
primers were designed: 5′-CCCTTCTACTGGGCACAATACA-3′ and
5′-GCATGGCAGGAATAGTATGCTTT-3′ for STEAP1;
5′-TTTTTCGGGTAGTGGAAAACC-3′ and 5′-GCAGTAGAAATACGGCTGCAC-3′ for
c-Myc; and 5′-GGCATCCTCACCCTGAAGTA-3′ and
5′-GAAGGTGTGGTGCCAGATTT-3′ for β-actin.
Western blotting
As previously described (19), cells were solubilized in
radioimmunoprecipitation assay lysis buffer (50 mM Tris-HCl, pH
7.5, 1% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA, 150 mM NaCl, 1 mM
EGTA, and protease inhibitor cocktail; Sigma-Aldrich; Merck KGaA),
and centrifuged at 12,000 × g for 10 min. The supernatants were
collected, and protein concentrations were determined using a
bicinchoninic acid Protein Assay Kit (Thermo Fisher Scientific,
Inc.). Equal amounts of protein were separated on MULTIGEL II mini
gels (Cosmo Bio Co., Ltd.) and transferred to polyvinylidene
fluoride membranes using a QBlot Kit (ATTO, Tokyo, Japan). The
blots were probed using the following primary antibodies:
anti-STEAP1 (sc25514; Santa Cruz Biotechnology),
anti-STEAP1 (#88677; Cell Signaling Technology), anti-cyclin
D1 (#2987; Cell Signaling Technology,), anti-p27 Kip1 (#3686; Cell
Signaling Technology), anti-c-Myc (OP10L; EMD Biosciences), and
anti-actin-horse radish peroxidase (HRP; sc-47778; Santa Cruz
Biotechnology).
Evaluation of cell proliferation
Hepatocellular carcinoma cells were seeded at a
density of 2×103 cells/well into 96-well plates. Control
siRNA or two independent siRNAs targeting human STEAP1 were
transfected 24 h after seeding. Cell viability was assessed at 0,
24, 48 and 72 h using a WST-1 assay (Premix WST-a Cell
Proliferation Assay; Takara Bio) and Infinite M1000 Pro microplate
reader (Tecan Japan). A growth curve was constructed by plotting
absorbance against time.
Cell cycle analysis
Liver cancer cells were seeded at a density of
3×105 cells/well into 6-well plates and cultured for 24
h. Subsequently, cells were transfected with control siRNA or an
siRNA targeting human STEAP1, and incubated for 72 h. After
incubation, floating cells in media were collected and adhesive
cells were washed, fixed in ethanol, and stained with propidium
iodide using a cell-cycle analysis kit (FxCycle PI/RNase Staining
Solution; Thermo Fisher Scientific), followed by analysis on a BD
FACS II (BD Biosciences) instrument using FACSDiva (BD Biosciences)
as previously described (20).
Apoptosis assay
Apoptosis was evaluated using an Annexin
V/7-amino-actinomycin (AAD) staining kit (BD Biosciences). Liver
cancer cells were seeded at a density of 3×105
cells/well into 6-well plates and cultured for 24 h. Subsequently,
cells were transfected with control siRNA or an siRNA targeting
human STEAP1, and incubated for 72 h. After incubation,
floating cells in media were collected and adhesive cells were
washed, stained with Annexin V and 7-AAD, and analyzed on a BD
FACSCanto II (BD Biosciences) instrument using FACSDiva (BD
Biosciences) as previously described (21).
PCR array
Total RNA was reverse-transcribed using an
RT2 First Strand Kit (Qiagen). PCR array was performed
using RT2 Profiler™ PCR Array Human MYC Targets
(PAHS-177Z; Qiagen) according to the manufacturer's protocol.
Statistical analysis
The significance of differences was determined by
Student's t-test, Mann-Whitney U test, log-rank test or one-way
ANOVA followed by Bonferroni's post-hoc test, as appropriate.
Pearsons correlation was used to perform the correlation analysis.
All statistical analyses were performed using EZR software version
1.33 (17). Statistical significance
was defined as P<0.05.
Results
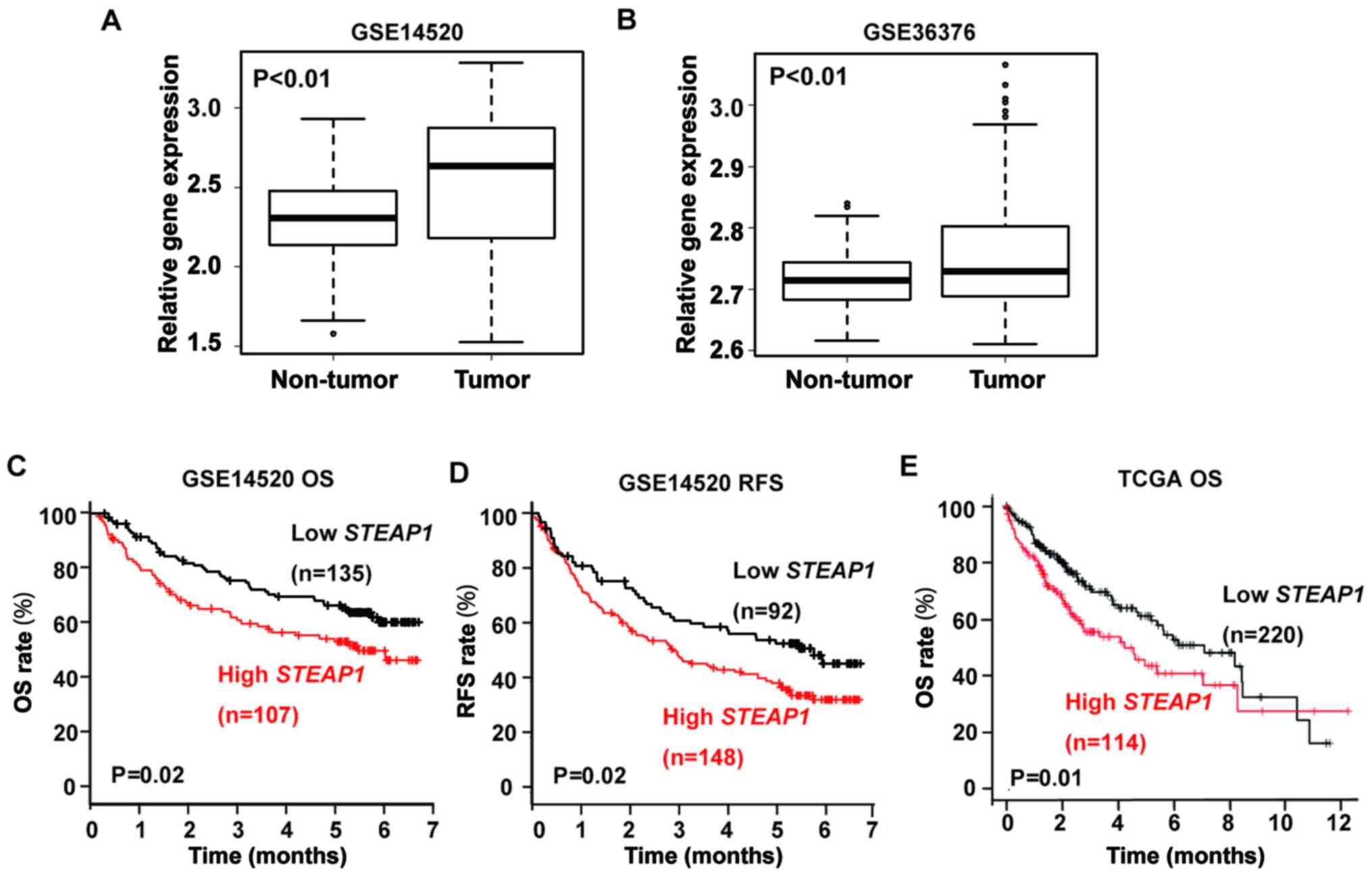
STEAP1 is up-regulated and
significantly associated with poor overall survival and
recurrence-free survival in liver cancer
We first investigated the expression of
STEAP1 in patients with liver cancer using publicly
accessible datasets (GSE14250 and GSE36376) from the Gene
Expression Omnibus. In both datasets, STEAP1 is
over-expressed in liver cancer tissues compared to non-cancerous
hepatic tissues (Fig. 1A and B).
Next, we evaluated the correlation between STEAP1 expression
and survival in patients with liver cancer using GSE14520 and TCGA
datasets. Patients with high STEAP1 expression presented
with significantly shorter overall survival (OS) and
recurrence-free survival (RFS) in GSE14520 and significantly
shorter OS in TCGA (Fig. 1C-E).
These data imply that STEAP1 may have oncogenic functions in liver
cancer.
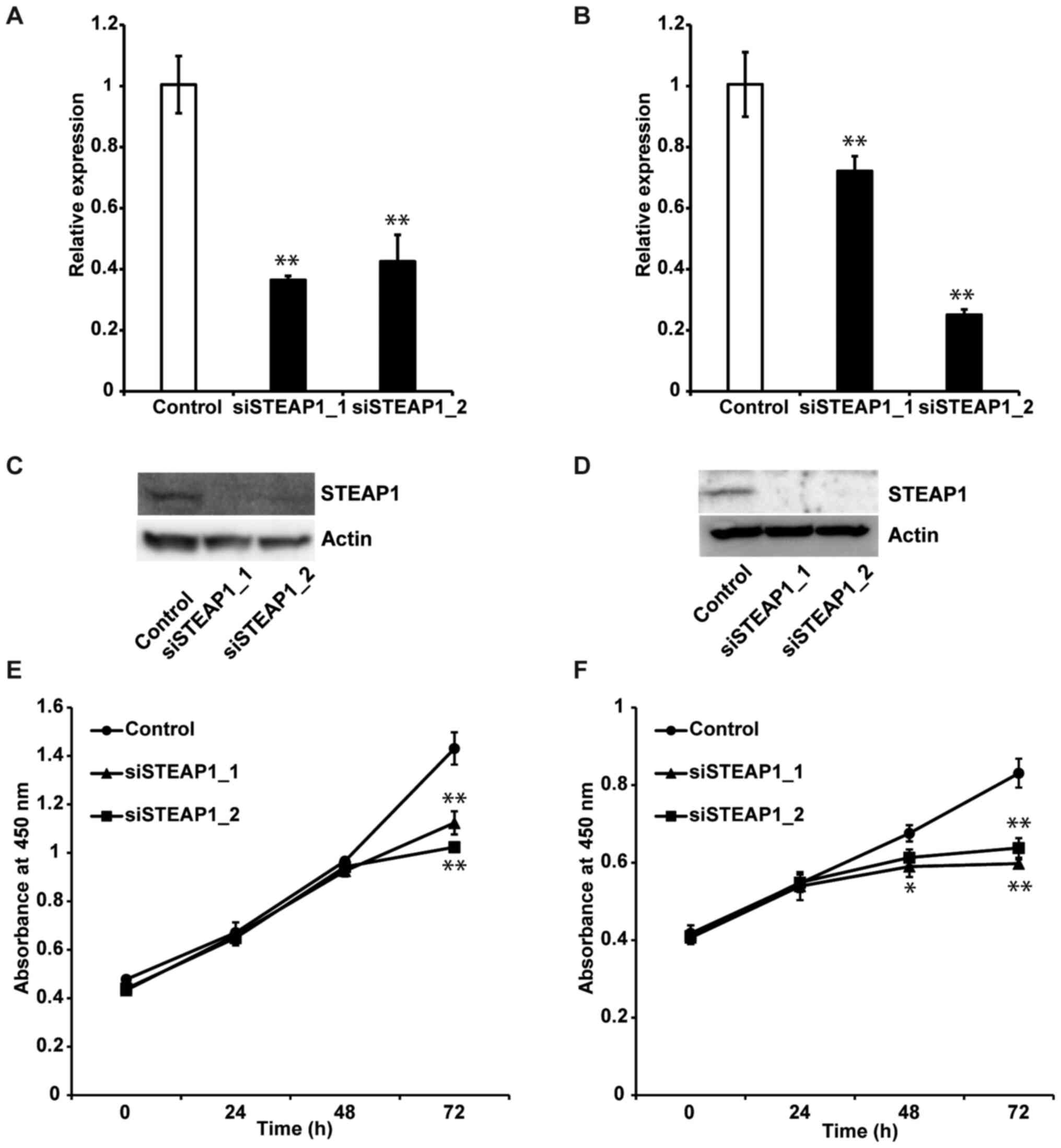
Knockdown of STEAP1 inhibits
proliferation of liver cancer cell lines
To evaluate the effect of STEAP1 on liver cancer, we
performed STEAP1 silencing using an RNA interference method
in two different liver cancer cell lines, HepG2 and Hep3B.
Knockdown efficiency was examined by RT-qPCR and western blot.
STEAP1 expression in these cell lines was significantly
down-regulated 72 h after transfection of two independent siRNAs
(Fig. 2A, B, D and E). We next
evaluated the impact of STEAP1 silencing on liver cancer
cell lines using WST-1 assays. STEAP1 silencing
significantly reduced proliferation in both cell lines (Fig. 2C and F). Based on these data, we
concluded that STEAP1 activated proliferation in liver cancer cell
lines.
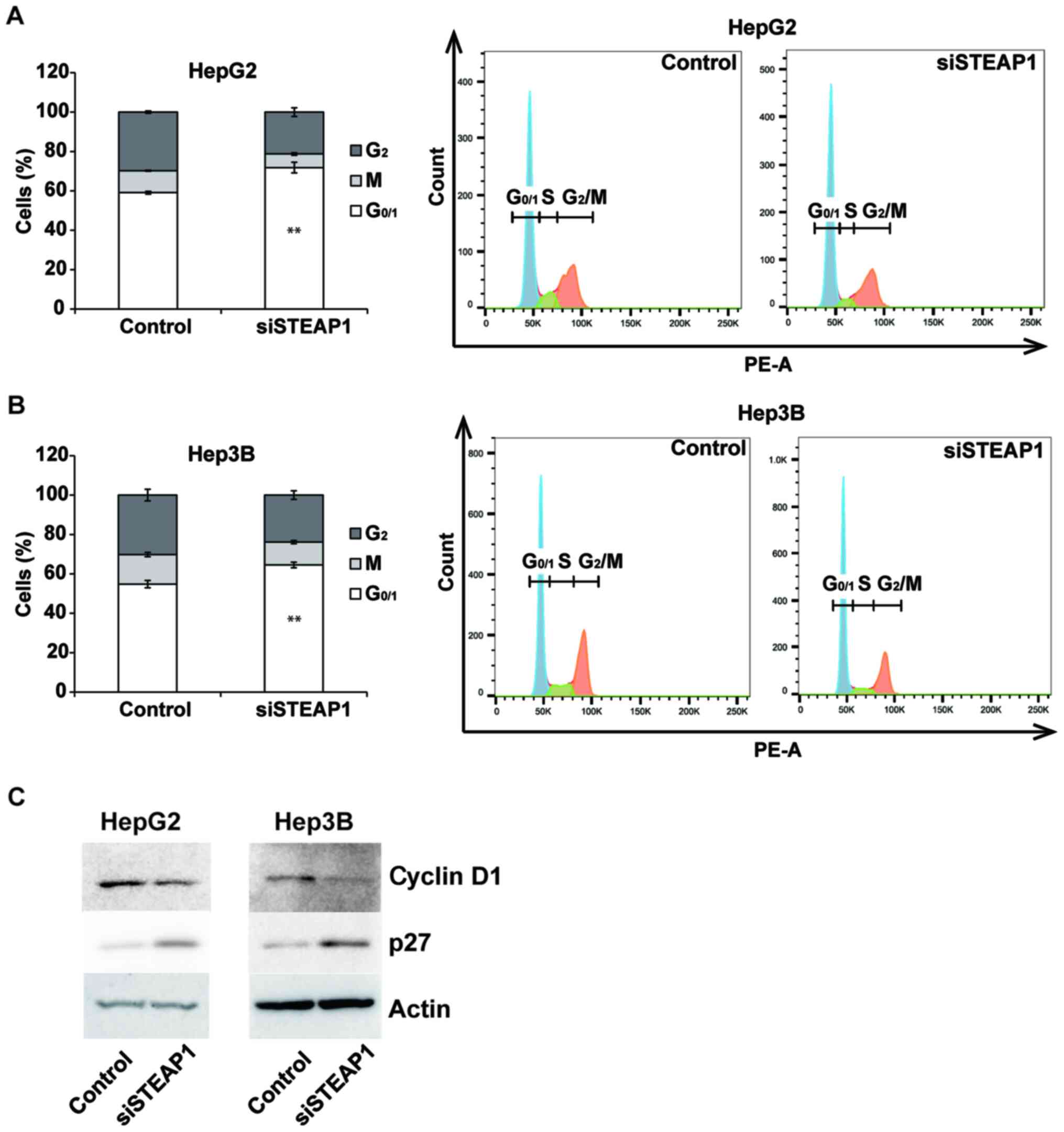
STEAP1 silencing promotes
G1 arrest in liver cancer cell lines
To evaluate the mechanism of decreasing
proliferation in response to the knockdown of STEAP1, we
examined the effects of STEAP1 silencing on the cell cycle
in the liver cancer cell lines, HepG2 and Hep3B. STEAP1
silencing significantly induced G1 arrest in both liver
cancer cell lines (Fig. 3A and B).
We also performed a flow cytometry analysis using Annexin V/7AAD
staining to evaluate the rate of apoptosis. However, an increased
percentage of apoptosis was not observed in STEAP1-silenced
liver cancer cell lines (Fig. S1A and
B). To analyze the mechanism of G1 arrest in HCC
cell lines induced by the knockdown of STEAP1, we evaluated
protein levels of several cell-cycle-related proteins in liver
cancer cell lines using western blot. The expression of the
G1 arrest-associated protein, cyclin D1, was decreased,
whereas the expression of p27, which promotes cell-cycle arrest,
was apparently increased (Fig.
3C).
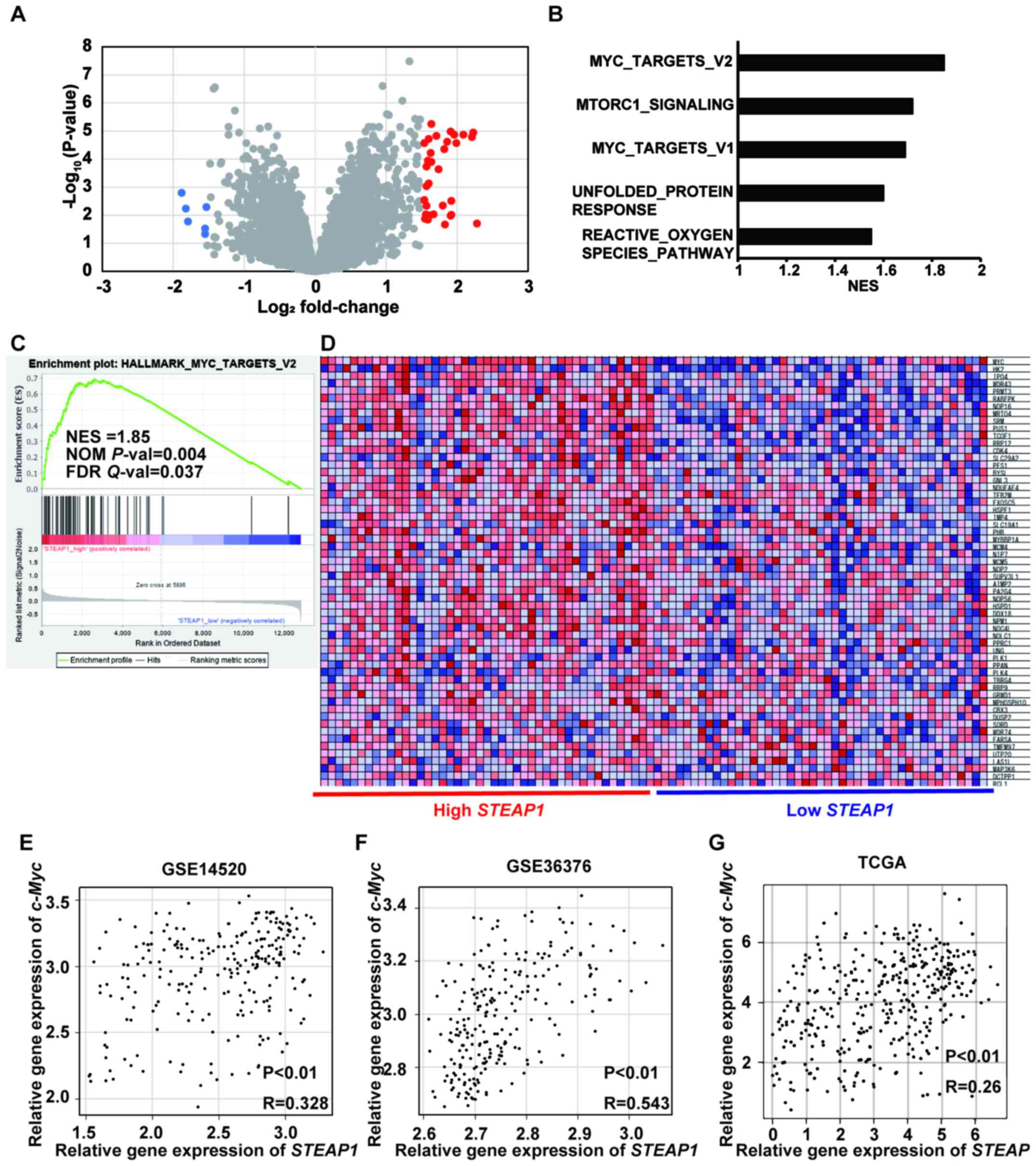
c-Myc target genes were significantly
enriched in patients with liver cancer showing high STEAP1
expression
To clarify the pathways related to STEAP1, we first
extracted DEGs between low and high STEAP1 liver cancer
samples in a publicly accessible dataset, GSE14520-GPL3921, using
GEO2R. The significant DEGs with |logFC| >1.5 and adjusted
P-value < 0.05 are highlighted in red and blue colors. Each gene
was represented as a volcano plot (Fig.
4A) and listed in a table (Table
I). Next, we conducted GSEA to explore the gene sets regulated
by STEAP1 in liver cancer and found five pathways which were
significantly enriched (NOM P-val <0.05 and FDR Q-val <0.25;
Fig. 4B, Fig. S2, and Table SI). The genes belonging to
MYC_TARGET_V2 were the most significantly enriched among these five
pathways (Fig. 4C and D). Based on
these findings, we hypothesized the existence of a relationship
between STEAP1 and c-Myc in liver cancer. To confirm this, we
evaluated their expression using the publicly accessible datasets,
GSE14250, GSE36376, and TCGA. Pearson's correlation coefficient
analysis revealed a significant positive relationship between
STEAP1 and c-Myc in all datasets (Fig. 4E-G).
 | Table I.List of significant DEGs in samples
with high and low STEAP1 expression in publicly accessible
gene expression profiling dataset, GSE14520-GPL3921. |
Table I.
List of significant DEGs in samples
with high and low STEAP1 expression in publicly accessible
gene expression profiling dataset, GSE14520-GPL3921.
| A, Upregulated
DEGs |
|---|
|
|---|
| Symbol | Gene name | log2
ratio | Adjusted
P-value |
|---|
| AFP | α fetoprotein | 2.28 | 0.0197 |
| SULT1C2 | Sulfotransferase
family 1C member 2 | 2.23 | 0.0000113 |
| MT1E | Metallothionein
1E | 2.09 | 0.0000135 |
| ABCB1 | ATP binding
cassette subfamily B member 1 | 1.99 | 0.0000268 |
| MT1G | Metallothionein
1G | 1.96 | 0.0000135 |
| GPX2 | Glutathione
peroxidase 2 | 1.92 | 0.00308 |
| C9 | Complement
component 9 | 1.92 | 0.00971 |
| MT1H | Metallothionein
1H | 1.91 | 0.0000105 |
| SPP1 | Secreted
phosphoprotein 1 | 1.91 | 0.0104 |
| MT1X | Metallothionein
1X | 1.86 | 0.0000241 |
| REG3A | Regenerating family
member 3α | 1.83 | 0.0215 |
| ROBO1 | Roundabout guidance
receptor 1 | 1.82 | 0.0000441 |
| LCN2 | Lipocalin 2 | 1.8 | 0.00455 |
| MYC | v-myc avian
myelocytomatosis viral oncogene homolog | 1.74 | 0.00023 |
| MT1M | Metallothionein
1M | 1.71 | 0.000015 |
| TSPAN8 | Tetraspanin 8 | 1.67 | 0.00928 |
| PLPPR1 | Phospholipid
phosphatase related 1 | 1.64 | 0.00000564 |
| MT1X | Metallothionein
1X | 1.64 | 0.000126 |
| MT1F | Metallothionein
1F | 1.63 | 0.0000604 |
| BCHE |
Butyrylcholinesterase | 1.61 | 0.0103 |
| MT1HL1 | Metallothionein
1H-like 1 | 1.6 | 0.0000192 |
| MTTP | Microsomal
triglyceride transfer protein | 1.6 | 0.000745 |
| SQSTM1 | Sequestosome 1 | 1.59 | 0.000114 |
| RELN | Reelin | 1.59 | 0.0144 |
| CXCL5 | C-X-C motif
chemokine ligand 5 | 1.57 | 0.000184 |
|
TRIM16L///TRIM16 | Tripartite motif
containing 16-like///tripartite motif containing 16 | 1.57 | 0.000923 |
| AKR1C4 | Aldo-keto reductase
family 1, member C4 | 1.57 | 0.00464 |
| CCL20 | C-C motif chemokine
ligand 20 | 1.56 | 0.00949 |
| COL2A1 | Collagen type II α
1 chain | 1.55 | 0.0134 |
| YBX3 | Y-box binding
protein 3 | 1.54 | 0.0000268 |
| IGF2BP3 | Insulin like growth
factor 2 mRNA binding protein 3 | 1.54 | 0.00289 |
|
| B, Downregulated
DEGs |
|
| Symbol | Gene
name | log2
ratio | Adjusted
P-value |
|
| SLPI | Secretory leukocyte
peptidase inhibitor | −3.17 |
3.31×10−08 |
| GNMT | Glycine
N-methyltransferase | −1.88 | 0.0016 |
| SPP2 | Secreted
phosphoprotein 2 | −1.82 | 0.00581 |
| LGALS4 | Galectin 4 | −1.79 | 0.0169 |
| CYP7A1 | Cytochrome P450
family 7 subfamily A member 1 | −1.55 | 0.0472 |
| SLC22A1 | Solute carrier
family 22 member 1 | −1.55 | 0.03 |
| PPP1R1A | Protein phosphatase
1 regulatory inhibitor subunit 1A | −1.53 | 0.00516 |
| CHI3L1 | Chitinase 3 like
1 | −1.52 | 0.12 |
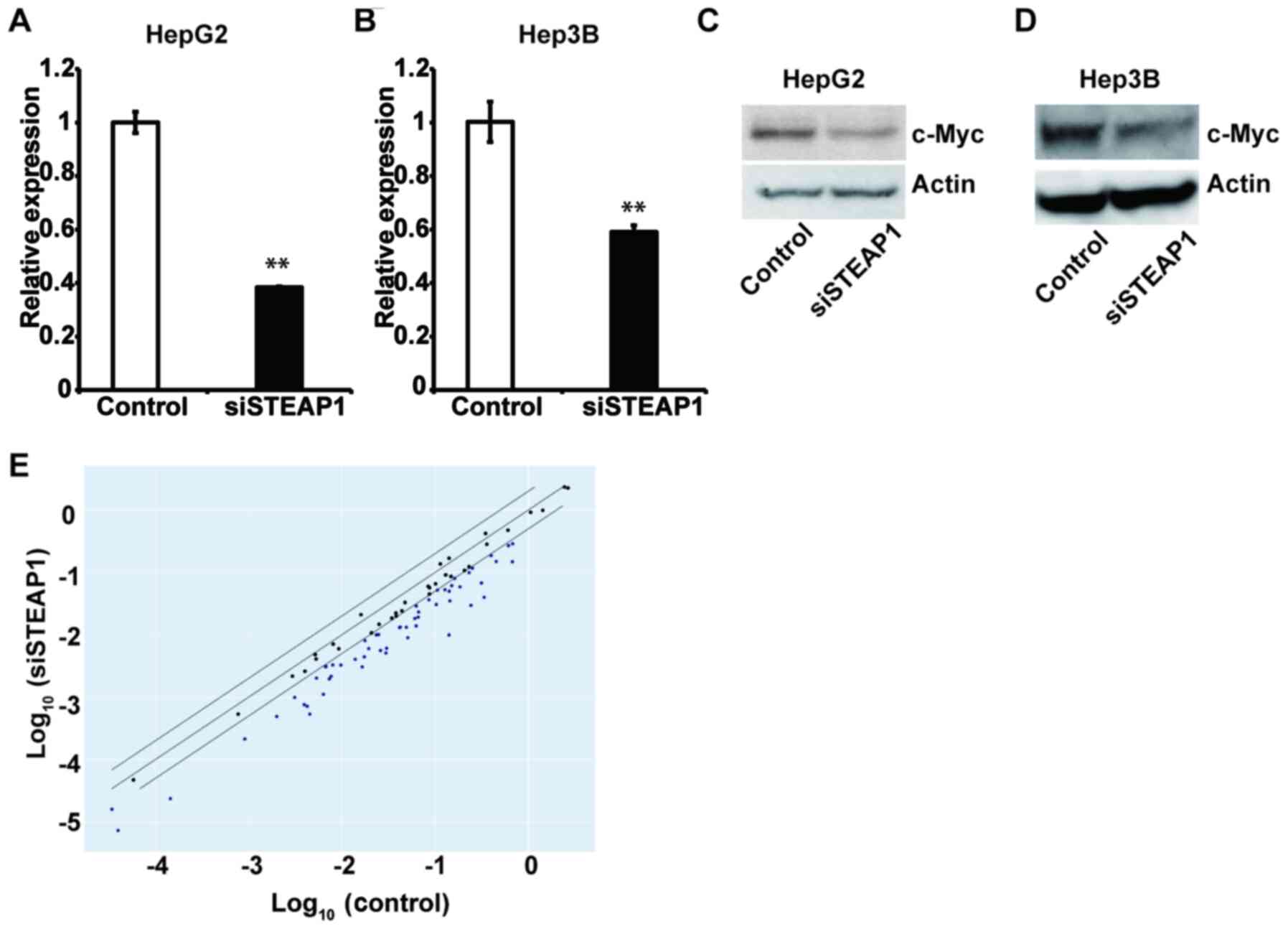
STEAP1 regulates c-Myc and its related
genes in liver cancer cell lines
To confirm the relationship between STEAP1 and c-Myc
in liver cancer, we evaluated the expression of c-Myc after
STEAP1 knockdown in HepG2 and Hep3B cell lines by RT-qPCR
and western blot. As we expected, downregulation of c-Myc was
observed in both cell lines when transfected with siRNA targeting
STEAP1 compared to non-targeting siRNA (Fig. 5A-D). Next, we conducted a PCR array
to analyze components of c-Myc-related genes; most were
significantly downregulated by STEAP1 silencing (Figs. 5E and S3). Taken together, our data suggest that
c-Myc lies downstream of STEAP1, and that the STEAP1-c-Myc pathway
promotes cell proliferation and cell-cycle progression in liver
cancer.
Discussion
Recently, treatment options for HCC have been
expanding as new drugs are approved (2–5).
However, unresectable HCC is an incurable disease; its median
overall survival remains around a year (22). Thus, the further exploration of novel
molecularly-based therapies is required to improve survival in
patients with advanced HCC. c-Myc is a high priority target of
liver cancer therapeutics because its pathological functions exist
in a subset of liver cancer cases. The structure of c-Myc has
hampered the development of c-Myc-specific inhibitors and
highlights the need for further investigations of novel c-Myc
signaling components as potential targets for liver cancer
therapeutics. The current study elucidated STEAP1 as a member of
the c-Myc signal transduction pathway using in vitro and
bioinformatic analyses. Inhibition of STEAP1 led to the
suppression of cell growth accompanied by G1 arrest in
liver cancer, encouraging the development of STEAP1 inhibitors as
therapeutics for STEAP1-c-Myc axis-driven liver cancer.
Additionally, STEAP1 is an attractive target for antibody drug
conjugates (ADC) in cancers because it is expressed on the plasma
membrane (11). In fact, DSTP3086S,
an ADC-targeting STEAP1, has been introduced for patients with
metastatic castration-resistant prostate cancer; it has been
evaluated as safe and shows promising therapeutics (23). Therefore, an ADC-targeting STEAP1 can
be used for patients with liver cancer, who, according to our data,
show the overexpression of STEAP1 in cancerous hepatic tissue
compared to adjacent non-cancerous parts (Fig. 1A and B).
In our previous work, we demonstrated that
STEAP1 knockdown led to apoptosis in colorectal cancer cells
in an NRF2-dependent fashion, corresponding to the increased
production of ROS (15). As shown in
Fig. S4, intracellular ROS levels
were increased by STEAP1 inhibition as found in our previous
work (Fig. S4A and B). Furthermore,
GSEA revealed an ROS-related pathway was significantly enriched in
patients with liver cancer showing upregulated STEAP1
(Fig. S2D). However, as mentioned
above, apoptotic cells were not increased by STEAP1
inhibition in liver cancer cells (Fig.
S1A and B). In addition, we found no statistical correlation
between STEAP1 and NRF2 in three individual datasets
(GSE14520, GSE36376 and TCGA; Fig.
S4C-E). Furthermore, previous studies reported that c-Myc
generates ROS in liver cancer cells (24,25).
However, the current study demonstrated that STEAP1 leads the
increased expression of c-Myc and reduced ROS production in liver
cancer cells. These results seem inconsistent, suggesting the
existence of an NRF2 or c-Myc independent ROS-related pathway in
the regulation of STEAP1-mediated cell growth. Additionally, others
have shown STEAP1 silencing induced cell growth inhibition,
which was associated with decreased levels of ROS in cases of Ewing
sarcoma (26). These results suggest
the existence of multiple pathways between STEAP1 and ROS in a
cancer-type specific manner. Accordingly, our next steps include
exploring the relationship between STEAP1 and ROS in STEAP1-driven
cancer cells.
In summary, this study provides a preclinical
concept for STEAP1 as a druggable target in liver cancer, an often
fatal cancer. The STEAP1-c-Myc axis has potential as an attractive
and promising therapeutic target in liver cancer, and its
manipulation will lead to the development of a novel strategy to
conquer this malignant disease.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Kei Yoneguchi
(Department of Medical Oncology, Sapporo Medical University School
of Medicine, Sapporo, Hokkaido, Japan) for her technical
assistance.
Funding
The present study was funded by a grant from Japan
Society for the Promotion of Science (grant no. 19K08397)
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Gene Expression Omnibus
repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173813.
Authors' contributions
KT, HN and KI were responsible for the conception
and design of the study and for confirming the authenticity of the
data. NH, TK, YU, SI, KM, MK and JK performed the analysis and
interpretation of data. HN and KT drafted the manuscript. JK
critically reviewed and revised the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al SHARP Investigators Study Group, : Sorafenib in advanced
hepatocellular carcinoma. N Engl J Med. 359:378–390. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bruix J, Qin S, Merle P, Granito A, Huang
YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, et al
RESORCE Investigators, : Regorafenib for patients with
hepatocellular carcinoma who progressed on sorafenib treatment
(RESORCE): A randomised, double-blind, placebo-controlled, phase 3
trial. Lancet. 389:56–66. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kudo M, Finn RS, Qin S, Han KH, Ikeda K,
Piscaglia F, Baron A, Park JW, Han G, Jassem J, et al: Lenvatinib
versus sorafenib in first-line treatment of patients with
unresectable hepatocellular carcinoma: A randomised phase 3
non-inferiority trial. Lancet. 391:1163–1173. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Finn RS, Qin S, Ikeda M, Galle PR, Ducreux
M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, et al IMbrave150
investigators, : Atezolizumab plus bevacizumab in unresectable
hepatocellular carcinoma. N Engl J Med. 382:1894–1905. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stine ZE, Walton ZE, Altman BJ, Hsieh AL
and Dang CV: MYC, metabolism, and cancer. Cancer Discov.
5:1024–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abou-Elella A, Gramlich T, Fritsch C and
Gansler T: c-myc amplification in hepatocellular carcinoma predicts
unfavorable prognosis. Mod Pathol. 9:95–98. 1996.PubMed/NCBI
|
|
8
|
Schaub FX, Dhankani V, Berger AC, Trivedi
M, Richardson AB, Shaw R, Zhao W, Zhang X, Ventura A, Liu Y, et al
Cancer Genome Atlas Network, : Pan-cancer alterations of the MYC
oncogene and its proximal network across the Cancer Genome Atlas.
Cell Syst. 6:282–300.e2. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duffy MJ and Crown J: Drugging
‘undruggable’ genes for cancer treatment: Are we making progress?
Int J Cancer. 148:8–17. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Massó-Vallés D and Soucek L: Blocking Myc
to treat cancer: Reflecting on two decades of omomyc. Cells.
9:8832020. View Article : Google Scholar
|
|
11
|
Hubert RS, Vivanco I, Chen E, Rastegar S,
Leong K, Mitchell SC, Madraswala R, Zhou Y, Kuo J, Raitano AB, et
al: STEAP: A prostate-specific cell-surface antigen highly
expressed in human prostate tumors. Proc Natl Acad Sci USA.
96:14523–14528. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moreaux J, Kassambara A, Hose D and Klein
B: STEAP1 is overexpressed in cancers: A promising therapeutic
target. Biochem Biophys Res Commun. 429:148–155. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gomes IM, Maia CJ and Santos CR: STEAP
proteins: From structure to applications in cancer therapy. Mol
Cancer Res. 10:573–587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oosterheert W and Gros P: Cryo-electron
microscopy structure and potential enzymatic function of human
six-transmembrane epithelial antigen of the prostate 1 (STEAP1). J
Biol Chem. 295:9502–9512. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nakamura H, Takada K, Arihara Y, Hayasaka
N, Murase K, Iyama S, Kobune M, Miyanishi K and Kato J:
Six-transmembrane epithelial antigen of the prostate 1 protects
against increased oxidative stress via a nuclear erythroid
2-related factor pathway in colorectal cancer. Cancer Gene Ther.
26:313–322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Menyhárt O, Nagy Á and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. R Soc Open Sci.
5:1810062018. View Article : Google Scholar
|
|
17
|
Kanda Y: Investigation of the freely
available easy-to-use software ‘EZR’ for medical statistics. Bone
Marrow Transplant. 48:452–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mani M, Carrasco DE, Zhang Y, Takada K,
Gatt ME, Dutta-Simmons J, Ikeda H, Diaz-Griffero F, Pena-Cruz V,
Bertagnolli M, et al: BCL9 promotes tumor progression by conferring
enhanced proliferative, metastatic, and angiogenic properties to
cancer cells. Cancer Res. 69:7577–7586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takada K, Zhu D, Bird GH, Sukhdeo K, Zhao
JJ, Mani M, Lemieux M, Carrasco DE, Ryan J, Horst D, et al:
Targeted disruption of the BCL9/β-catenin complex inhibits
oncogenic Wnt signaling. Sci Transl Med. 4:148ra1172012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hayasaka N, Takada K, Nakamura H, Arihara
Y, Kawano Y, Osuga T, Murase K, Kikuchi S, Iyama S, Emori M, et al:
Combination of eribulin plus AKT inhibitor evokes synergistic
cytotoxicity in soft tissue sarcoma cells. Sci Rep. 9:57592019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Matsuoka K, Koreth J, Kim HT, Bascug G,
McDonough S, Kawano Y, Murase K, Cutler C, Ho VT, Alyea EP, et al:
Low-dose interleukin-2 therapy restores regulatory T cell
homeostasis in patients with chronic graft-versus-host disease. Sci
Transl Med. 5:179ra432013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Llovet JM, Montal R, Sia D and Finn RS:
Molecular therapies and precision medicine for hepatocellular
carcinoma. Nat Rev Clin Oncol. 15:599–616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Danila DC, Szmulewitz RZ, Vaishampayan U,
Higano CS, Baron AD, Gilbert HN, Brunstein F, Milojic-Blair M, Wang
B, Kabbarah O, et al: Phase I study of DSTP3086S, an antibody-drug
conjugate targeting six-transmembrane epithelial antigen of
prostate 1, in metastatic castration-resistant prostate cancer. J
Clin Oncol. 37:3518–3527. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dolezal JM, Wang H, Kulkarni S, Jackson L,
Lu J, Ranganathan S, Goetzman ES, Bharathi SS, Beezhold K,
Byersdorfer CA, et al: Sequential adaptive changes in a
c-Myc-driven model of hepatocellular carcinoma. J Biol Chem.
292:10068–10086. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng K, Cubero FJ and Nevzorova YA:
c-MYC-making liver sick: Role of c-MYC in hepatic cell function,
homeostasis and disease. Genes (Basel). 8:1232017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grunewald TG, Diebold I, Esposito I, Plehm
S, Hauer K, Thiel U, da Silva-Buttkus P, Neff F, Unland R,
Müller-Tidow C, et al: STEAP1 is associated with the invasive and
oxidative stress phenotype of Ewing tumors. Mol Cancer Res.
10:52–65. 2012. View Article : Google Scholar : PubMed/NCBI
|