Introduction
Gastric cancer (GC) is one of the most commonly
occurring malignancies worldwide, and its incidence and mortality
rates rank fifth and second, respectively, among all types of
cancer according to the Global Cancer Statistics 2018 (1). The 5-year overall survival (OS) rate of
patients with GC is <20%, and this low survival rate is partly
due to a lack of effective early diagnostic methods and prognostic
indicators (2). In different types
of cancer, genomic instability is associated with chromosomal
aberrations, which affect numerous genes, further promoting tumor
progression (3). Comparative genomic
hybridization (CGH) data have shown that gains of DNA copy number
are observed frequently at chromosomes 1q, 3q, 5p, 7p, 7q, 8q, 11q,
17q, 20p and 20q, and losses of one copy are frequently detected at
chromosomes 3p, 4p, 4q, 8p, 9p, 18p and 18q in GC (4–9). Several
chromosomal aberrations, particularly amplification of 20q, have
been observed in various types of cancer, including pancreatic
(10), breast (11,12),
colon (13,14) and stomach cancer (15), implying that gain of 20q may have a
vital role in tumorigenesis.
Amplified genes on chromosome 20q contain various
functionally important genes involved in cell cycle regulation
(E2F1, TPX2, KIF3B, PIGT and B4GALT5), nuclear function (CSEL1),
viral replication (PSMA7 and LAMA5), methylation and chromatin
remodeling (ASXL1, AHCY and C20orf20), as well as transcription
regulation (TCEA2) (16,17). Amplification of chromosome 20q
deregulates several specific cancer-associated signaling pathways,
including the MAPK and p53 signaling pathways (13).
Long non-coding RNAs (lncRNAs), a type of non-coding
RNA, serve vital roles in the regulation of various biological
processes, including transcription, intracellular trafficking,
chromosome remodeling, cell proliferation, metastasis and migration
(18–20). Genome-wide association studies of
tumor samples have revealed that numerous lncRNAs associated with
the processes of tumorigenesis and metastasis become dysregulated
with expression alteration and mutations (21–23).
Multiple lncRNAs have been considered as either tumor suppressors
or oncogenes according to their genome-wide expression patterns and
tissue-specific expression characteristics (21,23).
Furthermore, lncRNAs have been suggested as novel biomarkers and
therapeutic targets for bladder (18), prostate (18), gastric (19,20),
pancreatic (24) and breast cancer
(25). Accumulating studies have
indicated that lncRNAs, such as TERRA, HOXA11-AS, AGAP2-AS1,
HOTAIR, CCAT1, MALAT1 and Xist, are dysregulated in gastric cancer
(GC) (21,26–30).
Investigating lncRNAs amplified on 20q may provide novel insights
into GC diagnosis and targeted therapy. Therefore, the present
study aimed to evaluate lncRNAs on amplified chromosome 20q13.33 in
GC tissues and cell lines.
Materials and methods
GC tissue specimens and cell
lines
A total of 60 paired samples of fresh frozen GC
tissues and corresponding adjacent non-tumor tissue samples (>5
cm away from tumor) were obtained from The Third Affiliated
Hospital of Harbin Medical University (Harbin, China) between March
2006 and March 2008. All cases were reviewed by pathologists and
histologically confirmed as GC. Patients were not subjected to
local or systemic treatment prior to the procedure. The age range
of the 60 patients (46 male and 14 female) was 40–76 years, with a
median age of 57 years. The study was approved by the Ethics
Committee of Southeast University affiliated to Zhongda Hospital
(Nanjing, China).
The normal gastric epithelial GES-1 cell line and
the GC AGS, MKN-45 and MKN-74 cell lines were purchased from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. The cells were cultured in RPMI-1640 medium containing
10% FBS (both Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin at 37°C in a humidified atmosphere with 5%
CO2. The culture medium was changed every 1–2 days and
subcultured when the cell confluence reached 80–90%.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the fresh GC tissues
and cell lines with TRIzol® reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
cDNA was synthesized using PrimeScript@ RT Reagent kit (Takara Bio,
Inc.) under standard conditions according to the manufacturer's
instructions. RT-qPCR was performed to determine the expression
levels of specific genes using a SYBR Premix Ex Taq kit (Takara
Bio, Inc.), and β-actin was used as the internal control for
normalization of the data. The thermocycling conditions for
amplification were as follows: 95°C for 5 min, followed by 40
cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, and
a final step at 72°C for 10 min. All the experiments were performed
on a StepOne Plus system (Applied Biosystems; Thermo Fisher
Scientific, Inc.), and the sequences of the primers for RT-qPCR are
shown in Table SI. Relative gene
expression compared with its control was assessed using the
2−ΔΔCq method (31). Each
sample was analyzed in triplicate.
Cell transfection
For in vitro assays, the small interfering
(si)RNA sequences and non-targeting negative controls (NCs) were
synthesized by Shanghai GenePharma Co., Ltd., and were as follows:
si-LINC00659-1, 5′-GGGACUUGGAUGCUUAACATT-3′; si-LINC00659-2,
5′-GCCGUGCUCUGGAUAUAUATT-3′; and si-NC,
5′-UUCUCCGAACGUGUCACGUTT-3′. Short hairpin (sh)RNA sequences were
designed according to the aforementioned siRNA sequences. Human
embryonic kidney 293T cells were used for lentiviral packing
(second-generation packaging system). 293T cells were provided by
Shanghai GeneChem Co., Ltd., and originally purchased from the
American Type Culture Collection. The cells were cultured in 10-cm
dishes for 2–3 days until they reached 90–95% confluency. The
recombinant virus plasmid GV112 (20 µg), which encoded
sh-LINC00659, and the control, together with packaging plasmids
pHelper 1.0 (15 µg) and pHelper 2.0 (10 µg), were co-transfected
into 293T cells using Lipofectamine® 2000 (all from
Shanghai GeneChem Co., Ltd.). After 48 h of transduction, the
lentiviral particles contained in the supernatant of 293T cells
were harvested and then concentrated by passing through a 0.45-µm
filter.
GC cells (1×105 cells/well) were seeded
on a 24-well plate, and cell transfection was performed when the
cell number reached ~2×105 cells/well. Fresh medium
containing 6 µg/ml polybrene was added for transfection at a
multiplicity of infection of 10 with control or sh-LINC00659
lentiviruses (Shanghai GeneChem Co., Ltd.) at 37°C after the cells
had been washed 3 times with PBS. After 8 h of incubation at 37°C,
fresh medium was added to the cells, and puromycin (2 µg/ml) was
added to screen the transfected cells on the 3rd day of
transfection. After 7 days, the cells were used for subsequent
experimentation.
Cell Counting Kit (CCK)-8 assay
The infected cells were suspended and seeded in a
96-well plate at a density of 2,000 cell/well. After 0, 24, 48, 72
and 96 h of incubation at 37°C, the culture medium was replaced
with 100 µl CCK-8 reagent (Dojindo Molecular Technologies, Inc.).
After 4 h of incubation, the cell proliferative ability was
determined by measuring the optical density at 450 nm.
Wound healing assay
The human GC AGS and MKN-74 cell lines, which were
stably transfected with sh-LINC00659, were seeded in 6-well plates
and reached a density of ~90% after one day. A 200-µl pipette tip
was used to scratch the cell culture surface. The cells were washed
three times with PBS and were then cultured in serum-free medium.
The cells were cultured at 37°C in a humidified atmosphere with 5%
CO2. Wound healing was recorded at 0 and 24 h using an
inverted light microscope (Olympus Corporation; magnification,
×200).
Cell migration and invasion assay
The cell invasive and migratory potential was
measured using Transwell chambers (EMD Millipore) containing a
polycarbonate membrane with 8.0-µm pores. The human GC AGS and
MKN-74 cell lines were stably transfected with sh-LINC00659 using a
lentivirus system, as aforementioned. After 48 h of transfection,
1.5×104 cells in serum-free medium were placed into the
upper chamber of an insert for migration, and the cells were
allowed to migrate for 18 h at 37°C. For the invasion assay,
2×104 cells in serum-free medium were seeded in
Matrigel-coated (Sigma-Aldrich; Merck KGaA) inserts and allowed to
invade for 36 h at 37°C. In both assays, the lower Transwell
chamber was filled with medium supplemented with 10% FBS. After
incubation, the cells that had migrated or invaded through the
membrane were stained with methanol and 0.1% crystal violet for 30
min at room temperature, imaged and counted using an IX71 inverted
light microscope (Olympus Corporation; magnification, ×200).
The Cancer Genome Atlas (TCGA) data
analysis
Processed TCGA expression data including tumor
(n=408) and normal samples (n=36) of patients with GC were
downloaded from TCGA database (http://cancergenome.nih.gov/), and the upper quantile
normalized fragments per kilobase per million values were collected
and converted into log2 data form. The data were used to analyze
the survival of patients with GC based on lncRNA expression with
Kaplan-Meier survival curves. The samples were first divided into
two groups (high and low LINC00659 groups) according to the median
expression levels (FPKM value, 37) of LINC00659. The 5-year
survival analysis was performed using the survival package
(https://cran.r-project.org/web/packages/survival)
in the R software.
Enrichment analysis
To explore the potential downstream pathways of
LINC00659, LINC00659 expression was categorized into two groups of
high and low expression, according to the median value (FPKM value,
37) of LINC00659 expression. Gene set enrichment analysis (GSEA)
2-2.2.3 (JAVA version) was downloaded from the GSEA website
(http://www.gsea-msigdb.org/gsea/downloads.jsp).
Subsequently, the downloaded dataset was imported using the GSEA
software. Gene sets associated with biological signal transduction
were identified on the Molecular Signatures Database (MSigDB;
http://software.broadinstitute.org/gsea/msigdb). The
simulation was repeated 1,000 times for each analysis, according to
the default weighted enrichment statistical method. Gene sets with
a false discovery rate <0.25 and P<0.05 were selected.
Target gene prediction
The correlation between the co-expression of
LINC00659 and all protein coding genes (PCGs) was determined by
calculating the Pearson correlation coefficients and z test. The
PCGs positively or negatively correlated with LINC00659 were
considered as LINC00659-associated PCGs (|Pearson correlation
coefficient|>0.4 and P<0.01).
Statistical analysis
All experiments were performed independently at
least three times and data are presented as the mean ± SD.
Statistical analyses were performed using SPSS 18.0 software (SPSS,
Inc.). Comparisons between two groups were made using the
independent sample t-test; unpaired Student's t-test was used to
analyze the differences between two groups, and paired Student's
t-test was used to compare the paired tissue samples. Comparisons
among >2 groups were made using ANOVA followed by Fisher's LSD
post-hoc test (for 3 groups) or Dunnett's post-hoc test (for >3
groups). Fisher's exact test or χ2 test was used to
analyze the association between LINC00659 expression and
clinicopathological parameters. Survival curves were plotted using
the Kaplan-Meier method, and the difference in OS survival rates
was assessed using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
LINC00659 expression is upregulated
and associated with a poor prognosis in patients with GC included
in TCGA database
Alteration of chromosomal duplication and gene
amplification are crucial for functional gain and overexpression of
oncogenes. Based on the CGH analysis, several chromosomal
aberrations, especially recurrent gain and amplification of the
long arm of chromosome 20 (20q13.33), have been observed in
numerous types of cancer, including GC (12–17). In
order to explore the contribution of lncRNAs located on amplified
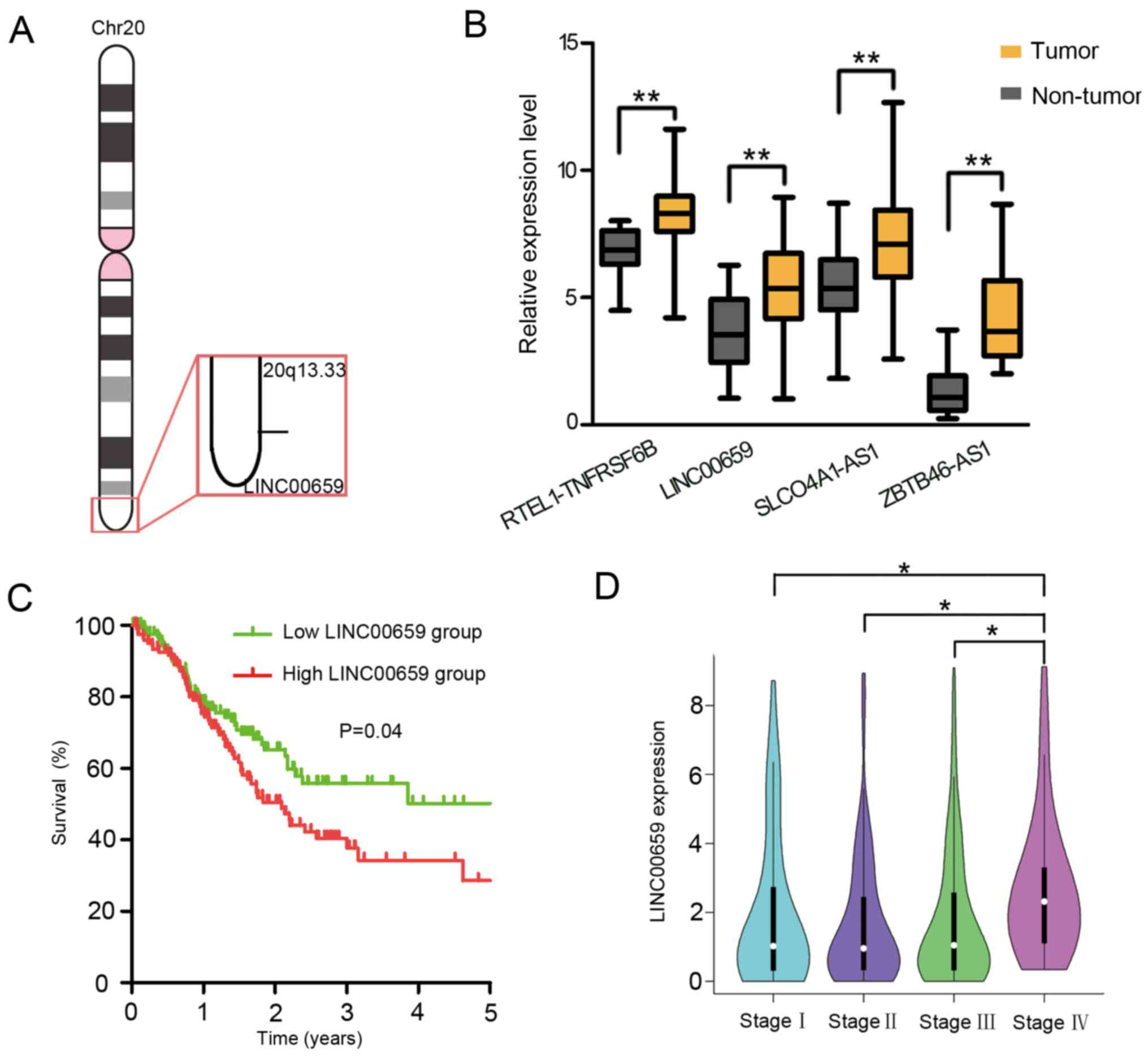
20q13.33 (Fig. 1A), TCGA database
was screened, and the expression levels of lncRNAs in GC were
analyzed. The expression levels of four lncRNAs, namely
RTEL1-TNFRSF6B, SLCO4A1-AS1, ZBTB46-AS1 and LINC00659, were found
to be significantly upregulated in gastric tumor tissues compared
with in non-tumor tissues (Fig. 1B).
Subsequently, Kaplan-Meier analysis using a log-rank test was
performed to investigate whether the expression levels of these
four lncRNAs were associated with the survival of patients with GC.
The results demonstrated that patients with a high level of
LINC00659 had a significantly poorer OS rate (P=0.04) compared with
patients with a low level of LINC00659 expression (Fig. 1C). However, the other three lncRNAs,
RTEL-TNFRSF6B, SLCO4A1-AS1 and ZBTB46-AS1, were not associated with
OS rate or tumor stage (Figs. S1
and S2). Additionally, the
expression levels of LINC00659 were significantly increased in
patients with advanced GC (stage IV) compared with in those with
stages I–III (Fig. 1D). These data
indicated that patients with GC with increased levels of LINC00659
expression may be prone to progress to a more advanced stage of the
disease.
LINC00659 expression is upregulated in
GC clinical tissues and cell lines
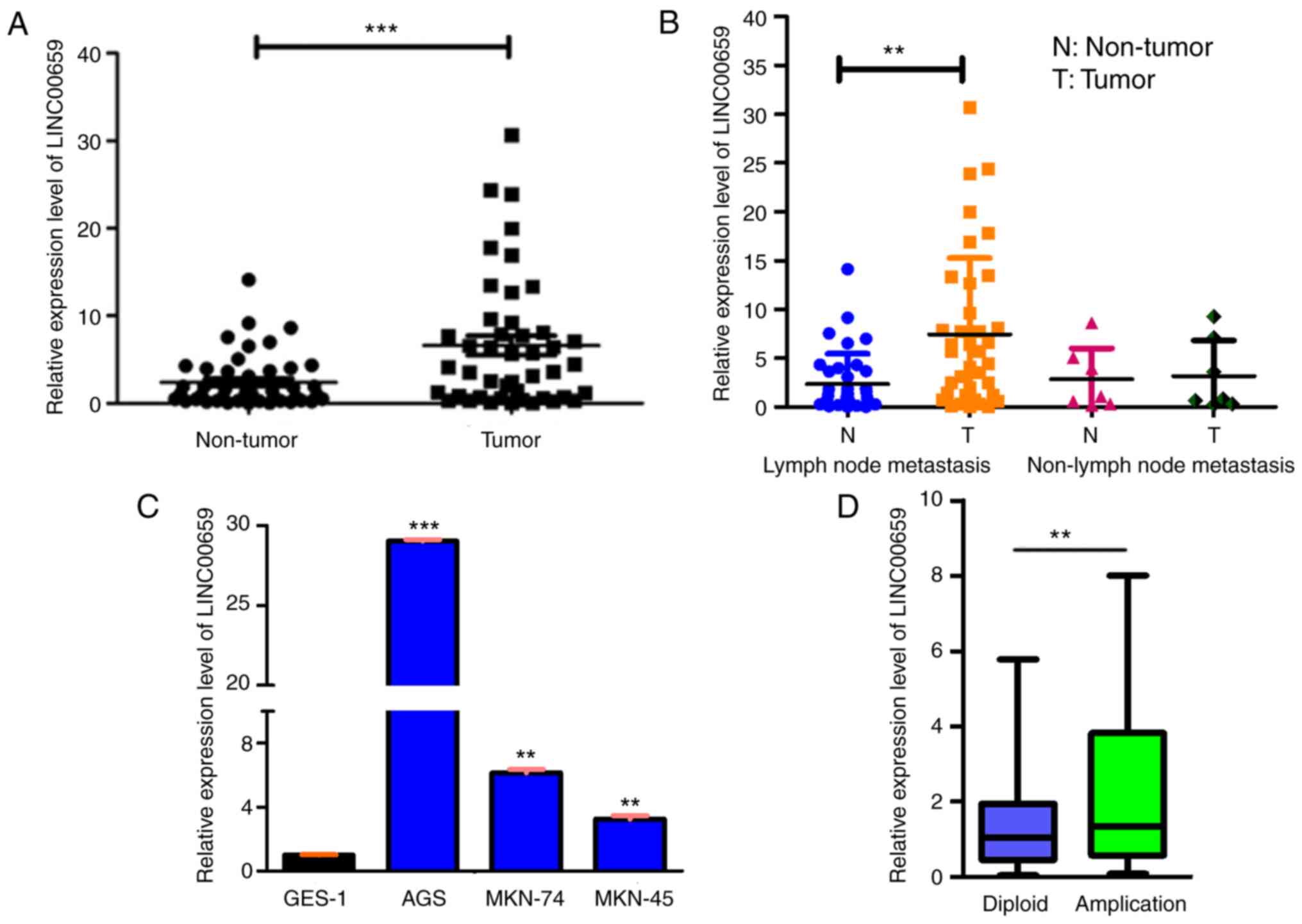
The relative expression levels of LINC00659 were
detected in 60 paired GC and adjacent non-cancerous tissues via
RT-qPCR. LINC00659 expression was significantly upregulated in the
cancerous tissues compared with in their non-cancerous counterparts
(Fig. 2A). Furthermore, the
association of LINC00659 expression with clinical features of GC
was examined, revealing that higher expression levels of LINC00659
were associated with differentiation and vessel invasion; however,
no association was observed with sex, age, tumor size, lymph node
metastasis and Helicobacter pylori infection (Table SII). Additionally, the expression
levels of LINC00659 were higher in cancer tissues than in adjacent
non-cancerous tissues in patients with lymph node metastasis
(Fig. 2B). Subsequently, LINC00659
expression was detected in three GC cell lines (AGS, MKN74 and
MKN-45) and the normal gastric epithelial GES-1 cell line via
RT-qPCR. As presented in Fig. 2C,
all three GC cell lines exhibited significantly higher expression
levels of LINC00659 compared with the normal gastric epithelial
cell line. Furthermore, LINC00659 was more highly expressed in the
copy number-amplified group than in the normal copy number group
(P<0.01; Fig. 2D). This finding
indicated that the number of amplifications of LINC00659 may be
closely associated with its expression. Overall, these results
indicated that upregulation of LINC00659 expression may have
important roles in GC development and progression.
Function of LINC00659 in GC cells
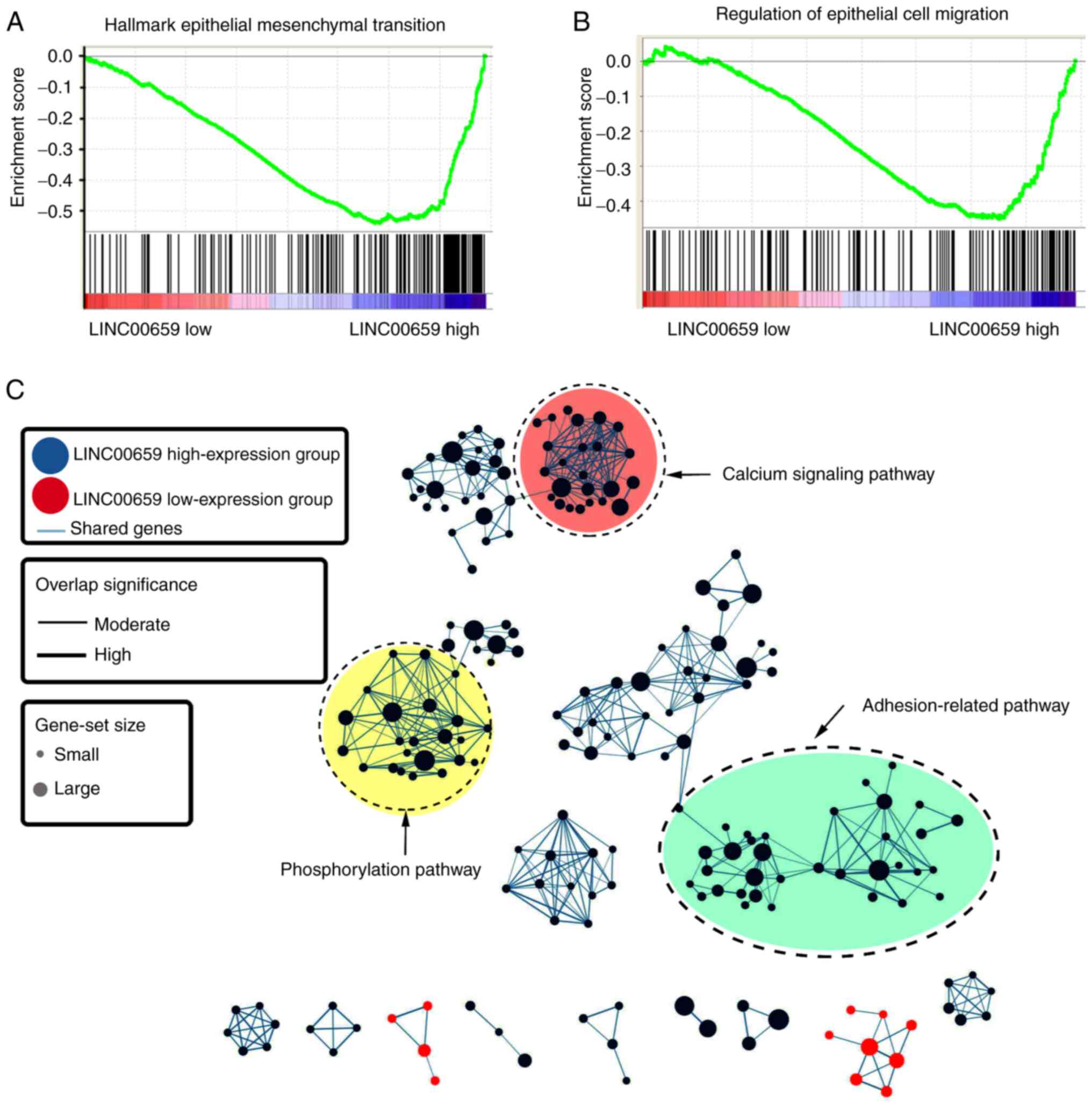
To explore the biological function of LINC00659,
GSEA analysis based on the mRNA expression levels of LINC00659 in
GC was performed using TCGA data. Enrichment plots of gene
expression signatures illustrated that epithelial cell migration
and epithelial-mesenchymal transition pathways were associated with
the expression levels of LINC00659 (Fig.
3A and B). With further analysis, the potential function of
LINC00659 was aggregated at a few functional clusters, including
calcium signaling pathway, phosphorylation pathway and
adhesion-related pathway (Fig. 3C).
These data implied that the function of LINC00659 may be associated
with cell migration and invasion involved in GC metastasis.
LINC00659 has no effect on cell
proliferation according to CCK-8 assay
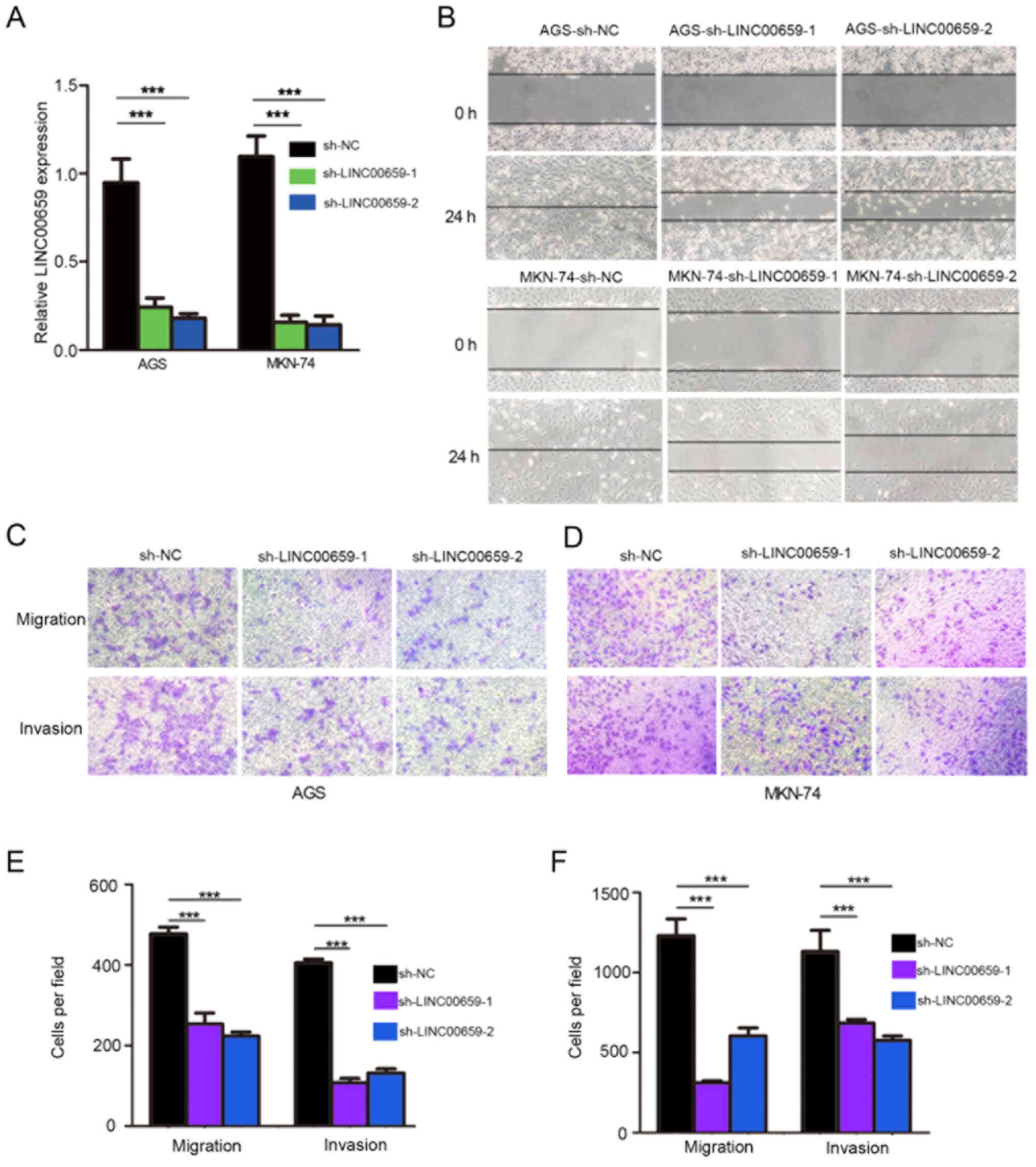
To assess the effects of LINC00659 expression on GC
cells, sh-LINC00659 or sh-NC were used to infect AGS and MKN-74
cells, which exhibited higher LINC00659 expression. The efficiency
of infection was confirmed by RT-qPCR, and a significant decrease
in LINC00659 expression was observed following transfection with
both sh-LINC00659 molecules (Fig.
4A), with sh-LINC00659-2 being chosen for subsequent
experiments. The CKK-8 assay revealed that LINC00659 inhibition by
sh-LINC00659-2 did not affect the proliferation of AGS and MKN-74
cells (Fig. S3).
LINC00659 promotes GC cell migration
and invasion in vitro
In order to clarify the function of LINC00659 in
terms of cell migration and invasion, the effects of
LINC00659-knockdown on cell migration and invasion were assessed.
The results of the wound-healing assays revealed that the migratory
ability of AGS and MKN-74 cells was decreased in the
LINC00659-knockdown group compared with in the NC group (Fig. 4B). Transwell migration assays
displayed that LINC00659-knockdown significantly suppressed the
migratory capabilities of both AGS and MKN-74 cancer cells compared
with the control group (Fig. 4C and
E). Moreover, GC cells with LINC00659-knockdown exhibited
significantly weaker abilities to invade through Matrigel compared
with the control group (Fig. 4D and
F). These results indicated that knocking down LINC00659 led to
a clear suppression of the migratory and invasive abilities of GC
cells.
IQ motif-containing GTPase activating
protein 3 (IQGAP3) and matrix metalloproteinase 15 (MMP15) are
potential downstream targets involved in LINC00659-induced tumor
metastasis
To investigate the underlying mechanism of LINC00659
in cell migration and invasion, the correlation between the
expression levels of LINC00659 and all PCGs was examined using
two-sided Pearson correlation coefficients and z-test. The PCGs
positively or negatively correlated with LINC00659 were considered
as LINC00659-associated PCGs (|Pearson correlation
coefficient|>0.4 and P<0.01). The expression levels of eight
genes, namely IQGAP3, MMP15, integrin subunit α6 (ITGA6), G
protein-coupled receptor class C group 5 member A (GPRC5A),
hematological and neurological expressed 1 (HN1), keratin 8 (KRT8),
engulfment and cell motility 3 (ELMO3), and TNF receptor associated
factor 4 (TRAF4), were found to be positively correlated with
LINC00659 expression (Fig. 5A-H). To
determine whether these eight genes were downstream targets of
LINC00659, their expression patterns were investigated in GC cells
where LINC00659 expression had been knocked down by sh-LINC00659-2.
As shown in Fig. 6A and B, the
expression levels of IQGAP3 and MMP15 were significantly
downregulated in LINC00659-knockdown GC cells, and ITGA6 expression
was significantly downregulated only in AGS cells. GPRC5A, HN1,
KRT8, ELMO3 and TRAF4 exhibited no change in expression when
LINC00659 was knocked down in AGS and MKN-74 cells (Fig. 6C). These results suggested that
LINC00659 may be associated with metastasis via IQGAP3 and MMP15 as
its downstream targets.
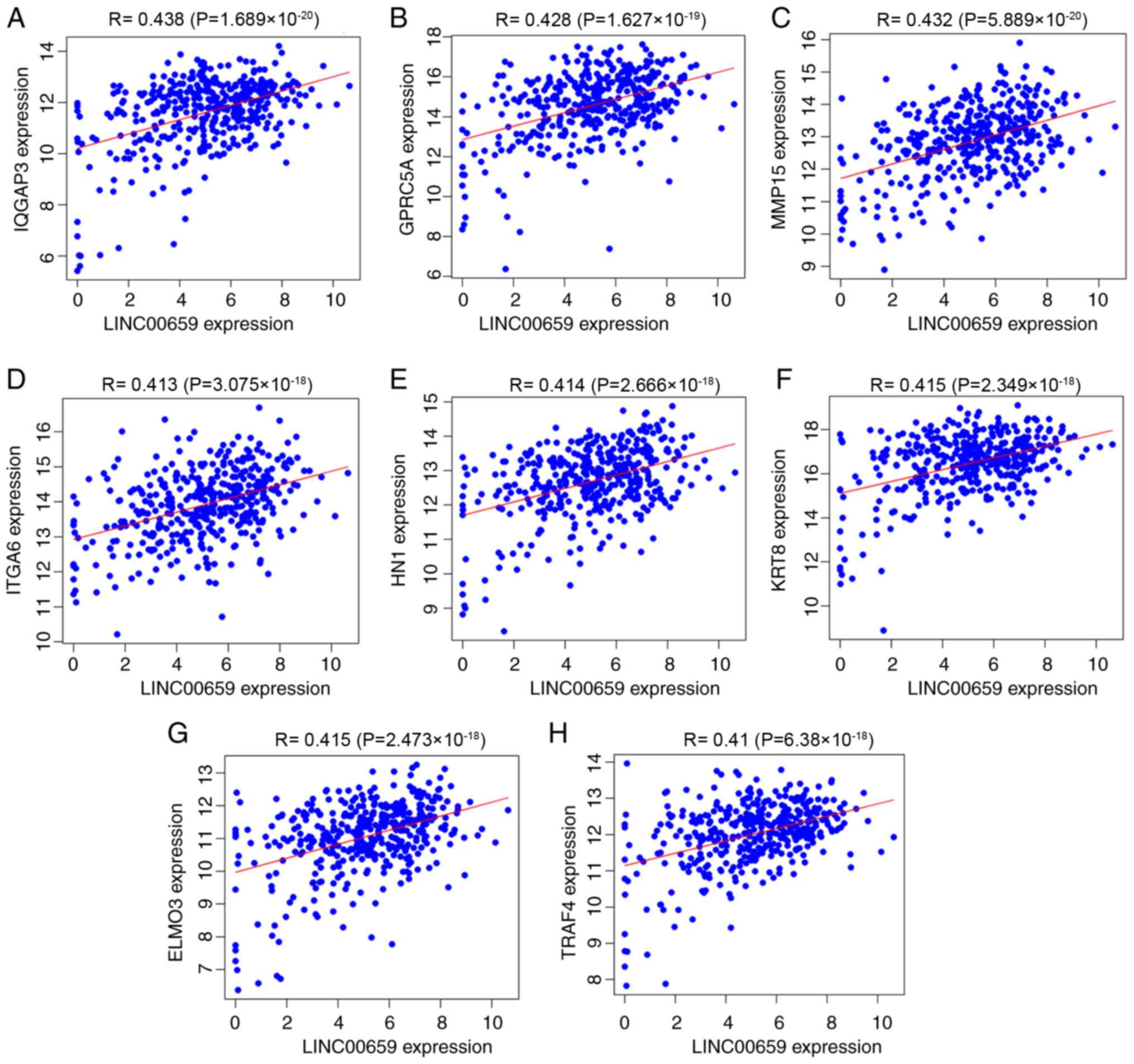 | Figure 5.Pearson correlation analysis of the
expression levels of eight protein-coding genes co-expressed with
LINC00659 expression in The Cancer Genome Atlas database.
Correlation of LINC00659 expression with (A) IQGAP3, (B) GPRC5A,
(C) MMP15, (D) ITGA6, (E), HN1, (F) KRT8, (G) ELMO3 and (H) TRAF4.
IQGAP3, IQ motif-containing GTPase activating protein 3; MMP15,
matrix metalloproteinase 15; ITGA6, integrin subunit α6; GPRC5A, G
protein-coupled receptor class C group 5 member A; HN1,
hematological and neurological expressed 1; KRT8, keratin 8; ELMO3,
engulfment and cell motility 3; TRAF4, TNF receptor associated
factor 4. |
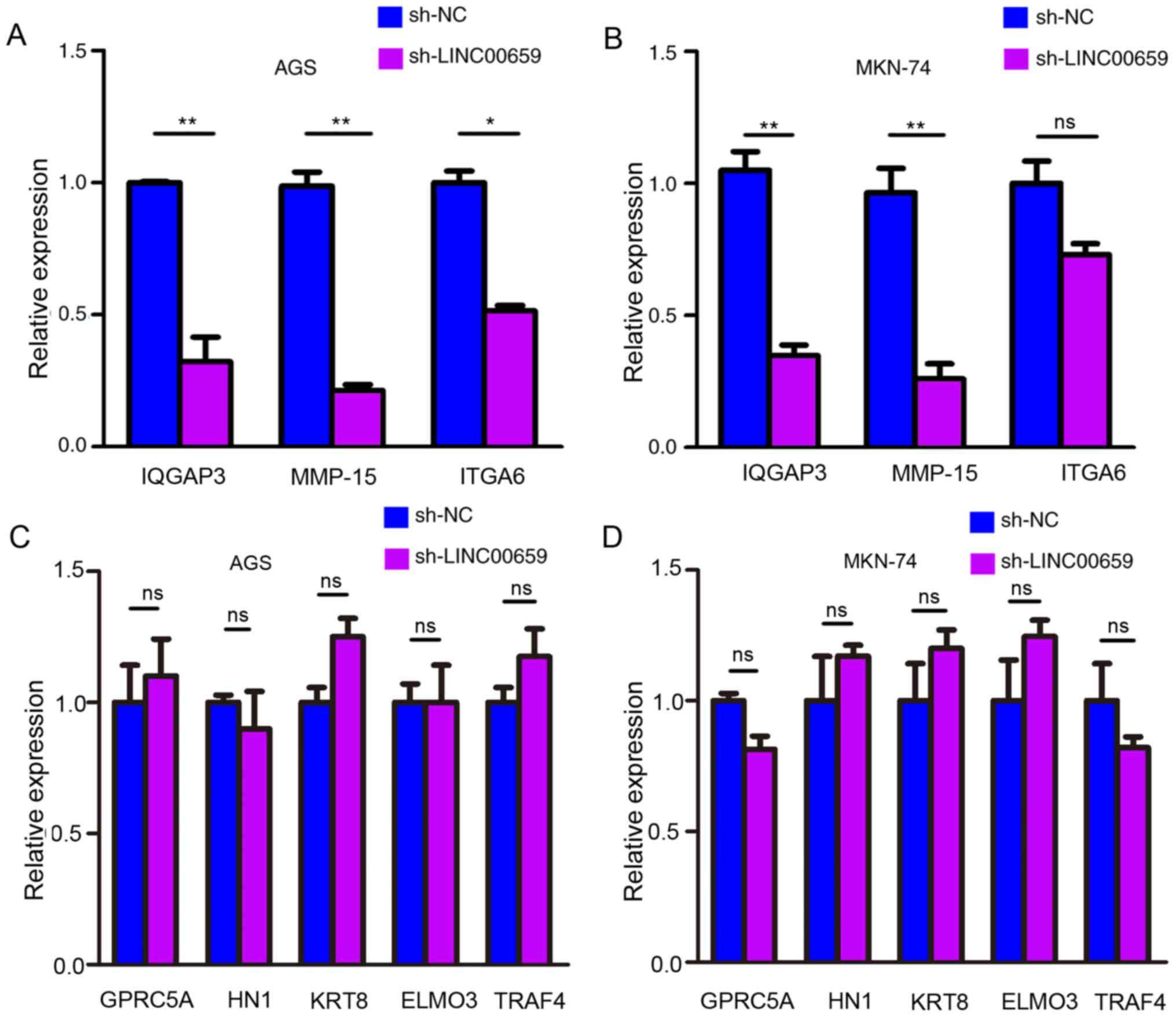 | Figure 6.IQGAP3 and MMP-15 are downstream
targets of LINC00659. Relative expression levels of potential
target genes (IQGAP3, MMP-15 and ITGA6) detected by reverse
transcription-quantitative PCR in (A) AGS and (B) MKN-74 cells
following transfection with sh-LINC00659. (C) Relative expression
levels of GPRC5A, HN1, KRT8, ELMO3 and TRAF4 following
LINC00659-knockdown in AGS. (D) Relative expression levels of
GPRC5A, HN1, KRT8, ELMO3 and TRAF4 following LINC00659-knockdown in
MKN-74. *P<0.05; **P<0.01 (Student's unpaired t-test). MMP15,
matrix metalloproteinase 15; IQGAP3, IQ motif-containing GTPase
activating protein 3; ITGA6, integrin subunit α6; GPRC5A, G
protein-coupled receptor class C group 5 member A; HN1,
hematological and neurological expressed 1; KRT8, keratin 8; ELMO3,
engulfment and cell motility 3; TRAF4, TNF receptor associated
factor 4; sh-NC, short hairpin RNA negative control; ns, not
significant. |
Discussion
An increasing number of studies has revealed that
lncRNAs serve a critical role in cancer (22,32,33), and
although a large number of lncRNAs have been identified in the
human genome, only a small number of them have been experimentally
validated and functionally annotated in GC (34). The initiation and progression of GC
involve deregulation of numerous lncRNAs (35). Some lncRNA functions have been
uncovered, so that they have become potential biomarkers and
therapeutic targets for the prognosis and treatment of GC (36). For example, the lncRNA HOXA11-AS
promotes GC cell proliferation and invasion via
serine/arginine-rich splicing factor 1, and may function as a
putative marker in GC (37).
Additionally, lncRNA antisense non-coding RNA in the INK4 locus may
have potential as a biomarker and therapeutic target for GC
prognosis and treatment (38).
LINC00337, MNX1-AS1 and MALAT1 have been shown to promote
proliferation, migration and invasion of GC cells (39–41).
Genomic amplifications are hallmarks of cancer, and investigation
of non-coding elements, such as lncRNAs, localized on amplicons is
often employed in research to delineate their roles in malignant
transformation (37–40,42). A
frequent expansion on chromosome 20 by CGH has often been
demonstrated (43), and the
chromosome has numerous GC susceptibility loci in 20q13.33
(44,45). The present study identified four
lncRNAs, namely RTEL1-TNFRSF6B, SLCO4A1-AS1, ZBTB46-AS1 and
LINC00659, in this amplified 20q13.33 region, which were
differentially expressed in GC tissues and para-cancerous tissues;
however, only LINC00659 was associated with the prognosis of
patients with GC. Additionally, genetic alterations of LINC00659
amplification were significantly associated with higher expression
levels of LINC00659.
A previous study has revealed that LINC00659 is a
novel oncogenic lncRNA involved in colon cancer cell proliferation
via modulating the cell cycle (46).
LINC00659-knockdown significantly suppresses colon cancer cell
proliferation by impairing cell cycle progression and suppressing
PI3K-AKT signaling (38). However,
to the best of our knowledge, the expression pattern and functional
significance of LINC00659 in GC cell migration and invasion have
remained elusive. In the present study, LINC00659 expression was
shown to be upregulated in GC tissues. The expression levels of
LINC00659 were associated with GC stage, and LINC00659 expression
was significantly increased in patients with advanced GC.
Additionally, the expression levels of LINC00659 were higher in
cancer tissues than in adjacent non-cancerous tissues in patients
with lymph node metastasis. After knocking down LINC00659, the AGS
and MKN-74GC cell lines displayed significantly attenuated
migratory and invasive abilities, although LINC00659-knockdown had
no influence on cell proliferation. IQGAP3 and MMP15 were
identified as potential downstream targets of LINC00659, with
crucial roles in GC metastasis. However, a limitation of the
present study is that the function of these two downstream targets
was not further analyzed in in vitro and in vivo
experiments. Wound healing, Transwell and tail vein injection
assays to observe the migration and invasion of GC cells following
IQGAP3- and MMP15-knockdown may provide more robust evidence for
the current findings.
In summary, the present study has identified a novel
lncRNA associated with a poor prognosis in patients with GC.
LINC00659 was identified as an oncogenic regulator that promoted GC
cell migration and invasion. The current findings suggested that
IQGAP3 and MMP15 may be putative targets for LINC00659, although
the mechanism requires further investigation. The present findings
indicated that LINC00659 may be an important molecular marker in GC
tumorigenesis, and may have the potential to be a novel promising
prognostic and therapeutic maker for patients with GC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81972664 and
81672414).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PG, YL, KZ and YX performed the experiments,
analyzed the data and wrote the manuscript. ML, XS, SY and YM
performed the experiments and analyzed the data. HF designed the
experiments, supervised the project and revised the manuscript. KZ,
YL and SY confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Southeast University affiliated to Zhongda
Hospital. Verbal informed consent was provided by all patients
included in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karimi P, Islami F, Anandasabapathy S,
Freedman ND and Kamangar F: Gastric cancer: Descriptive
epidemiology, risk factors, screening, and prevention. Cancer
Epidemiol Biomarkers Prev. 23:700–713. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Solomon E, Borrow J and Goddard AD:
Chromosome aberrations and cancer. Science. 254:1153–1160. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takada H, Imoto I, Tsuda H, Sonoda I,
Ichikura T, Mochizuki H, Okanoue T and Inazawa J: Screening of DNA
copy-number aberrations in gastric cancer cell lines by array-based
comparative genomic hybridization. Cancer Sci. 96:100–110. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hidaka S, Yasutake T, Kondo M, Takeshita
H, Yano H, Haseba M, Tsuji T, Sawai T, Nakagoe T and Tagawa Y:
Frequent gains of 20q and losses of 18q are associated with lymph
node metastasis in intestinal-type gastric cancer. Anticancer Res.
23:3353–3357. 2003.PubMed/NCBI
|
|
6
|
Oga A, Kong G, Ishii Y, Izumi H, Park CY
and Sasaki K: Preferential loss of 5q14-21 in intestinal-type
gastric cancer with DNA aneuploidy. Cytometry. 46:57–62. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kokkola A, Monni O, Puolakkainen P,
Larramendy ML, Victorzon M, Nordling S, Haapiainen R, Kivilaakso E
and Knuutila S: 17q12-21 amplicon, a novel recurrent genetic change
in intestinal type of gastric carcinoma: A comparative genomic
hybridization study. Genes Chromosomes Cancer. 20:38–43. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu MS, Chang MC, Huang SP, Tseng CC, Sheu
JC, Lin YW, Shun CT, Lin MT and Lin JT: Correlation of histologic
subtypes and replication error phenotype with comparative genomic
hybridization in gastric cancer. Genes Chromosomes Cancer.
30:80–86. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwon MJ, Kim RN, Song K, Jeon S, Jeong HM,
Kim JS, Han J, Hong S, Oh E, Choi JS, et al: Genes co-amplified
with ERBB2 or MET as novel potential cancer-promoting genes in
gastric cancer. Oncotarget. 8:92209–92226. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tabach Y, Kogan-Sakin I, Buganim Y,
Solomon H, Goldfinger N, Hovland R, Ke XS, Oyan AM, Kalland KH,
Rotter V and Domany E: Amplification of the 20q chromosomal arm
occurs early in tumorigenic transformation and may initiate cancer.
PLoS One. 6:e146322011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garcia-Murillas I, Sharpe R, Pearson A,
Campbell J, Natrajan R, Ashworth A and Turner NC: An siRNA screen
identifies the GNAS locus as a driver in 20q amplified breast
cancer. Oncogene. 33:2478–2486. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sana M and Malik HJ: Current and emerging
breast cancer biomarkers. J Cancer Res Ther. 11:508–513. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ptashkin RN, Pagan C, Yaeger R, Middha S,
Shia J, O'Rourke KP, Berger MF, Wang L, Cimera R, Wang J, et al:
Chromosome 20q amplification defines a subtype of microsatellite
stable, left-sided colon cancers with wild-type RAS/RAF and better
overall survival. Mol Cancer Res. 15:708–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sillars-Hardebol AH, Carvalho B, Belien
JA, de Wit M, Delis-van Diemen PM, Tijssen M, van de Wiel MA,
Ponten F, Fijneman RJ and Meijer GA: BCL2L1 has a functional role
in colorectal cancer and its protein expression is associated with
chromosome 20q gain. J Pathol. 226:442–450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hodgson JG, Chin K, Collins C and Gray JW:
Genome amplification of chromosome 20 in breast cancer. Breast
Cancer Res Treat. 78:337–345. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Snijders AM and Mao JH: Multi-omics
approach to infer cancer therapeutic targets on chromosome 20q
across tumor types. Adv Mod Oncol Res. 2:215–223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M and Murty VV: Identification of copy
number gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bolha L, Ravnik-Glavac M and Glavac D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:72439682017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng S, Xie X, Xiao YF, Tang B, Hu CJ,
Wang SM, Wu YY, Dong H, Li BS and Yang SM: Long noncoding RNA
LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress
gastric cancer progression. Cancer Lett. 412:179–187. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan L, Liang W, Fu M, Huang ZH, Li X,
Zhang W, Zhang P, Qian H, Jiang PC, Xu WR and Zhang X:
Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes
gastric cancer progression. J Cancer Res Clin Oncol. 143:991–1004.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Z, Chen Z, Fan R, Jiang B, Chen X,
Chen Q, Nie F, Lu K and Sun M: Over-expressed long noncoding RNA
HOXA11-AS promotes cell cycle progression and metastasis in gastric
cancer. Mol Cancer. 16:822017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Forrest ME and Khalil AM: Review:
Regulation of the cancer epigenome by long non-coding RNAs. Cancer
Lett. 407:106–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
ENCODE Project Consortium, ; Birney E,
Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qi F, Liu X, Wu H, Yu X, Wei C, Huang X,
Ji G, Nie F and Wang K: Long noncoding AGAP2-AS1 is activated by
SP1 and promotes cell proliferation and invasion in gastric cancer.
J Hematol Oncol. 10:482017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
YiRen H, YingCong Y, Sunwu Y, Keqin L,
Xiaochun T, Senrui C, Ende C, XiZhou L and Yanfan C: Long noncoding
RNA MALAT1 regulates autophagy associated chemoresistance via
miR-23b-3p sequestration in gastric cancer. Mol Cancer. 16:1742017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thorvaldsen JL, Duran KL and Bartolomei
MS: Deletion of the H19 differentially methylated domain results in
loss of imprinted expression of H19 and Igf2. Genes Dev.
12:3693–3702. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chu C, Zhang QC, da Rocha ST, Flynn RA,
Bharadwaj M, Calabrese JM, Magnuson T, Heard E and Chang HY:
Systematic discovery of Xist RNA binding proteins. Cell.
161:404–416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nasrollahzadeh-Khakiani M, Emadi-Baygi M,
Schulz WA and Nikpour P: Long noncoding RNAs in gastric cancer
carcinogenesis and metastasis. Brief Funct Genomics. 16:129–145.
2017.PubMed/NCBI
|
|
35
|
Sun M, Nie FQ, Wang ZX and De W:
Involvement of lncRNA dysregulation in gastric cancer. Histol
Histopathol. 31:33–39. 2016.PubMed/NCBI
|
|
36
|
Yuan L, Xu ZY, Ruan SM, Mo S, Qin JJ and
Cheng XD: Long non-coding RNAs towards precision medicine in
gastric cancer: Early diagnosis, treatment, and drug resistance.
Mol Cancer. 19:962020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu Y, Zhang YM, Ma FB, Pan SR and Liu BZ:
Long noncoding RNA HOXA11-AS promotes gastric cancer cell
proliferation and invasion via SRSF1 and functions as a biomarker
in gastric cancer. World J Gastroenterol. 25:2763–2775. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deng W, Zhang Y, Cai J, Zhang J, Liu X,
Yin J, Bai Z, Yao H and Zhang Z: LncRNA-ANRIL promotes gastric
cancer progression by enhancing NF-kB signaling. Exp Biol Med
(Maywood). 244:953–959. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu K, Ren Q and Zhao Y: lncRNA MALAT1
overexpression promotes proliferation, migration and invasion of
gastric cancer by activating the PI3K/AKT pathway. Oncol Lett.
17:5335–5342. 2019.PubMed/NCBI
|
|
40
|
Ma JX, Yang YL, He XY, Pan XM, Wang Z and
Qian YW: Long noncoding RNA MNX1-AS1 overexpression promotes the
invasion and metastasis of gastric cancer through repressing
CDKN1A. Eur Rev Med Pharmacol Sci. 23:4756–4762. 2019.PubMed/NCBI
|
|
41
|
Hu B, Wang X and Li L: Long noncoding RNA
LINC00337 promote gastric cancer proliferation through repressing
p21 mediated by EZH2. Am J Transl Res. 11:3238–3245.
2019.PubMed/NCBI
|
|
42
|
Li D, Chen Y, Mei H, Jiao W, Song H, Ye L,
Fang E, Wang X, Yang F, Huang K, et al: Ets-1 promoter-associated
noncoding RNA regulates the NONO/ERG/Ets-1 axis to drive gastric
cancer progression. Oncogene. 37:4871–4886. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nicolet C, Guerin E, Neuville A, Kerckaert
JP, Wicker N, Bergmann E, Brigand C, Kedinger M, Gaub MP and Guenot
D: Evidence for various 20q status using allelotyping, CGH arrays,
and quantitative PCR in distal CIN colon cancers. Cancer Lett.
282:195–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tanikawa C, Kamatani Y, Toyoshima O,
Sakamoto H, Ito H, Takahashi A, Momozawa Y, Hirata M, Fuse N,
Takai-Igarashi T, et al: Genome-wide association study identifies
gastric cancer susceptibility loci at 12q24.11-12 and 20q11.21.
Cancer Sci. 109:4015–4024. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jang SH, Park JW, Kim HR, Seong JK and Kim
HK: ADRM1 gene amplification is a candidate driver for metastatic
gastric cancers. Clin Exp Metastasis. 31:727–733. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ,
Hsu CW, Chen WS and Wang JH: Linc00659, a long noncoding RNA, acts
as novel oncogene in regulating cancer cell growth in colorectal
cancer. Mol Cancer. 17:722018. View Article : Google Scholar : PubMed/NCBI
|