Introduction
In the context of tumor biology, the six hallmarks
of cancer have been proposed to be associated with progressively
growing tumors and to be responsible for the complexity of
neoplastic diseases, and these are limitless replicative potential,
evading apoptosis, self-sufficiency in growth signals, sustained
angiogenesis, insensitivity to anti-growth signals, invasion and
metastasis (1). In the last decade,
two emerging hallmarks have been added to this list, namely
deregulating cellular energetics and evading immune destruction
(2). Evasion of immune destruction,
resulting in the formation of the tumor microenvironment through a
theory known as ‘cancer immune-editing’, remains a major concern
(2). This theory comprises three
distinct phases: Elimination, equilibrium and escape (2). Tumor cells induce the immune system,
and, in turn, tumor-infiltrating lymphocytes migrate to the tumor
site to eradicate the transformed cells (3). However, some of these transformed cells
can escape the immune destruction and can progressively grow and
give rise to a clinically apparent tumor (3). Therefore, the immune system is
considered as a dual weapon; it either suppresses tumor formation
or facilitates tumor progression by sculpting the immunogenicity of
the tumors (4). This has brought
immunotherapy to the forefront of oncology, aiming to inhibit tumor
growth and activate antitumor activity. Among the different
approaches of immunotherapy, immune checkpoints serve an important
role. Notably, programmed cell-death protein-1 (PD-1) is expressed
by T-lymphocytic cells (T-cells) during the effector phase to limit
its function via binding to its ligand, known as programmed
cell-death ligand-1 (PD-L1) on the surface of tumor cells, thus
leading to T-cell exhaustion (5).
Nivolumab and pembrolizumab are two anti-PD-1 immunotherapies that
have been approved for the treatment of melanoma and non-small cell
lung cancer (NSCLC), respectively (6,7).
Alternative oncogenic signaling pathways promote PD-L1 expression
in tumor cells, which is the ‘innate immune response’. The
induction of PD-L1 expression in response to IFN-γ is known as the
‘adaptive immune response’ (8).
Based on these important signaling pathways, the regulation of
PD-L1 expression is a broad area of investigation in several types
of cancer, including breast cancer (BC).
In 2018, BC was the most common type of cancer among
women worldwide and ranked first among Egyptian women (9). BC is associated with a poor prognosis
due to the strong heterogeneity of its pathogenesis. Disease
complexity has prompted researchers to investigate what is beyond
the genetic disruption of the disease. The results of these studies
revealed that the epigenetic regulation of the disease pathogenesis
and progression also serves an important role in BC. Emerging
evidence has suggested that the newly discovered non-coding RNAs
(ncRNAs) greatly contribute to carcinogenesis (10). microRNAs (miRNAs/miRs), a subtype of
ncRNAs, may lead to gene silencing via binding to the 3′
untranslated region (3′UTR) of target mRNAs, either through
translational repression or mRNA cleavage (11). Several miRNAs, such as Let-7a and
miR-145, have been reported to be tumor suppressors in BC,
resulting in decreased cellular proliferation and metastasis
(12). Another class of miRNAs that
contribute to cancer cell proliferation are oncomiRs, such as
miR-10b and miR-21 (13). The
present study focused on miR-182-5p, which has been reported to
serve as either an oncogene or tumor suppressor in numerous types
of cancer. Previous studies have demonstrated that the inhibition
of miR-182-5p attenuates cell proliferation and invasion in BC
(14), hepatocellular carcinoma
(HCC) (15) and oral squamous cell
carcinoma (16). Furthermore,
miR-182-5p suppresses renal cell carcinoma cell proliferation by
regulating the AKT/FOXO3a signaling pathway (17).
Another important group of ncRNAs are long
non-coding RNAs (lncRNAs). lncRNAs serve a pivotal role in gene
silencing and disease progression (18). The lncRNA X-inactive specific
transcript (XIST) is exclusively expressed from the X-inactivation
center of the inactive X chromosome and is essential for the
initiation and spread of X chromosome inactivation (19). A previous review article reported
that XIST exerts contradictory functions in different types of
cancer (20). For example, in
invasive pituitary adenoma, XIST acts as an oncogene (21). In addition, XIST could promote brain
metastasis following its silencing in BC (22). Our previous study demonstrated that
XIST combined with PD-L1 expression could serve as a potential
biomarker in patients with BC (23).
Furthermore, a recent study has supported the role of PD-L1 as a
useful biomarker for immunotherapy (24). Another study revealed that PD-L1
expression is positively associated with that of lncRNA T cell
leukemia/lymphoma 6 (TCL6) (25). In
addition, it has been reported that lncRNA TCL6 is associated with
a poor prognosis in patients with BC and increased immune cell
infiltration (25). Additionally,
lncRNA GATA binding protein 3 antisense RNA 1 induces the
deubiquitination of PD-L1, thus resulting in PD-L1 stabilization
and enhanced triple-negative breast cancer (TNBC) progression
(26). TSIX transcript, XIST
antisense RNA (TSIX) is considered to orchestrate the initiation of
X chromosome inactivation, thus determining which X chromosome
remains active by blocking the expression of the antisense XIST RNA
(27). Another lncRNA,
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1),
was originally identified as a prognostic marker for metastatic
lung cancer (28); however, it is
also associated with several other human tumors, such as HCC
(29) and glioblastoma (30). Therefore, the current study aimed to
identify potential ncRNAs regulating PD-L1 expression in TNBC cell
lines.
Materials and methods
Egyptian patients
The present study included 41 patients with BC
(4.88% males and 95.12% females) who underwent tumor resection
surgery between September 2016 and April 2018 at the following
hospitals: Demerdash, Cleopatra, Queens and Nozha Hospitals (Cairo,
Egypt). BC tissues biopsies as well as their adjacent non-cancerous
tissues together with their metastatic lymph nodes (LNs) were
removed. Tissues were subdivided into luminal BC (n=30; 73.1%),
TNBC (n=7; 17.07%) and HER-2-positive (n=4; 9.75%) subtype. The age
of patients ranged between 28 and 70 years with a mean of 49 years.
Pathological examination was performed to assess tumor grade and
stage (The Eighth Edition of the American Joint Committee Cancer
Staging Manual) using the TNM staging system (31). Immunohistochemistry was performed to
analyze receptors (estrogen receptor, progesterone receptor and
HER2) and Ki67. Tumor molecular subtyping was performed for all
tumor tissues by a pathologist during the surgical resection, it
was not performed at our laboratory. This was performed at Elia
Laboratory (Cleopatra Hospital, Heliopolis, Cairo, Egypt).
Furthermore, CEA and CA15-3 were analyzed before surgery. All human
biopsies were obtained with written informed consent. Patients were
subjected to clinical assessment as shown in Table I. The Ethical Committee of the German
University in Cairo and Ain Shams University (Cairo, Egypt)
approved the present study. The inclusion criteria were: All
molecular subtypes of BC, all ages and all types of treatment. The
exclusion criteria were: Male sex.
 | Table I.Characteristics of patients with
breast cancer. |
Table I.
Characteristics of patients with
breast cancer.
| No. | Size, cm | Type | Grade | Stage | Axillary lymph
node | Treatment | Duration of cancer
since diagnosis | Molecular
subtype | Ki67, % |
|---|
| 1 | 4.0 | IDC | 3 | 4 | Positive | N/A | 2 months | TNBC | 35 |
| 2 | 2.0 | IDC | 3 | 1 | Positive | N/A | 2 months | Luminal B,
HER2− | 40 |
| 3 | 2.4 | IDC | 2 | 2 | Positive | N/A | 6 months | Luminal B,
HER2+ | 40 |
| 4 | 4.0 | IDC | 3 | 2 | Positive | N/A | 6 months | Luminal B,
HER2− | 15 |
| 5 | 2.5 | IDC | 2 | 3 | Negative | Neoadjuvant
chemotherapy (6 cycles) | 8 months | Luminal B,
HER2+ | 25 |
| 6 | 2.5 | IDC | 2 | 2 | Positive | N/A | 2 months | Luminal B,
HER2− | 23 |
| 7 | 1.4 | IDC | 2 | 1 | Negative | N/A | 2 months | Luminal A | 12 |
| 8 | 2.1 | IDC | 2 | 2 | Negative | N/A | 4 months | Luminal B,
HER2− | 23 |
| 9 | 1.4 | IDC | 2 | 1 | Negative | N/A | 3 months | Luminal B,
HER2− | 30 |
| 10 | 3.5 | IDC | 2 | 2 | Negative | N/A | 1 year | Luminal A | 17 |
| 11 | 5.0 | IDC | 3 | 3 | Negative | N/A | 6 months |
HER2+ | 50 |
| 12 | 0.3 | IDC | 2 | 2 | Negative | Chemotherapy and
radiotherapy | 1 month | TNBC | 30 |
| 13 | 2.0 | IDC | 2 | 2 | Negative | N/A | 1 month | TNBC | 35 |
| 14 | 2.7 | IDC | 2 | 2 | Positive | N/A | 2 weeks | Luminal B,
HER2+ | 30 |
| 15 | 4.2 | IDC | 2 | 2 | Negative | N/A | 6 months |
HER2+ | 40 |
| 16 | 2.0 | IDC | 2 | 2 | Negative | N/A | 2 months | Luminal B,
HER2+ | 14 |
| 17 | 2.0 | IDC | 2 | 2 | Negative | Chemotherapy and
radiotherapy | 1 month | Luminal A | 18 |
| 18 | 4.0 | IDC | 2 | 2 | Positive | N/A | 8 months | Luminal B | 24 |
| 19 | 2.5 | IDC | 2 | 2 | Negative | N/A | 3 months | Luminal B,
HER2− | 50 |
| 20 | 3.0 | IDC | 2 | 2 | Positive | N/A | 4 months | TNBC | 85 |
| 21 | 0.3 | IDC | 2 | 1 | Positive | N/A | 1 month |
HER2+ | 30 |
| 22 | 3.0 | IDC | 2 | 2 | Positive | N/A | 4 months | Luminal A | 20 |
| 23 | 4.0 | ILC | 2 | 2 | Positive | N/A | 6 months | Luminal A | 18 |
| 24 | 1.5 | IDC | 1 | 1 | Negative | N/A | 2 years | Luminal A | 14 |
| 25 | 6.0×3.0 | IDC | 2 | 2 | Positive | N/A | 6 months | Luminal A | 5 |
| 26 | 2.0×2.5 | IDC | 3 | 3 | Positive | N/A | 10 months | TNBC | 18 |
| 27 | 2.5 | IDC | 2 | 2 | Negative | N/A | 2 years | Luminal A | 10 |
| 28 | 2.5×2.3 | IDC | 2 | 2 | Positive | N/A | 7 months | Luminal B | 35 |
| 29 | 2.5×2.0 | IDC | 2 | 3 | Positive | N/A | 8 months | Luminal B | 22 |
| 30 | 4.0 | IDC | 3 | 4 | Negative | N/A | 6 months | Luminal B | 60 |
| 31 | 1.0×1.0 | IDC | 3 | N/A | Positive | N/A | 6 months | Luminal B,
HER2− | 35 |
| 32 | 9.0 | IDC | 2 | N/A | Negative | N/A | 1 year | Luminal B | 50 |
| 33 | 4.2 | IDC | 2 | 2 | Positive | N/A | 1 month | Luminal B | 30 |
| 34 | 1.6 | ILC | 1 | 2 | Negative | N/A | 2 months | Luminal B | 22 |
| 35 | 2.5×2.0 | IDC | 2 | 2 | Positive | N/A | 6 months |
HER2+ | 35 |
| 36 | 1.5×1.0 | IDC | 2 | 1 | Positive | N/A | 4 months | Luminal A | 8 |
| 37 | 3.5×2.5 | IDC | 2 | 2 | Positive | N/A | 3 months | TNBC | 30 |
| 38 | 1.8 | IDC | 2 | 1 | Negative | N/A | 1 month | Luminal A | 7 |
| 39 | 2.0×1.5 | IDC | 2 | 2 | Positive | N/A | 4 months | TNBC | 30 |
| 40 | 2.5×2.0 | IDC | 2 | 2 | Positive | N/A | 2 months | Luminal A | 18 |
| 41 | 4.0×3.0 | IDC | 2 | 4 | Positive | N/A | 6 months | Luminal A | 15 |
Cell culture
MDA-MB-231 cells (Vacsera) were cultured and
maintained in DMEM (Lonza Group, Ltd.) supplemented with 4.5 g/l
glucose + L-Glutamine, 10% FBS (Lonza Group, Ltd.) and 1%
penicillin/streptomycin (Lonza Group, Ltd.) at 37°C in a 5%
CO2 atmosphere.
Bioinformatics
To detect the potential miRNAs targeting the 3′UTR
of PD-L1 mRNA, the TargetScan (release number, 7.2; http://www.targetscan.org/vert_72/)
bioinformatics target prediction algorithm was used. Based on
binding scores and number of hits, miRNAs with good scores were
selected. PD-L1 upstream targets were predicted. RNA22 software
version 2.0 (http://cm.jefferson.edu/rna22/Interactive/) competing
endogenous RNA (ceRNA; version 2.0; https://web.archive.org/web/20130922123437/http://starbase.sysu.edu.cn/mrnaCeRNA.php)
and TargetScan prediction software were used to analyze the
potential binding of miR-182-5p to lncRNAs XIST, MALAT1 and PD-L1
(position 1193–1199 in the UTR). Furthermore, lnCedb (Gencode 19
version; http://gyanxet-beta.com/lncedb/index.php) and Diana
tools software (version 7.0; http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/page&view=software)
were used to predict the potential binding of lncRNAs XIST and
MALAT1 to PD-L1.
Transfection of MDA-MB-231 cells using
miRNA and small interfering RNA (siRNA/si) oligonucleotides
MDA-MB-231 cells were transfected with 1 nmol mimics
(GeneGlobe, cat. no. 219600) and inhibitors (antagomiRs) of 1 nmol
miR-182-5p (GeneGlobe Id-MIN0000259;
5′-UUUGGCAAUGGUAGAACUCACACU-3′; cat. no. 219300; Qiagen GmbH) at
25°C for 1 h to examine the effect of miR-182-5p on PD-L1, MALAT1,
XIST and TSIX transcript expression. In addition, a parallel
experiment was carried out for transfection efficiency analysis.
This was followed by a series of transfection experiments using 5
nmol siRNAs (predesigned siRNA; Qiagen GmbH) for each lncRNA,
MALAT1 (NR_002819), XIST (NR_001564) and TSIX (NR_003255).
Co-transfection experiments were performed to examine the combined
effect of the upstream manipulators (miR-182-5p and lncRNAs XIST
and MALAT1) of PD-L1 on its expression levels. All transfection
experiments were carried out in quadruplicate using HiPerfect
Transfection Reagent (Qiagen GmbH) according to the manufacturer's
protocol. A group of scrambled (non-specific) siRNAs (cat. no.
1022076; Qiagen GmbH) and scrambled miRNA mimics and antagomirs:
Mixtures of mimics of miR-15a-5p (cat. no. 219600) and miR-122
(cat. no. 219600) for scrambled miRs and mixtures of
anti-miR-15a-5p and anti-miR-122 for scrambled anti-miRs
(hsa-miR-15a-5p; MIMAT0000068; 5′-UAGCAGCACAUAAUGGUUUGUG-3′; cat.
no. 219300; and hsa-miR-122-5p; MIMAT0000421;
5′-UGGAGUGUGACAAUGGUGUUUG-3′; cat. no. 219300; Qiagen GmbH) were
used as negative controls in gene knockdown and miRNA gain/loss of
function experiments, respectively. Cells that were only exposed to
transfection reagent were designated as mock cells, cells
transfected with miR-182-5p were referred to as miR-182-5p cells
and cells transfected with miR-182-5p inhibitor were referred to as
anti-miR-182-5p cells. Cells transfected with siRNAs of MALAT1,
XIST and TSIX were referred to as siMALAT1, siXIST and siTSIX,
respectively. The cells were transfected and incubated under normal
culture conditions (37°C with 5% CO2) for 48 h.
mRNA and miRNA extraction from breast
biopsies and MDA-MB-231 cells (TNBC cell lines)
Breast samples (healthy, cancerous and adjacent LN
tissues) were collected during surgery and were immediately
snap-frozen (−196°C) in liquid nitrogen. The specimens were
manually pulverized in liquid nitrogen. Subsequently, ~100 mg
tissue powder was used for large and small RNA extraction using
Biozol reagent (BioFlux) according to the manufacturer's protocol.
MDA-MB-231 cells were harvested 48 h after transfection according
to the HiPerfect Transfection Reagent protocol. RNA was isolated
using Biozol reagent, followed by cDNA synthesis using a
High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) at 37°C for 135 min. Subsequently,
RNA was quantified using reverse transcription-quantitative PCR
(RT-qPCR). Experiments were performed in quadruplicate.
miRNA and mRNA quantification
The extracted miRNAs were reverse transcribed into
single stranded cDNA using the TaqMan MicroRNA Reverse
Transcription Kit (Bio Basic, Inc.). mRNA was reverse transcribed
into cDNA using the high-capacity cDNA reverse transcription kit
(Bio Basic, Inc.) according to the manufacturer's protocol at 37°C
for 75 min. Relative expression levels of miR-182-5p and RNU6B
(housekeeping gene) were measured using specific primers for
hsa-miR-182-5p and RNU6B. Their assay IDs were 002334 and 001093,
respectively, as well as MALAT1, XIST, PD-L1, TSIX and β-2
microglobulin (as a housekeeping gene for normalization) were
quantified using TaqMan Real-Time Q-PCR (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The ABI Assay IDs for MALAT1, XIST,
PD-L1, TSIX and B2M were Hs00273907_m1, Hs01079824_m1,
Hs01125301_m1, Hs03299334_ml and Hs00187842_m1, respectively. A
StepOne™ System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) was used. The 2−ΔΔCq method was used for
quantification (32). The
thermocycling conditions were as follows: 25°C for 10 min, 37°C for
120 min, 85°C for 5 min, 4°C for infinity consisting of 40 cycles
of denaturation, annealing and extension, respectively.
Statistical analysis
All data are presented as the mean relative
quantitation ± SEM and repeated in quadruplicates. The statistical
method used for multiple groups was one-way ANOVA and multiple
comparisons were analyzed by Tukey's multiple comparison test (when
the mean of each column was compared with every other column) and
Dunnett's multiple comparison test (when the mean of each column
was compared with the mean of the control column). Analysis was
performed using GraphPad Prism 7.02 software (GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Screening of PD-L1 in breast
tissues
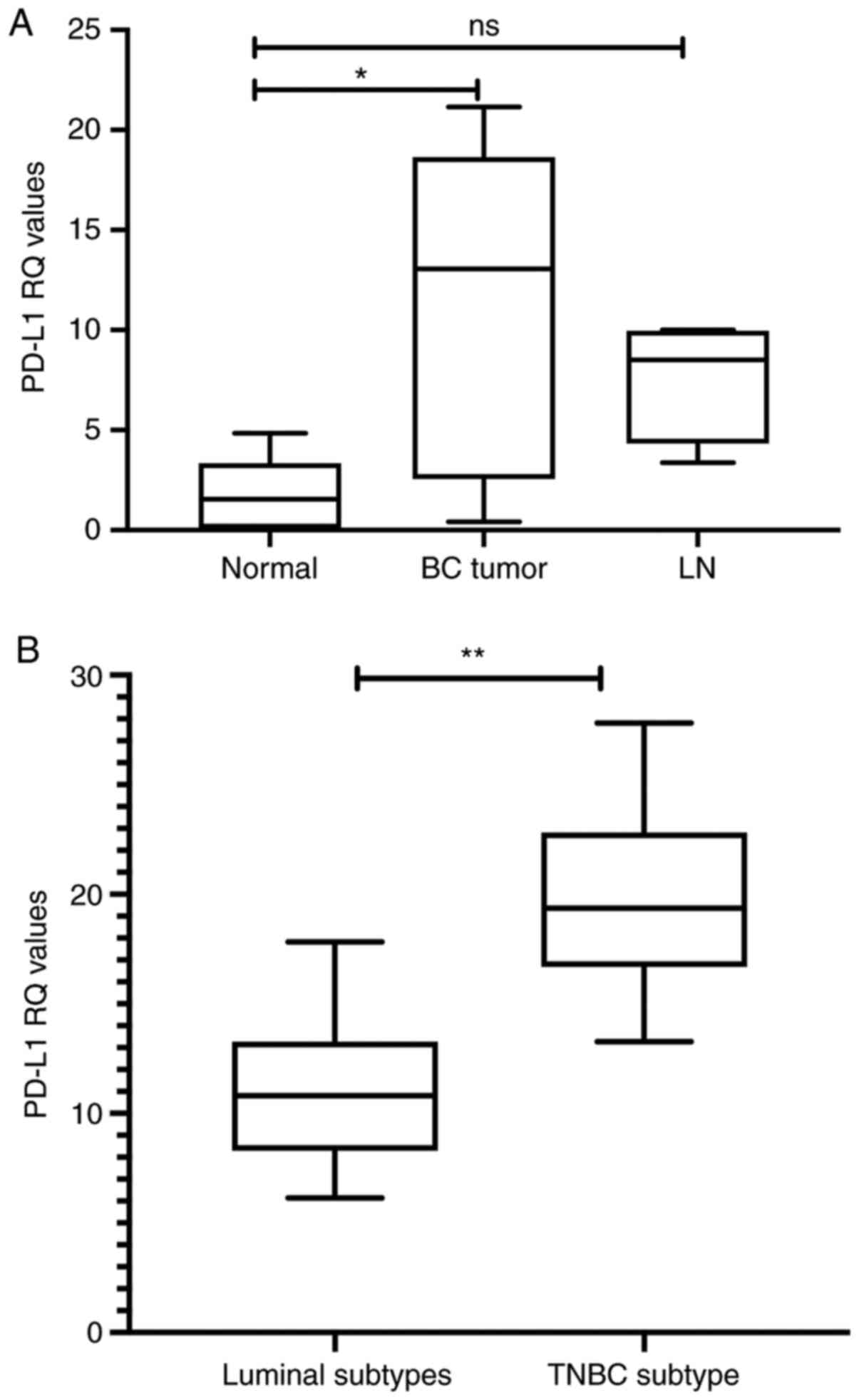
Statistically significant upregulation of PD-L1
transcript expression was observed in all BC subtype tissues
(P=0.0217) compared with healthy tissues, whereas this change was
not significant in the LNs compared with adjacent normal tissues
(P=0.0360; Fig. 1A). However, when
patients were categorized into luminal molecular subtypes and TNBC,
a marked difference in PD-L1 expression was observed. Patients with
TNBC exhibited significant upregulation of PD-L1 expression
(P=0.0037) compared with patients with luminal subtypes (Fig. 1B).
Selection of potential upstream
regulators of PD-L1 mRNA
In silico predictions were performed using
all aforementioned software. According to bioinformatics analysis,
miR-182-5p was predicted to target PD-L1, MALAT1 and XIST.
Additionally, MALAT1 and XIST were identified to target PD-L1
mRNA.
Screening of miR-182-5p, MALAT1 and
XIST expression in BC tissues
miR-182-5p expression was identified to be
upregulated in tumor tissues and LNs (P=0.0061 and P=0.0014,
respectively) compared with in healthy tissues (ANOVA P=0.0013;
Fig. 2A). A marked increase in
MALAT1 mRNA expression was observed in patients with all subtypes
of BC compared with normal adjacent tissue controls (P=0.00001),
whereas there was no significant difference observed for the
expression in LNs (ANOVA P<0.0001; Fig. 2B). In our previous study, XIST
expression was decreased in tissues of patients with BC and
adjacent LN samples from these patients, and markedly downregulated
in TNBC (23).
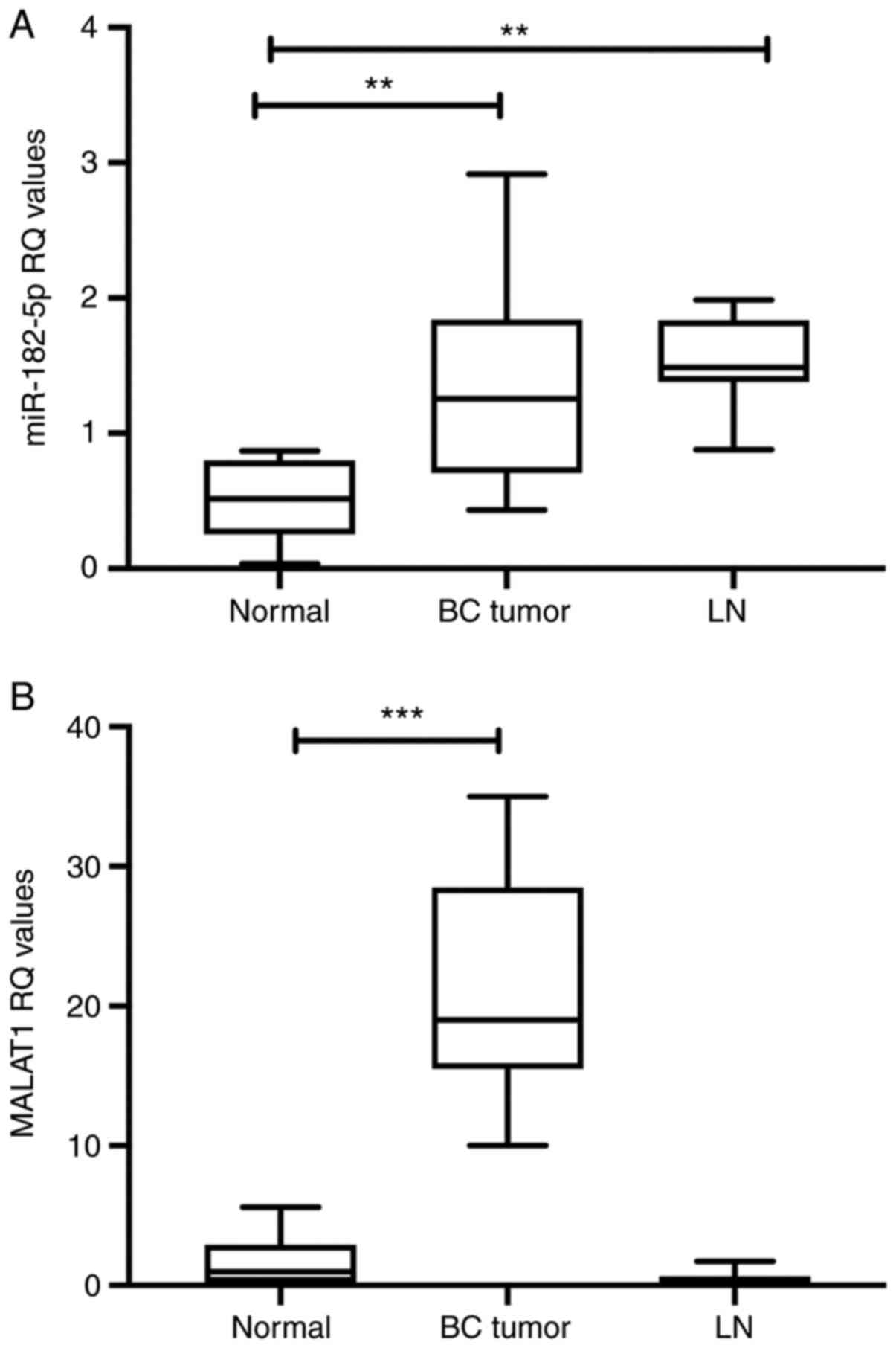 | Figure 2.miR-182-5p, MALAT1 expression in BC
tissues. (A) miR-182-5p was analyzed using RT-qPCR and normalized
to RNU6B as an endogenous control. BC samples, as well as LN
samples, exhibited upregulation of miR-182-5p expression. Analysis
was performed using one-way ANOVA for multiple groups and Tukey's
multiple comparison test. (B) lncRNA MALAT1 expression was analyzed
using RT-qPCR and normalized to B2M as an endogenous control.
MALAT1 expression was upregulated in BC tissues compared with
healthy tissues, whereas no significant difference in expression
was observed in LNs. Analysis was performed using one-way ANOVA for
multiple groups and Dunnett's multiple comparison test. Experiments
were performed in quadruplicate. **P<0.01, ***P<0.001. BC,
breast cancer; LN, lymph node; lncRNA, long non-coding RNA; MALAT1,
metastasis-associated lung adenocarcinoma transcript 1; miR,
microRNA; RQ, relative quantitation; RT-qPCR, reverse
transcription-quantitative PCR; XIST, X-inactive specific
transcript. |
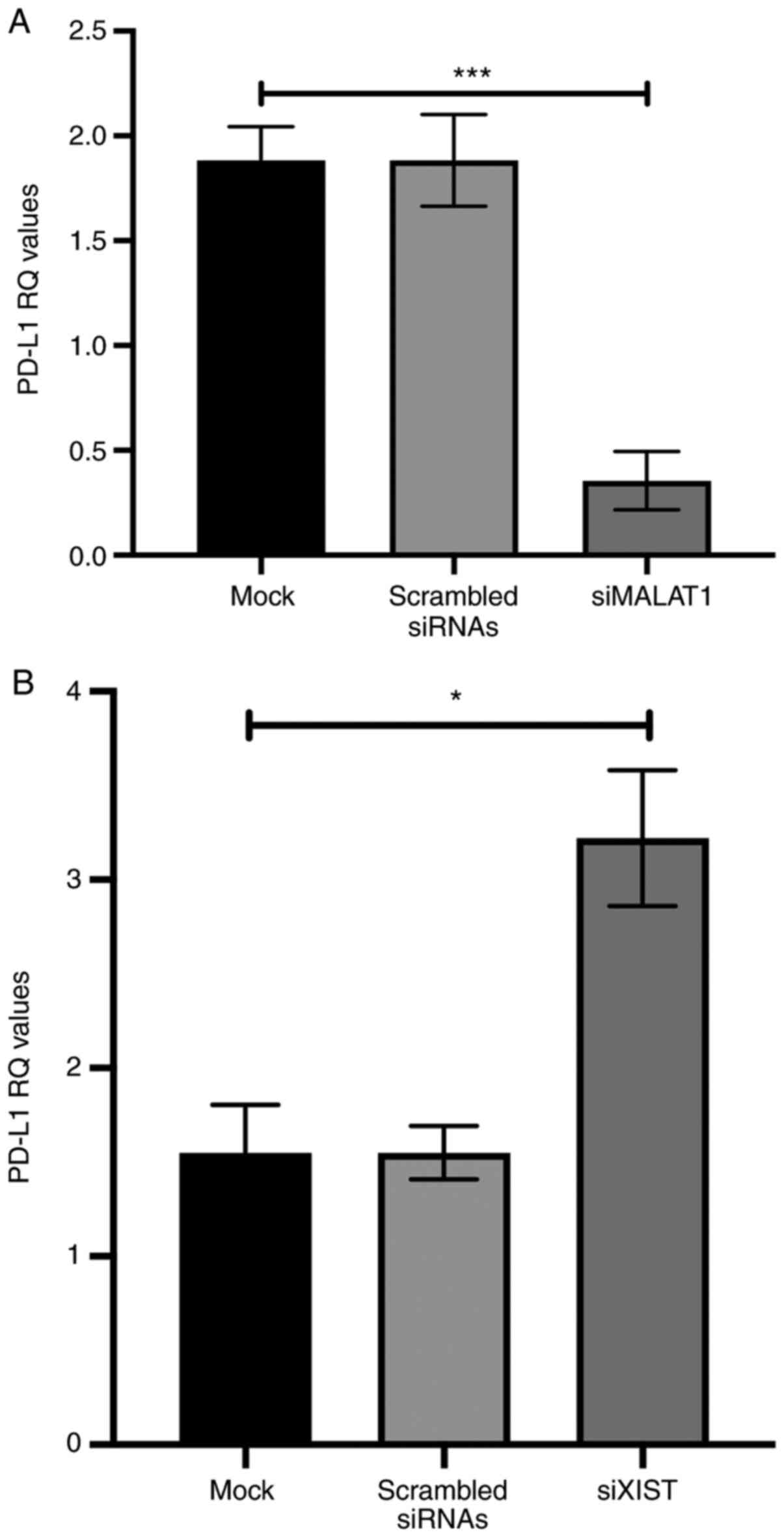
Transfection efficiency for gene
knockdown and miRNA ectopic expression
In order to assure successful transfection of
siRNAs, transfection efficiency was first assessed at 48 h after
transfection using RT-qPCR. The mRNA expression levels of MALAT1
(Fig. S1A), XIST (Fig. S1B) and Tsix (Fig. S1C) were markedly decreased in cells
transfected with their siRNAs compared with their respective mock
cells (P=0.0021, P=0.0056 and P=0.0051, respectively).
Additionally, the expression levels of miR-182-5p were assessed in
MDA-MB-231 cells. miR-182-5p expression was markedly increased in
miR-182-transfected cells compared with mock cells (P=0.0044;
Fig. S1D).
Effect of ectopic miR-182-5p
expression on its downstream targets in MDA-MB-231 cells
Ectopic miR-182-5p expression in MDA-MB-231 cells
was assessed. MDA-MB-231 cells transfected with miR-182-5p mimics
exhibited significant upregulation of PD-L1 expression, as well as
MALAT1 transript expression (P=0.0062 and P=0.0007, respectively),
compared with mock untransfected cells (ANOVA P=0.0065 and 0.0004,
respectively; Fig. 3A and B).
Inhibitors of miR-182-5p significantly decreased PD-L1 and MALAT1
transcript expression (P=0.034 and P=0.0053, respectively) compared
with miR-182-transfected cells (Fig. 3A
and B). However, transfection of mimics of miR-182-5p was
associated with a significant decrease in XIST expression
(P=0.0026) compared with that in the scrambled miRs group of cells,
while miR-182-5p antagomirs increased XIST expression compared with
that in cells transfected with mimics (P=0.0434; ANOVA P=0.0001;
Fig. 3C). Since Tsix is the
anti-sense of XIST, the expression levels of lncRNA Tsix were
examined. In MDA-MB-231 cells, overexpression of miR-182-5p
increased Tsix expression compared with that in mock cells
(P=0.0004; ANOVA P<0.0001; Fig.
3D).
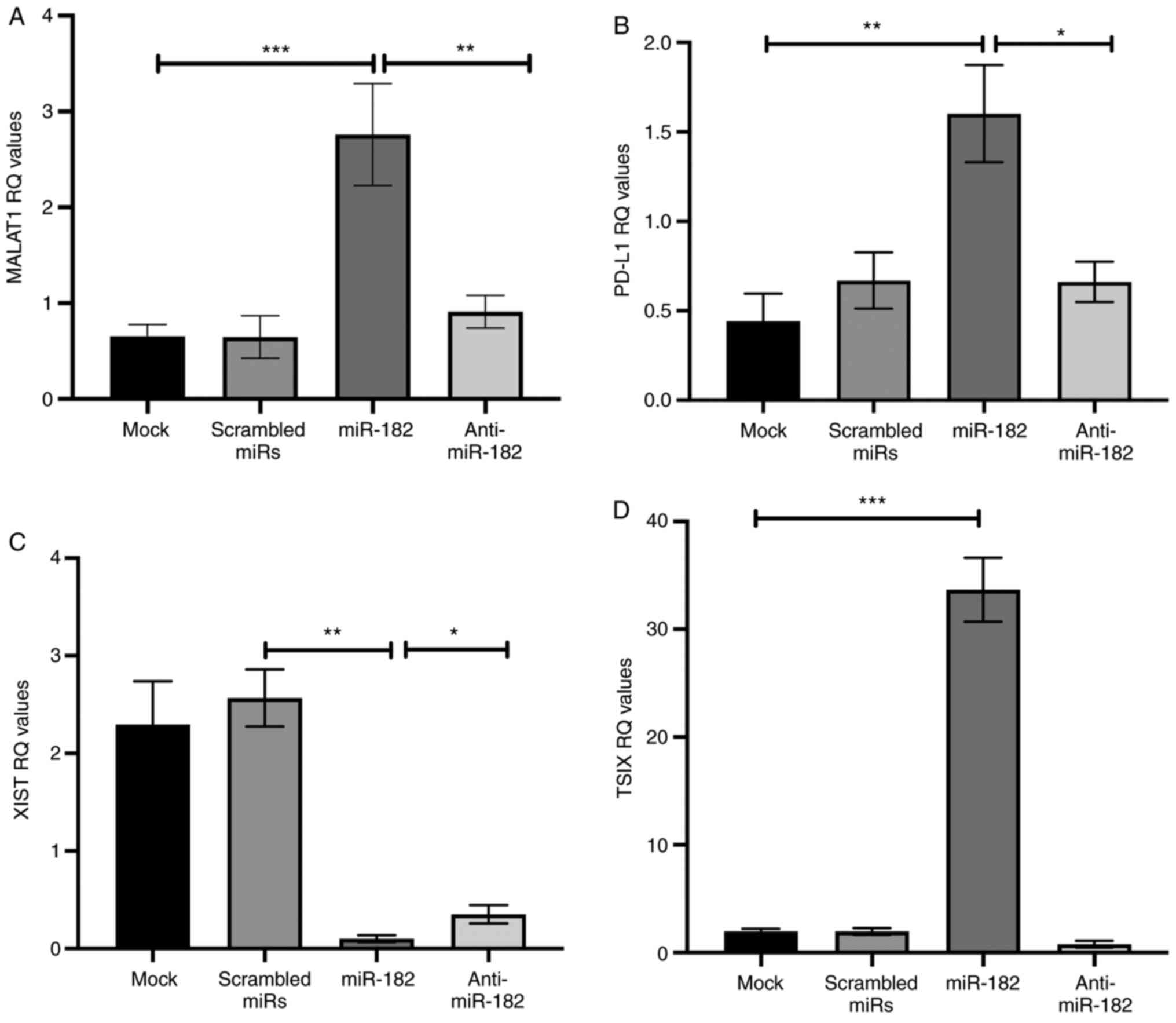 | Figure 3.Effect of ectopic miR-182-5p
expression on its downstream targets in MDA-MB-231 cells. (A)
miR-182-5p mimic transfection of MDA-MB-231 cells resulted in
significant upregulation of MALAT1 mRNA expression compared with
that in mock cells, whereas anti-miR-182-5p transfection decreased
MALAT1 expression compared with mimic-transfected cell lines. (B)
PD-L1 mRNA expression was assessed using reverse
transcription-quantitative PCR and normalized to B2M as an
endogenous control. miR-182-5p mimic transfection of MDA-MB-231
cells resulted in significant upregulation of PD-L1 mRNA expression
compared with that in mock cells, whereas anti-miR-182-5p
transfection decreased PD-L1 expression compared with that in
miR-182-transfected cells. (C) XIST mRNA expression was decreased
significantly in miR-182-transfected cells compared with scrambled
miRs. Furthermore, anti-miR-182-5p transfection resulted in an
increase in XIST expression compared with that in mimic-transfected
cells. (D) miR-182-5p mimic transfection resulted in significant
upregulation of TSIX expression compared with that in mock cells.
Analysis was performed using one-way ANOVA for multiple groups and
Tukey's multiple comparison test. Experiments were performed in
quadruplicate. *P<0.05, **P<0.01, ***P<0.001. MALAT1,
metastasis-associated lung adenocarcinoma transcript 1; miR,
microRNA; PD-L1, programmed cell-death ligand-1; RQ, relative
quantitation; XIST, X-inactive specific transcript; Tsix, TSIX
transcript, XIST antisense RNA. |
Effect of lncRNAs MALAT1 and XIST on
PD-L1 expression in MDA-MB-231 cells
At 48 h after transfection with specific siRNAs
against MALAT1 and XIST, PD-L1 expression was analyzed and
normalized to that of B2M. PD-L1 mRNA expression was decreased
significantly in MALAT1-silenced MDA-MB-231 cells (P=0.0007)
compared with mock cells (ANOVA P=0.0004; Fig. 4A). In contrast to overexpression of
PD-L1, in XIST-silenced MDA-MB-231 cells, PD-L1 mRNA expression was
increased significantly in BC cells (P=0.0178) compared with
untransfected mock cells (ANOVA P=0.0071; Fig. 4B).
Combined effect of the ncRNAs on PD-L1
expression in MDA-MB-231 cells
In three groups of MDA-MB-231 cells, co-transfection
of miR-182-5p mimics was performed once with siXIST and PD-L1 mRNA
expression was significantly increased compared with that of mock
cells (P=0.00004). In order to induce XIST expression in BC cells,
another group of cells was transfected with siRNAs of Tsix, a
negative regulator of XIST, combined with mimics of miR-182-5p.
miR-182-siTsix co-transfection resulted in a significant decrease
in PD-L1 mRNA expression in BC cells, compared with that in mock
cells (P=0.0357). Additionally, the third group of cells was
co-transfected with miR-182-5p mimics combined with siMALAT1, and
the expression levels of PD-L1 were significantly decreased
compared with those in mock cells (P=0.0331; ANOVA P=0.0003;
Fig. 5).
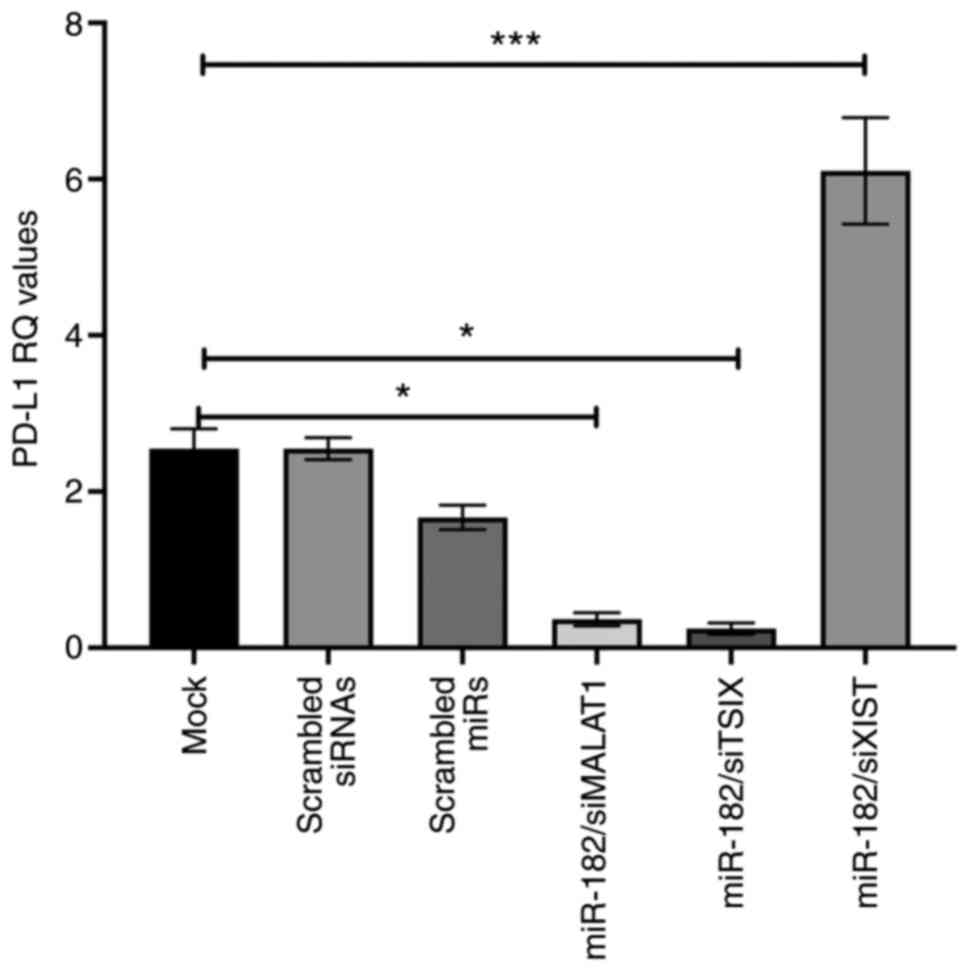 | Figure 5.Combined effect of the two upstream
regulators of the PD-L1 promoter (miR-182-5p and lncRNAs; XIST and
MALAT1) in MDA-MB-231 cells. Following transfection of miR-182-5p
mimics and silencing of XIST, PD-L1 expression was increased
compared with that of mock cells. However, following transfection
with miR-182-5p mimic and knockdown of Tsix, PD-L1 expression was
identified to be significantly downregulated, and PD-L1 expression
was revealed to be downregulated following transfection with
miR-182-5p mimic and silencing of MALAT1. Analysis was performed
using one-way ANOVA for multiple groups and Dunnett's multiple
comparison test. Experiments were performed in quadruplicate.
*P<0.05, ***P<0.001. MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; miR, microRNA; PD-L1, programmed
cell-death ligand-1; RQ, relative quantitation; si, small
interfering RNA; XIST, X-inactive specific transcript; Tsix, TSIX
transcript, XIST antisense RNA. |
Effect of combined knockdown of
lncRNAs on PD-L1 expression in MDA-MB-231 cells
PD-L1 expression was analyzed at 48 h after
transfection of MDA-MB-231 cells. Following transfection with
MALAT1 siRNAs combined with XIST siRNAs, a marked decrease in PD-L1
expression was observed compared with that of mock cells (P=0.006).
Furthermore, following silencing of MALAT1 combined with Tsix
knockdown, significant downregulation of PD-L1 mRNA expression was
observed compared with mock cells (P=0.0002; ANOVA P=0.0004;
Fig. 6).
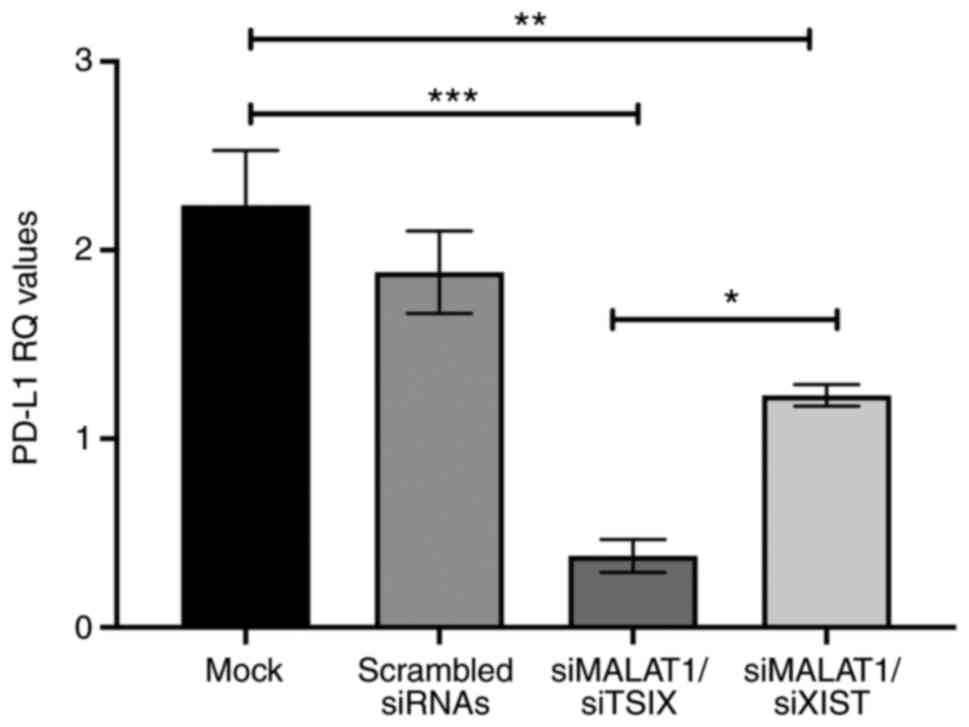 | Figure 6.Effect of combined knockdown of
lncRNAs on PD-L1 expression in MDA-MB-231 cells. The change in
PD-L1 expression as a response to co-transfection of the cells
using siRNAs of MALAT1 and XIST was assessed using TaqMan reverse
transcription-quantitative PCR and normalized to B2M. The results
revealed a marked decrease in PD-L1 expression compared with that
in mock cells, in addition to a significant decrease in PD-L1
expression following silencing of the oncogenic MALAT1 and
silencing of Tsix, the antisense of XIST. Analysis was performed
using one-way ANOVA for multiple groups and Tukey's multiple
comparison test. Experiments were performed in quadruplicate.
*P<0.05, **P<0.01, ***P<0.001. lncRNA, long on-coding RNA;
MALAT1, metastasis-associated lung adenocarcinoma transcript 1;
PD-L1, programmed cell-death ligand-1; RQ, relative quantitation;
si, small interfering RNA; XIST, X-inactive specific transcript;
Tsix, TSIX transcript, XIST antisense RNA. |
Effect of miR-182-5p mimic
transfection combined with lncRNAs on MALAT1 expression in
MDA-MB-231 cells
MALAT1 expression was evaluated at 48 h after
transfection of MDA-MB-231 cells with miR-182-5p mimics combined
with XIST siRNAs. Based on the results of RT-qPCR, transfection
with mimics of miR-182-5p and XIST siRNAs resulted in a significant
increase in MALAT1 expression compared with that of mock cells
(P=0.0004). Additionally, combined knockdown of Tsix and ectopic
miR-182-5p expression was associated with significant upregulation
of MALAT1 expression compared with that of mock cells (P<0.0001;
ANOVA P<0.0001; Fig. 7).
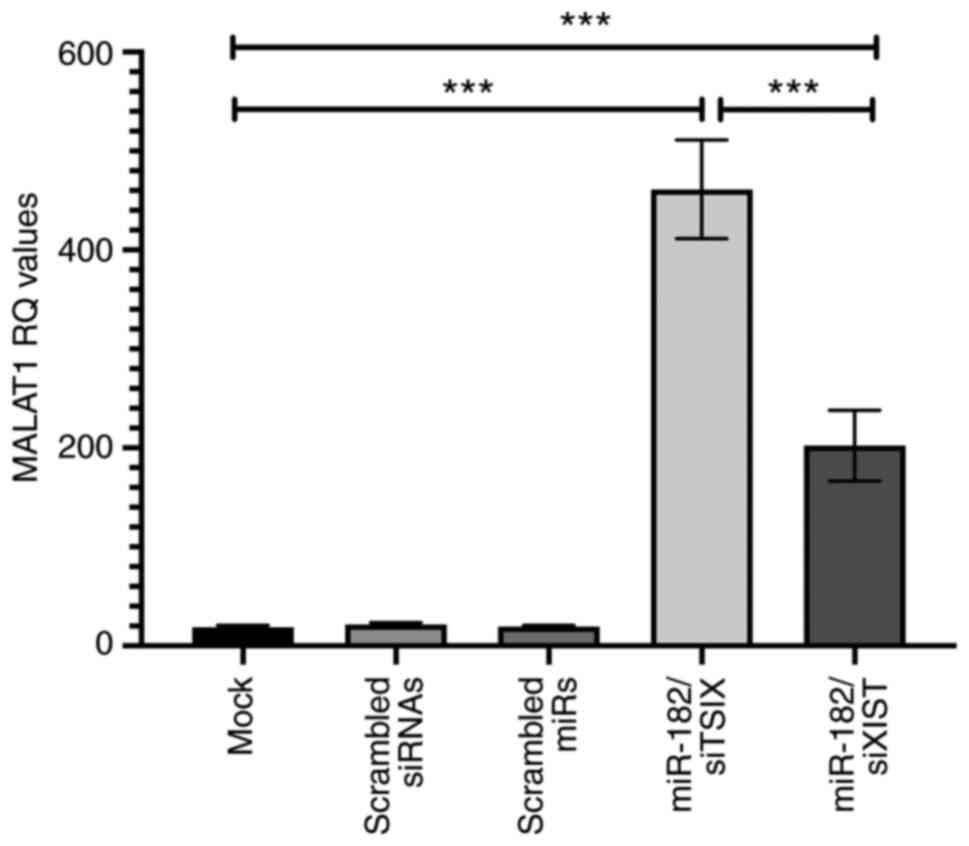 | Figure 7.Effect of miR-182-5p mimic
transfection combined with lncRNAs on MALAT1 regulation in
MDA-MB-231 cells. Using reverse transcription-quantitative PCR, the
relative expression levels of MALAT1 were analyzed and normalized
to B2M. A significant increase in MALAT1 expression was observed in
breast cancer cells when XIST siRNAs were co-transfected with
miR-182-5p mimics compared with mock cells. Notably, MALAT1 mRNA
expression was increased when breast cancer cells were
co-transfected with siTsix combined with miR-182-5p mimics compared
with that of mock cell. Analysis was performed using one-way ANOVA
for multiple groups and Tukey's multiple comparison test.
Experiments were performed in quadruplicate ***P<0.001. lncRNA,
long on-coding RNA; MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; miR, microRNA; RQ, relative
quantitation; si, small interfering RNA; XIST, X-inactive specific
transcript; Tsix, TSIX transcript, XIST antisense RNA. |
Discussion
In the last decade, significant progress and
previous advances in cancer immunology have provided novel
therapeutic approaches for the treatment of cancer (33). The clinical response observed in
patients treated with antibodies blocking the immune checkpoints,
namely the expression of cytotoxic T-lymphocyte-associated protein
4 and the PD-1/PD-L1 signaling pathway, led to their approval by
the Food and Drug Administration for the treatment of melanoma in
2011 and 2014, respectively (34).
The antibody against PD-1, nivolumab, was approved in 2015 for
squamous lung cancer treatment (6).
In addition, it has been reported that antibodies targeting PD-1 or
PD-L1 are effective and safe in treating several types of tumors,
including bladder cancer, Hodgkin's lymphoma and renal cell
carcinoma (35). The effects of
immune checkpoint inhibitors in different types of cancer have
promoted the targeting of these signaling pathways in other tumors,
such as BC (36). The association
between PD-L1 and the prognosis of patients with different types of
cancer has been a research topic of considerable interest. However,
the prognostic value of PD-L1 in patients with BC remains
controversial. Therefore, the current study aimed to investigate
the differential expression of lncRNAs in tissues derived from
patients with BC and the MDA-MB-231 cell line. In addition, the
miRNA-mediated regulation of PD-L1, XIST and MALAT1 expression
remains poorly investigated, particularly in BC. Therefore, the
present study also aimed to reveal novel miRNA/lncRNA interactions
in BC and their immune-modulatory effects on PD-L1 expression, in
order to provide novel possible immunotherapeutic targets for the
treatment of different subtypes of BC, particularly TNBC. The
results demonstrated that PD-L1 expression was upregulated in BC
tissues, with higher expression levels observed in TNBC compared
with the luminal subtypes. A previous study has demonstrated that
PD-L1 expression is associated with LN metastasis and TNBC,
suggesting that PD-L1 could serve as a promising biomarker for
monitoring prognosis in patients with BC and selecting the
appropriate immunotherapy (37).
Consistent with a previous study that demonstrated that PD-L1
expression is upregulated in the MDA-MB-468 cell line (38), the PD-L1 expression levels were
increased in MDA-MB-231 cells in the present study. Early clinical
trials have revealed that treatment with PD-1/PD-L1 inhibitors is
efficient against BC tumors, and particularly against TNBC
(39). Furthermore, a previous study
has demonstrated that patients with metastatic TNBC present a
positive clinical response to the blockade of PD-1 or PD-L1 with
specific antibodies, such as pembrolizumab or atezulizumab
(40). This finding was consistent
with the results of the present study. Notably, PD-L1 expression is
associated with several clinicopathological parameters, including
tumor size, grade and invasion (41). PD-L1 expression is associated with
higher tumor grade (42), shortened
survival rate and poor therapeutic outcome (43,44).
Since the present study demonstrated that PD-L1
expression was upregulated in TNBC, bioinformatics tools were
subsequently applied to identify the upstream modulators of its
expression. It has been reported that ncRNAs are involved in BC and
the regulation of gene expression. The majority of studies have
focused on determining the functions of miRNAs and lncRNAs, and
only a few have investigated how their expression is
transcriptionally regulated. Furthermore, numerous studies have
reported the targeting effect of miRNAs on lncRNAs in different
types of cancer. For example, a study demonstrated that miR-130a
could directly target FOS-like antigen 1, thus inhibiting cancer
cell migration and invasion in TNBC (45). In addition, lncRNAs can compete with
miRNAs for the same target-gene and they serve as precursors for
miRNAs. Emerging evidence has suggested that miRNAs are critical
key players in cancer immunotherapy as they act as crucial
regulators of immune responses under physiological and pathological
conditions (46). It has been
demonstrated that miRNAs are involved in cell transformation and
multiplication by acting as oncomiRs or tumor suppressors in
various types of cancer (47).
Several miRNAs have been identified as regulators of PD-L1
expression. Tumor suppressors, such as miR-15a and miR-16, are
predicted to target PD-L1, thus resulting in downregulation of
PD-L1 expression in malignant pleural mesothelioma (48). In the present study, miR-182-5p was
selected based on the results of the bioinformatics analysis, and
it was demonstrated that miR-182-5p exerted strong binding affinity
with PD-L1, MALAT1 and XIST mRNAs. The results revealed that
miR-182-5p expression was upregulated in TNBC, as well as in
luminal subtypes. This finding was consistent with previous
studies, showing that miR-182-5p expression is increased in TNBC
(49,50). Additionally, another study revealed
that treatment with miR-182-5p inhibitors attenuates cell apoptosis
and proliferation via regulation of CRISPR associated protein 9
expression in MCF-7 human BC cells (51). Additionally, miR-182-5p has been
identified to be upregulated in TNBC and luminal A breast tumors
(52). Furthermore, miR-182-5p was
highly expressed in a panel of human BC samples, highlighting its
role as a potential oncomiR in BC that could positively regulate
metastasis and promote cell colonization (53).
lncRNA MALAT1 serves an important oncogenic role in
different types of cancer. The present study demonstrated that
MALAT1 mRNA expression was significantly elevated in BC tissues.
These findings were in agreement with previous studies showing that
lncRNA MALAT1 could promote cell proliferation and invasion in TNBC
(54) and lung cancer (55). Additionally, miR-182-5p could
regulate MALAT1 expression in MDA-MB-231 cells. A previous study
revealed that MALAT1 overexpression is associated with poor
prognosis in patients with colorectal carcinoma (53). In addition, the expression levels of
MALAT1 are positively associated with the LN status, tumor stage
and histological grade in BC (54).
MALAT1 downregulation suppresses the progression of osteosarcoma
(56). Furthermore, miR-129-5p could
upregulate MALAT1, resulting in cell proliferation and the
progression of colon cancer (57).
Previous studies have demonstrated that MALAT1 serves a
protumorigenic role in pancreatic cancer (58), NSCLC (59) and ovarian cancer (60). Furthermore, accumulating evidence has
suggested that MALAT1 contributes to the initiation and progression
of bladder cancer via regulation of the expression levels of miRNAs
(61). A previous study suggested
that MALAT1 could serve as a therapeutic target or a novel
diagnostic biomarker for BC (62).
Consistent with the findings of the present study, another study
demonstrated that MALAT1 knockdown inhibits BC cell proliferation,
migration and invasion, and induces apoptosis (63). The present results revealed that
silencing of MALAT1 decreased PD-L1 expression. Furthermore, it has
been demonstrated that lysine demethylase 5B expression could
promote BC aggressiveness via MALAT1 overexpression and
downregulation of miR-448 (64). In
TNBC, MALAT1 expression is upregulated, and patients with increased
levels of MALAT1 exhibit poor overall survival (65). It has been reported that lncRNA XIST
serves an important role as a tumor suppressor or oncogene in
several types of cancer, such as prostate cancer where it acts as a
tumor suppressor (66). In the
present study, XIST expression was downregulated in MDA-MB-231
cells following miR-182-5p overexpression. Similarly, a previous
study revealed that XIST expression is downregulated in BC
(67). By contrast, another study
demonstrated that the expression levels of XIST were increased in
BRCA1-positive BC, suggesting that XIST expression could be used as
a marker to discriminate between BRCA1-positive and -negative
breast tumors (68). Additionally,
XIST upregulation promotes osteosarcoma (69), HCC (70) and bladder cancer (71) cell proliferation, while it acts as an
oncogene in NSCLC via regulation of the miR-37a/la-related protein
1 downstream signaling pathway (72).
Since PD-L1 and MALAT1 act as immune-modulatory
targets in the scope of the downstream signaling pathway, their
expression pattern was investigated following manipulation of
miR-182-5p expression. Ectopic miR-182-5p expression was assessed
in MDA-MB-231 cells using RT-qPCR and resulted in the upregulation
of PD-L1 and TSIX expression. In addition, elevated mRNA expression
levels of MALAT1 were observed in MDA-MB-231 cells transfected with
miR-182-5p mimics compared with mock cells. In contrast to renal
cancer, where miR-182-5p mimics decrease MALAT1 expression,
resulting in inhibition of cancer cell proliferation (73), the treatment with miR-182-5p
inhibitor reversed this effect. Transfection with miR-182-5p mimics
markedly downregulated XIST expression. Bioinformatics analysis
predicted that PD-L1 could strongly bind with MALAT1 and XIST.
Therefore, it was hypothesized that the regulation of PD-L1
expression could be mediated by lncRNAs, as upstream regulators,
rather than miR-182-5p. Therefore, the effect of each lncRNA on
PD-L1 expression was investigated. The expression levels of PD-L1
were determined in cells transfected with siMALAT1 or siXIST. PD-L1
expression was upregulated following transfection with siXIST;
however, it was downregulated following MALAT1 knockdown. This
finding was consistent with the results of another study,
demonstrating that the expression levels of MALAT1 are positively
associated with PD-L1 expression in NSCLC (59). Additionally, following treatment of
B-cell lymphoma human cell lines with short hairpin RNA MALAT1,
PD-L1 levels are decreased, resulting in inhibition of tumor cell
proliferation (74).
To investigate the regulatory association between
miR-182-5p and PD-L1, and to explore the combined effect of the two
upstream factors on the regulation of PD-L1 expression, the
expression levels of PD-L1 were assessed in cells co-transfected
with different modulators. PD-L1 expression was downregulated in
cells co-transfected with siMALAT1 and miR-182-5p mimics.
Furthermore, silencing of Tsix (a negative regulator of XIST) and
miR-182-5p overexpression in MDA-MB-231 cells decreased PD-L1
expression. However, the mRNA expression levels of PD-L1 were
increased in MDA-MB-231 cells following co-transfection with siXIST
and miR-182-5p mimics. Additionally, it was hypothesized that the
effect of miR-182-5p on PD-L1 expression was abolished in the
presence of lncRNAs. Therefore, the combined effect of lncRNA
expression on that of PD-L1 was further investigated. MDA-MB-231
cells were co-transfected with siMALAT1 and siXIST or siMALAT1
combined with siTsix to upregulate XIST expression. For both
co-transfection conditions, PD-L1 mRNA expression levels were
evaluated. The results revealed that following the silencing of
both MALAT1 and Tsix, PD-L1 expression was downregulated compared
with that of control cells. In addition, the expression levels of
PD-L1 were decreased in MDA-MB-231 cells transfected with siXIST
and siMALAT1. These opposing forces on the regulation of PD-L1
expression indicated that XIST could augment the inhibitory effect
of MALAT1 knockdown on PD-L1 expression. This hypothesis prompted
an investigation into the effect of the main lncRNAs, MALAT1 and
XIST, on PD-L1 expression. Therefore, the expression levels of
MALAT1 were determined in cells co-transfected with miR-182-5p
mimics and siXIST. The results revealed that MALAT1 expression was
upregulated in the aforementioned cells. Additionally, MALAT1
expression was upregulated in cells co-transfected with miR-182-5p
mimics and siTsix. Notably, PD-L1 expression was downregulated in
these cells, suggesting that XIST could be the dominant endogenous
competitor in the regulation of PD-L1 expression. Nevertheless, the
lack of experiments in additional TNBC cell lines should be
considered to be a potential limitation of the present study.
In conclusion, the present study introduced a novel
immune-modulatory miRNA-lncRNA interaction network in BC, namely
the MALAT1/XIST/miR-182-5p/PD-L1 axis. A previous study (75) has demonstrated that miR-182 acts as
an oncomiR, since its expression increases cell migration and
proliferation in vitro. In vivo assays in mice have
demonstrated that the expression of miR-182 significantly increases
tumor volume and enhances instant metastasis in the lungs (75). The results of the present study
suggested that the upregulation of miR-182-5p could act as an
oncomiR in BC tissues and MDA-MB-231 cells, and highlighted its
molecular effects on pivotal immunomodulatory signaling pathways by
promoting the upregulation of oncogenic lncRNAs PD-L1 and MALAT1 in
the MDA-MB-231 BC cell line. In addition, miR-182-5p downregulated
the expression of the tumor suppressor gene XIST in the same cells.
These findings supported the key role of the ceRNA network,
MALAT1/XIST, in regulating PD-L1 expression in BC, and suggested
their potential role as immunotherapeutic targets. Overall, both
molecules could be utilized as promising biomarkers in clinical
diagnosis and prognosis of aggressive BC tumors.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AS performed the practical work, data analysis,
writing and revision of the manuscript. RAT was the sample
provider. RAT was also involved in the acquisition of data, editing
and revising the manuscript, and contributed to the conception and
design of the study, and to obtaining materials and analysis tools,
as well as assessing the authenticity of all the raw data. HMET was
the principle investigator of the project who supervised the work.
HMET made substantial contributions to conception and design, in
addition to analysis and interpretation of data, was involved in
drafting the manuscript, assessing the authenticity of all the raw
data and revising it critically for important intellectual content
and gave final approval of the version to be published. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent for participation in the
study or use of their tissue was obtained from all participants.
The Ethical Committee of the German University in Cairo and Ain
Shams University (Cairo, Egypt) approved the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fouad YA and Aanei C: Revisiting the
hallmarks of cancer. Am J Cancer Res. 7:1016–1036. 2017.PubMed/NCBI
|
|
2
|
Mittal D, Gubin MM, Schreiber RD and Smyth
MJ: New insights into cancer immunoediting and its three component
phases-elimination, equilibrium and escape. Curr Opin Immunol.
27:16–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vaziri Fard E, Ali Y, Wang XI, Saluja K, H
Covinsky M, Wang L and Zhang S: Tumor-infiltrating lymphocyte
volume is a better predictor of disease-free survival than stromal
tumor-infiltrating lymphocytes in invasive breast carcinoma. Am J
Clin Pathol. 152:656–665. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Y, Wang J, Wang X, Li X, Song J, Fang
J, Liu X, Liu T, Wang D, Li Q, et al: Pik3ip1 is a negative immune
regulator that inhibits antitumor T cell immunity. Clin Cancer Res.
25:6180–6194. 2019.PubMed/NCBI
|
|
5
|
Alsaab HO, Sau S, Alzhrani R, Tatiparti K,
Bhise K, Kashaw SK and Iyer AK: PD-1 and PD-L1 checkpoint signaling
inhibition for cancer immunotherapy: Mechanism, combinations, and
clinical outcome. Front Pharmacol. 8:5612017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo L, Zhang H and Chen B: Nivolumab as
Programmed Death-1 (PD-1) inhibitor for targeted immunotherapy in
tumor. J Cancer. 8:410–416. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
McDermott J and Jimeno A: Pembrolizumab:
PD-1 inhibition as a therapeutic strategy in cancer. Drugs Today
(Barc). 51:7–20. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karachaliou N, Gonzalez-Cao M, Crespo G,
Drozdowskyj A, Aldeguer E, Gimenez-Capitan A, Teixido C,
Molina-Vila MA, Viteri S, De Los Llanos Gil M, et al: Interferon
gamma, an important marker of response to immune checkpoint
blockade in non-small cell lung cancer and melanoma patients. Ther
Adv Med Oncol. Jan 18–2018.(Epub ahead of print). doi:
10.1177/1758834017749748. View Article : Google Scholar
|
|
9
|
Azim HA and Ibrahim AS: Breast cancer in
Egypt, China and Chinese: Statistics and beyond. J Thorac Dis.
6:864–866. 2014.PubMed/NCBI
|
|
10
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin HY and Xiao C: MicroRNA mechanisms of
action: What have we learned from mice? Front Genet. 6:3282015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sempere LF, Christensen M, Silahtaroglu A,
Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S and Cole CN:
Altered MicroRNA expression confined to specific epithelial cell
subpopulations in breast cancer. Cancer Res. 67:11612–11620. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Asghari F, Haghnavaz N, Baradaran B,
Hemmatzadeh M and Kazemi T: Tumor suppressor microRNAs: Targeted
molecules and signaling pathways in breast cancer. Biomed
Pharmacother. 81:305–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao YS, Yang WC, Xin HW, Han JX and Ma
SG: MiR-182-5p knockdown targeting PTEN inhibits cell proliferation
and invasion of breast cancer cells. Yonsei Med J. 60:148–157.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cao MQ, You AB, Zhu XD, Zhang W, Zhang YY,
Zhang SZ, Zhang KW, Cai H, Shi WK, Li XL, et al: miR-182-5p
promotes hepatocellular carcinoma progression by repressing FOXO3a.
J Hematol Oncol. 11:122018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li N, Nan CC, Zhong XY, Weng JQ, Fan HD,
Sun HP, Tang S, Shi L and Huang SX: miR-182-5p promotes growth in
oral squamous cell carcinoma by inhibiting CAMK2N1. Cell Physiol
Biochem. 49:1329–1341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu X, Wu J, Li S, Hu Z, Xu X, Zhu Y, Liang
Z, Wang X, Lin Y, Mao Y, et al: Downregulation of microRNA-182-5p
contributes to renal cell carcinoma proliferation via activating
the AKT/FOXO3a signaling pathway. Mol Cancer. 13:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fang H, Disteche CM and Berletch JB: X
Inactivation and escape: Epigenetic and structural features. Front
Cell Dev Biol. 7:2192019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Samir A, Salama E and El Tayebi HM: The
long non-coding RNA XIST: A new cornerstone in carcinogenesis. J
Mol Genet Med. 12:22018.
|
|
21
|
Zhou K, Li S, Du G, Fan Y, Wu P, Sun H and
Zhang T: LncRNA XIST depletion prevents cancer progression in
invasive pituitary neuroendocrine tumor by inhibiting bFGF via
upregulation of microRNA-424-5p. Onco Targets Ther. 12:7095–7109.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo
YY, Feng J, Sanders S, Jin G, Singh R, et al: Loss of XIST in
breast cancer activates MSN-c-Met and reprograms microglia via
exosomal miRNA to promote brain metastasis. Cancer Res.
78:4316–4330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salama EA, Adbeltawab RE and El Tayebi HM:
XIST and TSIX: Novel cancer immune biomarkers in
PD-L1-overexpressing breast cancer patients. Front Oncol.
9:14592019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu Y, Zhang W, Li A, Chen Y, Ou Q, He Z,
Zhang Y, Liu R, Yao H and Song E: Association of long noncoding RNA
biomarkers with clinical immune subtype and prediction of
immunotherapy response in patients with cancer. JAMA Netw Open.
3:e2021492020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Li Z, Chen M, Chen H, Zhong Q,
Liang L and Li B: lncRNA TCL6 correlates with immune cell
infiltration and indicates worse survival in breast cancer. Breast
Cancer. 27:573–585. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang M, Wang N, Song P, Fu Y, Ren Y, Li Z
and Wang J: LncRNA GATA3-AS1 facilitates tumour progression and
immune escape in triple-negative breast cancer through
destabilization of GATA3 but stabilization of PD-L1. Cell Prolif.
53:e128552020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gayen S, Maclary E, Buttigieg E, Hinten M
and Kalantry S: A primary role for the Tsix lncRNA in maintaining
random X-chromosome inactivation. Cell Rep. 11:1251–1265. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao ZB, Chen F and Bai XF: Long noncoding
RNA MALAT1 regulates hepatocellular carcinoma growth under hypoxia
via sponging MicroRNA-200a. Yonsei Med J. 60:727–734. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Argadal OG, Mutlu M, Ak Aksoy S, Kocaeli
H, Tunca B, Civan MN, Egeli U, Cecener G, Bekar A, Taskapilioglu
MO, et al: Long noncoding RNA MALAT1 may be a prognostic biomarker
in IDH1/2 wild-type primary glioblastomas. Bosn J Basic Med Sci.
20:63–69. 2020.PubMed/NCBI
|
|
31
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC Cancer Staging Manual. 8th edition.
Springer International Publishing; 2017, View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mittendorf EA, Philips AV, Meric-Bernstam
F, Qiao N, Wu Y, Harrington S, Su X, Wang Y, Gonzalez-Angulo AM,
Akcakanat A, et al: PD-L1 expression in triple-negative breast
cancer. Cancer Immunol Res. 2:361–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Blackburn SD, Shin H, Haining WN, Zou T,
Workman CJ, Polley A, Betts MR, Freeman GJ, Vignali DA and Wherry
EJ: Coregulation of CD8+ T cell exhaustion by multiple inhibitory
receptors during chronic viral infection. Nat Immunol. 10:29–37.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Buchbinder EI and Desai A: CTLA-4 and PD-1
pathways: Similarities, differences, and implications of their
inhibition. Am J Clin Oncol. 39:98–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Linsley PS, Brady W, Urnes M, Grosmaire
LS, Damle NK and Ledbetter JA: CTLA-4 is a second receptor for the
B cell activation antigen B7. J Exp Med. 174:561–569. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
van der Merwe PA, Bodian DL, Daenke S,
Linsley P and Davis SJ: CD80 (B7-1) binds both CD28 and CTLA-4 with
a low affinity and very fast kinetics. J Exp Med. 185:393–403.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sasidharan Nair V, Toor SM, Ali BR and
Elkord E: Dual inhibition of STAT1 and STAT3 activation
downregulates expression of PD-L1 in human breast cancer cells.
Expert Opin Ther Targets. 22:547–557. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Monneur A, Gonçalves A and Bertucci F:
PD-L1 expression and PD-1/PD-L1 inhibitors in breast cancer. Bull
Cancer. 105:263–274. 2018.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Alegre ML, Frauwirth KA and Thompson CB:
T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol. 1:220–228.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang X, Teng F, Kong L and Yu J: PD-L1
expression in human cancers and its association with clinical
outcomes. Onco Targets Ther. 9:5023–5039. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Takahashi T, Tagami T, Yamazaki S, Uede T,
Shimizu J, Sakaguchi N, Mak TW and Sakaguchi S: Immunologic
self-tolerance maintained by CD25(+)CD4(+) regulatory T cells
constitutively expressing cytotoxic T lymphocyte-associated antigen
4. J Exp Med. 192:303–310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krummel MF and Allison JP: CTLA-4
engagement inhibits IL-2 accumulation and cell cycle progression
upon activation of resting T cells. J Exp Med. 183:2533–2540. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wing K, Onishi Y, Prieto-Martin P,
Yamaguchi T, Miyara M, Fehervari Z, Nomura T and Sakaguchi S:
CTLA-4 control over Foxp3+ regulatory T cell function. Science.
322:271–275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mao H, Zhang L, Yang Y, Zuo W, Bi Y, Gao
W, Deng B, Sun J, Shao Q and Qu X: New insights of CTLA-4 into its
biological function in breast cancer. Curr Cancer Drug Targets.
10:728–736. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Duraiswamy J, Kaluza KM, Freeman GJ and
Coukos G: Dual blockade of PD-1 and CTLA-4 combined with tumor
vaccine effectively restores T-cell rejection function in tumors.
Cancer Res. 73:3591–3603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hodi FS, O'Day SJ, McDermott DF, Weber RW,
Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel
JC, et al: Improved survival with ipilimumab in patients with
metastatic melanoma. N Engl J Med. 363:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kao SC, Cheng YY, Williams M, Kirschner
MB, Madore J, Lum T, Sarun KH, Linton A, McCaughan B, Klebe S, et
al: Tumor suppressor microRNAs contribute to the regulation of
PD-L1 expression in malignant pleural mesothelioma. J Thorac Oncol.
12:1421–1433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schütz F, Stefanovic S, Mayer L, von Au A,
Domschke C and Sohn C: PD-1/PD-L1 pathway in breast cancer. Oncol
Res Treat. 40:294–297. 2017. View Article : Google Scholar
|
|
50
|
Krishnan K, Steptoe AL, Martin HC, Wani S,
Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR,
Gabrielli B, et al: MicroRNA-182-5p targets a network of genes
involved in DNA repair. RNA. 19:230–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dong H, Strome SE, Salomao DR, Tamura H,
Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al:
Tumor-associated B7-H1 promotes T-cell apoptosis: A potential
mechanism of immune evasion. Nat Med. 8:793–800. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yee D, Shah KM, Coles MC, Sharp TV and
Lagos D: MicroRNA-155 induction via TNF-alpha and IFN-gamma
suppresses expression of programmed death ligand-1 (PD-L1) in human
primary cells. J Biol Chem. 292:20683–20693. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhan Y, Li X, Liang X, Li L, Cao B, Wang
B, Ma J, Ding F, Wang X, Pang D and Liu Z: MicroRNA-182 drives
colonization and macroscopic metastasis via targeting its
suppressor SNAI1 in breast cancer. Oncotarget. 8:4629–4641. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Robainas M, Otano R, Bueno S and
Ait-Oudhia S: Understanding the role of PD-L1/PD1 pathway blockade
and autophagy in cancer therapy. Onco Targets Ther. 10:1803–1807.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wu F, Yin Z, Yang L, Fan J, Xu J, Jin Y,
Yu J, Zhang D and Yang G: Smoking induced extracellular vesicles
release and their distinct properties in non-small cell lung
cancer. J Cancer. 10:3435–3443. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cha YJ and Shim HS: PD-L1 expression and
CD8+ tumor-infiltrating lymphocytes are associated with ALK
rearrangement and clinicopathological features in inflammatory
myofibroblastic tumors. Oncotarget. 8:89465–89474. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Peters S, Kerr KM and Stahel R: PD-1
blockade in advanced NSCLC: A focus on pembrolizumab. Cancer Treat
Rev. 62:39–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jia YH, et al: Long noncoding RNA
MALAT1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wei S, Wang K, Huang X and Zhao Z and Zhao
Z: LncRNA MALAT1 contributes to non-small cell lung cancer
progression via modulating miR-200a-3p/programmed death-ligand 1
axis. Int J Immunopathol Pharmacol. 33:20587384198596992019.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Gordon MA, Babbs B, Cochrane DR, Bitler BG
and Richer JK: The long non-coding RNA MALAT1 promotes ovarian
cancer progression by regulating RBFOX2-mediated alternative
splicing. Mol Carcinog. 58:196–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xie H, Liao X, Chen Z, Fang Y, He A, Zhong
Y, Gao Q, Xiao H, Li J, Huang W and Liu Y: LncRNA MALAT1 inhibits
apoptosis and promotes invasion by antagonizing miR-125b in bladder
cancer cells. J Cancer. 8:3803–3811. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ou X, Gao G, Bazhabayi M, Zhang K, Liu F
and Xiao X: MALAT1 and BACH1 are prognostic biomarkers for
triple-negative breast cancer. J Cancer Res Ther. 15:1597–1602.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xiping Z, Bo C, Shifeng Y, Feijiang Y,
Hongjian Y, Qihui C and Binbin T: Roles of MALAT1 in development
and migration of triple negative and Her-2 positive breast cancer.
Oncotarget. 9:2255–2267. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bamodu OA, Huang WC, Lee WH, Wu A, Wang
LS, Hsiao M, Yeh CT and Chao TY: Aberrant KDM5B expression promotes
aggressive breast cancer through MALAT1 overexpression and
downregulation of hsa-miR-448. BMC Cancer. 16:1602016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zheng L, Zhang Y, Fu Y, Gong H, Guo J, Wu
K, Jia Q and Ding X: Long non-coding RNA MALAT1 regulates BLCAP
mRNA expression through binding to miR-339-5p and promotes poor
prognosis in breast cancer. Biosci Rep. 39:BSR201812842019.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Du Y, Weng XD, Wang L, Liu XH, Zhu HC, Guo
J, Ning JZ and Xiao CC: LncRNA XIST acts as a tumor suppressor in
prostate cancer through sponging miR-23a to modulate RKIP
expression. Oncotarget. 8:94358–94370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jiang L, Yu X, Ma X, Liu H, Zhou S, Zhou
X, Meng Q, Wang L and Jiang W: Identification of transcription
factor-miRNA-lncRNA feed-forward loops in breast cancer subtypes.
Comput Biol Chem. 78:1–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Schouten PC, Vollebergh MA, Opdam M,
Jonkers M, Loden M, Wesseling J, Hauptmann M and Linn SC: High XIST
and Low 53BP1 expression predict poor outcome after high-dose
alkylating chemotherapy in patients with a BRCA1-like breast
cancer. Mol Cancer Ther. 15:190–198. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yang C, Wu K, Wang S and Wei G: Long
non-coding RNA XIST promotes osteosarcoma progression by targeting
YAP via miR-195-5p. J Cell Biochem. 119:5646–5656. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu WG and Xu Q: Long non-coding RNA XIST
promotes hepatocellular carcinoma progression by sponging
miR-200b-3p. Eur Rev Med Pharmacol Sci. 23:9857–9862.
2019.PubMed/NCBI
|
|
71
|
Zhou K, Yang J, Li X and Chen W: Long
non-coding RNA XIST promotes cell proliferation and migration
through targeting miR-133a in bladder cancer. Exp Ther Med.
18:3475–3483. 2019.PubMed/NCBI
|
|
72
|
Xu Z, Xu J, Lu H, Lin B, Cai S, Guo J,
Zang F and Chen R: LARP1 is regulated by the XIST/miR-374a axis and
functions as an oncogene in non-small cell lung carcinoma. Oncol
Rep. 38:3659–3667. 2017.PubMed/NCBI
|
|
73
|
Kulkarni P, Dasgupta P, Bhat NS, Shahryari
V, Shiina M, Hashimoto Y, Majid S, Deng G, Saini S, Tabatabai ZL,
et al: Elevated miR-182-5p associates with renal cancer cell
mitotic arrest through diminished MALAT-1 expression. Mol Cancer
Res. 16:1750–1760. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang QM, Lian GY, Song Y, Huang YF and
Gong Y: LncRNA MALAT1 promotes tumorigenesis and immune escape of
diffuse large B cell lymphoma by sponging miR-195. Life Sci.
231:1163352019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang X, Ma G, Liu J and Zhang Y:
MicroRNA-182 promotes proliferation and metastasis by targeting
FOXF2 in triple-negative breast cancer. Oncol Lett. 14:4805–4811.
2017. View Article : Google Scholar : PubMed/NCBI
|