Colorectal cancer (CRC) is responsible for ~10% of
all diagnosed cases of cancer and cancer-associated deaths
worldwide, with ~900,000 deaths annually (1,2). The
incidence and death rate of CRC has increased amongst individuals
aged <50 years old between 2000 and 2013 in the United States,
where the incidence rate has increased by 22% (3). Although the development of traditional
or novel treatment options, including endoscopy, surgery,
downstaging preoperative radiotherapy, systemic therapy, targeted
therapy and immunotherapy, has extended the overall survival of
patients with advanced stage disease to ~3 years, the cure rate of
patients with metastases remains low (2,4). Thus,
understanding the underlying biology of CRC progression may
highlight novel potential biomarkers and therapeutic targets for
assistance in the early diagnosis of CRC, or as treatment
targets.
In 1976, circular RNAs (circRNAs/circs) were first
identified in plant-based RNA viruses under an electron microscope
(5). However, for decades, circRNAs
were considered as functionless junk-RNA or by-products developed
from mRNA splicing (6). In 2013,
Hansen et al (7) revealed
that circRNAs can competitively bind to microRNAs (miRNAs/miRs) and
inhibit their expression, functioning as a miRNA ‘sponge’, and
in-turn increasing expression of the downstream miRNA target genes.
Since then, functional analysis of circRNAs has increased the
current understanding of several physiological and
pathophysiological processes. In recent years, due to the rapid
development of bioinformatics algorithms and experimental
techniques, such as high-throughput RNA sequencing and circRNA
microarray screening, thousands of circRNAs have been identified
and found to be involved in various disease processes. In
cardiovascular diseases, circRNAs regulate the activation of
endothelial cells, vascular smooth muscle cells and macrophages,
and thus function in the initiation and development of
atherosclerosis (8). Additionally,
emerging evidence from in vitro and in vivo
experimental studies have indicated that circRNAs can regulate
adipogenesis and obesity (9,10). Cerebellar degeneration-related
protein 1 antisense RNA (CDR1as, also known as CiRs-7), was the
first circRNA found to act as a sponge of a miRNA, miR-7 (7), serving important roles in Alzheimer's
disease and Parkinson's disease, amongst other neurodegenerative
diseases (11). In 2015,
Bachmayr-Heyda et al (12)
first reported a global reduction of circRNA abundance in CRC cell
lines and cancer tissues compared with in normal cells and tissues.
These results suggest that cells with high proliferative rates,
particularly tumors, universally trend towards exhibiting low
levels of circRNA expression. This may suggest that circRNAs are
not likely to be involved in cancer (13). However, the roles of several circRNAs
in different types of cancer have emerged in recent years. In the
present review, the association between circRNAs and CRC is
discussed. The biogenesis and functions of circRNAs are first
discussed, followed by a comprehensive summary of the role of
circRNAs in CRC biological processes, their association with
clinicopathological features, as well as their involvement in the
therapeutic response, highlighting their potential as CRC
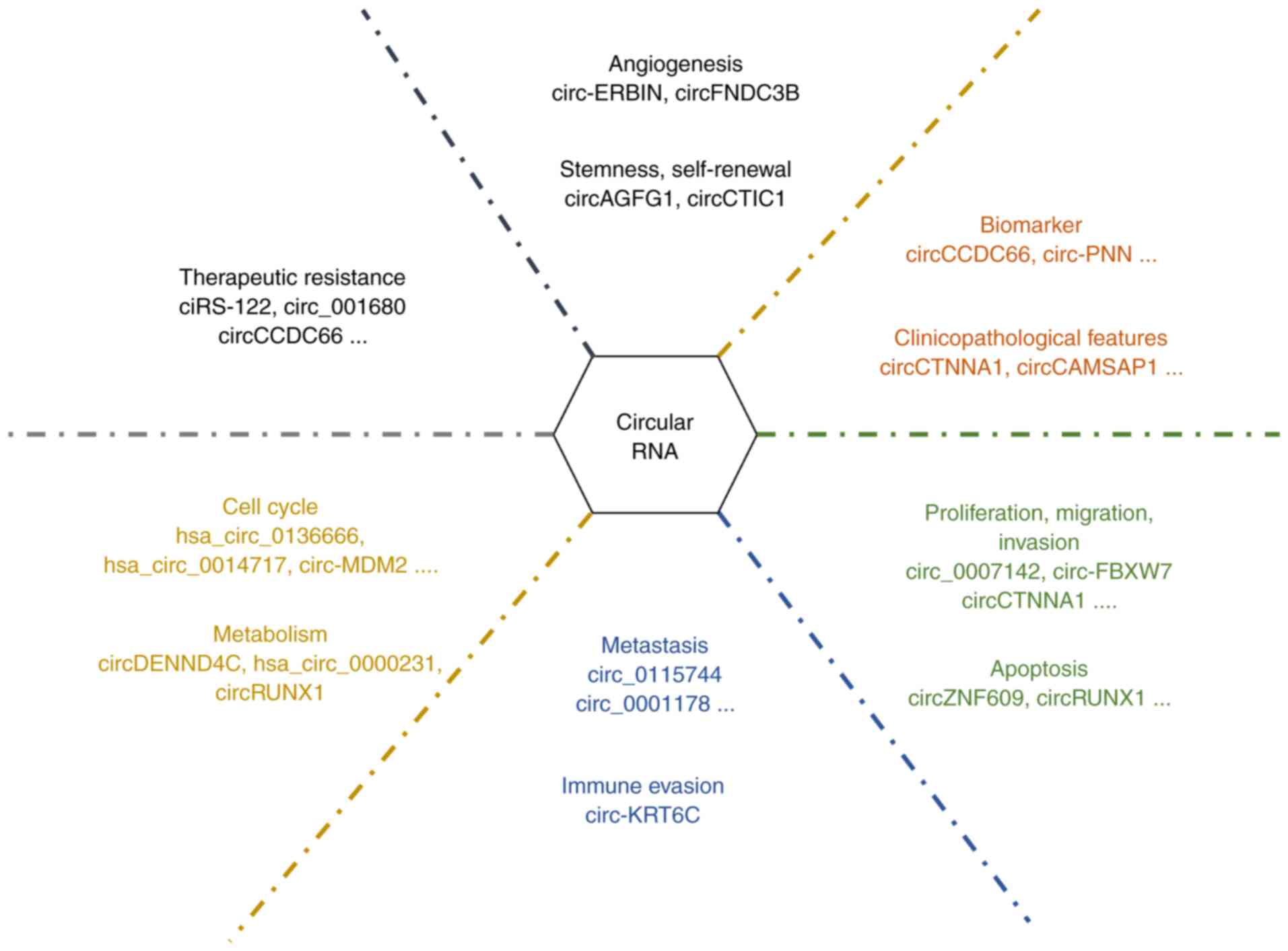
biomarkers in diagnosis, prognosis and treatment of CRC (Fig. 1).
circRNAs are a major type of non-coding RNA that are
produced by back-splicing of exons from pre-mRNA (14). During back-splicing, a downstream
splice-donor site is covalently linked to an upstream
splice-acceptor site (15), through
which a covalently closed RNA molecule is formed. Typically, mRNA
maturation consists of transcription, splicing, capping,
polyadenylation, export and final surveillance (16); however, in circRNA production, no
polyadenylation or capping is required (14). Notably, different circRNAs can be
produced from the same sequence through alternative back-splicing
events (17). Generally, according
to the different structures and cycling mechanisms, circRNA are
divided into four subtypes: Exonic circRNA, intronic circRNA,
exon-intron circRNA and intergenic circRNA, with exonic circRNAs
being the most common type (15).
Although back-splicing of exons takes place in the nucleus, most
circRNAs are localized to the cytoplasm by RNA helicase in a
length-dependent manner (18).
Compared with the linear mRNA counterpart, due to the presence of a
covalent bond joining the 3′ and 5′ end, circRNAs form a continuous
loop structure and are thus resistant to the degradation by RNA
exonucleases, as well as being highly stable, with a longer median
half-life ranging from 18.8-23.7 h (15,17). How
circRNAs are degraded remains poorly understood. Park et al
(19) demonstrated that
N6-methyladenosine (m6A)-containing circRNAs are
selectively cleaved by RNase P/MRP, which are essential
ribonucleoprotein complexes that function as endoribonucleases, and
engage in tRNA maturation and the cleavage of ribosomal RNAs, long
non-coding RNAs and mRNAs. Other possible mechanisms have been
reviewed elsewhere (17). The
characteristics of circRNAs can be summarized as universality,
diversity, stability and conservatism of evolution (20).
Numerous studies have shown that circRNAs function
as miRNA sponges, protein sponges, decoys, scaffolds, recruiters
and translation templates, and can promote transcription in
multiple biological processes.
RNA binding proteins (RBPs) participate in gene
transcription and translation, and interaction with RBPs is
regarded as a central role of circRNAs (22). circRNAs can interact with regulatory
RBPs, through which they act as protein sponges, decoys, scaffolds
or recruiters, and further affect their target mRNAs (23). For example, circular antisense
non-coding RNA in the INK4 locus (circANRIL) impairs pre-ribosomal
RNA processing and ribosome biogenesis by binding to pescadillo
homologue 1, an essential 60S pre-ribosomal assembly factor, in
human vascular smooth muscle cells and macrophages (24). As a result, circANRIL increases
nucleolar stress and p53 activation, which may improve the
atheroprotective effect by promoting the removal of
hyperproliferative cells from atherosclerotic plaques (24). circFOXO3 functions as a protein
scaffold and promotes MDM2-induced mutant p53 ubiquitination and
subsequent degradation, causing an overall reduction in p53 levels
(25). Another nuclear circRNA,
circ-potassium sodium-activated channel subfamily T member 2,
functions as a protein recruiter, and inhibits basic leucine zipper
ATF-like transcription factor (Batf) expression by recruiting the
nucleosome remodeling deacetylase complex onto the Batf promoter,
which then represses IL-17 expression, and thereby inactivates
group 3 innate lymphoid cells (ILC3), to promote resolution of
innate colitis (26). Certain
circRNAs possess dual roles in the regulation of protein
expression. For example, circ-mitochondrial ribosomal protein S35
(circMRPS35) functions as a protein scaffold to recruit the histone
acetyltransferase lysine acetyltransferase 7 to the promoters of
FOXO1 and FOXO3a genes, which leads to acetylation of H4K5 in their
promoters. circMRPS35 specifically and directly binds to the
FOXO1/3a promoter regions, significantly increasing their
transcription, and thus triggering activation of their downstream
target genes, including p21, p27, Twist1 and E-cadherin (27). Thus, circMRPS35 contributes to a
suppressive effect on cell proliferation and invasion.
Previously, circRNAs have been regarded as
non-coding RNAs due to their circular structure, which lacks 5′ and
3′ untranslated regions that are crucial for the initiation of
translation in eukaryotic cells (28). However, more recently, circRNAs have
been found to encode peptides, where an Internal Ribosome Entry
site and N6-methyladenosines mediated cap-independent
translation initiation were suggested as potential mechanisms
involved in the translation of circRNAs. Detailed mechanisms of
circRNA translation are reviewed elsewhere (28–30).
Legnini et al (31) provided
an example of translatable circRNAs in eukaryotes, suggesting that
circ-zinc finger protein 609 (circZNF609) was translated into
protein in a splicing-dependent and cap-independent manner, and
this was shown to be involved in regulating myogenesis. Translation
of circβ-catenin, another translatable circRNA, produces a novel
β-catenin isoform that can antagonize GSK3β-induced β-catenin
phosphorylation and degradation, and thus stabilize full-length
β-catenin, resulting in the activation of the Wnt signaling pathway
and promoting liver cancer cell proliferation (32).
In addition to the aforementioned functions,
nucleolar circRNAs promote transcription. Li et al (33) showed circRNAs that contain introns
that regulate gene transcription in cis by specifically interacting
with the U1 small nuclear ribonucleoprotein RNA (snRNA). The
intercommunication between U1 snRNA and the U1-binding sites of
exon-intron circRNAs, EIciEIF3J and EIciPAIP2, enhance eukaryotic
translation initiation factor 3 subunit J and poly(A) binding
protein interacting protein 2 transcription, respectively (33).
Using RNA-sequencing, microarray or other sequencing
techniques combined with reverse transcription-quantitative
(RT-q)PCR, differential expression levels of circRNAs in cancerous
vs. non-cancerous tissues have been previously detected (34,35).
With their special circular structure making them resistant to the
degradation of RNase (15), circRNAs
are considered as promising candidates for liquid biopsy, which is
a non-invasive tool to reflect the disease state using body fluids,
such as plasma or urine (36).
Studies have demonstrated that the variations in the expression
levels of circRNAs are significantly associated with several
clinicopathological features of patients with CRC, including
overall survival, prognosis, TNM stage, lymphovascular invasion and
lymph node metastasis (37–39). Thus, these circRNAs are likely to
serve as novels target genes for screening, diagnosis and
monitoring of CRC.
Three circRNAs (hsa_circ_0082182, hsa_circ_0000370
and hsa_circ_0035445) have been validated to be differentially
expressed (increased for hsa_circ_0082182 and hsa_circ_0000370, and
decreased for hsa_circ_0035445) in CRC plasma compared with in
normal plasma by microarray analysis, with area under the curves
(AUCs) of 0.815, 0.737, and 0.703, respectively (40). Moreover, the expression levels of
hsa_circ_0082182 and hsa_circ_0035445 were significantly different
between preoperative and postoperative stages (40). Lin et al (41) investigated the plasma levels of
circ-coiled-coil domain containing 66 (CCDC66), circ-ATP binding
cassette subfamily C member 1 and circ-STIL centriolar assembly
protein (STIL) by RT-qPCR, revealing that their plasma expression
levels were significantly decreased in patients with CRC (n=45)
compared with those in healthy controls (HC; n=61) (41). Receiver operating characteristic
(ROC) curve analysis demonstrated that the AUC of the three-circRNA
panel was 0.780, exceeding that of carcinoembryonic antigen (CEA;
AUC, 0.695) and carbohydrate antigen 19-9 (CA19-9; AUC, 0.678)
(41). Combining the circRNA panel
with CEA and CA19-9 further improved the accuracy of CRC diagnosis
(AUC, 0.855) (41). It has been also
found that circ-CCDC66 and circ-STIL may be used for the diagnosis
of early-stage CRC, and the three-circRNA panel may be useful in
diagnosing CEA-negative and CA19-9-negative CRC (41). However, using the same techniques,
Hsiao et al (42) verified
increased expression levels of circCCDC66 in polyps and colon
cancer using RT-qPCR (n=48) and demonstrated its association with a
poor prognosis. This controversy may be explained by the
heterogeneity of CRC; thus, large cohorts of patients of various
ethnicities, possibly through multicenter studies, are required for
further confirmation.
Overall, the aforementioned studies indicate the
potential value of circRNAs as diagnostic and prognostic
biomarkers, as well as therapeutic targets for CRC. Other circRNAs
with similar potential functions are presented in Table I (34,37–39,41–61).
Understanding the underlying mechanisms by which CRC
cells progress is key to the identification of novel therapeutic
targets. Emerging studies have shown the role of circRNAs in
numerous biological processes associated with the development
and/or progression of CRC. circRNAs exert their oncogenic or
suppressive roles by promoting or inhibiting CRC cell
proliferation, migration, invasion and apoptosis.
hsa_circ_0007142 is significantly upregulated in CRC
tissues compared with in neighboring para-cancerous tissues
(62). Bioinformatics analysis and
luciferase reporter assays have revealed that hsa_circ_0007142
sponges miR-103a-2-5p, and silencing of hsa_circ_0007142 using
small interfering (si)RNAs decreases the proliferation, migration
and invasion of HT-29 and HCT-116 cells (62). Yin et al (63) showed that knockdown of circ_0007142
decreased cell division cycle 25A expression by sponging
miR-122-5p, and repressed CRC cell proliferation, colony formation,
migration and invasion.
Thus, distinct mechanisms exerted by the same
circRNA and the effects of expression levels of the same circRNA
highlight the heterogeneity and complexity of circRNAs. A deeper
understanding of the biological behaviors of circRNAs is required
to reconcile these differences and other contrasting results.
EMT is a process in which dynamic changes occur in
the cellular organization transforming cells from an epithelial
phenotype to a mesenchymal phenotype, and this facilitates the
development of migratory and invasive cells (66). Metastasis or advanced CRC are the
major causes of cancer morbidity, mortality and tumor burden.
Several studies have shown that circRNAs regulate EMT and
metastasis in CRC.
Although the field of EMT research has seen
increased interest over the past two decades, particularly in the
past 5 years, there remain several unknowns (64). It is necessary to explore the various
roles of numerous circRNAs, either as oncogenic or suppressive
agents, to delay the progression of EMT and metastasis. Other
circRNAs involved in EMT and metastasis are summarized in Table II.
In addition to the aforementioned biological
functions, circRNAs can regulate other processes. circ-MDM2
(hsa_circ_0027492) is coded by the MDM2 gene, which is regarded as
a transcriptional target of p53 (70). Based on a previous study, which
revealed that MDM2 is crucial in suppressing p53 activity and p53
protein expression (71), Chaudhary
et al (72) knocked down
circ-MDM2 using siRNAs, resulting in an increase in basal p53
levels and growth defects, both in vitro and in vivo.
Further transcriptome profiling following knockdown of circMDM2
showed upregulation of several direct p53 targets, decreased
expression of retinoblastoma protein phosphorylation and
G1-S progression defects (72). Overall, a new role for the circRNA
derived from the MDM2 locus was identified in cell cycle
progression, which preceded the suppression of p53 levels (72). High expression levels of
hsa_circ_0136666 and hsa_circ_0014717 result in arrest of CRC cells
in the G0/G1 phase (73). Mechanistically, hsa_circ_0136666
increases SH2B adaptor protein 1 expression by sponging miR-136
(73), whereas hsa_circ_0014717
induces cell cycle G0/G1 phase arrest in
vitro partly by upregulating p16 expression, a cell cycle
inhibitory protein (74).
circRNAs can exert their roles in the complex
processes of cellular metabolism. circRNA differentially expressed
in normal cells and neoplasia domain containing 4C has been found
to accelerate proliferation and migration, as well as glycolysis,
in CRC cells by increasing glucose transporter 1 expression by
sponging miR-760 (75). Knockdown of
hsa_circ_0000231 blocks CRC glycolysis and progression via Myosin
VI downregulation by sponging miR-502-5p (76). During serum deprivation, circ-ACC1,
which derives from preACC1 mRNA, increases glycolysis and fatty
acid oxidation to adapt the metabolic change of HCT116 cells
(77). Another novel circRNA,
circRUNX1, has been found to promote glutamine metabolism and to
repress apoptosis by upregulating solute carrier family 38 member 1
SLC38A1 through miR-485-5p (78).
circ-001971 functions as an oncogenic ceRNA, which
aggravates the proliferation, invasion and angiogenesis of CRC by
relieving miR-29c-3p-induced inhibition of vascular endothelial
growth factor A (79). circ-ERBB2
interacting protein (ERBIN), which derives from exons 2 to 4 of the
ERBIN gene, promotes angiogenesis, proliferation, invasion and
migration of CRC cells by targeting miR-125a-5p and miR-138-5p;
this sponging effect increases eIF4E-binding protein 1 expression,
which then increases hypoxia inducible factor-1α (HIF-1α)
translation and activates the HIF-1α signaling pathway (80). Zeng et al (81) revealed significantly decreased
expression levels of circ-fibronectin type III domain containing 3B
(FNDC3B) in CRC tissues, cell lines and exosomes. Functional
experiments indicated that overexpression of circFNDC3B suppressed
CRC angiogenesis, which could be reversed by overexpression of
miR-937-5p (81). Furthermore, it
was demonstrated that tumor growth, angiogenesis and liver
metastasis were suppressed by overexpression of circFNDC3B or
circFNDC3B-exosome treatment (81).
Recent findings have indicated the role of circRNAs
in the self-renewal of CSCs and the maintenance of stemness in CRC.
Silencing of circRNA ArfGAP with FG repeats 1 (circAGFG1) markedly
suppresses CRC cell stemness and promotes apoptosis (82). Further experiments have revealed that
circAGFG 1 sponges miR-4262 and miR-185-5p, and promotes CTNNB1
gene (also known as β-catenin) transcription in CRC cells (82). Circular colon tumor initiating cells
1 (circCTIC1) is upregulated in colon TICs compared with in
non-TICs; depletion of this circRNA impairs the self-renewal
capacity of colon TICs, while its overexpression promotes colon TIC
self-renewal (83). Mechanistically,
circCTIC1 recruits the nuclear remodeling factor complex to the
c-Myc promotor and drives the initiation of c-Myc transcription
(83).
Immune evasion is a crucial problem in effective
anticancer therapeutic strategies (84). Emerging evidence has shown that
utilization of immune checkpoints by cancer cells is important for
immune evasion (85). Recently,
Jiang et al (86) revealed
the association between circRNAs and immune evasion in CRC. It was
demonstrated that circ-keratin 6C (KRT6C), which is encoded from
the KRT6C gene, functioned as a miR-485-3p sponge and promoted
immune evasion by upregulating programmed cell death receptor
ligand 1, which is the ligand for the immune check point programmed
cell death protein 1 (86). This
suggests the possible role of circRNAs in immune regulation and
immune evasion.
Additional potential biological functions of
circRNAs are now under exploration to provide a deeper
understanding of the roles of circRNAs in CRC progression.
Additionally, as a clearer picture of the complex network of
non-coding RNA regulation is built, the clinical value of circRNAs
has become more evident. The roles of other circRNAs in biological
processes in CRC are listed in Table
II (38,45,55–60,62–65,69,72–76,78–82,87–126).
In addition to surgical resection, chemotherapy and
radiotherapy constitute some of the primary therapeutic options
utilized for the treatment of CRC. However, escaping from
chemotherapy- or radiotherapy-induced cell death is one of the
characteristics of cancer cells, and numerous mechanisms contribute
to therapeutic resistance (127).
Although limited in number, some studies have investigated the role
of certain circRNAs in CRC therapeutic resistance, highlighting
potential targeted strategies to overcome or inhibit the
acquisition of resistance.
circCCDC66 (hsa_circ_0001313) has recently been
identified to be aberrantly upregulated in CC tissues (42). Wang et al (129) found that circCCDC66 was
significantly increased in CRC cells following radiation treatment,
whereas knockdown of circCCDC66 decreased cell viability and colony
formation rate, and increased caspase-3 activity. Another
circCCDC66 study conducted by Lin et al (130) indicated increased circCCDC66
expression in oxaliplatin-resistant CRC cells, and knockdown of
circCCDC66 decreased oxaliplatin-resistance. Notably, it was found
that phosphatidylinositol 3-kinase-related kinases-mediated
DExH-box helicase 9 phosphorylation, which favors oncogenic
circCCDC66 expression, was involved in the development of
oxaliplatin resistance (130). The
discovery of the roles of circRNAs in acquisition of therapeutic
resistance is an important avenue for future research. Other
circRNAs associated with acquisition of therapeutic resistance are
summarized in Table III (96,131–140).
In the present review, the role of circRNAs in CRC
was summarized, from their involvement in cellular processes to
their association with clinicopathological features and therapeutic
resistance. Additionally, their potential value as diagnostic,
prognostic and therapeutic targets in patients with CRC was
highlighted.
However, there are still significant challenges that
remain to be addressed before circRNAs can be considered in
clinical applications. First, despite the notable progress in the
field of circRNA research, relatively few circRNAs with biological
functions have been discovered, and the exact underlying molecular
mechanisms of circRNA generation, localization, degradation and
turnover process remain unclear. Further understanding of their
biology may demonstrate why circRNAs are dysregulated in tumors,
and thus accelerate their clinical utility. Second, there are
controversies amongst different studies on the same circRNA, such
as the expression levels of the same circRNA in different studies.
Large cohorts from multicenter studies are required for further
confirmation. Third, the biological functions of circRNAs are
complex. One circRNA can exert its function through multiple
pathways and targets. Thus, the roles of circRNAs and their
crosstalk with the tumor microenvironment requires further study.
Roles of circRNAs and their crosstalk with the tumor
microenvironment, cancer cell metabolism and therapeutic resistance
need further investigations. Finally, although certain circRNAs
have been suggested as promising diagnostic and prognostic
biomarkers, especially as non-invasive biomarkers, increasing their
sensitivity and specificity for clinical use is challenging.
Utilization of circRNAs in clinical practice has several hurdles to
overcome. Understanding how to block those with oncogenic
properties and magnify those with tumor suppressive effects may be
helpful. Notably, an artificial synthesized circRNA from linear RNA
molecule containing miR-21 binding sites using simple enzymatic
ligation steps has been proven to function as a miR-21 sponge and
to suppress the downstream cancer protein death domain-associated
tumor suppressor protein (141).
The artificial synthesis of circRNAs may be another effective tool
in clinical application. With the development of new technologies,
the crosstalk between circRNAs and tumor biogenesis will be further
explored, and this may lead to the development of promising
clinical approaches for the treatment of CRC.
Not applicable.
The present study was supported by Shenzhen Science
and Technology Innovation Commission Project (grant nos.
GJHZ20180420180754917, ZDSYS20190902092855097 and
KCXFZ20200201101050887) and Shenzhen Sanming Project (grant no.
SZSM201612041).
Not applicable.
MZ conceptualized and wrote the manuscript. SW
edited the manuscript. Data authentication is not applicable. All
authors have read and approved the final version of the
manuscript.
Not applicable.
Not applicable.
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dekker E, Tanis PJ, Vleugels JL, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RG, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Piawah S and Venook AP: Targeted therapy
for colorectal cancer metastases: A review of current methods of
molecularly targeted therapy and the use of tumor biomarkers in the
treatment of metastatic colorectal cancer. Cancer. 125:4139–4147.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16:582017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cao Q, Guo Z, Du S, Ling H and Song C:
Circular RNAs in the pathogenesis of atherosclerosis. Life Sci.
255:1178372020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zaiou M: The emerging role and promise of
circular RNAs in obesity and related metabolic disorders. Cells.
9:92020. View Article : Google Scholar
|
|
10
|
Zaiou M: circRNAs signature as potential
diagnostic and prognostic biomarker for diabetes mellitus and
related cardiovascular complications. Cells. 9:92020. View Article : Google Scholar
|
|
11
|
Ma Y, Liu Y and Jiang Z: CircRNAs: A new
perspective of biomarkers in the nervous system. Biomed
Pharmacother. 128:1102512020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation - exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Patop IL and Kadener S: circRNAs in
Cancer. Curr Opin Genet Dev. 48:121–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kristensen LS, Andersen MS, Stagsted LV,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nilsen TW and Graveley BR: Expansion of
the eukaryotic proteome by alternative splicing. Nature.
463:457–463. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen LL: The expanding regulatory
mechanisms and cellular functions of circular RNAs. Nat Rev Mol
Cell Biol. 21:475–490. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang C, Liang D, Tatomer DC and Wilusz
JE: A length-dependent evolutionarily conserved pathway controls
nuclear export of circular RNAs. Genes Dev. 32:639–644. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park OH, Ha H, Lee Y, Boo SH, Kwon DH,
Song HK and Kim YK: Endoribonucleolytic cleavage of m6A-containing
RNAs by RNase P/MRP complex. Mol Cell. 74:494–507.e8. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin
D, Liu J and Sun Z: The role of N6-methyladenosine (m6A)
modification in the regulation of circRNAs. Mol Cancer. 19:1052020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Holdt LM, Stahringer A, Sass K, Pichler G,
Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou
A, et al: Circular non-coding RNA ANRIL modulates ribosomal RNA
maturation and atherosclerosis in humans. Nat Commun. 7:124292016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu B, Ye B, Zhu X, Yang L, Li H, Liu N,
Zhu P, Lu T, He L, Tian Y, et al: An inducible circular RNA
circKcnt2 inhibits ILC3 activation to facilitate colitis
resolution. Nat Commun. 11:40762020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q,
Tang Q, Shan C, Lv Y, Zhang K, et al: CircMRPS35 suppresses gastric
cancer progression via recruiting KAT7 to govern histone
modification. Mol Cancer. 19:562020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong S, Tao M, Shen X and Ju S:
Translatable circRNAs and lncRNAs: Driving mechanisms and functions
of their translation products. Cancer Lett. 483:59–65. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prats AC, David F, Diallo LH, Roussel E,
Tatin F, Garmy-Susini B and Lacazette E: Circular RNA, the key for
translation. Int J Mol Sci. 21:212020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM, et al: Translation of the
circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ge J, Jin Y, Lv X, Liao Q, Luo C, Ye G and
Zhang X: Expression profiles of circular RNAs in human colorectal
cancer based on RNA deep sequencing. J Clin Lab Anal.
33:e229522019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ding B, Yao M, Fan W and Lou W:
Whole-transcriptome analysis reveals a potential
hsa_circ_0001955/hsa_circ_0000977-mediated miRNA-mRNA regulatory
sub-network in colorectal cancer. Aging (Albany NY). 12:5259–5279.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: The new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen HY, Li XN, Ye CX, Chen ZL and Wang
ZJ: Circular RNA circHUWE1 is upregulated and promotes cell
proliferation, migration and invasion in colorectal cancer by
sponging miR-486. OncoTargets Ther. 13:423–434. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX
and Yang L: Circular RNA circVAPA is up-regulated and exerts
oncogenic properties by sponging miR-101 in colorectal cancer.
Biomed Pharmacother. 112:1086112019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Li X, Lu L, He L, Hu H and Xu Z:
Circular RNA hsa_circ_0000567 can be used as a promising diagnostic
biomarker for human colorectal cancer. J Clin Lab Anal.
32:e223792018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ye DX, Wang SS, Huang Y and Chi P: A
3-circular RNA signature as a noninvasive biomarker for diagnosis
of colorectal cancer. Cancer Cell Int. 19:2762019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin J, Cai D, Li W, Yu T, Mao H, Jiang S
and Xiao B: Plasma circular RNA panel acts as a novel diagnostic
biomarker for colorectal cancer. Clin Biochem. 74:60–68. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xie Y, Li J, Li P, Li N, Zhang Y, Binang
H, Zhao Y, Duan W, Chen Y, Wang Y, et al: RNA-Seq profiling of
serum exosomal circular RNAs reveals Circ-PNN as a potential
biomarker for human colorectal cancer. Front Oncol. 10:9822020.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pan B, Qin J, Liu X, He B, Wang X, Pan Y,
Sun H, Xu T, Xu M, Chen X, et al: Identification of serum exosomal
hsa-circ-0004771 as a novel diagnostic biomarker of colorectal
cancer. Front Genet. 10:10962019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen P, Yao Y, Yang N, Gong L, Kong Y and
Wu A: Circular RNA circCTNNA1 promotes colorectal cancer
progression by sponging miR-149-5p and regulating FOXM1 expression.
Cell Death Dis. 11:5572020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhou C, Liu HS, Wang FW, Hu T, Liang ZX,
Lan N, He XW, Zheng XB, Wu XJ, Xie D, et al: circCAMSAP1 Promotes
tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol
Ther. 28:914–928. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang W, Yang S, Liu Y, Wang Y, Lin T, Li
Y and Zhang R: Hsa_circ_0007534 as a blood-based marker for the
diagnosis of colorectal cancer and its prognostic value. Int J Clin
Exp Pathol. 11:1399–1406. 2018.PubMed/NCBI
|
|
48
|
Zhang R, Xu J, Zhao J and Wang X:
Silencing of hsa_circ_0007534 suppresses proliferation and induces
apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci.
22:118–126. 2018.PubMed/NCBI
|
|
49
|
Ji W, Qiu C, Wang M, Mao N, Wu S and Dai
Y: Hsa_circ_0001649: A circular RNA and potential novel biomarker
for colorectal cancer. Biochem Biophys Res Commun. 497:122–126.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang Y, Li Z, Xu S and Guo J: Novel
potential tumor biomarkers: Circular RNAs and exosomal circular
RNAs in gastrointestinal malignancies. J Clin Lab Anal.
34:e233592020.PubMed/NCBI
|
|
51
|
Zhuo F, Lin H, Chen Z, Huang Z and Hu J:
The expression profile and clinical significance of circRNA0003906
in colorectal cancer. OncoTargets Ther. 10:5187–5193. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ruan H, Deng X, Dong L, Yang D, Xu Y, Peng
H and Guan M: Circular RNA circ_0002138 is down-regulated and
suppresses cell proliferation in colorectal cancer. Biomed
Pharmacother. 111:1022–1028. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S, et al: Decreased expression of
hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025.
2015.PubMed/NCBI
|
|
54
|
Li J, Ni S, Zhou C and Ye M: The
expression profile and clinical application potential of
hsa_circ_0000711 in colorectal cancer. Cancer Manag Res.
10:2777–2784. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yuan Y, Liu W, Zhang Y, Zhang Y and Sun S:
CircRNA circ_0026344 as a prognostic biomarker suppresses
colorectal cancer progression via microRNA-21 and microRNA-31.
Biochem Biophys Res Commun. 503:870–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang Z, Su M, Xiang B, Zhao K and Qin B:
Circular RNA PVT1 promotes metastasis via miR-145 sponging in CRC.
Biochem Biophys Res Commun. 512:716–722. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ge Z, Li LF, Wang CY, Wang Y and Ma WL:
CircMTO1 inhibits cell proliferation and invasion by regulating
Wnt/β-catenin signaling pathway in colorectal cancer. Eur Rev Med
Pharmacol Sci. 22:8203–8209. 2018.PubMed/NCBI
|
|
58
|
Ren C, Zhang Z, Wang S, Zhu W, Zheng P and
Wang W: Circular RNA hsa_circ_0001178 facilitates the invasion and
metastasis of colorectal cancer through upregulating ZEB1 via
sponging multiple miRNAs. Biol Chem. 401:487–496. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen Z, Ren R, Wan D, Wang Y, Xue X, Jiang
M, Shen J, Han Y, Liu F, Shi J, et al: Hsa_circ_101555 functions as
a competing endogenous RNA of miR-597-5p to promote colorectal
cancer progression. Oncogene. 38:6017–6034. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li C and Zhou H: Circular RNA
hsa_circRNA_102209 promotes the growth and metastasis of colorectal
cancer through miR-761-mediated Ras and Rab interactor 1 signaling.
Cancer Med. 9:6710–6725. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL and
Yang Y: RNA sequencing reveals the expression profiles of circRNA
and indicates that circDDX17 acts as a tumor suppressor in
colorectal cancer. J Exp Clin Cancer Res. 37:3252018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhu CL, Sha X, Wang Y, Li J, Zhang MY, Guo
ZY, Sun SA and He JD: Circular RNA hsa_circ_0007142 is upregulated
and targets miR-103a-2-5p in colorectal cancer. J Oncol.
2019:98368192019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yin W, Xu J, Li C, Dai X, Wu T and Wen J:
Circular RNA circ_0007142 facilitates colorectal cancer progression
by modulating CDC25A expression via miR-122-5p. OncoTargets Ther.
13:3689–3701. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wu L, Xia J, Yang J, Shi Y, Xia H, Xiang X
and Yu X: Circ-ZNF609 promotes migration of colorectal cancer by
inhibiting Gli1 expression via microRNA-150. J BUON. 23:1343–1349.
2018.PubMed/NCBI
|
|
65
|
Zhang X, Zhao Y, Kong P, Han M and Li B:
Expression of circZNF609 is down-regulated in colorectal cancer
tissue and promotes apoptosis in colorectal cancer cells by
upregulating p53. Med Sci Monit. 25:5977–5985. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang J, Antin P, Berx G, Blanpain C,
Brabletz T, Bronner M, Campbell K, Cano A, Casanova J, Christofori
G, et al EMT International Association (TEMTIA), : Guidelines and
definitions for research on epithelial-mesenchymal transition. Nat
Rev Mol Cell Biol. 21:341–352. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xu H, Wang C, Song H, Xu Y and Ji G:
RNA-Seq profiling of circular RNAs in human colorectal Cancer liver
metastasis and the potential biomarkers. Mol Cancer. 18:82019.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ma X, Lv L and Xing C: Circ_ 0115744 acts
as miR-144 sponge to promote and predict the metastasis of
colorectal cancer. Aging (Albany NY). 13:5892–5905. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Xiao H and Liu M: Circular RNA
hsa_circ_0053277 promotes the development of colorectal cancer by
upregulating matrix metallopeptidase 14 via miR-2467-3p
sequestration. J Cell Physiol. 235:2881–2890. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Slack A, Chen Z, Tonelli R, Pule M, Hunt
L, Pession A and Shohet JM: The p53 regulatory gene MDM2 is a
direct transcriptional target of MYCN in neuroblastoma. Proc Natl
Acad Sci USA. 102:731–736. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Haupt Y, Maya R, Kazaz A and Oren M: Mdm2
promotes the rapid degradation of p53. Nature. 387:296–299. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chaudhary R, Muys BR, Grammatikakis I, De
S, Abdelmohsen K, Li XL, Zhu Y, Daulatabad SV, Tsitsipatis D,
Meltzer PS, et al: A circular RNA from the MDM2 locus controls cell
cycle progression by suppressing p53 levels. Mol Cell Biol.
40:402020. View Article : Google Scholar
|
|
73
|
Jin C, Wang A, Liu L, Wang G and Li G:
Hsa_circ_0136666 promotes the proliferation and invasion of
colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol.
234:7247–7256. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang F, Wang J, Cao X, Xu L and Chen L:
Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits
tumor growth by promoting p16 expression. Biomed Pharmacother.
98:775–782. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang ZJ, Zhang YH, Qin XJ, Wang YX and Fu
J: Circular RNA circDENND4C facilitates proliferation, migration
and glycolysis of colorectal cancer cells through miR-760/GLUT1
axis. Eur Rev Med Pharmacol Sci. 24:2387–2400. 2020.PubMed/NCBI
|
|
76
|
Liu Y, Li H, Ye X, Ji A, Fu X, Wu H and
Zeng X: Hsa_circ_0000231 knockdown inhibits the glycolysis and
progression of colorectal cancer cells by regulating
miR-502-5p/MYO6 axis. World J Surg Oncol. 18:2552020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R,
Thorne RF, Zhang XD, Hu W and Wu M: CircACC1 regulates assembly and
activation of AMPK complex under metabolic stress. Cell Metab.
30:157–173.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yu J, Chen X, Li J and Wang F: CircRUNX1
functions as an oncogene in colorectal cancer by regulating
circRUNX1/miR-485-5p/SLC38A1 axis. Eur J Clin Invest. Mar
26–2021.(Epub ahead of print). doi: 10.1111/eci.13540. View Article : Google Scholar
|
|
79
|
Chen C, Huang Z, Mo X, Song Y, Li X, Li X
and Zhang M: The circular RNA 001971/miR-29c-3p axis modulates
colorectal cancer growth, metastasis, and angiogenesis through
VEGFA. J Exp Clin Cancer Res. 39:912020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Chen LY, Wang L, Ren YX, Pang Z, Liu Y,
Sun XD, Tu J, Zhi Z, Qin Y, Sun LN, et al: The circular RNA
circ-ERBIN promotes growth and metastasis of colorectal cancer by
miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α
translation. Mol Cancer. 19:1642020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zeng W, Liu Y, Li WT, Li Y and Zhu JF:
CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit
colorectal cancer progression. Mol Oncol. 14:2960–2984. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang L, Dong X, Yan B, Yu W and Shan L:
CircAGFG1 drives metastasis and stemness in colorectal cancer by
modulating YY1/CTNNB1. Cell Death Dis. 11:5422020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhan W, Liao X, Wang Y, Li L, Li J, Chen
Z, Tian T and He J: circCTIC1 promotes the self-renewal of colon
TICs through BPTF-dependent c-Myc expression. Carcinogenesis.
40:560–568. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Vinay DS, Ryan EP, Pawelec G, Talib WH,
Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara HM, et
al: Immune evasion in cancer: Mechanistic basis and therapeutic
strategies. Semin Cancer Biol. 35 (Suppl 1):S185–S198. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Su R, Dong L, Li Y, Gao M, Han L,
Wunderlich M, Deng X, Li H, Huang Y, Gao L, et al: Targeting FTO
suppresses cancer stem cell maintenance and immune evasion. Cancer
Cell. 38:79–96.e11. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Jiang Z, Hou Z, Liu W, Yu Z, Liang Z and
Chen S: Circ-KRT6C promotes malignant progression and immune
evasion of colorectal cancer through miR-485-3p/PDL1 axis. J
Pharmacol Exp Ther. Mar 26–2021.(Epub ahead of print). doi:
10.1124/jpet.121.000518. View Article : Google Scholar
|
|
87
|
Lu C, Fu L, Qian X, Dou L and Cang S:
Knockdown of circular RNA circ-FARSA restricts colorectal cancer
cell growth through regulation of miR-330-5p/LASP1 axis. Arch
Biochem Biophys. 689:1084342020. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lu H, Yao B, Wen X and Jia B: FBXW7
circular RNA regulates proliferation, migration and invasion of
colorectal carcinoma through NEK2, mTOR, and PTEN signaling
pathways in vitro and in vivo. BMC Cancer. 19:9182019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Chen LY, Zhi Z, Wang L, Zhao YY, Deng M,
Liu YH, Qin Y, Tian MM, Liu Y, Shen T, et al: NSD2 circular RNA
promotes metastasis of colorectal cancer by targeting
miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol.
248:103–115. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chen ZL, Li XN, Ye CX, Chen HY and Wang
ZJ: Elevated levels of circRUNX1 in colorectal cancer promote cell
growth and metastasis via miR-145-5p/IGF1 signalling. OncoTargets
Ther. 13:4035–4048. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Cui W, Dai J, Ma J and Gu H:
circCDYL/microRNA-105-5p participates in modulating growth and
migration of colon cancer cells. Gen Physiol Biophys. 38:485–495.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Du H, He Z, Feng F, Chen D, Zhang L, Bai
J, Wu H, Han E and Zhang J: Hsa_circ_0038646 promotes cell
proliferation and migration in colorectal cancer via
miR-331-3p/GRIK3. Oncol Lett. 20:266–274. 2020.PubMed/NCBI
|
|
93
|
Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W,
Wu C, Zhu D, Wu C and Jiang J: Hsa_circ_0009361 acts as the sponge
of miR-582 to suppress colorectal cancer progression by regulating
APC2 expression. Clin Sci (Lond). 133:1197–1213. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Han K, Wang FW, Cao CH, Ling H, Chen JW,
Chen RX, Feng ZH, Luo J, Jin XH, Duan JL, et al: CircLONP2 enhances
colorectal carcinoma invasion and metastasis through modulating the
maturation and exosomal dissemination of microRNA-17. Mol Cancer.
19:602020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
He JH, Li YG, Han ZP, Zhou JB, Chen WM, Lv
YB, He ML, Zuo JD and Zheng L: The CircRNA-ACAP2/Hsa-miR-21-5p/
Tiam1 regulatory feedback circuit affects the proliferation,
migration, and invasion of colon cancer SW480 cells. Cell Physiol
Biochem. 49:1539–1550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Jian X, He H, Zhu J, Zhang Q, Zheng Z,
Liang X, Chen L, Yang M, Peng K, Zhang Z, et al: Hsa_circ_001680
affects the proliferation and migration of CRC and mediates its
chemoresistance by regulating BMI1 through miR-340. Mol Cancer.
19:202020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Jin Y, Yu LL, Zhang B, Liu CF and Chen Y:
Circular RNA hsa_circ_0000523 regulates the proliferation and
apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med
Biol Res. 51:e78112018. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Li H, Jin X, Liu B, Zhang P, Chen W and Li
Q: CircRNA CBL.11 suppresses cell proliferation by sponging
miR-6778-5p in colorectal cancer. BMC Cancer. 19:8262019.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Li R, Wu B, Xia J, Ye L and Yang X:
Circular RNA hsa_circRNA_102958 promotes tumorigenesis of
colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res.
11:6887–6893. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Li W, Xu Y, Wang X, Cao G, Bu W, Wang X,
Fang Z, Xu Y, Dong M and Tao Q: circCCT3 modulates vascular
endothelial growth factor A and Wnt signaling to enhance colorectal
cancer metastasis through sponging miR-613. DNA Cell Biol.
39:118–125. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C, et al: A novel protein encoded by a
circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Zhao H, Chen S and Fu Q: Exosomes from
CD133+ cells carrying circ-ABCC1 mediate cell stemness
and metastasis in colorectal cancer. J Cell Biochem. 121:3286–3297.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang Y, Zhang Z, Yi Y, Wang Y and Fu J:
CircNOL10 acts as a sponge of miR-135a/b-5p in suppressing
colorectal cancer progression via regulating KLF9. OncoTargets
Ther. 13:5165–5176. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang XL, Xu LL and Wang F:
Hsa_circ_0020397 regulates colorectal cancer cell viability,
apoptosis and invasion by promoting the expression of the miR-138
targets TERT and PD-L1. Cell Biol Int. 41:1056–1064. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang X, Xu Y, Yamaguchi K, Hu J, Zhang L,
Wang J, Tian J and Chen W: Circular RNA circVAPA knockdown
suppresses colorectal cancer cell growth process by regulating
miR-125a/CREB5 axis. Cancer Cell Int. 20:1032020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang Q, Zhang C, Ma JX, Ren H, Sun Y and
Xu JZ: Circular RNA PIP5K1A promotes colon cancer development
through inhibiting miR-1273a. World J Gastroenterol. 25:5300–5309.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Shen T, Cheng X, Liu X, Xia C, Zhang H,
Pan D, Zhang X and Li Y: Circ_0026344 restrains metastasis of human
colorectal cancer cells via miR-183. Artif Cells Nanomed
Biotechnol. 47:4038–4045. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yong W, Zhuoqi X, Baocheng W, Dongsheng Z,
Chuan Z and Yueming S: Hsa_circ_0071589 promotes carcinogenesis via
the miR-600/EZH2 axis in colorectal cancer. Biomed Pharmacother.
102:1188–1194. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yang Z, Zhang J, Lu D, Sun Y, Zhao X, Wang
X, Zhou W, He Q and Jiang Z: Hsa_circ_0137008 suppresses the
malignant phenotype in colorectal cancer by acting as a
microRNA-338-5p sponge. Cancer Cell Int. 20:672020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Yang L, Sun H, Liu X, Chen J, Tian Z, Xu
J, Xiang B and Qin B: Circular RNA hsa_circ_0004277 contributes to
malignant phenotype of colorectal cancer by sponging miR-512-5p to
upregulate the expression of PTMA. J Cell Physiol. Jan
21–2020.(Epub ahead of print). doi: 10.1002/jcp.29484.
|
|
112
|
Li X, Wang J, Zhang C, Lin C, Zhang J,
Zhang W, Zhang W, Lu Y, Zheng L and Li X: Circular RNA circITGA7
inhibits colorectal cancer growth and metastasis by modulating the
Ras pathway and upregulating transcription of its host gene ITGA7.
J Pathol. 246:166–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Yang G, Zhang T, Ye J, Yang J, Chen C, Cai
S and Ma J: Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1,
suppressing colorectal cancer proliferation. Cancer Manag Res.
11:6499–6509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yang B, Du K, Yang C, Xiang L, Xu Y, Cao
C, Zhang J and Liu W: CircPRMT5 circular RNA promotes proliferation
of colorectal cancer through sponging miR-377 to induce E2F3
expression. J Cell Mol Med. 24:3431–3437. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Xu XW, Zheng BA, Hu ZM, Qian ZY, Huang CJ,
Liu XQ and Wu WD: Circular RNA hsa_circ_000984 promotes colon
cancer growth and metastasis by sponging miR-106b. Oncotarget.
8:91674–91683. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Xian ZY, Hu B, Wang T, Cai JL, Zeng JY,
Zou Q and Zhu PX: CircABCB10 silencing inhibits the cell
ferroptosis and apoptosis by regulating the miR-326/CCL5 axis in
rectal cancer. Neoplasma. 67:1063–1073. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wang X, Ren Y, Ma S and Wang S: Circular
RNA 0060745, a novel circRNA, promotes colorectal cancer cell
proliferation and metastasis through miR-4736 sponging. OncoTargets
Ther. 13:1941–1951. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Wang J, Luo J, Liu G and Li X: Circular
RNA hsa_circ_0008285 inhibits colorectal cancer cell proliferation
and migration via the miR-382-5p/PTEN axis. Biochem Biophys Res
Commun. 527:503–510. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Wang DK, Chong RF, Song BL, Fan KF and Liu
YF: Circular RNA circ-SMAD7 is downregulated in colorectal cancer
and suppresses tumor metastasis by regulating epithelial
mesenchymal transition. Eur Rev Med Pharmacol Sci. 24:1736–1742.
2020.PubMed/NCBI
|
|
120
|
Pei FL, Cao MZ and Li YF: Circ_0000218
plays a carcinogenic role in colorectal cancer progression by
regulating miR-139-3p/RAB1A axis. J Biochem. 167:55–65. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Ma Z, Han C, Xia W, Wang S, Li X, Fang P,
Yin R, Xu L and Yang L: circ5615 functions as a ceRNA to promote
colorectal cancer progression by upregulating TNKS. Cell Death Dis.
11:3562020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lu X, Yu Y, Liao F and Tan S: Homo sapiens
circular RNA 0079993 (hsa_circ_0079993) of the POLR2J4 gene acts as
an oncogene in colorectal cancer through the microRNA-203a-3p.1 and
CREB1 axis. Med Sci Monit. 25:6872–6883. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Li YF, Pei FL and Cao MZ: CircRNA_101951
promotes migration and invasion of colorectal cancer cells by
regulating the KIF3A-mediated EMT pathway. Exp Ther Med.
19:3355–3361. 2020.PubMed/NCBI
|
|
124
|
Li Y, Li C, Xu R, Wang Y, Li D and Zhang
B: A novel circFMN2 promotes tumor proliferation in CRC by
regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond).
133:2463–2479. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Chen C, Yuan W, Zhou Q, Shao B, Guo Y,
Wang W, Yang S, Guo Y, Zhao L, Dang Q, et al:
N6-methyladenosine-induced circ1662 promotes metastasis of
colorectal cancer by accelerating YAP1 nuclear localization.
Theranostics. 11:4298–4315. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Liu X, Qin Y, Tang X, Wang Y, Bian C and
Zhong J: Circular RNA circ_0000372 contributes to the
proliferation, migration and invasion of colorectal cancer by
elevating IL6 expression via sponging miR-495. Anticancer Drugs.
32:296–305. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Wang X, Zhang H, Yang H, Bai M, Ning T,
Deng T, Liu R, Fan Q, Zhu K, Li J, et al: Exosome-delivered circRNA
promotes glycolysis to induce chemoresistance through the
miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 14:539–555.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang L, Peng X, Lu X, Wei Q, Chen M and
Liu L: Inhibition of hsa_circ_0001313 (circCCDC66) induction
enhances the radio-sensitivity of colon cancer cells via tumor
suppressor miR-338-3p: Effects of cicr_0001313 on colon cancer
radio-sensitivity. Pathol Res Pract. 215:689–696. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Lin YC, Yu YS, Lin HH and Hsiao KY:
Oxaliplatin-Induced DHX9 phosphorylation promotes oncogenic
circular RNA CCDC66 expression and development of chemoresistance.
Cancers (Basel). 12:122020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Chen H, Pei L, Xie P and Guo G: Circ-PRKDC
contributes to 5-fluorouracil resistance of colorectal cancer cells
by regulating miR-375/FOXM1 axis and Wnt/β-catenin pathway.
OncoTargets Ther. 13:5939–5953. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
He X, Ma J, Zhang M, Cui J and Yang H:
Circ_0007031 enhances tumor progression and promotes 5-fluorouracil
resistance in colorectal cancer through regulating miR-133b/ABCC5
axis. Cancer Biomark. 29:531–542. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Wang Y, Wang H, Zhang J, Chu Z, Liu P,
Zhang X, Li C and Gu X: Circ_0007031 serves as a sponge of miR-760
to regulate the growth and chemoradiotherapy resistance of
colorectal cancer via regulating DCP1A. Cancer Manag Res.
12:8465–8479. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Xiong W, Ai YQ, Li YF, Ye Q, Chen ZT, Qin
JY, Liu QY, Wang H, Ju YH, Li WH, et al: Microarray analysis of
circular RNA expression profile associated with
5-fluorouracil-based chemoradiation resistance in colorectal cancer
cells. BioMed Res Int. 2017:84216142017. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Xu Y, Qiu A, Peng F, Tan X, Wang J and
Gong X: Exosomal transfer of circular RNA FBXW7 ameliorates the
chemoresistance to oxaliplatin in colorectal cancer by sponging
miR-18b-5p. Neoplasma. 68:108–118. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Abu N, Hon KW, Jeyaraman S, Yahaya A,
Abdullah NM, Mustangin M, Sulaiman SA, Jamal R and Ab-Mutalib NS:
Identification of differentially expressed circular RNAs in
chemoresistant colorectal cancer. Epigenomics. 11:875–884. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Ren TJ, Liu C, Hou JF and Shan FX:
CircDDX17 reduces 5-fluorouracil resistance and hinders
tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1
axis. Eur Rev Med Pharmacol Sci. 24:1743–1754. 2020.PubMed/NCBI
|
|
138
|
Zhang W, Wang Z, Cai G and Huang P:
Downregulation of Circ_0071589 suppresses cisplatin resistance in
colorectal cancer by regulating the miR-526b-3p/KLF12 axis. Cancer
Manag Res. 13:2717–2731. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zhao K, Cheng X, Ye Z, Li Y, Peng W, Wu Y
and Xing C: Exosome-mediated transfer of circ_0000338 enhances 5-FU
resistance in colorectal cancer through regulating miR-217 and
miR-485-3p. Mol Cell Biol. 41:e00517–20. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Xi L, Liu Q, Zhang W, Luo L, Song J, Liu
R, Wei S and Wang Y: Circular RNA circCSPP1 knockdown attenuates
doxorubicin resistance and suppresses tumor progression of
colorectal cancer via miR-944/FZD7 axis. Cancer Cell Int.
21:1532021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Liu X, Abraham JM, Cheng Y, Wang Z, Wang
Z, Zhang G, Ashktorab H, Smoot DT, Cole RN, Boronina TN, et al:
Synthetic circular RNA functions as a miR-21 sponge to suppress
gastric carcinoma cell proliferation. Mol Ther Nucleic Acids.
13:312–321. 2018. View Article : Google Scholar : PubMed/NCBI
|