Introduction
Sarcoma has numerous subtypes and typically develops
at a relatively young age compared with other cancers (1). Osteosarcoma (OS), the most common
primary sarcoma of bone, preferentially develops at 10–14 years of
age, which is a period of accelerated bone growth (2,3). Ewing's
sarcoma (EWS) is a small round sarcoma and the second most common
sarcoma of bone in children (1).
Liposarcoma (LPS), a common subtype of soft tissue sarcoma (STS),
accounts for 20% of STS cases (4).
LPS is classified into 4 categories, which are well differentiated
liposarcoma (WDLPS), dedifferentiated liposarcoma (DDLPS), myxoid
liposarcoma and pleomorphic liposarcoma (1,4). DDLPS
accounts for ~20% of these and shares a genetic background with
WDLPS and occurs focally in the WDLPS lesion (1,5).
microRNAs (miRNAs/miRs) are non-coding RNAs with a
length of 17–25 base pairs. miRNAs regulate gene expression at the
transcriptional level by binding to target mRNAs (6). Over a period of 10 years of miRNA
research, numerous functions of miRNAs have been identified,
including their involvement in cancer (7). Exosomes are small vesicles of ~100 nm
in size that function as an active transport system in cells
(8). Exosomes are enclosed in a
lipid bilayer membrane and incorporate mRNAs, miRNAs and proteins
(8). Exosomes are released from most
cell types and function as intercellular communication tools
(8). Cancer cells interact with the
microenvironment through exosomes, and tumor-derived exosomes
induce premetastatic niches (9).
There are currently no effective biomarkers for
sarcomas, to the best of our knowledge. In recent years,
circulating miRNAs have attracted attention as candidate biomarkers
in several cancers, and these miRNAs are released into the
extracellular space by small vesicles, particularly exosomes
(7).
Several circulating miRNAs have been identified as
candidate markers for OS. The expression levels of miR-195, -Let7A,
−9 and −21 are increased in the serum or plasma of patients with OS
compared with healthy controls (10–13).
miR-135b, −148a, −27a and −9 levels are increased in the exosomes
of the serum of patients with OS with a good response to
chemotherapy, and these miRNAs can predict the therapeutic efficacy
(14).
DDLPS is difficult to detect because it often occurs
in the retroperitoneum and trunk (1,15,16).
Therefore, it is often discovered after it has grown and is often
difficult to resect. When comparing miRNAs in formalin-fixed
paraffin-embedded LPS and adipose tissue samples, miR-155 and −21
are upregulated in LPS samples (17,18).
High expression of miR-155 and −26a-2 is correlated with a poor
prognosis in DDLPS (18,19). miR-25-3p and −92a-3p are highly
expressed in peripheral blood plasma vesicles derived from human
LPS patient samples. These miRNAs are secreted from LPS cell lines
through extracellular vesicles and transferred to macrophages
(20).
In previous work from our group, it was identified
that serum miRNAs that can serve as biomarkers for certain types of
cancer, such as sarcoma, bladder cancer and ovarian cancer
(21–23). However, whether the identified miRNAs
are released from the tumor site remains unclear. In the present
study, the relevance of the identified miRNAs to sarcoma was
determined by comparing miRNA expression levels between serum and
tissue samples from the same patient based on the microarray
data.
Materials and methods
Clinical samples
All clinical serum samples were obtained from
patients undergoing tumor resection at the National Cancer Center
Hospital (Tokyo, Japan) (NCCH) between January 2007 and December
2013. The patients that became inoperable during preoperative
treatment and were judged to be inappropriate as participants by
the doctor or refused to participate in this study were excluded.
The clinicopathological data were collected and TNM staging was
performed according to French Fédération Nationale des Centres de
Lutte Contre Le Cancer (FNCLCC) system described in American Joint
Committee on Cancer (AJCC) system, 7th edition (1). Serum samples were stored at 4°C for 1
week and then stored at −20°C until further use. The serum samples
were collected before the operation or preoperative treatment at
the NCCH. Tissue samples were obtained from surgical specimens of
patients undergoing surgery at the NCCH and stored at −80°C in the
NCC Biobank. The study included 22 OS samples, 17 DDLPS samples and
three EWS samples (Table
SI,Table SII,Table SIII). All of these samples were used
by Asano et al (21). The
present study was approved by the NCCH Institutional Review Board
(Tokyo, Japan; approval nos. 2004-050, 2013-111 and 2015-266).
Written informed consent was obtained from each participant on the
first visit. When the patient was under 20 years old, the informed
consent was obtained from their parents, relatives or guardians of
minors.
miRNA expression array of clinical
serum and tissue samples
Serum RNA was extracted from 300 µl of serum using
the 3D-Gene® RNA extraction reagent (Toray Industries,
Inc.). Fresh-frozen tissues were crushed to a powder using a
Multibead Shocker (Yasui Kikai Corporation) under liquid nitrogen
(−196°C). Total RNA was extracted from frozen tumor tissue powder
using the miRNeasy Mini kit (cat. no. 217004; Qiagen GmbH).
Comprehensive miRNA expression analysis was performed using the
3D-Gene® miRNA Labeling kit and the 3D-Gene®
Human miRNA Oligo Chip (both Toray Industries, Inc.), which was
designed to detect 2,588 miRNA sequences registered in miRBase
release 21 database (http://www.mirbase.org/). Microarray experiments were
performed by Kamakura Techno-Science Inc. miRNAs with a signal
intensity >26 were considered detected miRNAs.
Principal component analysis (PCA) map and heatmap were generated
by Genomics Suite version 6.6 (Partek Inc.).
Cell culture
Human DDLPS cell lines (LP6 and LPS12) were
previously established and kindly provided by Dr Andrew J. Wagner
(Dana Farber Cancer Institute; USA). Human adipose-derived stem
cells (ADSCs) were purchased from Invitrogen; Thermo Fisher
Scientific, Inc. (cat. R7788115). LP6 cells were cultured in
RPMI-1640 medium (cat. 11875093; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; cat. 10270-106;
Thermo Fisher Scientific, Inc.). LPS12 cells were cultured in
DMEM/F12 (cat. 21331020; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS and GlutaMAX (cat. 35050061; Thermo
Fisher Scientific, Inc.). ADSCs were cultured in MesenPRO RS™
medium (cat. 12746012; Thermo Fisher Scientific, Inc.). All cell
cultures were treated with 1% antibiotic-antimycotic solution (cat.
15240062; Thermo Fisher Scientific, Inc.) and cultured at 37°C in
5% CO2.
Cell proliferation assay
LP6 cells were seeded at a density of
5×103 cells per well into a 96-well dish at 37°C in 5%
CO2. After 1 day of seeding, the medium was replaced by
miRNA mimics (hsa-miR-1246, ID MC13182; 5′AAUGGAUUUUUGGAGCAGG-3′;
hsa-miR-4532, ID MC21908; 5′-CCCCGGGGAGCCCGGCG-3′; hsa-miR-4454,
ID: MC21186; 5′-GGAUCCGAGUCACGGCACCA-3′; hsa-miR-619-5p, ID:
MC28761; 5′-GCUGGGAUUACAGGCAUGAGCC-3′; and hsa-miR-6126, ID:
MCMC25200; 5′-GUGAAGGCCCGGCGGAGA-3′; all Thermo Fisher Scientific,
Inc.) in DharmaFECT reagent-1 (cat. no. T-2001-02; GE Healthcare
Dharmacon, Inc.) for miRNA transfection. Negative Control #1 (NC)
(cat. no. 4464058; Thermo Fisher Scientific, Inc.) was used as a
non-targeting negative control. The concentrations of miRNA mimics
and NC were 2 µM in the original stocks. The transfection
efficiency was estimated (n=4) based the average using HiLyte
Fluor488 negative control miRNA mimic (Nippon Gene Co., Ltd.) and
evaluated using reverse transcription-quantitative PCR with
miRNA-specific primers. Each miRNA was added to three wells. For
all miRNA mimic transfection, final concentrations were 10 nM. The
time of transfection was 8 h at 37°C in 5% CO2. After
transfection (24, 48 and 72 h), the number of living cells was
counted using the Cell Counting Kit-8 (Dojindo Molecular
Technologies, Inc.) for 3 days, and measurements (Gen5 Synergy H4;
BioTek Instruments, Inc.) were performed three times. The average
value of day 2 and 3 was normalized to that of day 1. Proliferation
curves were drawn with the average value, and the standard
deviation was calculated. Images were captured using a fluorescent
microscope (BZ-X700; Keyence Corporation).
Purification and analysis of
exosomes
Exosomes were prepared as previously described
(24). Briefly, to avoid the
contamination of exosomes from FBS, cells were washed with PBS and
the culture medium was replaced with advanced RPMI medium (cat.
12633012; Thermo Fisher Scientific, Inc.) with 2 mM L-glutamine
(cat. 25030149; Thermo Fisher Scientific, Inc.) for LP6 cells and
advanced DMEM/F12 medium (cat. 12491015; Thermo Fisher Scientific,
Inc.) with 2 mM L-glutamine for LPS12 cells. The ADSC culture
medium was replaced by StemPro™ MSC SFM medium (cat. A1033201;
Thermo Fisher Scientific, Inc.). The LP6, LPS12 and ADSC cells were
incubated for 48 h at 37°C in 5% CO2. After 48 h of
incubation, the conditioned medium (CM) was collected and
centrifuged at 2,000 × g for 10 min at 4°C. The supernatant was
filtered through a 0.22-µm filter (EMD Millipore). The filtered CM
was ultracentrifuged at 110,000 × g using a SW41Ti rotor for 70 min
at 4°C using an Optima XPN-100 Ultracentrifuge (Beckman Coulter,
Inc.). The supernatant was discarded, and the pellet was washed by
adding PBS, ultracentrifuged at 35,000 rpm 110,000 × g using the
SW41Ti rotor for 70 min at 4°C, and resuspended in PBS. For
determination of the size distribution of exosomes, nanoparticle
tracking analysis was performed using the NanoSight system
(NanoSight; Malvern Panalytical, Ltd.) in samples diluted
500–1,000-fold with PBS for analysis (24).
RNA extraction and RT-qPCR
miRNAs were isolated from the LP6, LPS12 and ADSC
cells and exosomes using the miRNeasy® Mini kit (cat.
no. 217004; Qiagen GmbH), and cDNA was produced using the miScript
II RT kit (cat. no. 218160; Qiagen GmbH) according to the
manufacturer's instructions. The cDNA samples with miScript primers
were subjected to quantitative PCR using a miScript®
SYBR® Green PCR kit (cat. no. 218075; Qiagen GmbH).
Reactions were performed three times on the StepOnePlus Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The following specific primers were used: Hs_miR-1246_1 (cat. no.
MS00014224), Hs_miR-4532_1 (cat. no. MS00040705), Hs_miR-4454_1
(cat. no. MS00037597), Hs_miR-619-5p_1 (cat. no. MS00046032) and
Hs_miR-6126_1 (cat. no. MS00045416) (all Qiagen GmbH). The
expression levels were normalized to those of RNU6-2_11 (cat. no.
MS00033740; Qiagen GmbH), and relative expression was calculated
using the 2−ΔΔCq method (25). The expression levels of each miRNA in
LP6 and LPS12 were compared with those in ADSCs to calculate the
fold-changes. The average values and standard deviations were
calculated.
Immunoblotting
In total, 1 µg of exosomes were extracted by Sample
Buffer Solution (2-Mercaptoethanol-) (FUJIFILM Wako Pure Chemical
Corporation) and 1 µg of proteins were loaded per lane onto 4–15%
Mini-PROTEAN TGX gels (Bio-Rad Laboratories, Inc.) and
electrotransferred (100 V, 30 mA) (26). The proteins were transferred to
polyvinylidene difluoride membranes (EMD Millipore). The membranes
were blocked with Blocking One solution (cat. 03953-95; Nacalai
Tesque) on shacking machine for 1 h at room temperature and then
incubated for 1 h at room temperature with primary antibodies:
Anti-CD63 (1:1,000; cat. no. 12A12; Cosmo Bio Co., Ltd.) and
anti-CD9 (1:1,000; cat. no. 8A12; Cosmo Bio Co., Ltd.). After
washing, the membranes were incubated for 1 h at room temperature
with secondary antibodies (horseradish peroxidase-linked anti-mouse
IgG; cat. no. NA931 or horseradish peroxidase-linked anti-rabbit
IgG; cat. no. Na934; both 1:5,000; GE Healthcare). After washing,
the membranes were then exposed to ImmunoStar LD for development
(cat. 292-69903; FUJIFILM Wako Pure Chemical Corporation).
Statistical analysis
miRNA expression levels were analyzed using
Pearson's correlation, with the vertical axis representing tissues
and the horizontal axis representing serum. The expression levels
of miRNAs in cells and exosomes were compared by calculating the
average values and the standard deviation. The data are presented
as mean ± SD of 3 or 4 biological replicates. A total of 3
biological replicates were performed for the experiments of miRNA
extraction from cell lines and exosomes and cell proliferation. A
total of 4 biological replicates were performed of the
transfections of fluorescently-labeled miRNA mimic. The
significance of the average values was analyzed using one-way ANOVA
with a Tukey's HSD post hoc test. P<0.05 was considered to
indicate a statistically significant difference. Statistical
analyses were performed using Excel 2019 (Microsoft Corporation)
and online One-way ANOVA with post-hoc Tukey HSD Test Calculator
for comparing multiple treatments (https://astatsa.com/OneWay_Anova_with_TukeyHSD/).
Results
Expression of miRNAs in the tissues
and serum of patients with sarcoma
A total of 22 OS, 17 DDLPS and three EWS samples
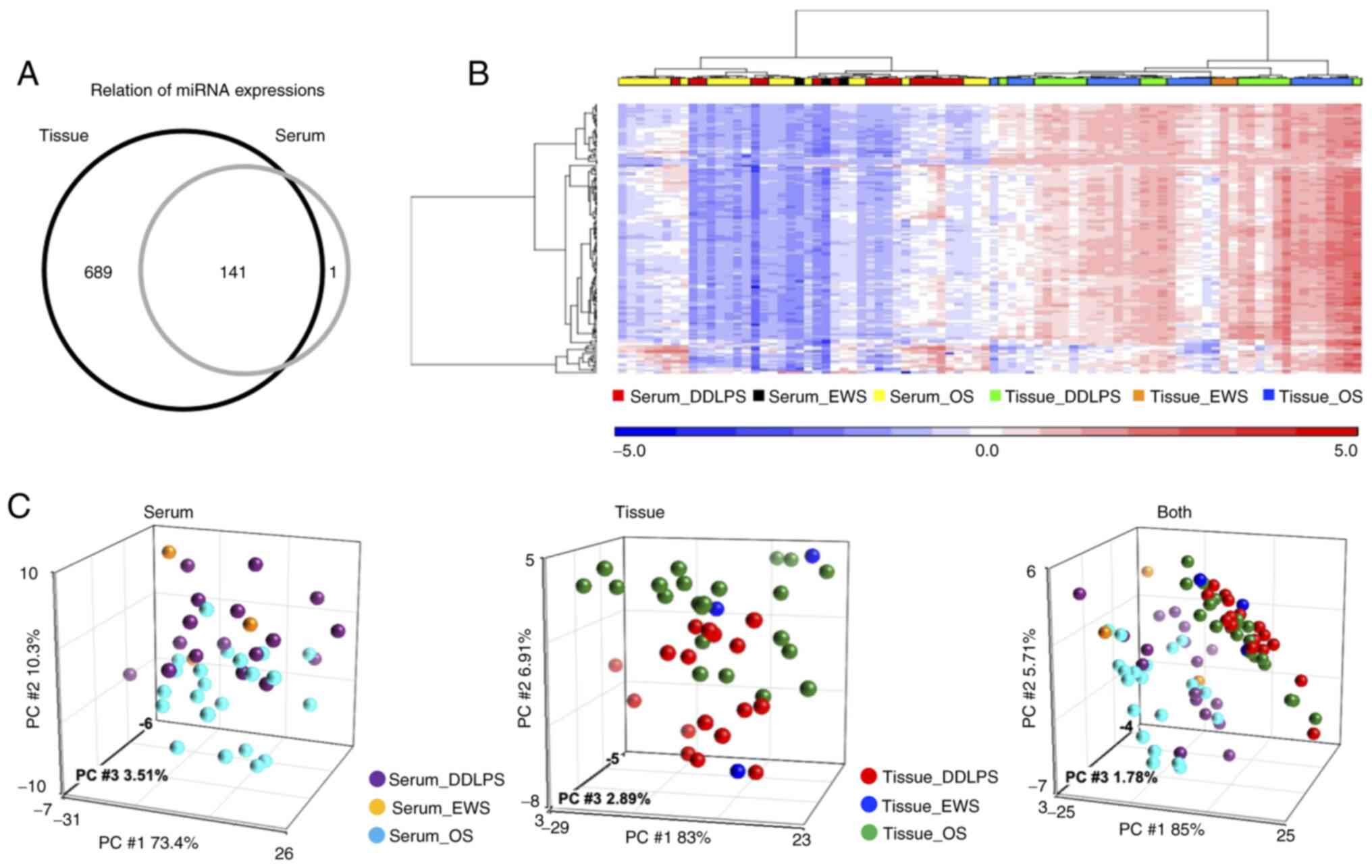
were analyzed. Overall, 830 miRNAs were detected in tissues and 142
miRNAs in serum (Fig. 1A), and 141
miRNAs were expressed both in tissues and serum. Fig. 1B shows the results of hierarchical
unsupervised clustering with miRNAs on the vertical axis and
clinical cases on the horizontal axis. The expression of miRNAs was
generally higher in tissues compared with in serum. Based on the
PCA map (Fig. 1C), DDLPS and OS
samples were separated in both serum and tissue samples. The
expression profiles in tissues and serum were separated in the PCA
map (Fig. 1C, right panel).
Different trends were observed in miRNA expression patterns for
each tumor type and miRNAs that were highly expressed in both tumor
and serum miRNAs were found.
Correlation coefficients of sarcoma
samples and DDLPS samples
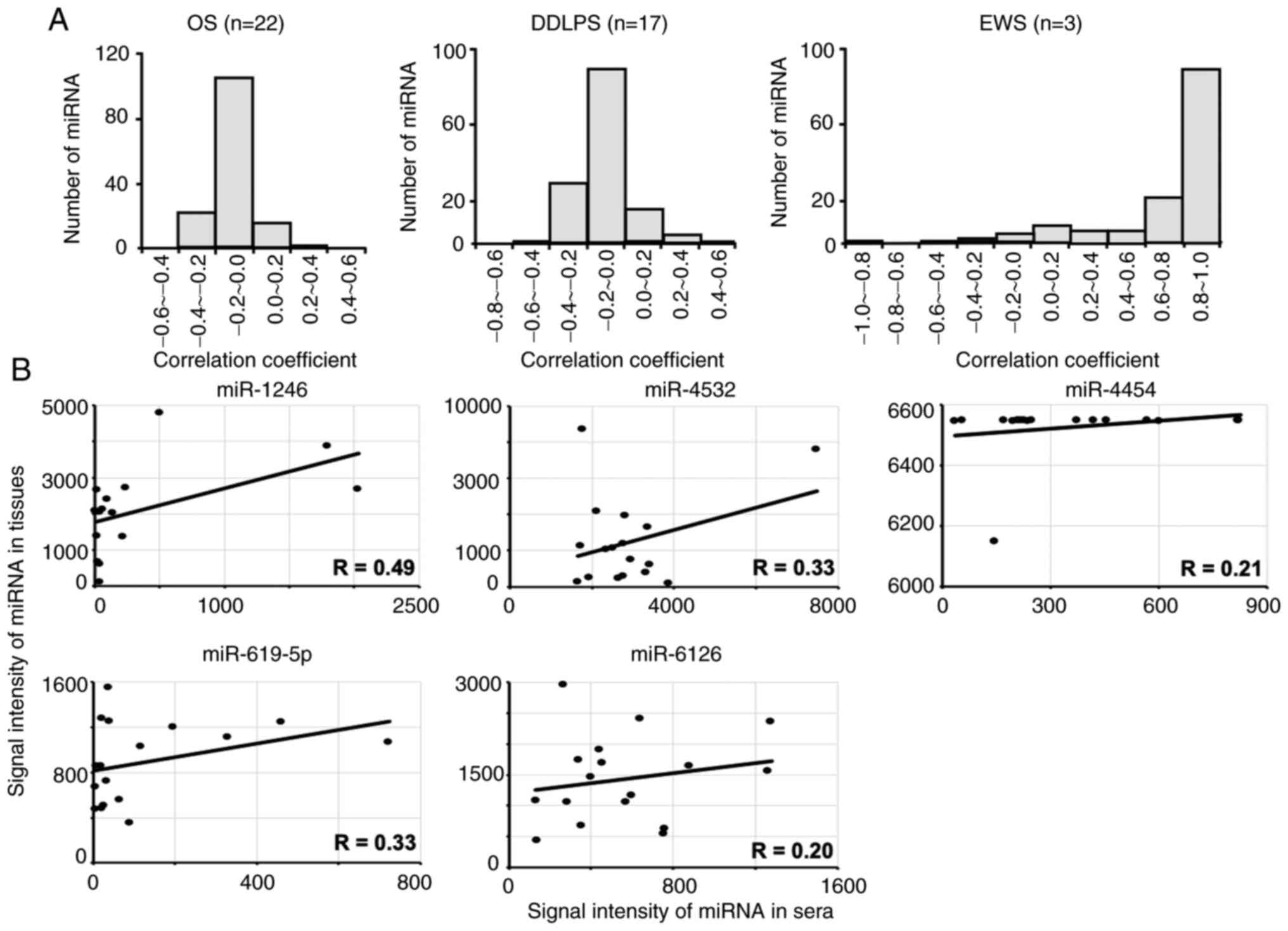
The Pearson's correlation coefficients in miRNA
expression between the serum and tissues were calculated (Fig. 2A). In the histograms of Fig. 2A, the distribution of Pearson
correlation coefficient (R) was shown. In OS and DDLPS, there were
a few miRNA which showed positive correlation between the serum and
the tissues (Fig. 2A). Although
miRNAs in EWS samples were expressed at high levels, the number of
detected miRNAs was too low (n=3) to determine statistical
significance. However, several miRNAs in DDLPS were expressed at
relatively high levels, including miR-1246, −4532, −4454, −619-5p
and −6126. The correlation these miRNAs in tissues and sera in
DDLPS is presented in Fig. 2B. The
expression levels of certain miRNAs, such as −1246, −4532 and
−619-5p, were weakly correlated between serum and tissue samples
(R=0.33–0.49), suggesting that these serum miRNAs could be derived
from tumors in patients with DDLPS.
miRNA expression in DDLPS cell lines
and effect on cell proliferation
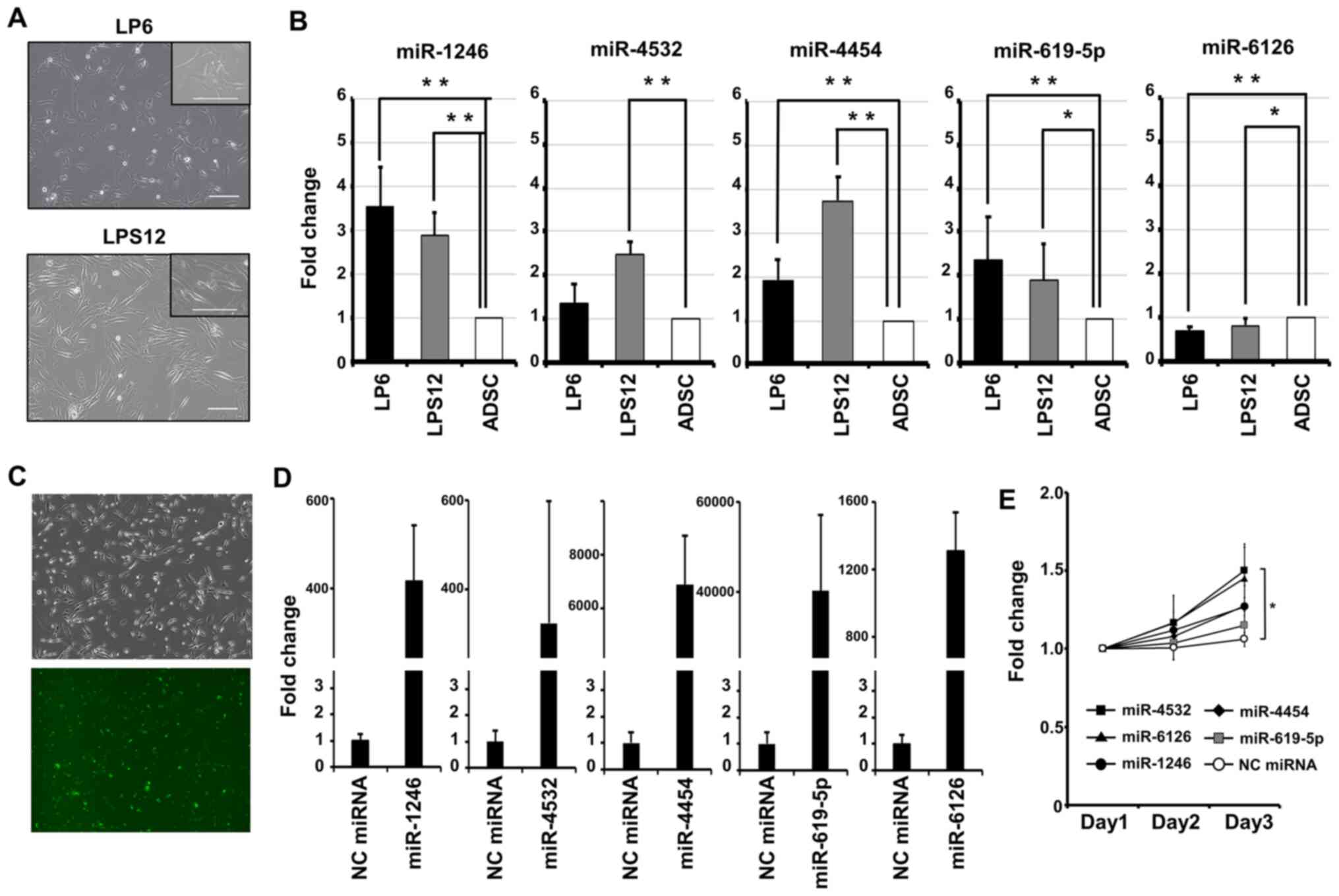
It was examined whether the identified miRNAs were
released from the human DDLPS cell lines LP6 and LPS12 (Fig. 3A). For measurement of the expression
levels of the miRNAs in DDLPS cells, total RNA was extracted from
LP6 and LPS12 cells and ADSCs as controls and subjected to
quantitative PCR. The expression of miR-1246, −4532, −4454 and
−619-5p was higher in LP6 and LPS12 cells compared with in ADSCs
(Fig. 3B). The ratio of the
expression level of miR-1246 was 3.52±0.89 (P<0.01) in LP6 and
2.87±0.52 (P<0.01) in LPS12 cells. That of miR-4532 was
1.36±0.43 (P=0.29) in LP6 and 2.48±0.28 (P<0.01) in LPS12 cells.
That of miR-4454 was 1.90±0.47 (P<0.01) in LP6 and 3.72±0.52
(P<0.01) in LPS12 cells. That of miR-619-5p was 2.36±0.99
(P<0.01) in LP6 and 1.88±0.84 (P=0.02) in LPS12 cells. That of
miR-6126 was 0.62±0.12 (P<0.01) in LP6 and 0.83±0.15 (P=0.03) in
LPS12. To examine the effects of these miRNAs on DDLPS cell
proliferation, the transfection efficiency of miRNA in LP6 cells
was examined using a fluorescently-labeled miRNA mimic negative
control. Based on the fluorescence rate, efficiency was 92.5±3.3%
(Fig. 3C). Then, LP6 cells were
transiently transfected with miRNA mimics, and the overexpression
of each miRNA was confirmed using RT-qPCR (Fig. 3D). The cell proliferation rates were
monitored on days 1, 2 and 3 after transfection (Fig. 3E). The cells transfected with the
miRNAs exhibited a higher proliferative capacity compared with the
negative control. miR-4532 had a significant effect on promoting
cell proliferation in LP6 cells (P<0.05). miR-1246, −4454 and
−619-5p were significantly highly expressed in both DDLPS cell
lines. This result indicated the possibility that these miRNAs are
also highly expressed in DDLPS tissues.
DDLPS cells release exosomes
containing specific miRNAs
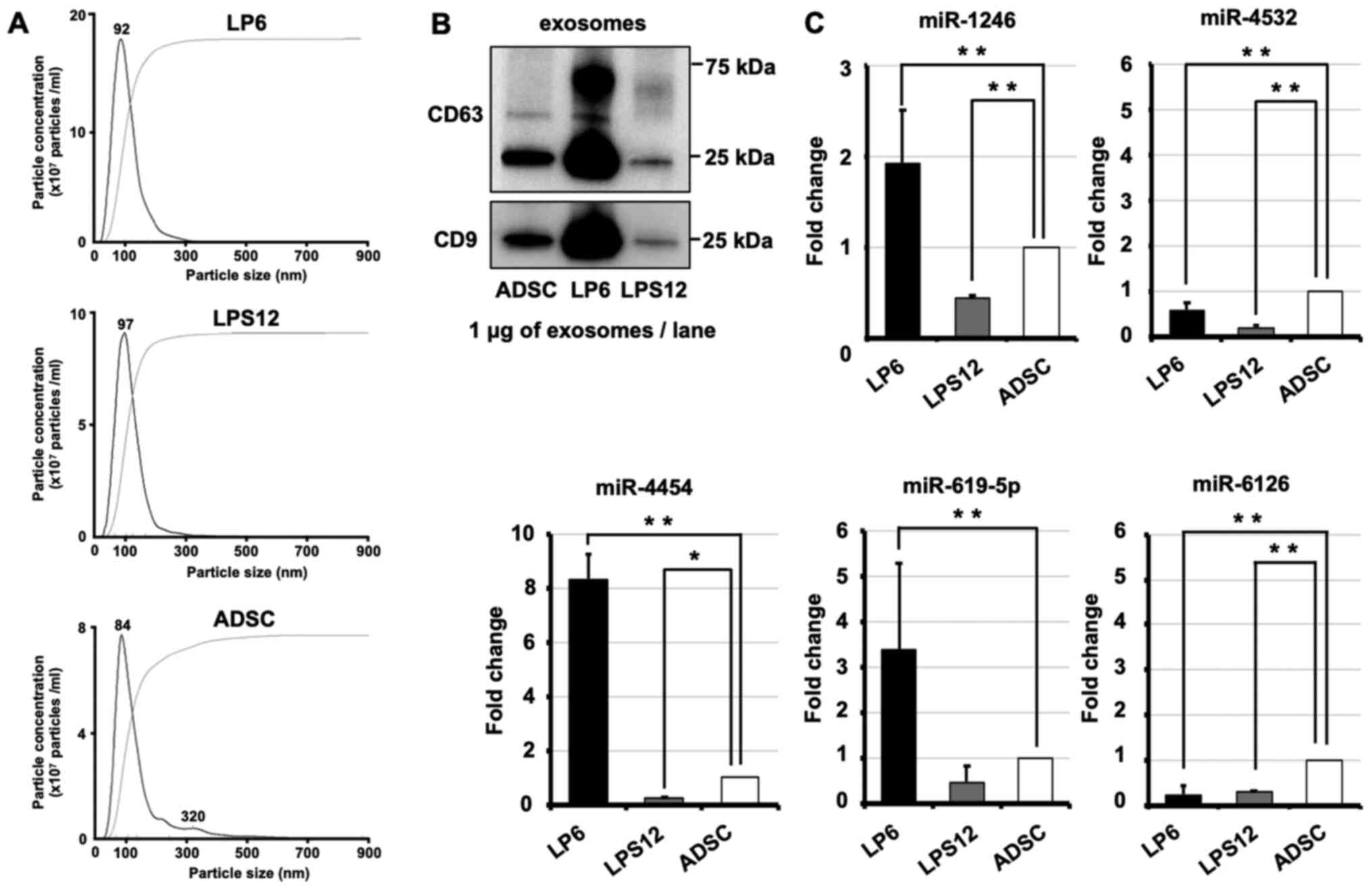
Exosomes were isolated from LP6 cells, LPS12 cells
and ADSCs by ultracentrifugation. Isolated exosomes were confirmed
using the NanoSight system (Fig.
4A). Typical exosome markers were confirmed with immunoblotting
(Fig. 4B). CD63 and CD9 were
expressed in the exosomes from LP6, LPS12 and ADSCs. Also, we
previously confirmed the isolation of exosomes from ADSC by
transmission electron microscopy (27). Total RNA was extracted from exosomes,
and the expression levels of miRNAs were examined using RT-qPCR
(Fig. 4C). miRNA expression levels
were lower in exosomes from LPS12 cells compared with those from
LP6 cells. The results of RT-qPCR analysis showed that miR-1246,
−4454 and −619-5p were highly expressed in LP6 exosomes. The ratio
of the expression level of exosomal miR-1246 was 1.92±0.89
(P<0.01) in LP6 and 0.42±0.52 (P<0.01) in LPS12 cells. That
of miR-4532 was 0.56±1.91 (P<0.01) in LP6 and 0.18±0.37
(P<0.01) in LPS12 cells. That of miR-4454 was 8.35±0.16
(P<0.01) in LP6 and 0.28±0.03 (P=0.02) in LPS12 cells. That of
miR-619-5p was 3.36±0.58 (P<0.01) in LP6 and 0.46±0.04 (P=0.67)
in LPS12 cells. That of miR-6126 was 0.23±0.88 (P<0.01) in LP6
and 0.28±0.02 (P<0.01) in LPS12 cells. These results indicated
that miR-1246, −4454 and −619-5p detected in the serum were derived
from sarcoma tissues. Exosomal miR-1246, −4454 and −619-5p were
detected in LP6 cells. This result indicated the possibility that
these miRNAs are present in DDLPS serum.
Discussion
The use of multidrug chemotherapy in combination
with surgery strongly improves the outcomes of patients with OS,
resulting in a 5-year overall survival rate of ~64-77% globally
(28–31). However, the 5-year survival rate of
patients with OS with metastatic disease and relapse at the first
visit is <20% globally (1,32). The
mortality rate of DDLPS is ~60% globally, which can be partly
attributed to tumors that are too large to be resected at the time
of diagnosis (33). The present
study identified miRNAs with potential value as biomarkers for
DDLPS. A correlation between the serum and tissue expression of
specific miRNAs [miR-1246 (R=0.49), miR-619-5p (R=0.33), miR-4532
(R=0.33), miR-4454 (R=0.21) and miR-6126 (R=0.20)] and measured the
expression levels of these miRNAs in DDLPS cell lines (LP6 and
LPS12) and released exosomes.
Most of the miRNAs showing high levels of expression
in the serum were also highly expressed in tissues in the present
study, suggesting that serum miRNAs could be derived from tumor
tissues. Although the number of EWS samples was too small to draw
firm conclusions, the number of OS and DDLPS samples was considered
to be sufficient for the analysis. There were differences in the
miRNA expression between serum and tissues. When these miRNAs were
evaluated individually, miR-1246 showed the highest correlation
coefficient. To determine whether miRNAs were secreted from DDLPS
cells, the miRNA expression levels between cells and exosomes were
compared. miR-1246, −4454 and −619-5p were highly expressed in both
DDLPS cell lines. miR-1246, −4454 and −619-5p were highly expressed
in LP6 exosomes, even though the expression levels of miRNAs were
generally low in LPS12 cells. The different results of these two
cell lines may be because the cell line was isolated from part of
one sample and may not have the characteristics of the original
tumor. These data indicated that miR-1246, −4454 and −619-5p may be
candidate miRNAs for the diagnosis of DDLPS. However, these miRNAs
were not candidates for other LPS subtypes (well differentiated,
myxoid/round cell or pleomorphic liposarcomas).
There are few studies on miRNA biomarkers of
sarcomas, but several studies have reported candidates for OS
biomarkers (10–13,34).
miR-195-5p, −199a-3p, −320a and −374a-5p were significantly
increased in the plasma of patients with OS compared with healthy
controls (34). The expression of
these four miRNAs decreased after tumor resections which were 83
extremities and 7 trunks and the expression of miR-195-5p and
−199a-3p was significantly increased in patients with metastasis
(34). miR-9 and −21 are upregulated
in the blood samples of patients with OS, but −195, -Let7A,
−199a-3p and −143 are downregulated (10–13).
These studies differed from the present report in that they
compared blood samples of patients with OS with those of healthy
controls. Moreover, Fujiwara et al (35) identified miR-25-3p as a diagnostic
and prognostic biomarker of OS in vitro, in vivo and in
clinical samples.
miR-155 was previously reported as a biomarker for
the diagnosis of DDLPS. miR-155 is highly expressed in the tissues
and plasma (36). Similarly, the
high expression of miR-155 in tissues is correlated with poor
prognosis (18). The present study
examined the correlation between miRNA expression in the serum and
tissue and serum miRNA levels may be used for predicting patient
with DDLPS prognosis.
The present assessment of the effect of miRNAs on
cell proliferation showed that miR-4532 and −6126 significantly
promoted DDLPS cell proliferation. This finding suggested that
these miRNAs may promote the progression of DDLPS. Meanwhile, other
papers have reported that miR-4532 promotes tumor progression and
that miR-6126 suppresses tumor progression. Breast cancer cells
overexpressing miR-4532 have enhanced cell viability upon
administration of adriamycin (37).
miR-4532 targeting hypermethylation in cancer 1 (HIC-1), and
overexpression of HIC-1 in breast cancer cells suppresses cell
invasion in Transwell assays (37).
Conversely, enhanced expression of miR-6126 suppresses ovarian
cancer progression. Ovarian cancer cells transfected with miR-6126
mimic have decreased cell migration and invasion (38). The reason for the discrepancy between
these previous studies and the present results is the different
cancer types. In addition, the present evaluation was insufficient
because only cell proliferation was evaluated. Additional
experiments are required to identify direct targets of miR-4532 and
miR-6126 in DDLPS.
The present study identified specific miRNAs that
were highly expressed in both the serum and tissues from patients
with DDLPS, and in vitro experiments suggested that certain
miRNAs were secreted from DDLPS cells. Taken together, the present
results suggested that the identified miRNAs could be of value as
biomarkers in DDLPS.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms Tomomi Fukuda
(Division of Molecular and Cellular Medicine, National Cancer
Center Research Institute), Dr Hiroko Tadokoro (Division of
Molecular and Cellular Medicine, National Cancer Center Research
Institute), Mr Tatsuya Suzuki (Division of Molecular and Cellular
Medicine, National Cancer Center Research Institute), Ms Makiko
Ichikawa (New Frontiers Research Institute, Toray Industries), Mr
Junpei Kawauchi (New Frontiers Research Institute, Toray
Industries) and Mr Satoshi Kondou (New Frontiers Research
Institute, Toray Industries) for technical assistance. The authors
thank Ms Noriko Abe (Clinical Laboratory, National Cancer Center
Research Institute) and Ms Michiko Ohori (Department of Biobank and
Tissue Resources, National Cancer Center Research Institute) for
collecting samples from the freezing room and Dr Kazuki Sudo
(Department of Breast and Medical Oncology, National Cancer Center
Hospital) for independent confirmation of participant eligibility.
The authors thank Dr Hitoshi Ichikawa (Department of Clinical
Genomics, National Cancer Center Research Institute) for mediating
the transfer of DDLPS cells.
Funding
The study was funded by The National Cancer Center
Research and Development Fund (grant no. 29-A-1) and through a
Development of Diagnostic Technology for Detection of miRNA in Body
Fluids grant from the Japan Agency for Medical Research and
Development.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
IK, NA, JM, YY, TY and RT performed the experiments,
analyzed data and wrote the manuscript. EK, HC, AK and TO conceived
the study, and analyzed and interpreted the data. ST, HS, KK and HF
contributed to data analysis. IK, JM and YY confirmed the
authenticity of raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by The National Cancer Centre
Hospital Institutional Review Board (Tokyo, Japan; approval nos.
2004-050, 2013-111 and 2015-266). The reason for several approval
numbers was that the data from several studies used for reanalysis
in the present study. Written informed consent was obtained from
each participant. When the patient was under 20 years old, the
informed consent was obtained from their parents, relatives or
guardians of minors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fletcher CDM, Bridge JA, Hogendoorn PCW
and Mertens F: WHO classification of soft tissue tumours. 4th
edition. IARC; Lyon: 2013
|
|
2
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kansara M, Teng MW, Smyth MJ and Thomas
DM: Translational biology of osteosarcoma. Nat Rev Cancer.
14:722–735. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mack TM: Sarcomas and other malignancies
of soft tissue, retroperitoneum, peritoneum, pleura, heart,
mediastinum, and spleen. Cancer. 75 (Suppl 1):S211–S244. 1995.
View Article : Google Scholar
|
|
5
|
Dalal KM, Kattan MW, Antonescu CR, Brennan
MF and Singer S: Subtype specific prognostic nomogram for patients
with primary liposarcoma of the retroperitoneum, extremity, or
trunk. Ann Surg. 244:381–391. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu R, Rai A, Chen M, Suwakulsiri W,
Greening DW and Simpson RJ: Extracellular vesicles in
cancer-implications for future improvements in cancer care. Nat Rev
Clin Oncol. 15:617–638. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoshino A, Costa-Silva B, Shen TL,
Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di
Giannatale A, Ceder S, et al: Tumour exosome integrins determine
organotropic metastasis. Nature. 527:329–35. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai H, Zhao H, Tang J and Wu H: Serum
miR-195 is a diagnostic and prognostic marker for osteosarcoma. J
Surg Res. 194:505–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hua J, Liu D, Cao L, Wang D, Wu T, Lin F,
Su P, Niu Y and Sun Y: Diagnostic and prognostic values of blood
microRNA-Let7A for osteosarcoma. J Bone Oncol. 12:65–68. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fei D, Li Y, Zhao D, Zhao K, Dai L and Gao
Z: Serum miR-9 as a prognostic biomarker in patients with
osteosarcoma. J Int Med Res. 42:932–937. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ouyang L, Liu P, Yang S, Ye S, Xu W and
Liu X: A three-plasma miRNA signature serves as novel biomarkers
for osteosarcoma. Med Oncol. 30:3402013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu JF, Wang YP, Zhang SJ, Chen Y, Gu HF,
Dou XF, Xia B, Bi Q and Fan SW: Exosomes containing differential
expression of microRNA and mRNA in osteosarcoma that can predict
response to chemotherapy. Oncotarget. 8:75968–75978. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vos M, Boeve WC, van Ginhoven TM, Sleijfer
S, Verhoef C and Grünhagen DJ: Impact of primary tumor location on
outcome of liposarcoma patients, a retrospective cohort study. Eur
J Surg Oncol. 45:2437–2442. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jour G, Gullet A, Liu M and Hoch BL:
Prognostic relevance of Fédération Nationale des Centres de Lutte
Contre le Cancer grade and MDM2 amplification levels in
dedifferentiated liposarcoma: A study of 50 cases. Mod Pathol.
28:37–47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vincenzi B, Iuliani M, Zoccoli A, Pantano
F, Fioramonti M, De Lisi D, Frezza AM, Rabitti C, Perrone G, Onetti
Muda A, et al: Deregulation of dicer and mir-155 expression in
liposarcoma. Oncotarget. 6:10586–10591. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kapodistrias N, Mavridis K, Batistatou A,
Gogou P, Karavasilis V, Sainis I, Briasoulis E and Scorilas A:
Assessing the clinical value of microRNAs in formalin-fixed
paraffin-embedded liposarcoma tissues: Overexpressed miR-155 is an
indicator of poor prognosis. Oncotarget. 8:6896–6913. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee DH, Amanat S, Goff C, Weiss LM, Said
JW, Doan NB, Sato-Otsubo A, Ogawa S, Forscher C and Koeffler HP:
Overexpression of miR-26a-2 in human liposarcoma is correlated with
poor patient survival. Oncogenesis. 2:e472013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Casadei L, Calore F, Creighton CJ,
Guescini M, Batte K, Iwenofu OH, Zewdu A, Braggio DA, Bill KL,
Fadda P, et al: Exosome-derived miR-25-3p and miR-92a-3p stimulate
liposarcoma progression. Cancer Res. 77:3846–3856. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Asano N, Matsuzaki J, Ichikawa M, Kawauchi
J, Takizawa S, Aoki Y, Sakamoto H, Yoshida A, Kobayashi E, Tanzawa
Y, et al: A serum microRNA classifier for the diagnosis of sarcomas
of various histological subtypes. Nat Commun. 10:12992019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Usuba W, Urabe F, Yamamoto Y, Matsuzaki J,
Sasaki H, Ichikawa M, Takizawa S, Aoki Y, Niida S, Kato K, et al:
Circulating miRNA panels for specific and early detection in
bladder cancer. Cancer Sci. 110:408–419. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka
Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda
T, et al: Integrated extracellular microRNA profiling for ovarian
cancer screening. Nat Commun. 9:43192018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa
M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F
and Ochiya T: Malignant extracellular vesicles carrying MMP1 mRNA
facilitate peritoneal dissemination in ovarian cancer. Nat Commun.
8:144702017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshioka Y, Konishi Y, Kosaka N, Katsuda
T, Kato T and Ochiya T: Comparative marker analysis of
extracellular vesicles in different human cancer types. J Extracell
Vesicles. 18:22013.PubMed/NCBI
|
|
27
|
Zhou Y, Yamamoto Y, Takeshita F, Yamamoto
T, Xiao Z and Ochiya T: Delivery of miR-424-5p via extracellular
vesicles promotes the apoptosis of MDA-MB-231 TNBC cells in the
tumor microenvironment. Int J Mol Sci. 22:8442021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iwamoto Y, Tanaka K, Isu K, Kawai A,
Tatezaki S, Ishii T, Kushida K, Beppu Y, Usui M, Tateishi A, et al:
Multiinstitutional phase II study of neoadjuvant chemotherapy for
osteosarcoma (NECO study) in Japan: NECO-93J and NECO-95J. J Orthop
Sci. 14:397–404. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bacci G, Bertoni F, Longhi A, Ferrari S,
Forni C, Biagini R, Bacchini P, Donati D, Manfrini M, Bernini G and
Lari S: Neoadjuvant chemotherapy for high-grade central
osteosarcoma of the extremity. Histologic response to preoperative
chemotherapy correlates with histologic subtype of the tumor.
Cancer. 97:3068–3075. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bacci G, Briccoli A, Ferrari S, Longhi A,
Mercuri M, Capanna R, Donati D, Lari S, Forni C and DePaolis M:
Neoadjuvant chemotherapy for osteosarcoma of the extremity:
Long-term results of the Rizzoli's 4th protocol. Eur J Cancer.
37:2030–2039. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bajpai J, Chandrasekharan A, Talreja V,
Simha V, Chandrakanth MV, Rekhi B, Khurana S, Khan A, Vora T, Ghosh
J, et al: Outcomes in non-metastatic treatment naive extremity
osteosarcoma patients treated with a novel non-high
dosemethotrexate-based, dose-dense combination chemotherapy regimen
‘OGS-12’. Eur J Cancer. 85:49–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chou AJ, Geller DS and Gorlick R: Therapy
for osteosarcoma: Where do we go from here? Paediatr Drugs.
10:315–27. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Henricks WH, Chu YC, Goldblum JR and Weiss
SW: Dedifferentiated liposarcoma: A clinicopathological analysis of
155 cases with a proposal for an expanded definition of
dedifferentiation. Am J Surg Pathol. 21:271–281. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lian F, Cui Y, Zhou C, Gao K and Wu L:
Identification of a plasma four-microRNA panel as potential
noninvasive biomarker for osteosarcoma. PLoS One. 10:e01214992015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fujiwara T, Uotani K, Yoshida A, Morita T,
Nezu Y, Kobayashi E, Yoshida A, Uehara T, Omori T, Sugiu K, et al:
Clinical significance of circulating miR-25-3p as a novel
diagnostic and prognostic biomarker in osteosarcoma. Oncotarget.
8:33375–33392. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Boro A, Bauer D, Born W and Fuchs B:
Plasma levels of miRNA-155 as a powerful diagnostic marker for
dedifferentiated liposarcoma. Am J Cancer Res. 6:544–552.
2016.PubMed/NCBI
|
|
37
|
Feng F, Zhu X, Wang C, Chen L, Cao W, Liu
Y, Chen Q and Xu W: Downregulation of hypermethylated in cancer-1
by miR-4532 promotes adriamycin resistance in breast cancer cells.
Cancer Cell Int. 18:1272018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kanlikilicer P, Rashed MH, Bayraktar R,
Mitra R, Ivan C, Aslan B, Zhang X, Filant J, Silva AM,
Rodriguez-Aguayo C, et al: Ubiquitous release of exosomal tumor
suppressor miR-6126 from ovarian cancer cells. Cancer Res.
76:7194–7207. 2016. View Article : Google Scholar : PubMed/NCBI
|