Introduction
Spinal cord glioma accounts for >80% of all
spinal intramedullary tumors (1,2). Due to
the high recurrence and poor survival rates of patients with spinal
cord glioma, its treatment is considered challenging (3,4). Studies
have suggested that the abnormal invasion, proliferation, migration
and apoptosis of spinal cord glioma cells are the main obstacles to
improving prognosis (5,6). Although progress has been made in
conventional treatments, their therapeutic effects are not
satisfactory (7). Therefore, the
identification of novel therapeutic targets and the development of
mechanism-based approaches are urgently required.
MicroRNAs (miRNAs/miRs) are endogenous non-coding
single-stranded RNAs 20–25 nucleotides in length, which have
regulatory functions and are present in the genomes of most
eukaryotes (8). According to the
principle of complementary base pairing, the 3′-untranslated region
(3′-UTR) of a target mRNA is identified by a miRNA, and the
silencing complex is guided to degrade or inhibit the translation
of the target mRNA. Certain miRNA precursors can form two stably
expressed mature miRNA, with the difference that ‘-3p’ or ‘-5p’ is
added to the end of the name depending on where it advances
(9). Recently, increasing evidence
has demonstrated that miRNAs serve an important role in specific
cellular processes, such as cell differentiation, morphology and
tumor formation (10,11).
During tumorigenesis and tumor development, miRNAs
may act as oncogenes or tumor suppressor genes, and are involved in
the regulation of tumor-associated genes (12). Studies have suggested that miR-21
(13), miR-10b (14), miR-95-3p (15), miR-106a-5p (16), miR-615-3p (17) and miR-155 (18) are important tumor growth factors. The
different expression profiles between glioma and non-cancerous
tissues may partially elucidate the roles of different miRNAs in
tumorigenesis, and may be therefore used to predict new therapeutic
targets (18).
In the present study, the expression levels of
miR-21, miR-10b, miR-95-3p, miR-106a-5p, miR-615-3p and miR-155
were compared between spinal cord glioma and non-cancerous spinal
cord tissues by reverse transcription-quantitative PCR (RT-qPCR).
Since miR-106a-5p was highly expressed in spinal cord glioma
tissues compared with in non-cancerous tissues, it was hypothesized
that miR-106a-5p may serve as a gene mediating the growth of spinal
cord glioma. To verify this hypothesis, the effects of miR-106a-5p
on cell proliferation, migration, invasion and apoptosis of spinal
cord glioma cells were evaluated. Furthermore, the results revealed
an inverse association between CUGBP Elav-like family member 2
(CELF-2) and miR-106a-5p expression in human spinal cord glioma
samples. Therefore, the effect of CELF-2 overexpression on the
biological activity of spinal cord glioma cells was evaluated in
vitro and in vivo.
Materials and methods
Cell culture and transfection
Human spinal cord glioma and non-cancerous spinal
cord tissues (normal adjacent tissues) were collected from the
Department of Neurosurgery of The First Affiliated Hospital of
University of Science and Technology of China (Hefei, China). All
21 patients (admitted between February 2017 and July 2018; age
range, 16–60 years; median age, 46 years) provided written informed
consent prior to enrolment. The present study was approved by the
Institutional Review Board of The First Affiliated Hospital of
University of Science and Technology of China. Patient 02–13 was
randomly selected for the isolation of spinal cord glioma cells
(0213SCG cells). Fresh human spinal cord glioma tissue was cut into
1-mm3 pieces and washed 2–3 times with PBS supplemented
with 1% penicillin/streptomycin solution (Gibco; Thermo Fisher
Scientific, Inc). PBS was removed and 0.25% trypsin was added to
digest the tumor tissue mass at 37°C in a water bath for 30 min.
The digested cell fluid was passed through a 100-mesh steel
strainer to prepare a tumor cell suspension, which was in turn
transferred to a centrifuge tube followed by centrifugation at 241
× g for 3 min at room temperature. Subsequently, the cells were
resuspended in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin/streptomycin solution, and were cultured at 37°C
in a 5% CO2 incubator. For tracking transplanted spinal
cord glioma cells in vivo, 0213SCG cells were transduced
with a self-inactivating lentiviral vector carrying an ubiquitin
promoter driving the expression of firefly luciferase and enhanced
green fluorescence protein (Fluc-eGFP) at a multiplicity of
infection (MOI) of 10. After 48 h, fluorescence images of eGFP in
0213SCG cells were observed at ×20 magnification under a
fluorescence microscope (Carl Zeiss AG), and luciferase expression
was identified. For CELF-2 overexpression in 0213SCG cells, the
pcDNA3.1-CELF-2-flag expression vector and blank-vector
(pCMV6-entry) were purchased from OriGene Technologies, Inc.
0213SCG cells were transfected with 2 µg of the
pcDNA3.1-CELF-2-flag plasmid or blank-vector for 24 h at 37°C in
serum-free Opti-MEM I (Invitrogen; Thermo Fisher Scientific, Inc.)
using Lipofectamine® 2000 (Guangzhou RioboBio Co., Ltd.)
according to the manufacturers instructions. Following transfection
for 24 h, the culture medium was replaced with fresh complete
medium containing 600 µg/ml G418. The positive 0213SCG cells were
obtained after 7 days of screening by G418. Finally, CELF-2
expression was determined by western blot analysis.
Virus particles production
For the production of virus, 293T cells (American
Type Culture Collection) were seeded into 6-well culture plates.
After 24 h, 5 µg of CMV-eGFP-T2A-Luciferase plasmid (GM-0220IV02;
Genomeditech; Jiman Biotechnology (Shanghai) Co., Ltd.), 3 µg of
package plasmid pCMV.R8.2 and 1 µg of envelope plasmid pMD.G were
transfected into 293T cells using the standard calcium phosphate
(Invitrogen; Thermo Fisher Scientific, Inc.) method (19). The transfected cells were incubated
in a 37°C incubator with 5% CO2 for 12 h and then the
medium was replaced with fresh medium. After 48 h from
transfection, virus-containing medium was harvested and centrifuged
at 845 × g for 5 min at room temperature. The virus-containing
medium was ultracentrifuged at 50,000 × g for 150 min at 4°C to
obtain the high-titer concentrated virus particles.
Cell transfection with miRNA
inhibitors/mimics
0213SCG cells were transfected with 20 nM Homo
sapiens miR-106a-5p inhibitors, miR-106a-5p mimics or miRNA
negative controls (NCs) using Lipofectamine® 2000
(Guangzhou RioboBio Co., Ltd.) for 48 h at 37°C in Opti-MEM (Gibco;
Thermo Fisher Scientific, Inc.) using Lipofectamine®
2000. Following transfection for 48 h, cells were collected for
subsequent investigation. Additionally, the TargetScan database
(www.targetscan.org) was utilized to
predict the potential target mRNAs of miR-106a-5p. The sequences of
all oligonucleotides used for transfection were as follows:
Mimics-control sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-mimics sense,
5′-AAAAGUGCUUACAGUGCAGGUAG-3′ and antisense,
5′-ACCUGCACUGUAAGCACUUUUUU-3′; inhibitor-control,
5′-CAGUACUUUUGUGUAGUACAA-3′; and miR-inhibitor,
5′-CUACCUGCACUGUAAGCACUUUU-3′.
Cell proliferation assay
For the cell proliferation assay, 0213SCG cells were
seeded into 96-well plates at a density of 8×103
cells/well. To detect the effects of miR-106a-5p inhibitors on cell
proliferation, 0213SCG cells were transfected with miR-106a-5p
inhibitors or NC inhibitors, as aforementioned. Cell proliferation
was determined after 24 and 48 h by MTT assay using dimethyl
sulfoxide as the solvent according to the manufacturers
instructions (Sigma-Aldrich; Merck KGaA). Furthermore, to evaluate
the effect of CELF-2 overexpression on 0213SCG cell proliferation,
0213SCG cells and 0213SCG cells overexpressing CELF-2 were seeded
into 96-well plates at a density of 8×103 cells/well.
Cell proliferation was then determined at 24 and 48 h by MTT assay,
as aforementioned. The absorbance at 490 nm was measured to
calculate cell proliferation using a multi-function enzyme-linked
analyzer (BioTek Instruments, Inc.; Agilent Technologies,
Inc.).
Western blot analysis
For CELF-2 protein expression analysis, the cells or
tissues were lysed on ice with radio-immunoprecipitation assay
buffer supplemented with a protease inhibitor cocktail
(Sigma-Aldrich; Merck KGaA). The tumor tissues were specifically
lysed for 30 min. BCA reagent (Thermo Fisher Scientific, Inc.) was
used to quantify the protein concentration. Total cell or tissue
proteins (50 µg/lane) were separated by 10% SDS-PAGE and
transferred onto PVDF membranes (EMD Millipore). Following blocking
with 5% skimmed dry milk in Tris-buffer containing 0.05%-Tween-20
(TBST) for 2 h at room temperature, the membranes were incubated
with primary antibodies at 4°C overnight. The membranes were washed
thrice with TBST for 5 min each time, and were then incubated with
HRP-conjugated secondary antibodies (1:10,000; cat. no. VA001;
Vicmed Biotech Co., Ltd.) at room temperature for 2 h. Following
washing with TBST for three times, the blots were visualized using
the SuperSignal West Dura Extended Duration Substrate (Thermo
Fisher Scientific, Inc). The primary antibodies used were against
GAPDH (1:1,000; cat. no. ab9485; Abcam) and CELF-2 (1:400; cat. no.
PA1-4130; Thermo Fisher Scientific, Inc.). GAPDH served as the
internal control. The grey levels of western blot analysis were
measured and quantified using ImageJ software (version ImageJ2;
National Institutes of Health).
Tumor model and bioluminescence
imaging (BLI)
Male nu/nu Nude mice (age, 6–8 weeks; weight 22 g;
n=40) were purchased from the Beijing Weitonglihua Experimental
Animal Co., Ltd., and were housed under standard laboratory
conditions (temperature, 20–26°C; relative humidity, 40–60%; noise,
<60 dB; ventilation degree, 10–20 times/h; light/dark cycle,
12/12 h; adequate food and drinking water.). All experimental
procedures were conducted in accordance with the institutional
guidelines of the University of Science and Technology of China for
The Care and Use of Laboratory Animals and conformed to the
National Institutes of Health Guide for Care and Use of Laboratory
Animals [approval no. 2019-N (A)-011]. Mice received subcutaneous
injection of 100 µl of the cell suspension containing
1×106 0213SCG cells or CELF-2-overexpressing 0213SCG
cells (Fluc-eGFP) into the right forelimb armpit. In addition, the
following humane endpoints were established in the present study:
i) The tumor burden should not exceed 5% of the normal body weight
of the mouse; ii) the tumor should not grow to a position that
seriously affects the normal function of the mouse, and the growth
of the tumor should not cause pain in the mouse; iii) the weight
loss of mice should not exceed 20% of the body weight of normal
mice; iv) there should be no ulcers or infections at the growth
point of the tumor; and v) mice should not exhibit self-harming
behavior. Animals were monitored every 3 days to evaluate tumor
progression. To monitor tumor development, BLI was applied using
the Imaging System IVIS Lumina (Xenogen Corp.). To detect Fluc
expression at days 1, 4, 7, 10, 17 and 20, mice were
intraperitoneally injected with D-luciferin (150 mg/kg body weight;
J&K Scientific Ltd.). The dose of isoflurane used to induce and
maintain anesthesia was 2%. For in vitro BLI, 0213SCG cells
(Fluc-eGFP) were seeded into 6-well plates at a density of 2, 4, 6
and 8×105 cells/well, and D-luciferin was added into
each well following incubation for 0.5 h at room temperature. On
day 20, the mice were euthanized by cervical dislocation, and tumor
samples were harvested and fixed with 4% paraformaldehyde for 48 h
at 4°C. The largest tumor volume observed in the study was 916
mm3.
Cell migration and invasion assay
Cell migration and invasion assays were performed
using Transwell chambers (8-µm pore; EMD Millipore) pre-coated with
(invasion assay) or without (migration assay) Matrigel (cat. no.
356234; Corning, Inc.). Transwell chambers for invasion assays were
coated with Matrigel at 37°C for 30 min. A total of
1×104 0213SCG cells transfected with miR-106a-5p
inhibitors or NC inhibitors were seeded into the inserts and
cultured with serum-free RPMI-1640 medium, and RPMI-1640 medium
containing 10% FBS was added to the lower chambers. Cell migration
and invasion were also determined in CELF-2 overexpressing 0213SCG
cells. Following culture for 24 h at 37°C, non-migrating or
non-invading cells on the upper side of the filter were removed
with a cotton swab. The migrated or invasive cells were stained
with crystal violet for 15 min at room temperature and counted at 6
randomly selected fields at ×20 magnification under an optical
microscope (Carl Zeiss AG).
Immunofluorescence staining and TUNEL
assay
Tumor tissues formed after transplanting 0213SCG
cells were harvested and fixed with 4% paraformaldehyde for 48 h at
4°C and cut into 5-µm-thick frozen sections at −20°C. The prepared
frozen sections were used for immunofluorescence and TUNEL
experiments. To evaluate CELF-2 expression in transplanted tumor
tissues, immunofluorescence staining was performed using a rabbit
anti-human CELF-2 antibody (1:400; cat. no. PA1-4130; Thermo Fisher
Scientific, Inc). In addition, apoptosis and proliferation in tumor
tissues were detected using the cleaved capase-3 (cat. no. ab49822)
and Ki67 (cat. no. ab15580) antibodies (both 1:400; Abcam),
respectively. Briefly, the frozen sections were blocked with 5% BSA
(Roche Diagnostics GmbH) for 2 h at room temperature, and then
incubated with primary antibody overnight at 4°C. The primary
antibodies were detected by Alexa Fluor 647-labeled secondary
antibody (1:1,000; cat. no. A-31573; Invitrogen; Thermo Fisher
Scientific, Inc.). Furthermore, the apoptosis in tumor tissues and
adherent cells, was evaluated using a TUNEL assay kit (DeadEnd™
Fluorometric TUNEL System; Promega Corporation) according to the
manufacturers protocol. Tissue sections and apoptotic cells were
counterstained with 10 µg/ml DAPI for 20 min at room temperature
and observed at ×40 magnification under a fluorescence microscope
(Carl Zeiss AG). The positive area and positive cells were measured
using Image-Pro Plus software (version 6.0; Media Cybernetics,
Inc.).
Flow cytometric analysis
Flow cytometry was utilized to determine the
apoptotic rate of 0213SCG cells transfected with miR-106a-5p
inhibitors and respective NC. Following transfection with
miR-106a-5p inhibitors/NCs for 24 h, 0213SCG cells were harvested
and stained using an Annexin V and PI kit (BD Bioscience) according
to the manufacturers protocol. Subsequently, the mean fluorescence
of the cells was measured and analyzed using the Beckman Coulter
FC500 Flow Cytometry system with CXP software (Beckman Coulter,
Inc.).
RT-qPCR
Total RNA was isolated from tissues or cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturers protocol. Subsequently,
first-strand cDNA was synthesized using reverse transcriptase and
oligo(dT) primers of an RT kit (cat. no. AE101-02; TransGen Biotech
Co., Ltd.) following the manufacturers protocol. Real-time PCR was
performed with SYBR-Green on the Bio-Rad Real-Time PCR System
(Bio-Rad Laboratories, Inc.). The expression levels of miR-21,
miR-10b, miR-155, miR-106a-5p, miR-95-3p, miR-615-3p and CELF-2
were quantified using SYBR-Green qPCR (TransGen Biotech Co., Ltd.).
The 2−∆∆Cq method (20)
was used to analyze the relative mRNA fold-changes. Primers are
listed in Table SI. In the
experiments, GAPDH was used as an internal control for mRNA, and U6
was used as an internal control for miRNA. The thermocycling
conditions were as follows: 94°C for 5 min, followed by 40 cycles
at 94°C for 30 sec, 60°C for 25 sec and 72°C for 25 sec.
Statistical analysis
All statistical analysis was performed using
GraphPad Prism 5.0 (GraphPad Software, Inc.). All data were
expressed as the mean ± SEM. Mixed two-way ANOVA was used to
evaluate tumor growth at different time points in the control and
CELF-2 OE groups, and the Bonferroni test was used as the post-hoc
test. Paired Students t-test was used to analyze the differences
between cancerous and non-cancerous tissues. The correlation
between Fluc fluorescence intensity and cell number was analyzed by
unary linear regression in 0213SCG cells transfected with
Fluc-eGFP. For the other datasets, statistically significant
differences between two or multiple groups were assessed by
unpaired Students t-test or one-way ANOVA with Tukeys post-hoc
test, respectively. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-106a-5p expression is upregulated
in spinal cord glioma tissues
It has been reported that miR-21, miR-10b,
miR-95-3p, miR-106a-5p, miR-615-3p and miR-155 are significant
regulators of tumor growth (13–18).
Therefore, the expression levels of the aforementioned six miRNAs
were determined in spinal cord glioma and non-cancerous tissues
(normal adjacent tissues) from patient 02–13. The results
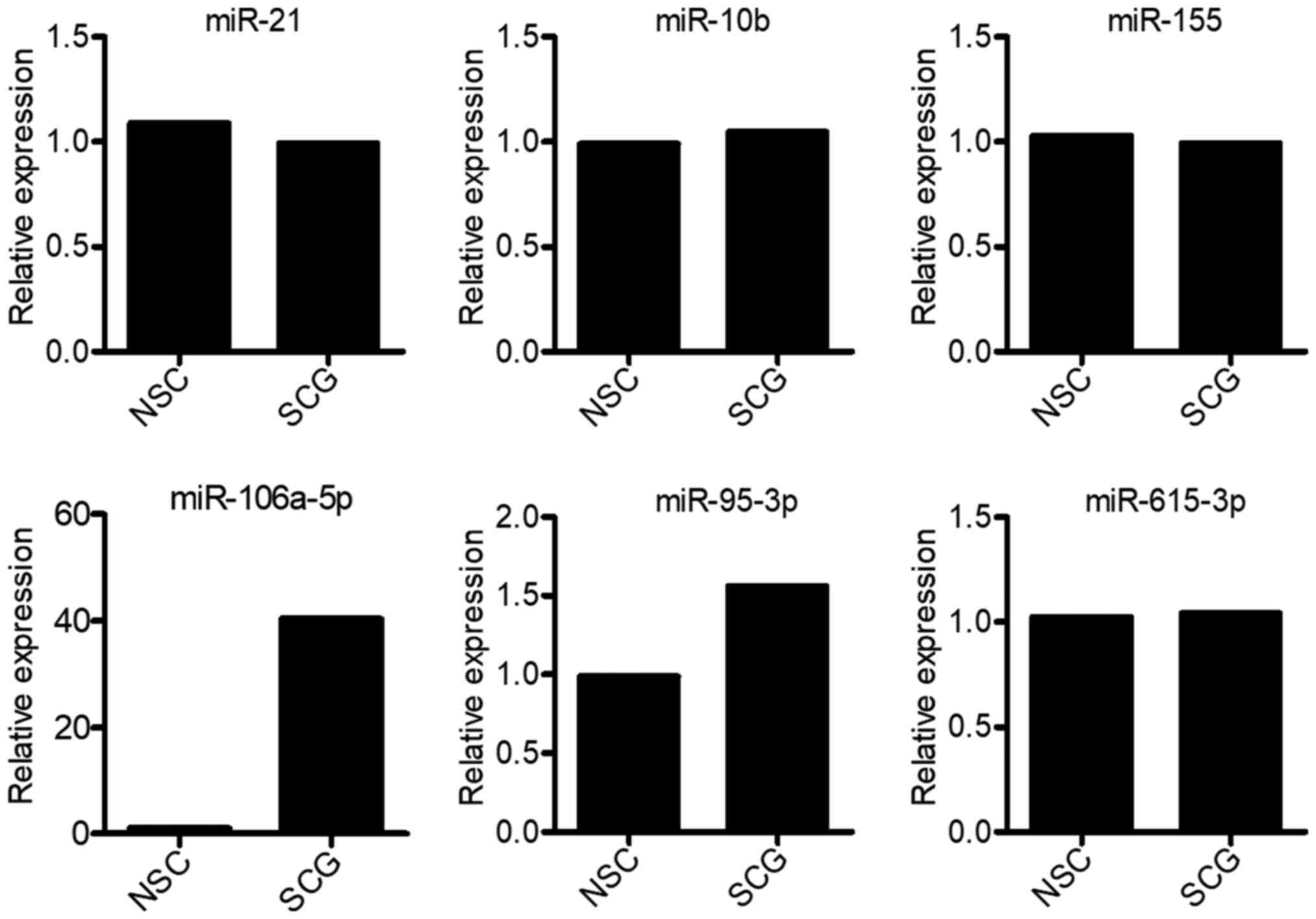
demonstrated that only miR-106a-5p and miR-95-3p expression was
upregulated in spinal cord glioma tissues compared with in
non-cancerous tissues (Fig. 1).
Compared with normal tissues, miR-106a-5p expression was markedly
upregulated in spinal cord glioma tissues, whereas miR-95-3p
expression was not markedly altered (Fig. 1). In addition, spinal glioma tissue
samples from another 20 patients with high-grade glioma were used
as the experimental group, while the non-cancerous spinal cord
tissue samples from the corresponding patients served as control.
The characteristics of the spinal cord tissue samples are listed in
Table SII. The RT-qPCR results
indicated that miR-106a-5p was markedly upregulated in spinal
glioma tissue samples compared with in normal tissues in all
patients (Fig. S1A and B).
Therefore, the current findings suggested that miR-615-3p may play
a carcinogenic role in spinal cord glioma.
Tumor-promoting effect of
miR-106a-5p
Emerging evidence has suggested that miR-106a-5p
expression is upregulated or downregulated in different tumor
tissues (21–25). However, to the best of our knowledge,
the effect of miR-106a-5p in spinal cord glioma has not been
previously investigated. To explore the effect of miR-106a-5p on
0213SCG cells isolated from patients with 02–13 spinal cord glioma,
cells were transiently transfected with miR-106a-5p inhibitors or
mimics or the corresponding NCs (inCON or miCON, respectively). The
expression levels of miR-106a-5p in transiently transfected 0213SCG
cells were determined by RT-qPCR, demonstrating that 0213SCG cells
were efficiently transfected with miR-106a-5p inhibitors or mimics
(Fig. S2A and B). Subsequently,
cell proliferation was evaluated in 0213SCG cells transfected with
miR-106a-5p inhibitors or NC inhibitors for 24 and 48 h by MTT
assays. Therefore, compared with the PBS or inCON groups, the
proliferation of 0213SCG cells was significantly attenuated in the
miR-106a-5p inhibitor group (Fig.
2A), while the proliferation of 0213SCG cells was significantly
increased in the miR-106a-5p mimics group compared with in the PBS
and miCON groups (Fig. S2C).
Furthermore, cell migration and invasion were evaluated by
Transwell assays. Compared with the PBS and inCON groups, 0213SCG
cells transfected miR-106a-5p inhibitors exhibited a significantly
attenuated ability to penetrate through the membrane pores
pre-coated with or without Matrigel (Fig. 2B-E). Additionally, the apoptosis of
0213SCG cells was analyzed by flow cytometry and TUNEL assay. The
results revealed that 0213SCG cells transfected with miR-106a-5p
inhibitors exhibited a significantly increased apoptosis rate
compared with those treated with inCON and PBS (Fig. 2F and G). However, compared with the
cells in the miCON and PBS groups, the apoptosis rate was not
markedly changed in 0213SCG cells transfected with miR-106a-5p
mimics (Fig. S2D). Overall, the
aforementioned results suggested that miR-95-3p inhibition affected
the proliferation, migration, invasion and apoptosis of 0213SCG
cells.
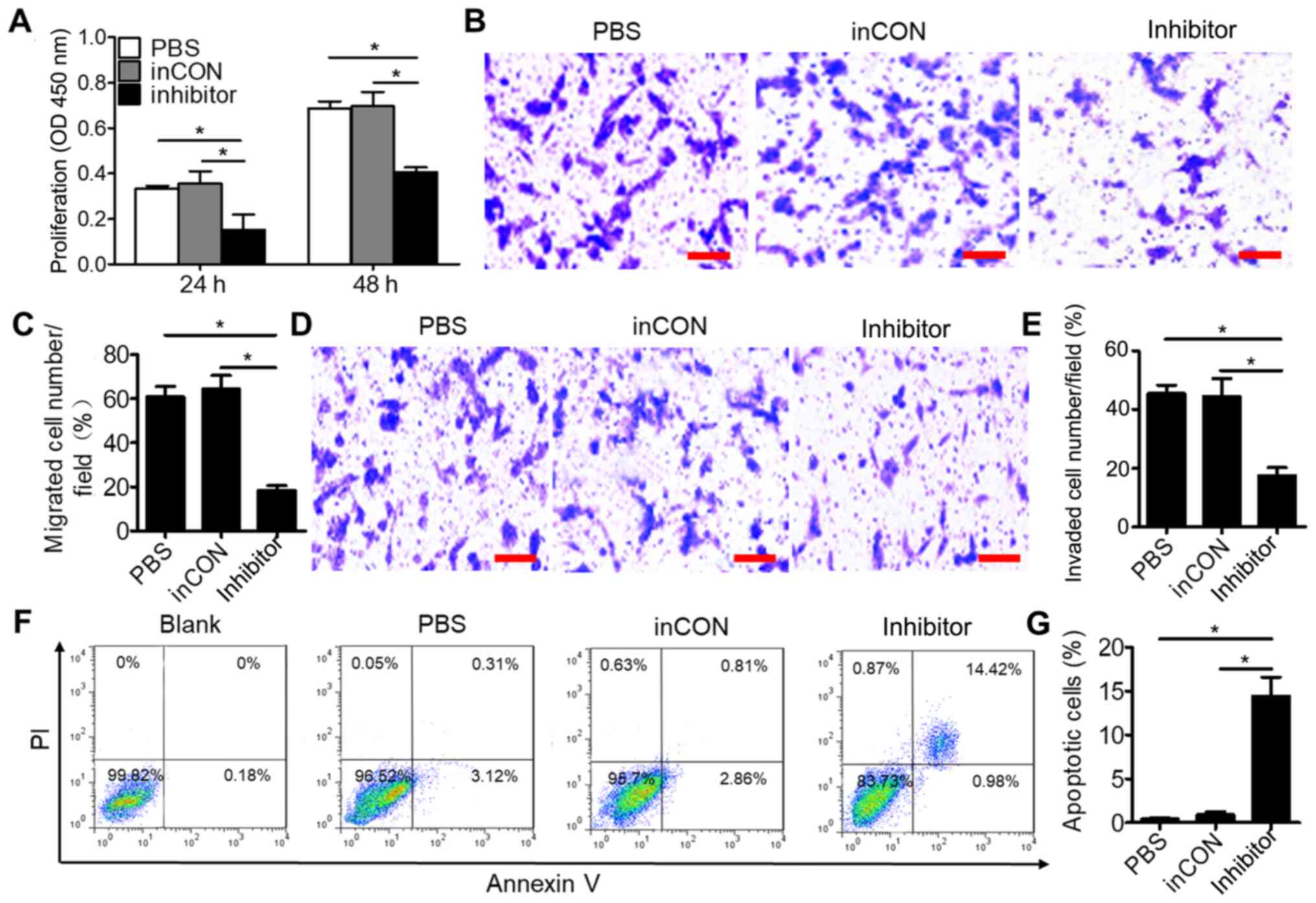 | Figure 2.Downregulated miR-106a-5p inhibits the
proliferation, migration and invasion, and promotes the apoptosis
of 0213SCG cells. (A) MTT assays result indicated that
downregulation of miR-106a-5p inhibited the proliferation of
0213SCG cells. (B) Representative photographs of Transwell
migration assay of 0213SCG cells treated with PBS, inCON and
miR-106a-5p inhibitor. Scale bar, 100 µm. (C) Quantitative analysis
results revealed that the migration of 0213SCG cells was
significantly suppressed by the miR-106a-5p inhibitor. (D)
Representative photographs of Transwell invasion assay of 0213SCG
cells treated with PBS, inCON and miR-106a-5p inhibitor. Scale bar,
100 µm. (E) Quantitative analysis results revealed that the
invasion of 0213SCG cells was significantly suppressed by the
miR-106a-5p inhibitor. (F) Flow cytometry was used to analyze the
apoptosis of 0213SCG cells treated with PBS, inCON and miR-106a-5p
inhibitor. (G) Quantitative analysis results revealed that
miR-106a-5p inhibitors significantly increased the apoptosis of
0213SCG cells. Data are expressed as the mean ± SEM. *P<0.05.
All experiments were performed in triplicate. 0213SCG cells, spinal
cord glioma cells isolated from patient 0213; inCON, miR-106a-5p
inhibitor negative control; miR, microRNA; OD, optical density. |
miR-106a-5p suppresses CELF-2
expression in spinal cord glioma cells
Previous studies have revealed that CELF-2 is a
tumor suppressor and is inversely associated with cancer cell
migration, invasion, proliferation and apoptosis (26–28).
However, the effect of CELF-2 in spinal cord glioma has not been
previously reported. To evaluate whether miR-106a-5p could regulate
the occurrence of spinal cord glioma via regulating CELF-2, the
TargetScan database was utilized to predict the potential target
mRNAs of miR-106a-5p. Among all the predicted target-mRNAs, the
analysis revealed that CELF-2 could be a putative target gene of
miR-106a-5p, since its potential binding site was identified
(Figs. 3A and S3). To verify that miR-106a-5p could
directly target CELF-2, total RNA and proteins were isolated from
transplanted tumor tissues, 0213SCG cells treated with PBS and
0213SCG cells transfected with miR-106a-5p inhibitors or NC.
Subsequently, RT-qPCR and western blot analyses were performed to
determine the expression levels of CELF-2. The results revealed
that compared with the NSC group, the gene and protein expression
levels of CELF-2 were downregulated in the PBS and NC groups;
however, cell transfection with miR-106a-5p inhibitors
significantly increased the gene and protein expression levels of
CELF-2 compared with the PBS and NC groups (Fig. 3B-D).
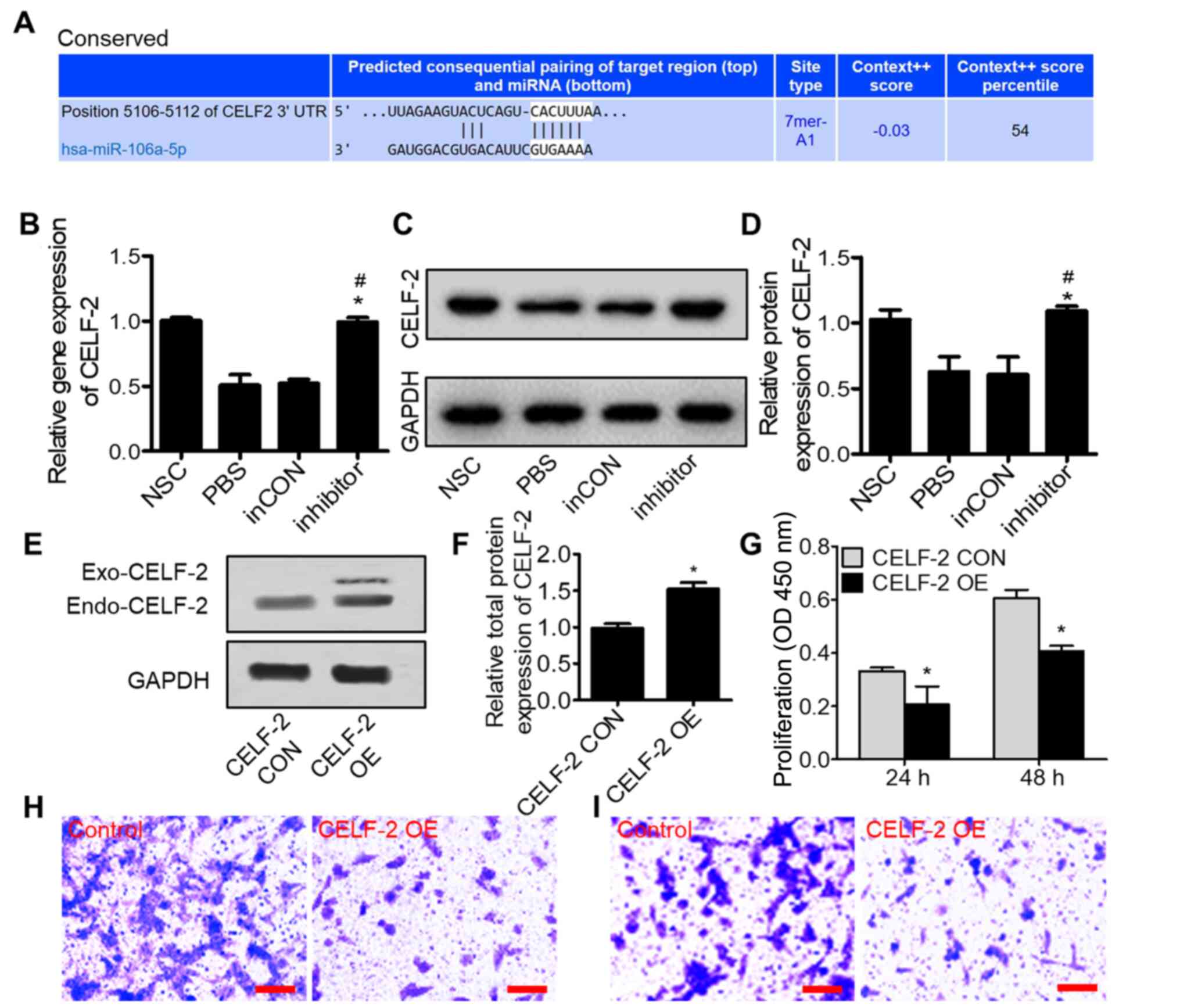 | Figure 3.miR-106a-5p promotes tumor progression
by inhibiting CELF-2 expression in 0213SCG cells. (A) Predicted
binding site of miR-106a-5p in the CELF-2 3′-UTR region using
TargetScan. (B) Reverse transcription-quantitative PCR assays
showed that the miR-106a-5p inhibitor significantly increased the
expression levels of CELF-2 in 0213SCG cells. (C) Western blot
results showed the CELF-2 protein expression in 0213SCG cells
treated with PBS, inCON and miR-106a-5p inhibitor. (D) Quantitative
analysis results revealed that CELF-2 protein expression was
significantly increased by the miR-106a-5p inhibitor. Data are
expressed as the mean ± SEM. *P<0.05 vs. PBS;
#P<0.05 vs. inCON. (E) Western blot analysis of
CELF-2 protein expression in CELF-2 OE and CELF-2 CON 0213SCG
cells. (F) Quantitative analysis results showed upregulated
expression levels of CELF-2 in CELF-2 OE 0213SCG cells. (G) MTT
assays results revealed that the overexpression of CELF-2
suppressed 0213SCG cell proliferation at 24 and 48 h. Data are
expressed as the mean ± SEM. *P<0.05 vs. CON. Representative
images of Transwell (H) migration and (I) invasion assays in CELF-2
OE and control 0213SCG cells. Scale bar, 100 µm. All experiments
were performed in triplicate. 0213SCG, spinal cord glioma cells
isolated from patient 0213; inCON, miR-106a-5p inhibitor negative
control; miR, microRNA; CELF-2, CUGBP Elav-like family member 2;
OE, overexpression; 3′-UTR, 3′-untranslated region; CON, control;
NSC, normal spinal cord tissue adjacent to tumor tissue; OD,
optical density; Exo-CELF-2, exogenous CELF-2 derived from the
pcDNA3.1-CELF-2-flag plasmid; Endo-CELF-2, endogenous CELF-2. |
To determine whether miR-106a-5p promoted the
development of spinal cord gliomas via targeting CELF-2, a 0213SCG
cell line transiently overexpressing CELF-2 was established. CELF-2
overexpression was verified by western blot analysis (Fig. 3E and F). Subsequently, to reveal
whether CELF-2 overexpression could affect the proliferation of
0213SCG cells, MTT assay was performed. The results indicated that,
compared with control 0213SCG cells, the proliferation of
CELF-2-overexpressing 0213SCG cells was significantly attenuated
(Fig. 3G). In addition, CELF-2
overexpression markedly decreased the migration and invasion of
0213SCG cells (Fig. 3H and I). The
current findings suggested that CELF-2 may be a functional
downstream target of miR-106a-5p regulating the development of
spinal cord glioma.
Overexpression of CELF-2 exerts
tumor-suppressive effects
To determine the effect of CELF-2 overexpression on
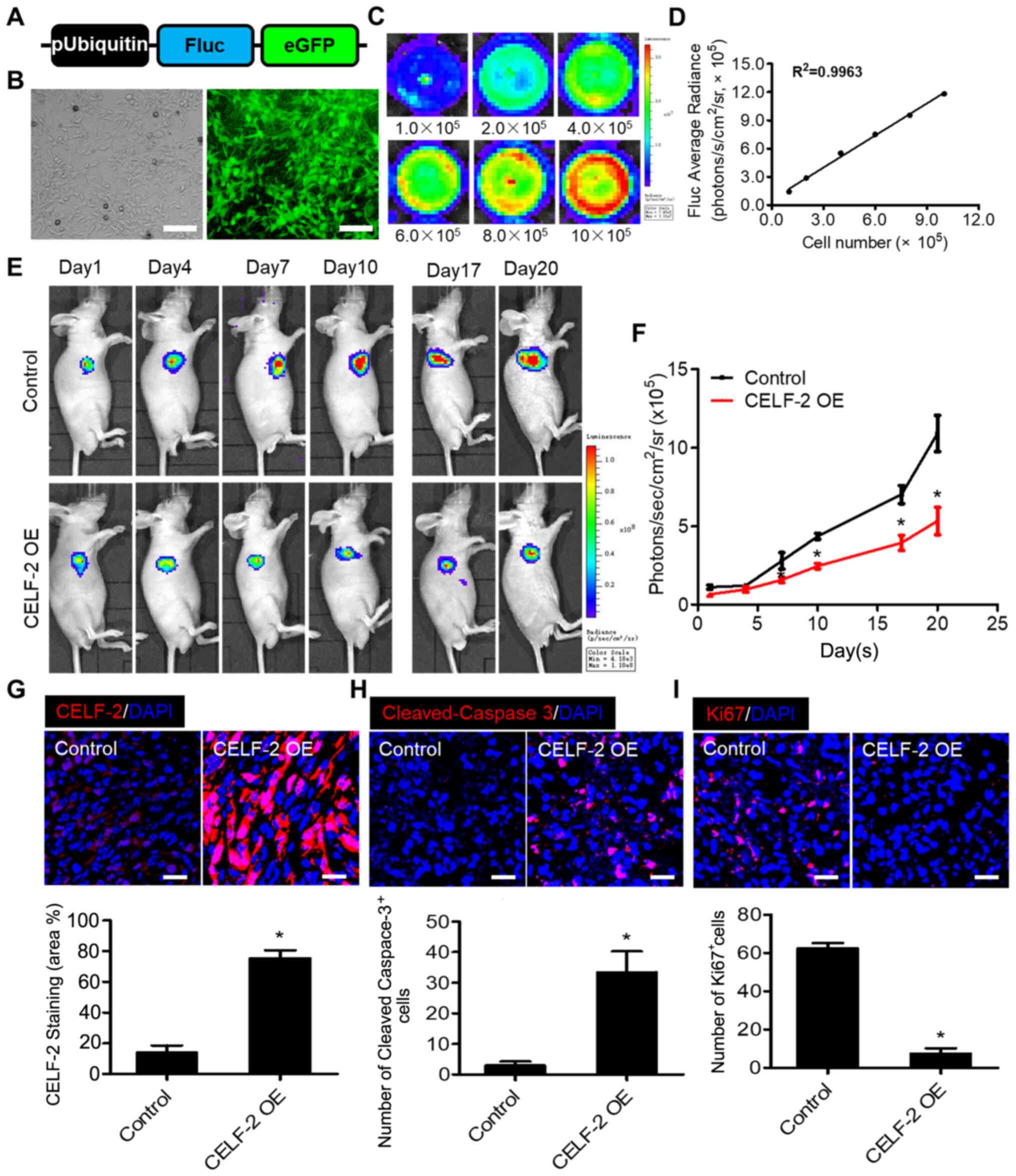
spinal cord glioma growth, Fluc imaging was performed to monitor
tumor progression. First, to monitor transplanted cells in
vivo, an imaging assay was developed using corresponding
reporter genes. Transduction of 0213SCG cells was performed with
double fusion (DF) (Fig. 4A).
Fluorescence microscopy images revealed that eGFP was robustly
expressed in 0213SCG cells (Fig.
4B). In addition, a strong association between Fluc activity
and 0213SCG cell number was revealed, suggesting that tumor growth
may be assessed in vivo by analyzing firefly signal
intensity (Fig. 4C and D). A spinal
cord glioma model was established in nu/nu nude mice by
subcutaneously injecting 1×105 0213SCG cells or
CELF-2-overexpressing 0213SCG cells labeled with DF reporter gene
into the right forelimb armpit. BLI of Fluc was performed to
monitor the development of tumors. Fluc signals were detected at
different time points (days 1, 4, 7, 10, 17 and 20). The results
indicated that the tumor growth was significantly attenuated in the
CELF-2 overexpression group compared with in the control group
(Fig. 4E and F). Compared with the
control group, immunofluorescence staining revealed that CELF-2 was
highly expressed in the tumors of the CELF-2 overexpression groups
(Fig. 4G). TUNEL staining assay
demonstrated a high number of apoptotic CELF-2-overexpressing
0213SCG cells on day 20 (Fig. S4A and
B). Furthermore, the results of cleaved-caspase-3 and Ki67
staining indicated that apoptosis was enhanced, whereas cell
proliferation was inhibited in the CELF-2-overexpressing 0213SCG
cell group (Fig. 4H and I). The
aforementioned results suggested that CELF-2 may partially
alleviate spinal cord glioma growth.
Discussion
Several miRNAs have been identified as regulators of
multiple biological processes in cancer (29). Herein, the differentially expressed
miRNAs (miR-106a-5p, miR-21, miR-10b, miR-155, miR-95-3p and
miR-615-3p) were screened in patients with spinal cord glioma by
RT-qPCR. The analysis revealed that miR-106a-5p expression was
upregulated in spinal cord glioma tissues. Previously studies have
suggested that miR-106a-5p may both promote and inhibit tumor
progression (21–25). Currently, the effect of miR-106a-5p
in spinal cord glioma has not been reported. Therefore, the present
study aimed to perform functional experiments on multiple aspects
of spinal cord glioma development, including proliferation,
migration, invasion and apoptosis. To establish an experimental
group, 0213SCG cells isolated from spinal cord glioma tissues were
transfected with miR-106a-5p inhibitor to knock down miR-106a-5p
expression. MTT assay revealed that transfection with miR-106a-5p
inhibitor alleviated the proliferation of 0213SCG cells. Since
tumor cell migration and invasion are key factors in tumor
metastasis (30), Transwell assays
were performed to investigate the effect of miR-106a-5p on these
processes. As expected, the number of migrating and invading
miR-106a-5p inhibitor-transfected cells was decreased compared with
the other groups. Apoptosis analysis revealed that the apoptosis
rate was significantly enhanced in the miR-106a-5p inhibitor
group.
Subsequently, the current study aimed to uncover the
mechanism of miR-106a-5p in spinal cord glioma. The TargetScan
online database was used to predict the targets of miR-106a-5p.
Among all predicted target mRNAs, CELF-2 was selected as a putative
target of miR-106a-5p, since a potential binding site for
miR-106a-5p was predicted in the 3′-UTR of CELF-2. Emerging
evidence has suggested that CELF-2 acts as a tumor suppressor
(26–28). Online miRNA target prediction tools
have predicted several potential miRNA targets (www.targetscan.org). However, the regulation of CELF-2
by miRNAs has been poorly studied. It has been reported that
miR-95-3p targets CELF-2 in glioma (31). Therefore, miR-95-3p downregulation
may inhibit cell proliferation and invasion and promote apoptosis
(31). Therefore, the present study
hypothesized that CELF-2 may also be associated with the occurrence
of spinal cord glioma. RT-qPCR and western blot analysis revealed a
negative association between CELF-2 and miR-106a-5p expression in
spinal cord glioma tissues. Additionally, CELF-2 may be directly
targeted by miR-106a-5p.
A previous study has demonstrated that CELF-2
hypermethylation is associated with shorter overall survival of
patients with breast cancer in the clinical setting, while CELF-2
restoration exerted an inhibitory effect on breast cancer growth
(32). Therefore, it has been
suggested that the epigenetic loss of CELF-2 may enhance breast
cancer growth and may be associated with worse outcomes in patients
with breast cancer (32). Therefore,
the current study hypothesized that CELF-2 may be a potential novel
therapeutic target for cancer. Herein, CELF-2 overexpression was
shown to attenuate the proliferation, migration and invasion of
spinal cord glioma cells in vitro. Furthermore, when
CELF-2-overexpressing spinal cord glioma cells were transplanted
into mice, BLI revealed that the proliferation of spinal cord
glioma cells was alleviated. Additionally, an increased number of
apoptotic cells was observed in CELF-2-overexpressing spinal cord
glioma tissues. The current findings further supported that CELF-2
may affect the proliferation of spinal cord glioma cells via
promoting apoptosis. However, the specific molecular mechanisms
underlying the effects of CELF-2 should be further
investigated.
Overall, the present study demonstrated that
miR-106a-5p expression was abnormally upregulated in spinal cord
glioma samples, while miR-106a-5p downregulation inhibited the
proliferation, migration and invasion, and promoted apoptosis of
spinal cord glioma cells via upregulating CELF-2. In addition, the
expression levels of miR-106a-5p and CELF-2 may be associated with
the clinical characteristics of patients, so there may be potential
applications in clinical practice, although this requires further
investigation. In summary, it is important to first find the common
characteristics of heterogeneous tumors, and secondly, verify this
common point in other samples one by one. Through the systematic
analysis of multiple samples, the goal of discovering universal
phenomena may be finally achieved.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fundamental
Research Funds for the Central Universities (grant no.
WK9110000126).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors contributions
HX and FW performed the experiments and data
analysis. HX and LW conceived and designed the study. HX and LW
assessed the raw data to ensure its legitimacy. WL reviewed and
edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of University of Science
and Technology of China (Hefei, China). Written informed consent
was obtained from all patients. The legal guardians of minors or
subjects restricted from participating in this study provided
informed consent on their behalf. All animal experimental
procedures were conducted in accordance with the institutional
guidelines of the University of Science and Technology of China for
The Care and Use of Laboratory Animals and conformed to the
National Institutes of Health Guide for Care and Use of Laboratory
Animals [approval no. 2019-N (A)-011].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ma CC, Xiong Z, Zhu GN, Wang C, Zong G,
Wang HL, Bian EB and Zhao B: Long non-coding RNA ATB promotes
glioma malignancy by negatively regulating miR-200a. J Exp Clin
Cancer Res. 35:902016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramaswamy V and Taylor MD: CAR T cells for
childhood diffuse midline gliomas. Nat Med. 24:534–535. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tobias A, Ahmed A, Moon KS and Lesniak MS:
The art of gene therapy for glioma: A review of the challenging
road to the bedside. J Neurol Neurosurg Psychiatry. 84:213–222.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zheng YJ, Liang TS, Wang J, Zhao JY, Yang
DK and Liu ZS: Silencing lncRNA LOC101928963 inhibits proliferation
and promotes apoptosis in spinal cord glioma cells by binding to
PMAIP1. Mol Ther Nucleic Acids. 18:485–495. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alvi MA, Ida CM, Paolini MA, Kerezoudis P,
Meyer J, Barr Fritcher EG, Goncalves S, Meyer FB, Bydon M and
Raghunathan A: Spinal cord high-grade infiltrating gliomas in
adults: Clinico-pathological and molecular evaluation. Mod Pathol.
32:1236–1243. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ropper AE, Zeng X, Haragopal H, Anderson
JE, Aljuboori Z, Han I, Abd-El-Barr M, Lee HJ, Sidman RL, Snyder
EY, et al: Targeted treatment of experimental spinal cord glioma
with dual gene-engineered human neural stem cells. Neurosurgery.
79:481–491. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
An T, Fan T, Zhang XQ, Liu YF, Huang J,
Liang C, Lv BH, Wang YQ, Zhao XG, Liu JX, et al: Comparison of
alterations in miRNA expression in matched tissue and blood samples
during spinal cord glioma progression. Sci Rep. 9:91692019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gebert LF and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wessels HH, Lebedeva S, Hirsekorn A,
Wurmus R, Akalin A, Mukherjee N and Ohler U: Global identification
of functional microRNA-mRNA interactions in Drosophila. Nat
Commun. 10:16262019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moradimotlagh A, Arefian E, Valojerdi RR,
Ghaemi S, Adegani FJ and Soleimani M: MicroRNA-129 inhibits glioma
cell growth by targeting CDK4, CDK6, and MDM2. Mol Ther Nucleic
Acids. 19:759–764. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu SJ, Hu HT, Li HL and Chang S: The role
of miRNAs in immune cell development, immune cell activation, and
tumor immunity: With a focus on macrophages and natural Killer
cells. Cells. 8:11402019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tang L, Chen HY, Hao NB, Tang B, Guo H,
Yong X, Dong H and Yang SM: microRNA inhibitors: Natural and
artificial sequestration of microRNA. Cancer Lett. 407:139–147.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abels ER, Maas SL, Nieland L, Wei Z, Cheah
PS, Tai E, Kolsteeg CJ, Dusoswa SA, Ting DT, Hickman S, et al:
Glioblastoma-associated microglia reprogramming is mediated by
functional transfer of extracellular miR-21. Cell Rep.
28:3105–3119.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ouyang H, Gore J, Deitz S and Korc M:
Erratum: microRNA-10b enhances pancreatic cancer cell invasion by
suppressing TIP30 expression and promoting EGF and TGF-β actions.
Oncogene. 36:4952. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ye J, Yao Y, Song Q, Li S, Hu Z, Yu Y, Hu
C, Da X, Li H, Chen Q and Wang QK: Up-regulation of miR-95-3p in
hepatocellular carcinoma promotes tumorigenesis by targeting p21
expression. Sci Rep. 6:340342016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Wang B, Shi Y, Xu C, Xiao HL, Ma
LN, Xu SL, Yang L, Wang QL, Dang WQ, et al: Oncogenic miR-20a and
miR-106a enhance the invasiveness of human glioma stem cells by
directly targeting TIMP-2. Oncogene. 34:1407–1419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan T, Ooi WF, Qamra A, Cheung A, Ma D,
Sundaram GM, Xu C, Xing M, Poon L, Wang J, et al: HoxC5 and
miR-615-3p target newly evolved genomic regions to repress hTERT
and inhibit tumorigenesis. Nat Commun. 9:1002018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bayraktar R and Van Roosbroeck K: miR-155
in cancer drug resistance and as target for miRNA-based
therapeutics. Cancer Metastasis Rev. 37:33–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miyoshi H, Blömer U, Takahashi M, Gage FH
and Verma IM: Development of a self-inactivating lentivirus vector.
J Virol. 72:8150–8157. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luan W, Zhou Z, Ni X, Xia Y, Wang J, Yan Y
and Xu B: Long non-coding RNA H19 promotes glucose metabolism and
cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J
Cancer Res Clin Oncol. 144:531–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He QY, Wang GC, Zhang H, Tong DK, Ding C,
Liu K, Ji F, Zhu X and Yang S: miR-106a-5p suppresses the
proliferation, migration, and invasion of osteosarcoma cells by
targeting HMGA2. DNA Cell Biol. 35:506–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pan YJ, Wei LL, Wu XJ, Huo FC, Mou J and
Pei DS: miR-106a-5p inhibits the cell migration and invasion of
renal cell carcinoma through targeting PAK5. Cell Death Dis.
8:e3155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li D, Wang Z, Chen Z, Lin L, Wang Y,
Sailike D, Luo K, Du G, Xiang X and Jiafu GD: MicroRNA-106a-5p
facilitates human glioblastoma cell proliferation and invasion by
targeting adenomatosis polyposis coli protein. Biochem Biophys Res
Commun. 481:245–250. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hoey C, Ray J, Jeon J, Huang X, Taeb S,
Ylanko J, Andrews DW, Boutros PC and Liu SK: miRNA-106a is a novel
regulator of radiation resistance through targeting LITAF and ATM
in prostate cancer. Mol Oncol. 12:1324–1341. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pencheva N and Tavazoie SF: Control of
metastatic progression by microRNA regulatory networks. Nat Cell
Biol. 15:546–554. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Liu L, Sun Y, Xue Y, Qu J, Pan S,
Li H, Qu H, Wang J and Zhang J: miR-615-3p promotes proliferation
and migration and inhibits apoptosis through its potential target
CELF2 in gastric cancer. Biomed Pharmacother. 101:406–413. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yeung YT, Fan S, Lu B, Yin S, Yang S, Nie
W, Wang M, Zhou L, Li T, Li X, et al: CELF2 suppresses non-small
cell lung carcinoma growth by inhibiting the PREX2-PTEN
interaction. Carcinogenesis. 41:377–389. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Madsen CD, Hooper S, Tozluoglu M,
Bruckbauer A, Fletcher G, Erler JT, Bates PA, Thompson B and Sahai
E: STRIPAK components determine mode of cancer cell migration and
metastasis. Nat Cell Biol. 17:68–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fan B, Jiao BH, Fan FS, Lu SK, Song J, Guo
CY, Yang JK and Yang L: Downregulation of miR-95-3p inhibits
proliferation, and invasion promoting apoptosis of glioma cells by
targeting CELF2. Int J Oncol. 47:1025–1033. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Piqué L, Martinez de Paz A, Piñeyro D,
Martínez-Cardús A, Castro de Moura M, Llinàs-Arias P, Setien F,
Gomez-Miragaya J, Gonzalez-Suarez E, Sigurdsson S, et al:
Epigenetic inactivation of the splicing RNA-binding protein CELF2
in human breast cancer. Oncogene. 38:7106–7112. 2019. View Article : Google Scholar
|