Introduction
Retinoblastoma (RB) is the most common primary
intraocular malignant tumor in children, with a constant incidence
worldwide of 1:15,000-1:20,000 live births (1). At present, the treatment of RB mainly
includes surgical enucleation, radiotherapy and chemotherapy
(1). Due to its limited sensitivity
to treatment, the prognosis of RB is often poor (1). RB is a disease caused by gene
mutations. It has been demonstrated that the mutation of the RB1
gene located on 13q14 plays a vital role in the development of RB.
The RB1 gene participates in tumor inhibition, maintenance of gene
stability and epigenetic inheritance. Therefore, unlimited
proliferation of cells occurs when the RB1 gene is mutated
(2).
Regarded as a catabolic process, autophagy is a
response to cell nutrient deficiency, environmental stress and
energy shortage, and plays an important role in clearing damaged
organelles and aggregation of denatured proteins, as well as in
recycling amino acids in the starvation state (3). Through various mechanisms, including
degradation of damaged intracellular organelles and protection of
DNA stability, autophagy maintains the normal function of cells and
ensures gene stability to inhibit the occurrence of tumors
(4,5). However, in tumor cells, such as in RB,
autophagy is the main resistance mechanism towards adverse factors,
including an external anoxic environment or drug therapy, leading
to the high invasion ability of tumors, an anti-anoxic environment
and multidrug resistance (6,7).
The present review describes the current research
progress on the association between autophagy and RB. The role and
regulatory mechanism of autophagy in the occurrence, progression
and treatment of RB are reported, and new strategies and targets
for the treatment of RB are provided.
Concept, classification, biological
characteristics and regulation of autophagy
Autophagy is associated with a variety of
pathological and physiological processes. Generally, according to
different mechanisms of action, autophagy is divided into three
types, namely large autophagy, small autophagy and
chaperon-mediated autophagy (8,9). Large
autophagy is what the present review calls autophagy. Under
particular conditions, such as lack of nutrients or energy, or
oxygen deficiency, autophagy is induced to degrade cytoplasmic
substances into metabolites in cells, thereby maintaining the
normal survival of cells (10). In a
previous study, the autophagy level was significantly decreased in
mouse acinar cells lacking autophagy-related gene 7 (ATG7)
(11), and further experimentation
conclusively showed that the protein synthesis ability of the cells
was markedly decreased, accompanied by a deficiency in the rough
endoplasmic reticulum, leading to progressive loss of pancreatic
function, development of pancreatic inflammation and cancer
recurrence (11). When ATG7 is
specifically knocked out in mouse nerve cells, abnormal protein
accumulation and motor function defects appear (12). Autophagy plays an important role in
growth and development, and it is the main form of innate immunity
in cells, since it plays a role in the defense against the invasion
of extracellular microorganisms (13). Furthermore, autophagy is an important
mechanism enabling immune cells to survive and serve their roles in
tumor microenvironments, which are characterized by poor metabolic
conditions and high levels of immunosuppression (14). Autophagy interacts with apoptosis,
promoting the mitosis and specific degradation of pro-apoptotic
proteins, thereby inhibiting apoptosis. By contrast, after
apoptosis is activated, necessary autophagy-related proteins (ARPs)
are inactivated or autophagy proteins are converted into apoptotic
proteins under certain conditions (15).
Autophagy includes five stages: Induction,
nucleation, expansion and containment, fusion and degradation.
Mammalian target of rapamycin receptor complex 1 (mTORC1) inhibits
autophagy (16). The initiation of
cell autophagy requires a variety of mTORC1 inhibition pathways to
be activated to reverse the upstream inhibition of autophagy
(17,18). The class III phosphatidylinositol
3-kinase (PI3K) complex constituting of ATG proteins plays an
important role in the recruitment and positioning of ATG18 and
ATG2, which in turn recruits ATG8, ATG9 and ATG12 to form the
pre-autophagosomal structure (19,20).
This complex process causes the cargo to gather and nucleate,
laying the foundation for the subsequent expansion of the membrane,
in which two connected systems participate. The connection between
ATG5 and ATG12 requires the participation of the ubiquitin sample
connection system (21). This
connection product is the complex required for the binding between
phosphoethanolamine and microtubule-associated protein 1 light
chain 3 (LC3). Finally, the mature autophagosome membrane is fused
with the lysosome membrane structure to release the encapsulated
cargo into the lysosome (22).
Regulation at the post-translational level and the
regulation of transcription factors are involved in the regulation
of autophagy. In the absence of amino acids, activation of general
control nonderepressible (Gcn)2 (a kinase) and Gcn4 (a
transcription factor) promotes ATG gene expression in yeast cells
(23). In mammals, the inactivation
of mTORC1 promotes the dephosphorylation and translocation of
transcription factor EB, and the transcription of various genes
associated with the autophagy process (24).
In the absence of energy, autophagy is activated via
several pathways, including the AMP-activated protein kinase A
pathway, which directly triggers autophagy (25). In addition, it has been found that
spermidine inhibits autophagy by inhibiting various acetyl
transferases (Fig. 1) (26).
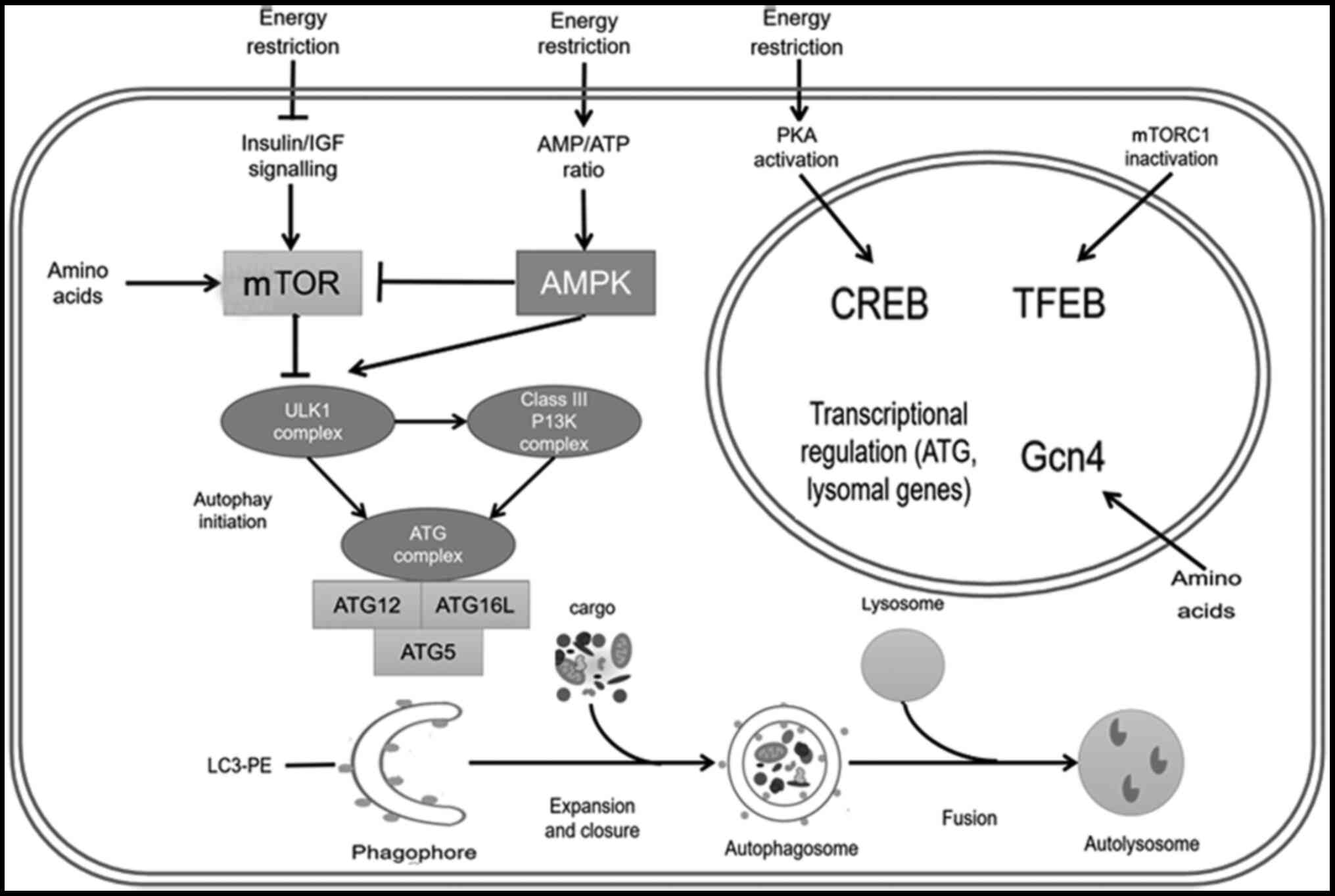 | Figure 1.Process and regulation of autophagy.
Energy restriction inhibits mTOR and promotes autophagy. Under
energy constraints, the increased AMP/ATP ratio activates the AMPK
pathway, which either directly promotes autophagy or promotes
autophagy by inhibiting the mTOR pathway. Energy restriction also
activates CREB protein through the protein kinase A signaling
pathway and activates transcription factor EB by inhibiting mTORC1.
These autophagy transcription factors will promote the
transcription of autophagy-related proteins and lysosomal proteins.
Amino acid deficiency activates mTOR and Gcn4 signaling pathways,
and promotes autophagy. mTOR inhibition and AMPK activation
promotes the activation of the ULK1 complex upstream of autophagy.
The ULK1 complex promotes the formation of the PI3K and ATG
complexes, and ultimately promotes the occurrence of autophagy.
After the initiation of autophagy, through a series of cascade
reactions, the process of expansion and closure occurs, followed by
consecutive encircle and fusion steps, which finally leads to the
formation of the autolysosome. ULK1, Unc-51-like autophagy
activating kinase 1; AMPK, AMP-activated protein kinase; CREB,
cAMP-response element binding; PKA, protein kinase A; mTORC1,
mammalian target of rapamycin receptor complex 1; Gcn4, general
control nonderepressible 4; ATG, autophagy-related gene; TFEB,
transcription factor EB; PE, phosphoethanolamine; LC3,
microtubule-associated protein 1 light chain 3. |
Autophagy and tumors
Autophagy prevents genes from being altered by
reactive oxygen species (ROS) and mitochondria (4), and inhibits the expression of
anti-apoptotic proteins to suppress tumorigenesis (27). Autophagy can also restrain
tumorigenesis by repressing mitochondrial swelling, p62
accumulation, oxidative stress and genome damage, particularly in
mouse liver tumors (5). Nuclear
factor-erythroid 2-related factor 2 (NRF2), which can be activated
by p62, is mainly responsible for the transcription of
anti-oxidation defensive genes, and for facilitating cell survival
and tumorigenesis (28). A previous
study found that autophagy deficiency by ATG7 deletion promoted p62
accumulation and NRF2 activation. In mouse hepatocarcinoma with
autophagy deficiency, knockdown of p62 partially suppressed tumor
progression (29), while knockdown
of Parkinson disease 2 (a E3 ubiquitin ligase that is responsible
for coding the E3 ligase parkin, a multifaceted protein involved in
the signaling pathway of mitochondrial autophagy) promoted liver
cancer (28,30). Additionally, systemic ATG5
elimination and ATG7 knockdown contribute to the formation of
hepatocarcinoma, particularly in the liver of mice (29). In summary, autophagy is associated
with the malignant transformation of hepatocytes in mice, and it
has been demonstrated to be important for stabilizing cellular
homeostasis in tumor suppression.
As a double-edged sword, autophagy has promotive
effects on tumorigenesis and tumor growth. It has been found that
autophagy is associated with high viability and resistance to
antitumor drugs. ROS stimulates autophagy through the Ras (a gene
involved in the regulation of various cell signaling pathways)/Raf1
(a serine/threonine kinase)/MEK1/2/ERK1/2 pathway, while it
restrains autophagy via activation of the PI3K and AKT (a protein
kinase)/mTOR pathway (31,32). Based on the role of autophagy in the
regulation of cell genetics and nutrient metabolism, enhanced
autophagy may relieve hypoxia and nutrient deficiency during the
overgrowth of cancer cells (33). A
previous study showed that both ROS and BRAF simultaneously
stimulated tumor growth and improved the levels of autophagy
(28). In cancer cells with KRAS
activation, inhibition of autophagy triggers mitochondrial
oxidative phosphorylation and increases ROS production, thus
exerting antitumor efficacy (34,35).
Deficiency of the autophagy-suppressor gene ATG7 restricts tumor
progression and promotes the accumulation of mitochondrial
dysfunction through BRAF activation (36). A previous study demonstrated that an
autophagy-deficient non-small cell lung cancer (NSCLC)-derived cell
line in mice with p53 deficiency was highly dependent on glutamine
from the environment for the normal functioning of the mitochondria
to support the metabolism and survival in starvation (37). Resistance to vemurafenib, a tumor
inhibitor, is linked with active autophagy in patients with
melanoma, while the resistance of melanoma to vemurafenib can be
reversed by hydroxychloroquine, which was discovered in a drug
stimulation trial in a melanoma cell line (38). Furthermore, clinical trials
evaluating the potential of autophagy inhibition protocols on brain
cancer have indicated that the combined use of chloroquine and
vemurafenib prevents the formation of drug resistance (39). Furthermore, autophagy has been found
to maintain the intracellular redox balance of tumor stem cells
(40), preserve the formation and
function of tumor stem cells (41),
promote vascular hyperplasia via activation of the receptors of
vascular endothelial growth factor (42) and participate in the antigen
presentation process of T helper cells (14). These findings indicate that autophagy
is beneficial for the survival of tumors, and autophagic inhibition
blocks the development of tumors (Fig.
2).
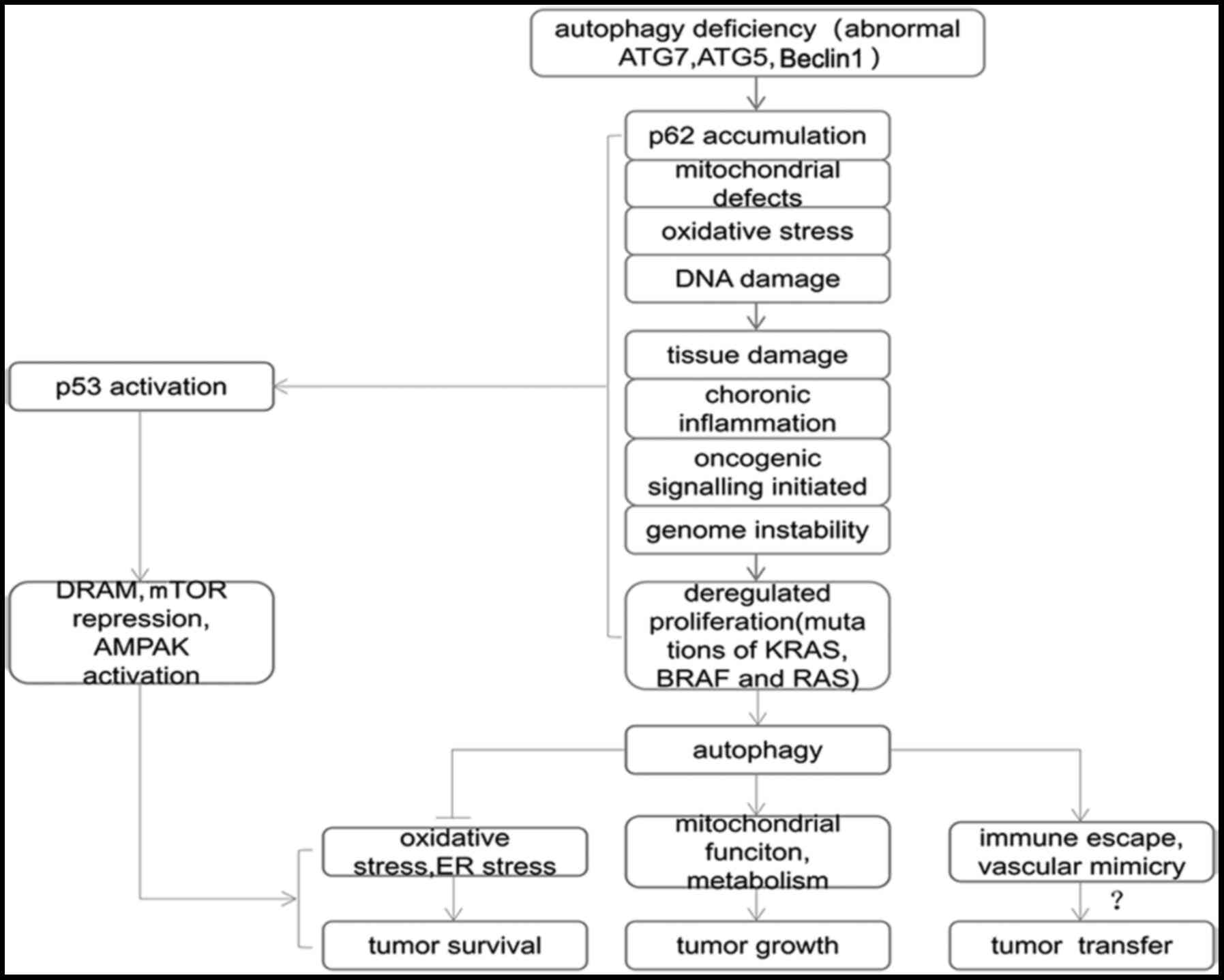 | Figure 2.Association between autophagy and
tumors. Gene abnormalities lead to autophagy defects in cells by
inducing p62 aggregation, mitochondrial defects, oxidative stress,
DNA damage and activation of other pathways', which cause tissue
damage, chronic inflammation, tumor gene signaling activation and
decreased gene stability, which are closely associated with the
occurrence of tumors. DNA damage, tumor gene activation and other
conditions promote the activation of anti-p53 and other related
genes to induce autophagy. Autophagy is used by tumors to promote
tumor survival, and may even be a potential mechanism of tumor
metastasis. ATG, autophagy-related gene; mTOR, mammalian target of
rapamycin; AMPK, AMP-activated protein kinase; ER, endoplasmic
reticulum. |
Association between autophagy and RB
Autophagy and the transformation and
occurrence of RB
Autophagy is involved in the malignant
transformation and gene mutation process of tumors, and the lack of
autophagy will lead to the instability of tumor genes, thus
promoting the occurrence of tumors (43). RB originates from normal cells, and
has been described as a multistep gene variation process of M1,
M2or M3 to Mn, with the RB1 allele variation belonging to the M1 to
M2 variation process (44,45). It has been clinically found that the
RB lesions of certain patients contain a benign tissue region
called a retinocytoma (RC). Furthermore, the cell regions of RC
have higher genetic stability than those of RB (46). RC may be an intermediate transition
state of RB or RB degradation, which provides a novel idea for the
transformation and occurrence of RB. As a benign lesion of RB, RC
has a stronger tolerance to chemotherapy, which was initially
speculated to be caused by the different roles of autophagy in RC
and RB. A study on the role of autophagy in RC and RB indicated
that the microtubule-associated protein LC3B was present at a high
level in the RC region (RCR) and in retinal cells, while its levels
were markedly lower in the adjacent undifferentiated RB region
(47). A large number of autophagic
vacuoles were found in RCR cells by electron microscopy, but not in
the undifferentiated RB region, which confirmed that autophagy is a
more common phenomenon in RCR (47).
Similar to RB, RC has the same mutation in the RB1 allele (48). Mutation of the RB1 gene leads to the
inhibition of the E2 transcription factor. When the RB1 gene was
transduced into human cancer cells deficient in RB protein, the
levels of autophagy were increased (49). A previous study reported that
silencing RB tumor repression proteins inhibited autophagy induced
by etoposide, leading to increased double-stranded DNA damage and
tumor cell death (50). RB1 gene
mutation reduces the level of autophagy by changing the expression
of RB tumor repression proteins, thus inducing cell gene damage or
cell death, which suggests that RC is prone to further M2 to Mn
gene mutations, which may be a benign intermediate transition form
of RB. Therefore, the decrease in autophagy levels may be one of
the critical mechanisms of RB formation. Due to the rarity of RC,
the majority of ophthalmologists do not recommend enucleation of
the eyeball for patients with RC, and experimental interventional
studies on RC are still relatively scarce. Further studies on the
role of RC in the regulation of autophagy may provide important
insights into understanding the pathogenesis of RB.
Autophagy and the invasion and
metastasis of RB
A previous study suggested a possible link between
autophagy and the invasion and metastasis of RB (51). It is recognized that the
sesquiterpenoid nootkatone induces the production of endogenous ROS
in RB to induce cytotoxicity and inhibit the migration of tumor
cells, while it promotes autophagy by promoting the expression of
ARPs in RB cells (52). Zhang et
al (53) found that p62 and LC3B
were highly expressed in the majority of RB tumor cells, and that
p53 was expressed in the cytoplasm of certain RB tumor cells. It
was also found that high protein levels of p62 and LC3B were
significantly associated with late-stage TNM and optic nerve
invasion in RB, while low levels of p53 were significantly
associated with calcification of RB tumors and optic nerve invasion
(53). It is known that p53 is an
autophagy inhibitor and LC3B is a marker of autophagy levels; thus,
the decreased level of cytoplasmic p53 and increased level of LC3B
can reflect high autophagy levels in RB cells. p62 is degraded by
autophagy, and inhibition of autophagy leads to the aggregation of
p62, which is not caused by the abnormal expression of p62; thus,
p62 can indirectly reflect the level of autophagy (54). Consequently, it was hypothesized that
high levels of autophagy may be one of the mechanisms of RB
invasion and metastasis. However, the molecular mechanism of
autophagy involved in tumor metastasis and invasion is unclear, and
further research is needed.
Autophagy and the resistance and
survival of RB
Previous studies have shown that autophagy mediates
drug resistance and has a certain protective effect in RB, which
indicates the close association between drug resistance and
autophagy (55,56). It was found that microRNA
(miRNA/miR)-34a regulated apoptosis and autophagy in RB cells by
targeting high mobility group box 1 (HMGB1), and it was
demonstrated that miR-34a inhibited the expression of HMGB1, thus
inhibiting autophagy and inducing apoptosis (57). Further results showed that inhibition
of autophagy enhanced chemotherapy treatment for DNA damage in RB
cells. X-inactive specific transcript (XIST), a 17-kb long
non-coding RNA (lncRNA) located on the X chromosome, accelerates
tumor progression in certain types of human cancer. Previous
studies have demonstrated that the expression of XIST increases in
RB tissues and cell lines, and that silencing XIST weakens RB
proliferation and autophagy, and enhances the sensitivity to
vincristine (VCR) (58,59). Further studies found that XIST
sponged miR-204-5p, and that the promoting effect of XIST on RB
proliferation and autophagy was reversed by miR-204-5p (59). Thus, it was speculated that decreased
miR-204-5p in RB may be associated with drug resistance and
proliferation in RB. LINC00152 is increased in RB, and enhances the
invasiveness of RB and its resistance to carboplatin and Adriamycin
through the sponging of miR-613; however, silencing LINC00152
inhibits the proliferation, invasiveness and autophagy of RB, and
then significantly promotes apoptosis (60,61). The
increased level of RB autophagy may be one of the mechanisms of
resistance to apoptosis and drug resistance. Huang et al
(61) found that
metastasis-associated lung adenocarcinoma transcript 1, as a
lncRNA, promoted the autophagy of RB cells by targeting miR-124,
thus providing a theoretical basis for drug resistance induced by
autophagy. Liu et al (62)
applied the autophagy blocker 3-methyladenine (3-MA) to explore the
effect of autophagy on RB resistance, and found that the inhibition
rate of cisplatin at different concentrations was higher than that
observed in tumor cells treated with cisplatin alone, and that the
transcription level of drug-resistant genes in cisplatin combined
with autophagy inhibitor 3-MA group was markedly downregulated
(62). CD24 is a glycosyl
phosphatidyl inositol-anchored protein highly expressed in RB
tissues and cell lines, and studies have shown that it activates
autophagy through the PTEN/AKT/mTORC1 signaling pathway, thereby
inactivating the sensitivity of RB to VCR (63). The aforementioned studies suggest
that autophagy is a crucial participant in the drug resistance
mechanism of RB.
Autophagy is not only associated with chemotherapy
resistance in RB, but it may also be an important mechanism for RB
survival. Under normal conditions, the expression levels of hypoxia
inducible factor-1 (HIF-1) and miR-320 in RB cells are increased
compared with those in normal cells. A previous study showed that
HIF-1α was a downstream target of miR-320, and that decreasing
miR-320 or HIF-1α expression inhibited autophagy in RB cells. LC3
aggregation was significantly increased under hypoxia, indicating a
high level of autophagy, while inhibition of miR-320 or HIF-1
caused a significant decrease in LC3 aggregation (64). Therefore, it was speculated that
increased expression of miR-320 promotes the expression of HIF-1,
thereby regulating the expression of ARPs and enhancing RB
autophagy. These results suggested that autophagy may be a
protective mechanism for RB to promote its survival in the hypoxic
environment of the center ischemic region of the RB. Studies have
found that miR-512-3p promotes RB cell apoptosis induced by
endoplasmic reticulum stress; however, this process is associated
with the inhibition of autophagy (65). Additionally, it was found that
inhibiting autophagy and inducing cytotoxicity in RB cells promoted
their apoptosis when using a suicide gene therapy with herpes
simplex virus type 1 thymidine kinase/ganciclovir (66).
The present review has so far presented a summary of
the various protective effects of autophagy in RB at the gene and
protein levels. Autophagy can also regulate the intracellular
environment of RB, which allocates available materials for
decomposition and utilization under conditions of starvation and
lack of nutrients, and also helps to maintain the homeostasis of
the cytoplasm against stress caused by adverse external
environments. To date, several studies have reported conflicting
results. Ginsenoside Rh2 inhibited RB proliferation but promoted
autophagy and apoptosis by downregulating miR-638, while
methylbutanol led to potential anti-RB effects by inducing
autophagy and cell cycle arrest, and inhibiting the PI3K/mTOR/Akt
signaling pathway (67,68). In these studies, the antitumor effect
of anticancer drugs or substances may have been accompanied by an
increase in the autophagy level, but such studies could not
demonstrate whether an elevated level of autophagy is associated
with RB repression. On the contrary, the aforementioned drugs with
anti-RB effects may induce autophagy and lead to important drug
resistance, similar to numerous anti-RB drugs such as etoposide and
cisplatin. A number of studies reached the opposite conclusion, and
found that dimethyldibutanol 2-methyl-2-butanol (MBT) led to RB
cell apoptosis, while the use of autophagy inhibitors blocked the
action of this drug (69). These
results indicate that the anti-RB effect of MBT may be associated
with an increase in the autophagy level, while the specific
mechanism remains unknown. Therefore, further studies on the
association between autophagy and RB will help us to understand the
mechanism of drug resistance in RB (Fig.
3).
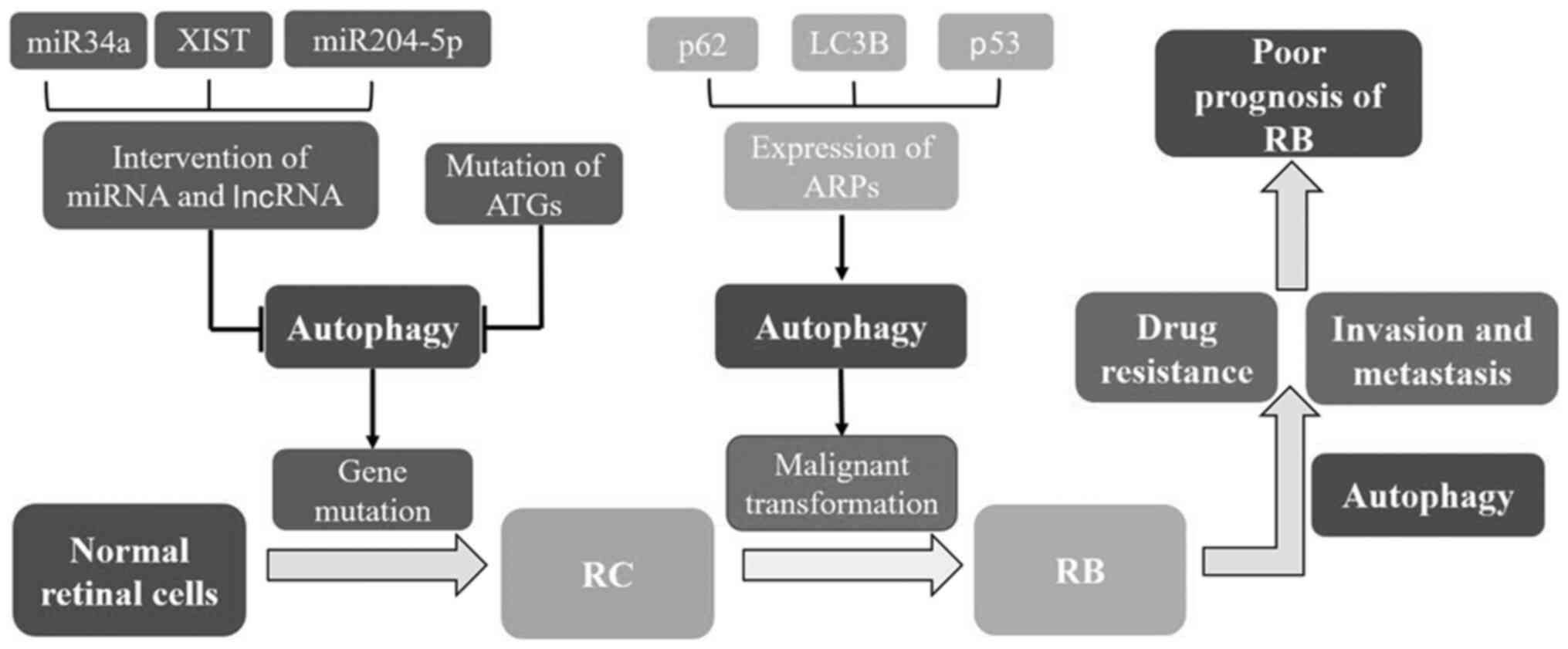 | Figure 3.Association between autophagy and RB.
Autophagy plays an important role in the occurrence and development
of RB via the regulation of various factors such as
autophagy-related genes, proteins, various microRNAs and long
non-coding RNAs. Inhibition of autophagy promotes gene damage and
mutation, thus inducing the formation of retinocytoma (an
intermediate transition form). Further genetic variations will lead
to malignant transformation and ultimately RB formation. Autophagy
is utilized by RB as one of the important mechanisms for drug
resistance and invasion, which may be the reason for the poor
prognosis of RB. RB, retinoblastoma; RC, retinocytoma; miR,
microRNA; XIST, X-inactive specific transcript; lncRNA, long
non-coding RNA; ATG, autophagy-related gene; ARP, autophagy-related
protein; LC3, microtubule-associated protein 1 light chain 3. |
Review and summary
Autophagy, a catabolic process conserved by
evolution, plays a vital role in maintaining the homeostasis of the
internal and external environments of the cell, and in enhancing
the adaptability of the cell to the external environment. The
present study reviewed relevant studies on the association between
autophagy and RB, and concluded that autophagy plays a
‘housekeeping’ role in normal, RB and other tumor cells by
protecting the cells, promoting the cell fight against external
adverse stimuli and improving cell viability. Autophagy degrades
damaged intracellular organelles in time and protects DNA stability
to maintain normal cell functions in normal cells. Contrary to the
situation in normal cells, in tumor cells, autophagy becomes the
main mechanism for RB cells to fight against external adverse
factors, leading to drug resistance, anti-hypoxia and high tumor
invasiveness of these cells. A marked decrease in autophagy in both
normal and RB cells has serious consequences. Decreased levels of
autophagy lead to apoptosis or promotion of cancer in normal cells,
whereas in RB or other tumor cells, they lead to cytotoxic damage
and decrease the drug resistance of tumor cells, eventually leading
to tumor cell death. Therefore, inhibition of autophagy in RB cells
may be an effective target for treatment or may serve as an adjunct
therapy to enhance the efficacy of anticancer drugs. The method to
specifically target RB tissues or cells without significantly
affecting autophagy in normal cells needs to be clarified.
Numerous studies have found that lncRNA and miRNA
have significant regulatory effects on autophagy in RB, and various
lncRNAs and miRNAs exert significant anti-RB effects by inhibiting
autophagy. In addition, the expression of certain lncRNAs and
miRNAs in RB showed obvious specificity. These findings suggest
that gene therapy targeting lncRNA and miRNA may become a new
effective method for RB therapy by regulating autophagy. However,
the molecular mechanism of the interaction between autophagy and RB
is still unknown, and further research will provide more reliable
theories and novel ideas for the clinical treatment of RB.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 82060250),
the Guangxi Science and Technology Plan Project (grant no.
AD20238035), the Open Project of Guangxi Key Laboratory of Brain
and Cognitive Neuroscience (grant no. GKLBCN-20190105-02) and the
Guangxi Zhuang Autonomous Region Students' Innovation and
Entrepreneurship Training Program of China (grant no.
202010601050).
Availability of data and materials
Not applicable.
Authors' contributions
TW and MF wrote the manuscript. ZW, XG and SZ
revised it critically for important intellectual content and gave
important advice. All authors read and approved the manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fabian ID, Onadim Z, Karaa E, Duncan C,
Chowdhury T, Scheimberg I, Ohnuma SI, Reddy MA and Sagoo MS: The
management of retinoblastoma. Oncogene. 37:1551–1560. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mallipatna A, Marino M and Singh AD:
Genetics of Retinoblastoma. Asia Pac J Ophthalmol (Phila).
5:260–264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Doherty J and Baehrecke EH: Life, death
and autophagy. Nat Cell Biol. 20:1110–1117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartsch K, Knittler K, Borowski C, Rudnik
S, Damme M, Aden K, Spehlmann ME, Frey N, Saftig P, Chalaris A, et
al: Absence of RNase H2 triggers generation of immunogenic
micronuclei removed by autophagy. Hum Mol Genet. 26:3960–3972.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takamura A, Komatsu M, Hara T, Sakamoto A,
Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K and Mizushima N:
Autophagy-deficient mice develop multiple liver tumors. Genes Dev.
25:795–800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kimmelman AC and White E: Autophagy and
tumor metabolism. Cell Metab. 25:1037–1043. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Poillet-Perez L and White E: Role of tumor
and host autophagy in cancer metabolism. Genes Dev. 33:610–619.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eskelinen EL: Autophagy: Supporting
cellular and organismal homeostasis by self-eating. Int J Biochem
Cell Biol. 111:1–10. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou S, Chen X, Xue R, Zhou Q, Hu P,
Ouyang X, Dai T, Zhu W and Tian S: Autophagy is involved in the
pathogenesis of experimental autoimmune neuritis in rats.
Neuroreport. 27:337–344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Parzych KR and Klionsky DJ: An overview of
autophagy: Morphology, mechanism, and regulation. Antioxid Redox
Signal. 20:460–473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Antonucci L, Fagman JB, Kim JY, Todoric J,
Gukovsky I, Mackey M, Ellisman MH and Karin M: Basal autophagy
maintains pancreatic acinar cell homeostasis and protein synthesis
and prevents ER stress. Proc Natl Acad Sci USA. 112:E6166–E6174.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Honda S, Arakawa S, Yamaguchi H, Torii S,
Tajima Sakurai H, Tsujioka M, Murohashi M and Shimizu S:
Association between Atg5-independent alternative autophagy and
neurodegenerative diseases. J Mol Biol. 432:2622–2632. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Keller MD, Torres VJ and Cadwell K:
Autophagy and microbial pathogenesis. Cell Death Differ.
27:872–886. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rybstein MD, Bravo-San Pedro JM, Kroemer G
and Galluzzi L: The autophagic network and cancer. Nat Cell Biol.
20:243–251. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mariño G, Niso-Santano M, Baehrecke EH and
Kroemer G: Self-consumption: The interplay of autophagy and
apoptosis. Nat Rev Mol Cell Biol. 15:81–94. 2014. View Article : Google Scholar
|
|
16
|
Laplante M and Sabatini DM: mTOR signaling
in growth control and disease. Cell. 149:274–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nazio F, Strappazzon F, Antonioli M,
Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J,
Piacentini M, Fimia GM, et al: mTOR inhibits autophagy by
controlling ULK1 ubiquitylation, self-association and function
through AMBRA1 and TRAF6. Nat Cell Biol. 15:406–416. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Z, Tian X, Ji X, Wang J, Chen H, Wang D
and Zhang X: ULK1-ATG13 and their mitotic phospho-regulation by
CDK1 connect autophagy to cell cycle. PLoS Biol. 18:e30002882020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Araki Y, Ku WC, Akioka M, May AI, Hayashi
Y, Arisaka F, Ishihama Y and Ohsumi Y: Atg38 is required for
autophagy-specific phosphatidylinositol 3-kinase complex integrity.
J Cell Biol. 203:299–313. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kotani T, Kirisako H, Koizumi M, Ohsumi Y
and Nakatogawa H: The Atg2-Atg18 complex tethers pre-autophagosomal
membranes to the endoplasmic reticulum for autophagosome formation.
Proc Natl Acad Sci USA. 115:10363–10368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hanada T, Noda NN, Satomi Y, Ichimura Y,
Fujioka Y, Takao T, Inagaki F and Ohsumi Y: The Atg12-Atg5
conjugate has a novel E3-like activity for protein lipidation in
autophagy. J Biol Chem. 282:37298–37302. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo H, Chitiprolu M, Roncevic L, Javalet
C, Hemming FJ, Trung MT, Meng L, Latreille E, Tanese de Souza C,
McCulloch D, et al: Atg5 Disassociates the V1V0-ATPase to Promote
exosome production and tumor metastasis independent of canonical
macroautophagy. Dev Cell. 43:716–730.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
B'Chir W, Maurin AC, Carraro V, Averous J,
Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P and Bruhat
A: The eIF2α/ATF4 pathway is essential for stress-induced autophagy
gene expression. Nucleic Acids Res. 41:7683–7699. 2013. View Article : Google Scholar
|
|
24
|
Settembre C, Di Malta C, Polito VA, Garcia
Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D,
Colella P, et al: TFEB links autophagy to lysosomal biogenesis.
Science. 332:1429–1433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Russell RC, Yuan HX and Guan KL: Autophagy
regulation by nutrient signaling. Cell Res. 24:42–57. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pietrocola F, Lachkar S, Enot DP,
Niso-Santano M, Bravo-San Pedro JM, Sica V, Izzo V, Maiuri MC,
Madeo F, Mariño G, et al: Spermidine induces autophagy by
inhibiting the acetyltransferase EP300. Cell Death Differ.
22:509–516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma
K, Li X, Wang L, Wang J, Zhang H, et al: Autophagy regulates
chromatin ubiquitination in DNA damage response through elimination
of SQSTM1/p62. Mol Cell. 63:34–48. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang T, Song X, Yang Y, Wan X, Alvarez
AA, Sastry N, Feng H, Hu B and Cheng SY: Autophagy and Hallmarks of
Cancer. Crit Rev Oncog. 23:247–267. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takamura A, Komatsu M, Hara T, Sakamoto A,
Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K and Mizushima N:
Autophagy-deficient mice develop multiple liver tumors. Genes Dev.
25:795–800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Poulogiannis G, McIntyre RE, Dimitriadi M,
Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams
DJ, et al: PARK2 deletions occur frequently in sporadic colorectal
cancer and accelerate adenoma development in Apc mutant mice. Proc
Natl Acad Sci USA. 107:15145–15150. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li L, Ishdorj G and Gibson SB: Reactive
oxygen species regulation of autophagy in cancer: Implications for
cancer treatment. Free Radic Biol Med. 53:1399–1410. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pattingre S, Bauvy C and Codogno P: Amino
acids interfere with the ERK1/2-dependent control of macroautophagy
by controlling the activation of Raf-1 in human colon cancer HT-29
cells. J Biol Chem. 278:16667–16674. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
White E: The role for autophagy in cancer.
J Clin Invest. 125:42–46. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang S, Wang X, Contino G, Liesa M, Sahin
E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, et al:
Pancreatic cancers require autophagy for tumor growth. Genes Dev.
25:717–729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo JY, Chen HY, Mathew R, Fan J,
Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM,
Karantza V, et al: Activated Ras requires autophagy to maintain
oxidative metabolism and tumorigenesis. Genes Dev. 25:460–470.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Strohecker AM and White E: Autophagy
promotes BrafV600E-driven lung tumorigenesis by preserving
mitochondrial metabolism. Autophagy. 10:384–385. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo JY, Karsli-Uzunbas G, Mathew R, Aisner
SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et
al: Autophagy suppresses progression of K-ras-induced lung tumors
to oncocytomas and maintains lipid homeostasis. Genes Dev.
27:1447–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ma XH, Piao SF, Dey S, McAfee Q,
Karakousis G, Villanueva J, Hart LS, Levi S, Hu J, Zhang G, et al:
Targeting ER stress-induced autophagy overcomes BRAF inhibitor
resistance in melanoma. J Clin Invest. 124:1406–1417. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Levy JM, Thompson JC, Griesinger AM, Amani
V, Donson AM, Birks DK, Morgan MJ, Mirsky DM, Handler MH, Foreman
NK, et al: Autophagy inhibition improves chemosensitivity in
BRAF(V600E) brain tumors. Cancer Discov. 4:773–780. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Perera RM, Stoykova S, Nicolay BN, Ross
KN, Fitamant J, Boukhali M, Lengrand J, Deshpande V, Selig MK,
Ferrone CR, et al: Transcriptional control of autophagy-lysosome
function drives pancreatic cancer metabolism. Nature. 524:361–365.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu K, Lee J, Kim JY, Wang L, Tian Y, Chan
ST, Cho C, Machida K, Chen D and Ou JJ: Mitophagy controls the
activities of tumor suppressor p53 to regulate hepatic cancer stem
cells. Mol Cell. 68:281–292.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu HB, Yang S, Weng HY, Chen Q, Zhao XL,
Fu WJ, Niu Q, Ping YF, Wang JM, Zhang X, et al: Autophagy-induced
KDR/VEGFR-2 activation promotes the formation of vasculogenic
mimicry by glioma stem cells. Autophagy. 13:1528–1542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Amaravadi RK, Kimmelman AC and Debnath J:
Targeting autophagy in cancer: Recent advances and future
directions. Cancer Discov. 9:1167–1181. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Soliman SE, Racher H, Zhang C, MacDonald H
and Gallie BL: Genetics and Molecular Diagnostics in Retinoblastoma
- An Update. Asia Pac J Ophthalmol (Phila). 6:197–207.
2017.PubMed/NCBI
|
|
45
|
Laurie NA, Donovan SL, Shih CS, Zhang J,
Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al:
Inactivation of the p53 pathway in retinoblastoma. Nature.
444:61–66. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dimaras H, Khetan V, Halliday W, Orlic M,
Prigoda NL, Piovesan B, Marrano P, Corson TW, Eagle RC Jr, Squire
JA, et al: Loss of RB1 induces non-proliferative retinoma:
Increasing genomic instability correlates with progression to
retinoblastoma. Hum Mol Genet. 17:1363–1372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu Y, Wan ST, Zhang P, Zhang WX, Zheng
JL, Lin JX and Li YP: Expression levels of autophagy related
proteins and their prognostic significance in retinocytoma and
retinoblastoma. Int J Ophthalmol. 7:594–601. 2014.PubMed/NCBI
|
|
48
|
Indovina P, Acquaviva A, De Falco G, Rizzo
V, Onnis A, Luzzi A, Giorgi F, Hadjistilianou T, Toti P, Tomei V,
et al: Downregulation and aberrant promoter methylation of
p16INK4A: A possible novel heritable susceptibility marker to
retinoblastoma. J Cell Physiol. 223:143–150. 2010.PubMed/NCBI
|
|
49
|
Jiang H, Martin V, Gomez-Manzano C,
Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N and
Fueyo J: The RB-E2F1 pathway regulates autophagy. Cancer Res.
70:7882–7893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Biasoli D, Kahn SA, Cornélio TA, Furtado
M, Campanati L, Chneiweiss H, Moura-Neto V and Borges HL:
Retinoblastoma protein regulates the crosstalk between autophagy
and apoptosis, and favors glioblastoma resistance to etoposide.
Cell Death Dis. 4:e7672013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen N and Karantza-Wadsworth V: Role and
regulation of autophagy in cancer. Biochim Biophys Acta.
1793:1516–1523. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhu X, Li X and Chen Z: Inhibition of
anticancer growth in Retinoblastoma cells by naturally occurring
sesquiterpene nootkatone is mediated via autophagy, endogenous ROS
production, cell cycle arrest and inhibition of NF-κB signalling
pathway. J BUON. 25:427–431. 2020.PubMed/NCBI
|
|
53
|
Zhang M, Zhou YF, Gong JY, Gao CB and Li
SL: Expression of autophagy-related protein LC3B, p62, and
cytoplasmic p53 in human retinoblastoma tissues. Eur Rev Med
Pharmacol Sci. 20:3152–3160. 2016.PubMed/NCBI
|
|
54
|
Komatsu M and Ichimura Y: Physiological
significance of selective degradation of p62 by autophagy. FEBS
Lett. 584:1374–1378. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye
WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in
cancer. Chin J Cancer. 36:522017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yoshida GJ: Therapeutic strategies of drug
repositioning targeting autophagy to induce cancer cell death: From
pathophysiology to treatment. J Hematol Oncol. 10:672017.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cai CL, Jin L, Lang XL and Li BL: Long
noncoding RNA XIST regulates cardiomyocyte apoptosis by targeting
miR-873-5p/MCL1 axis. Eur Rev Med Pharmacol Sci. 24:12878–12886.
2020.PubMed/NCBI
|
|
58
|
Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang
R, Cao L, Tang D and Duan X: MIR34A regulates autophagy and
apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy.
10:442–452. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yao L, Yang L, Song H, Liu TG and Yan H:
Silencing of lncRNA XIST suppresses proliferation and autophagy and
enhances vincristine sensitivity in retinoblastoma cells by
sponging miR-204-5p. Eur Rev Med Pharmacol Sci. 24:3526–3537.
2020.PubMed/NCBI
|
|
60
|
Wang Y, Xin D and Zhou L: LncRNA LINC00152
increases the aggressiveness of human retinoblastoma and enhances
carboplatin and adriamycin resistance by regulating
miR-613/Yes-associated protein 1 (YAP1) axis. Med Sci Monit.
26:e920886–1-e920886-17. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Huang J, Yang Y, Fang F and Liu K: MALAT1
modulates the autophagy of retinoblastoma cell through
miR-124-mediated stx17 regulation. J Cell Biochem. 119:3853–3863.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu Y, Su J and Wang LH: Role of autophagy
in cisplatin resistance of retinoblastoma Y79 cells and its
mechanism. Zhongliu Fangzhi Yanjiu. 45:517–522. 2018.PubMed/NCBI
|
|
63
|
Sun J, Feng D, Xi H, Luo J, Zhou Z, Liu Q,
Chen Y and Shao Q: CD24 blunts the sensitivity of retinoblastoma to
vincristine by modulating autophagy. Mol Oncol. 14:1740–1759. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Liang Y, Chen X and Liang Z: MicroRNA-320
regulates autophagy in retinoblastoma by targeting hypoxia
inducible factor-1α. Exp Ther Med. 14:2367–2372. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kong M, Han Y, Zhao Y and Zhang H:
miR-512-3p overcomes resistance to cisplatin in retinoblastoma by
promoting apoptosis induced by endoplasmic reticulum stress. Med
Sci Monit. 26:e923817–1-e923817-10. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yi QY, Bai ZS, Cai B, Chen N, Chen LS,
Yuan T and Mao JH: HSV-TK/GCV can induce cytotoxicity of
retinoblastoma cells through autophagy inhibition by activating
MAPK/ERK. Oncol Rep. 40:682–692. 2018.PubMed/NCBI
|
|
67
|
Li M, Zhang D, Cheng J, Liang J and Yu F:
Ginsenoside Rh2 inhibits proliferation but promotes apoptosis and
autophagy by down-regulating microRNA-638 in human retinoblastoma
cells. Exp Mol Pathol. 108:17–23. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yin L, Sun Z, Ren Q, Su X and Zhang D:
Methyl eugenol induces potent anticancer effects in RB355 human
retinoblastoma cells by inducing autophagy, cell cycle arrest and
inhibition of PI3K/mTOR/Akt signalling pathway. J BUON.
23:1174–1178. 2018.PubMed/NCBI
|
|
69
|
Li X, Zhu X, Xu C and Wu J: 2-Methyl
2-butanol suppresses human retinoblastoma cells through cell cycle
arrest and autophagy. Braz J Med Biol Res. 51:e68892018. View Article : Google Scholar : PubMed/NCBI
|

















