Introduction
Hepatocellular carcinoma is the most common primary
liver cancer and the fourth leading cause of cancer-related deaths
worldwide, accounting for more than 782,000 deaths in 2018
(1–3). Due to challenges associated with
diagnosis, the majority of patients are diagnosed at later stages
when surgical resection is not feasible (3). Therefore, the prognosis of liver cancer
remains extremely poor (2).
Clarifying the underlying molecular mechanisms may help develop
more effective pharmacological therapies.
A recent study reported that RNA-binding protein 24
(RBM24) has a tumor-suppressive role in prostate cancer,
suppressing cellular proliferation, migration, and invasion
(4,5). RBM24 exhibits a tumor-suppressive role,
and RNA-binding proteins (RBPs) are known to play a key role in
post-transcriptional regulation, including mRNA stabilization and
translation (6–8). Moreover, several oncogenes and tumor
suppressor genes control RBPs in various cancer cell lines
(9). RBM24, an RBP with a single
conserved RNA recognition motif domain, is involved in
post-transcriptional regulation and has been shown to influence the
proliferation and motility of various cancer cells (4,5,10). Previous studies have demonstrated
that RBM24 may regulate the stability of p21 and p63 mRNA
transcripts in different human cancer cell lines (11,12).
Conversely, RBM24 has also been shown to destabilize tumor protein
63 (TP63) mRNA by binding to its 3′-UTR (12).
Ruptier et al report that TP63 expression is
regulated via the Wnt/β-catenin pathway in human hepatocellular
carcinoma and squamous cell carcinoma cell lines (13), suggesting that the activation of the
β-catenin (CTNNB1) pathway may contribute to TP63 overexpression
during tumor progression in a cell type-specific manner. It has
also been reported that TP63 overexpression is related to
oncogenesis, cell migration, and epithelial-to-mesenchymal
transition (EMT)-related features of cancer (14,15).
However, the exact function of RBM24 in liver cancer tumorigenesis
and progression remains largely unknown.
In the present study, we demonstrate that RBM24 acts
as a tumor suppressor in liver cancer cells through regulation of
CTNNB1 and TP63. We suggest that RBM24 plays a critical role in
liver cancer progression and anticancer drug resistance.
Materials and methods
Cell culture and drug treatment
A total of 293 cells and human liver cells (Huh7,
Hep3B and HepG2) were purchased from the Korean Cell Line Bank
(KCLB, Seoul, Korea). LX-2 cells were kindly provided by Dr Park
(Yonsei University, Seoul, Korea). L-O2 cells (an immortalized
normal liver cell line) were provided by Dr Shin (Sung-Kyun-Kwan
University, Gyeonggi, Korea). The cell lines were cultured in a
growth medium consisting of Dulbecco's modified Eagle's medium
(DMEM; HyClone; Cytiva) and MEM supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), and 1%
antibiotics (Gibco; Thermo Fisher Scientific, Inc.) in a 5%
CO2-humidified incubator. The cell groups were treated
with various concentrations of sorafenib (0, 0.1, 0.5, 1, 5, and 10
µM) in 10% FBS-supplemented growth media for 4 days. Sorafenib
concentration referred to Liang et al reports (16).
Cloning of RBM24 in an expression
vector and siRNA knockdown
We constructed an RBM24 overexpressing DNA plasmid
(6.7 kb) using pcDNA3.1 (Addgene, Inc.). The cDNA of RBM24 (720 bp
fragment of RBM24 transcript variant 1 mRNA, GenBank accession no.
NM_001143942.1) was ligated to the mammalian expression vector
pcDNA3.1 (Addgene, Inc.), amplified in Escherichia coli (E.
coli) DH5a, identified by restriction analysis (BamHI
and EcoRI), and sequenced. RBM24-pcDNA contains CMV
promoter/enhancer sequences that control the expression of the gene
of interest in multiple cloning sites. This vector includes the Col
E1 origin of replication and the E. coli Ampr
gene for propagation and antibiotic selection in bacteria. The SV40
promoter controls the expression of the neomycin resistance gene
(Neor) that allows antibiotic selection in eukaryotic
cells. A total of 3 µg of RBM24 overexpression vector was
transfected into three human liver cancer cell lines (Huh7, Hep3B
and HepG2) using Lipofectamine 3000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C CO2
incubator. After 72 h transfected liver cancer cells were
G418-selected for two weeks. Scramble siRNA and the siRNAs against
human CTNNB1 (On-TARGETplus human CTNNB1-SMART pool,
L-003482-00-0005; GE Healthcare Dharmacon, Inc.) were purchased
from GE Healthcare Dharmacon, Inc. Cells were transfected with each
100 nM siRNA using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C CO2 incubator for 72 h.
Cell viability, spheroid formation,
and MCTS (multicellular tumor spheroid model) assay
Cell viability was assayed using the Cell Titer-Blue
Cell Viability Assay kit (Promega Corporation) according to the
manufacturer's protocol. The spheroids were trypsinized for 10 min
and incubated with Cell Titer-Blue fluorescence for 2 h and an
additional hour at ambient temperature. The fluorescence intensity
(555–585 nm, gain: 57) was measured using Varioskan Flash
Fluorescent Microplate Fluorometer. To generate spheroids, cells
were suspended in a complete medium and seeded as a series of cell
seeding/well density in a 94-well ultra-low attachment plate
(Corning, Inc.). The plates were incubated for 4 days at 37°C in a
humidified atmosphere supplied with 5% CO2. MCTS assay
was performed using the liver cancer cells and LX2 cells
co-cultured in spheroids at a 7:3 ratio. Data were analyzed using
SigmaPlot software (Systat Software, Inc.) to evaluate the logistic
three parameters and determine the IC50 of the
chemotherapeutic agents.
Quantitative PCR analysis
For RT-qPCR analysis of the target genes, total RNA
was isolated using the TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Reverse transcription was performed using
TOPscript™ RT DryMIX (Enzynomics). RT-qPCR was performed using the
iQ SYBR-Green supermix (Bio-Rad Laboratories, Inc.) and the CFX96™
Real-Time system [Bio-Rad Laboratories (Singapore)]. The
amplification conditions consisted of an initial denaturation at
95°C for 2 min, then 40 cycles of denaturation at 95°C for 5 sec,
and annealing at 58°C for 30 sec according to the manufacturer's
instructions. The relative amounts of target genes were normalized
to those of GAPDH. The RT-qPCR primer sets were summarized in
Table I. The 2−ΔΔCq
method was adopted to determine expression fold-changes (control
vs. sample) (17).
 | Table I.Reverse transcription-quantitative PCR
primers. |
Table I.
Reverse transcription-quantitative PCR
primers.
| Gene | 5′ primer
sequence | 3′ primer
sequence |
|---|
| RBM24 |
5′-GTGAACCTGGCATACTTAGGAGC-3′ |
5′-GCACAAAAGCCTGCGGATAGAC-3′ |
| CTNNB1 |
5′-CACAAGCAGAGTGCTGAAGGTG-3′ |
5′-GATTCCTGAGAGTCCAAAGACAG-3′ |
| TP63 |
5′-CAGGAAGACAGAGTGTGCTGGT-3′ |
5′-AATTGGACGGCGGTTCATCCCT-3′ |
| GAPDH |
5′-GTCTCCTCTGACTTCAACAGCG-3′ |
5′-ACCACCCTGTTGCTGTAGCCAA-3′ |
Immunoprecipitation and immunoblotting
assay
Cells were harvested and lysed with
immunoprecipitation lysis buffer (REF87787; Thermo Fisher
Scientific, Inc.). Transfer the lysate to a microcentrifuge tube
and centrifuge at ~13,000 × g for 10 min to pellet the cell debris
at 4°C. Protein concentrations were determined using a Bradford
protein assay kit (#5000006; Bio-Rad Laboratories, Inc.). For
immunoprecipitation, the isolated protein was transferred to a
clean Falcon tube and incubated with anti-His-tag (1:100; sc-8036;
Santa Cruz Biotechnology, Inc.), 2 µg of anti-Rbm24, or anti-rabbit
IgG antibodies overnight at 4°C. A total of 5 µl (0.25 mg) of
magnetic beads (Pierce™ Protein A/G Magnetic Beads, 88802; Thermo
Fisher Scientific, Inc.) conjugated with pre-treated protein were
incubated at room temperature for 1 h with continuous mixing. The
target protein was eluted from the conjugated magnetic beads with
elution buffer (0.1 M glycine, pH 2.0) and neutralization buffer (1
M Tris, pH 7.5-9). Equivalent amounts (30 µg) of protein from each
lysate were separated using 10% sodium dodecyl sulfate
(SDS)-polyacrylamide gel electrophoresis (PAGE) (Bio-Rad
Laboratories, Inc.) and transferred to nitrocellulose membranes
(#1620115; Bio-Rad Laboratories, Inc.) for immunoblotting. The
membranes were washed three times with phosphate-buffered saline
(PBS; Welgene, Inc.) containing 0.1% Tween-20 (PBST; Sigma-Aldrich;
Merck KGaA). After blocking with PBST containing 1% BSA (BSAS0.1;
Bovogen) for 1 h at room temperature, the membranes were incubated
with the appropriate primary antibody in PBST containing 1% BSA at
4°C overnight. All primary antibodies were diluted to an
appropriate concentration using PBST containing 1% BSA. After
treatment with antibodies against RBM24 (diluted 1:1,000; ab94567;
Abcam), CTNNB1 (diluted 1:1,000; 8480S; Cell Signaling Technology,
Inc.), p63 (diluted 1:1,000; sc-25268; Santa Cruz Biotechnology,
Inc.), caspase 3 (diluted 1:1,000; sc-7272; Santa Cruz
Biotechnology, Inc.), cleaved caspase 3 (diluted 1:1,000; 9661S;
Cell Signaling Technology, Inc.), lamin B1 (diluted 1:1,000;
ab16048; Abcam), or β-actin (diluted 1:1,000; sc-4778; Santa Cruz
Biotechnology, Inc.), the membranes were washed three times with
PBST for 30 min, followed by incubation with goat anti-rabbit
IgG-horseradish peroxidase-conjugated (diluted 1:1;000; #7074S;
Cell Signaling Technology, Inc.), or anti-mouse IgG-horseradish
peroxidase-conjugated secondary antibodies (diluted 1:1,000;
#7076S; Cell Signaling Technology, Inc.) for 2 h at room
temperature, and washed three times with TBST for 30 min. The
membranes were developed using ECL Buffer (REF34580; Thermo Fisher
Scientific, Inc.).
Immunocytochemistry
To examine CTNNB1 translocation, cells were
incubated with 25 mM LiCl (Sigma-Aldrich; Merck KGaA) for 24 h.
After fixing with 4% paraformaldehyde for 5 min, the cells were
incubated for 10 min with PBS containing 0.25% Triton X-100
(Sigma-Aldrich; Merck KGaA) at room temperature. The cells were
then washed three times with PBS, incubated with blocking solution
(1% PBS containing 1% BSA and 0.1% Tween-20) followed by primary
antibodies against CTNNB1 (1:100; 8480S; Cell Signaling Technology,
Inc.) and p63 (diluted 1:1,000; sc-25268; Santa Cruz Biotechnology,
Inc.) at 4°C for 24 h. Before incubation with primary antibodies,
the membranes were blocked with a blocking solution at room
temperature for 1 h. The cells were incubated with donkey
anti-mouse IgG conjugated with Alexa Fluor 594 (1:200; A21203;
Thermo Fisher Scientific, Inc.) and donkey anti-rabbit IgG Alexa
Fluor 488 (1:200; A21206; Thermo Fisher Scientific, Inc.) at room
temperature for 2 h. All secondary antibodies were diluted in an
appropriate concentration of the blocking solution. Nuclei were
stained with DAPI containing mounting solution (H-1200; Vector
Laboratories, Inc.). The cells were then visualized using an
Axiovert 200 fluorescence microscope (Carl Zeiss AG).
Statistical analysis
Statistical significance was analyzed using
SigmaPlot v12.5 software (Systat Software, Inc.). The densitometry
value was normalized to β-actin value using ImageJ v1.45s software.
All experiments were performed in triplicate, and the data are
presented as the mean ± standard deviation. Unpaired Student's
t-test was used to compare differences between two groups. A
one-way ANOVA test followed by a post hoc test using the Holm-Sidak
method was used to compare multiple groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
RBM24 inhibits liver cancer cell
growth
To assess whether RBM24 exhibits a tumor-suppressive
function, we compared RBM24 expression in three liver cancer cells
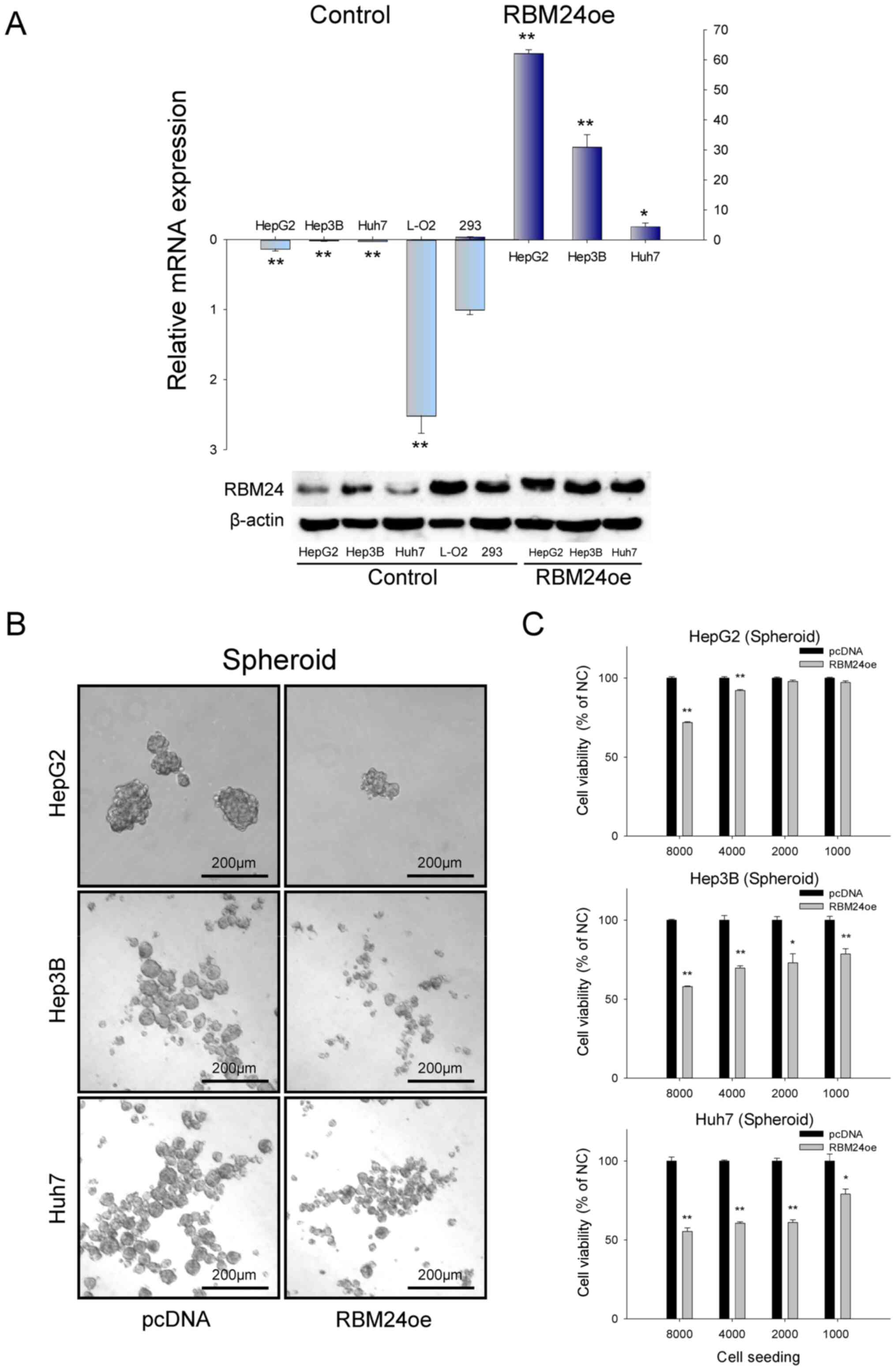
and generated RBM24 overexpression liver cancer cells. As shown in
Fig. 1A, RBM24 expression was
significantly downregulated in all three liver cancer cell lines
compared to those of the normal human fibroblast 293 cells.
However, normal liver cells (L-02) showed higher expression of
RBM24. To address the function of RBM24 in liver cancer cells, we
established RBM24 overexpression systems using a pcDNA expression
vector in the liver cancer cell lines of HepG2, Hep3B, and Huh7
cells. Then, we evaluated the effect of the overexpression of RBM24
in the spheroid culture models. We demonstrated that RBM24
overexpression could significantly reduce the efficiency of sphere
formation in all the three liver cancer cell lines, from an average
of 100% to 72% (HepG2), 58% (Hep3B), and 55% (Huh7), respectively,
under 8000 seeding conditions (Fig.
1B). These results confirm that RBM24 has a tumor-suppressive
function, suppressing sphere formation of liver cancers.
RBM24 inhibits liver cancer cell
progression and induces sorafenib sensitivity
Recently, a MCTS model has emerged as a powerful
tool for simulating tumor complexity and enhancing heterogeneity in
anticancer research, recapitulating the interplay between cancer
cells and their microenvironments (18). Hence, we investigated the
tumor-suppressive function of RBM24 in the MCTS for liver cancer
cells. We developed liver cancer MCTS by co-culturing the liver
cancer cells with a liver stellate cell lines LX-2. Using this
system, we could demonstrate that overexpression of RBM24 could
inhibit the viability of liver cancer cells of Hep3B (100% to 70%)
and Huh7 (100% to 72%), although the viability of HepG2 cells was
not inhibited by RBM24 overexpression (100% to 92%) (Fig. 2A).
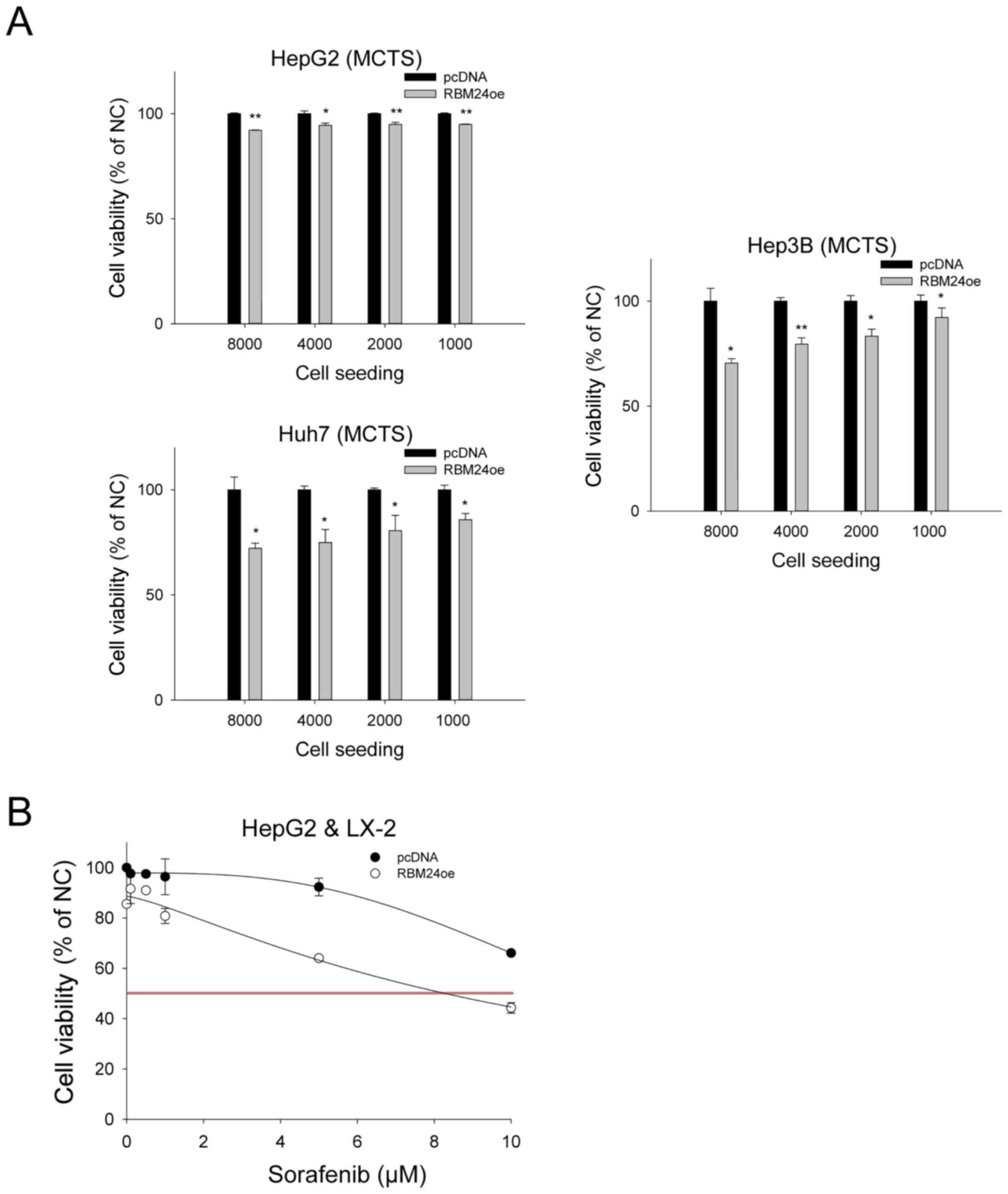 | Figure 2.RBM24-overexpression decreases tumor
progression in three liver cancer MCTS, and increases sensitivity
to sorafenib. All values were calculated based on cell viability
analyses and are depicted in the chart. (A) Using ultra-low
attachment plate analysis, three liver cancer cells and RBM24oe
liver cancer cells were analyzed for tumor suppression in liver
cancer-MCTS co-cultured with LX-2. (B) Sorafenib sensitivity was
assessed using RBM24-overexpression in HepG2-MCTS at 0, 0.1, 0.5,
1, 5 and 10 µM sorafenib. The values indicate the mean and standard
deviation of three independent experiments. *P<0.01,
**P<0.001 vs. pcDNA. MCTS, multicellular tumor spheroids; RBM24,
RNA-binding protein 24; RBM24oe, RBM24-overexpressing; pcDNA,
control plasmid; LX-2, stellate cells. |
In addition, liver cancer MCTS have shown strong
resistance against sorafenib treatment (19). When we evaluated the effect of the
RBM24 expression and sorafenib treatment in the spheroid growths of
liver cancer cells, we could observe that the RBM24 overexpression
could significantly reduce the spheroid progression of cancer cells
(Fig. 2B). Moreover, we could
demonstrate that the RBM24 expressing HepG2-MCTS cells exhibited
decreased sorafenib resistance (IC50: 8.2 vs. 99 µM).
Unfortunately, Hep3B-MCTS and HuH7-MCTS cells did not alter the
sorafenib sensitivity by RBM24 overexpression (data not shown).
Taken together, we suggest that RBM24 expression can increase the
sorafenib sensitivity, at least in HepG2 cells.
TP63 expression is suppressed by
interaction of RBM24 and CTNNB1
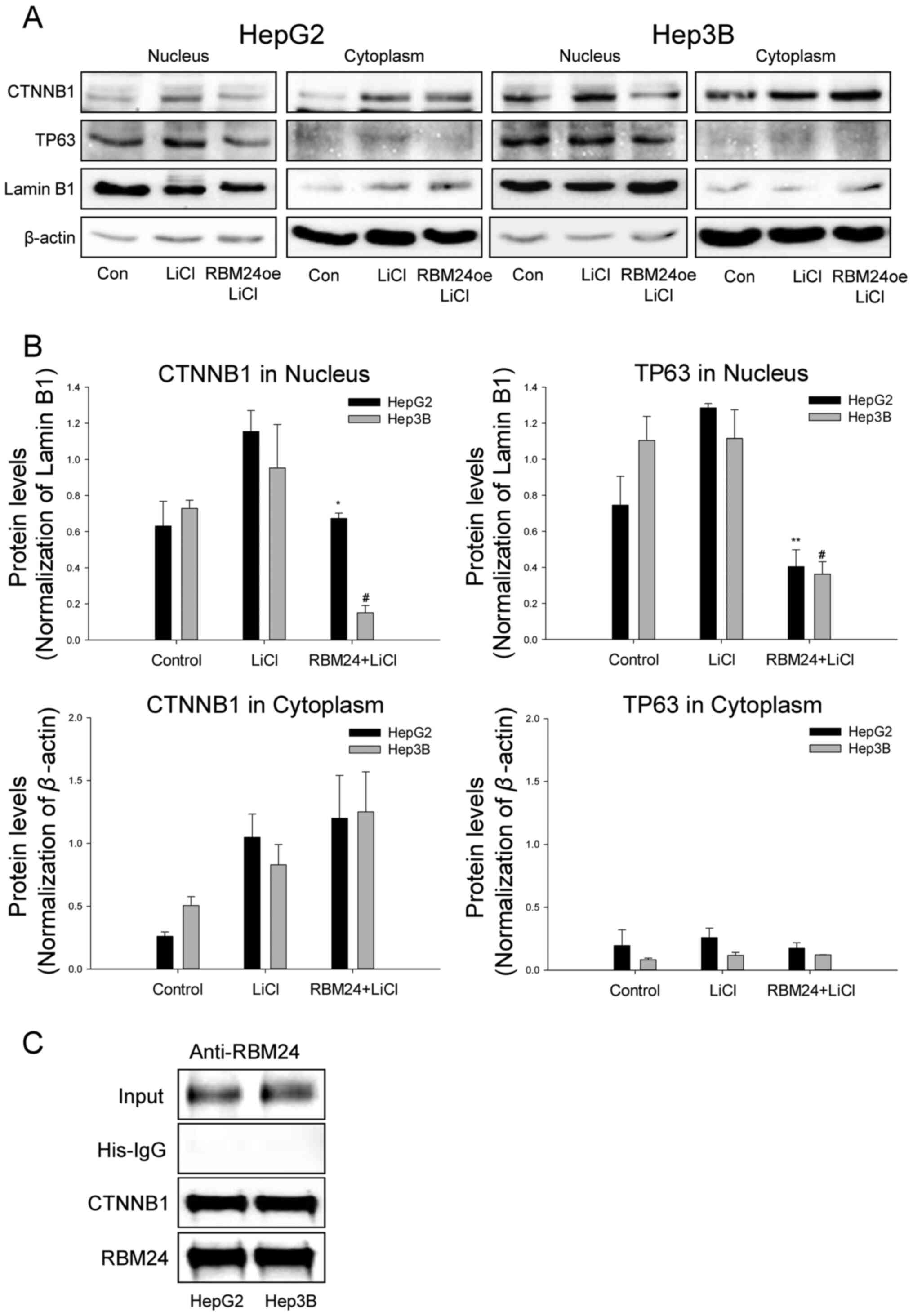
To investigate whether RBM24 interacts with CTNNB1
and impacts TP63 expression in the signaling pathway, we performed
protein expression analysis in the three liver cancer cell lines
overexpressing RBM24 using immunoblotting and RT-qPCR. The results
indicated that RBM24 overexpression in liver cancer cells
specifically decreased TP63 expression and induced a slight
reduction in CTNNB1 expression at the protein level (Fig. 3A and B). Additionally, RBM24
overexpression in the liver cancer cells could increase the
cleavaged-caspase3 levels, thereby inhibiting cell proliferation
and promoting apoptosis. To further confirm that the altered TP63
expression is associated with the interaction between RBM24 and
CTNNB1, we demonstrated that TP63 mRNA expression was inhibited by
RBM24 overexpression or by knockdown of CTNNB1 using RT-qPCR
(Fig. 3C) and immunoblotting
(Fig. 3D), respectively. These
results suggest that TP63 expression is regulated by RBM24 and
CTNNB1.
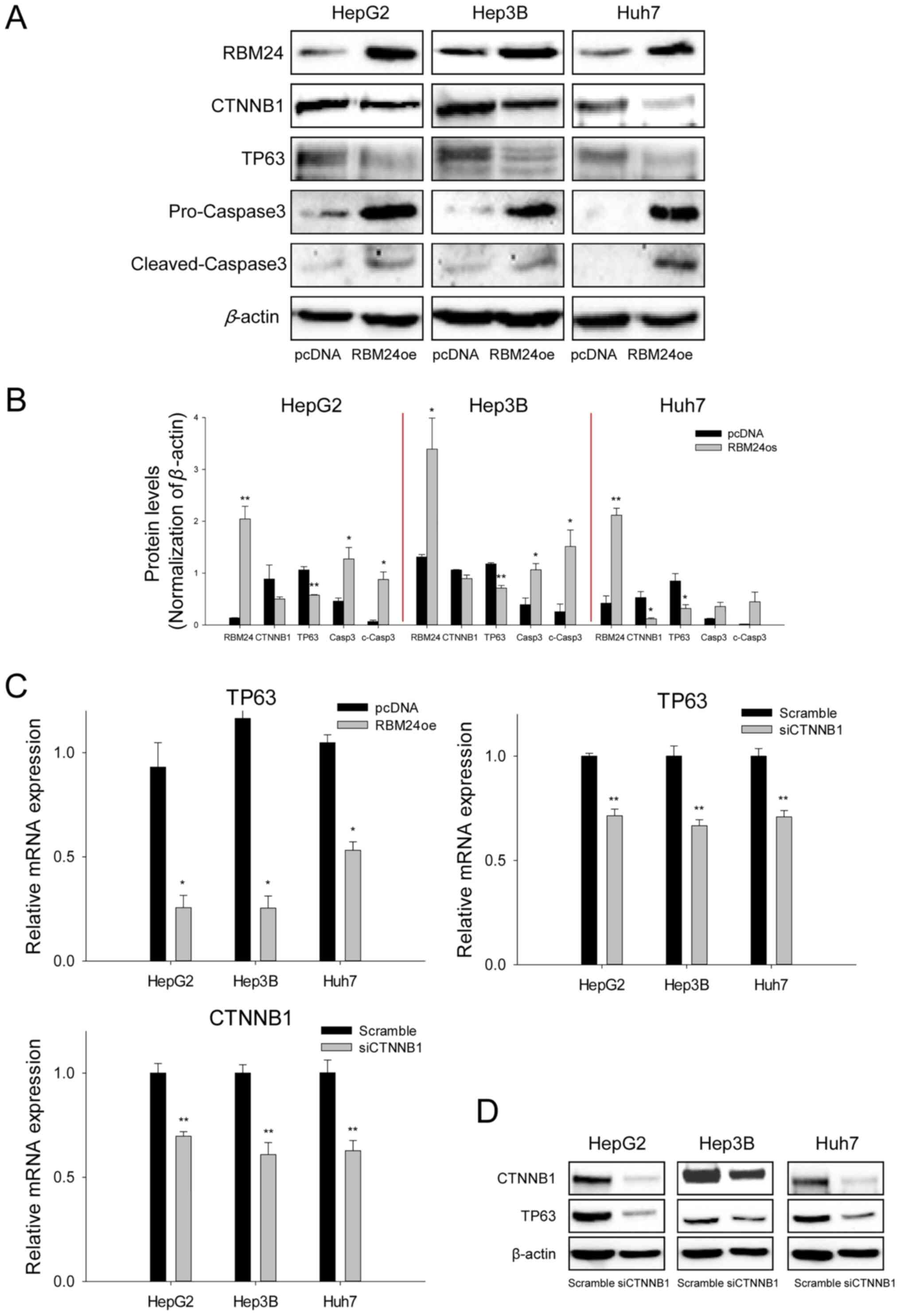 | Figure 3.RBM24 reduces TP63 expression by
downregulating CTNNB1 expression. (A) Immunoblot analysis of RBM24,
CTNNB1, TP63, c-Casp3 and Casp-3 expression in HepG2, Hep3B and
Huh7 cells transfected with the RBM24oe or pcDNA. (B) Quantified
immunoblotting results indicate the densitometry value, normalized
to β-actin value. (C) Reverse transcription-quantitative PCR
analysis of TP63 and CTNNB1 expression in three liver cancer cell
lines transfected with RBM24oe, siCTNNB1, pcDNA or scrambled
siRNAs. (D) Immunoblotting assay of TP63 and CTNNB1 expression in
three liver cancer cell lines transfected with siCTNNB1 or
scrambled siRNAs. The values indicate the mean and standard
deviation of three independent experiments. *P<0.05, **P<0.01
vs. pcDNA or Scramble. RBM24, RNA-binding protein 24; RBM24oe,
RBM24-overexpressing; pcDNA, control plasmid; TP63, tumor protein
63; CTNNB1, β-catenin; c-Casp3, cleaved-caspase 3; Casp-3,
pro-caspase-3; si-, small interfering. |
RBM24 inhibits nuclear translocation
of CTNNB1in liver cancer cells
Next, to further evaluate the subcellular
localization of CTNNB1 upon RBM24 overexpression, we performed
immunoblot analysis of CTNNB1 on both nuclear and cytoplasmic
subcellular fractions. Notably, RBM24 overexpression marginally
inhibited the CTNNB1 expression in HepG2 and Hep3B cells but
suppressed the TP63 expression (Fig. 4A
and B). In addition, we could observe that the treatment of
LiCl induced CTNNB1 translocation and increased TP63 expression in
liver cancer cells. Moreover, overexpression of RBM24 could
suppress the LiCl-induced CTNNB1 translocation and the nuclear TP63
expression. This finding the interaction between RBM24 and CTNNB1
could be further confirmed by immunoprecipitation (IP) analyses
using an anti-RBM24 antibody (Fig.
4C).
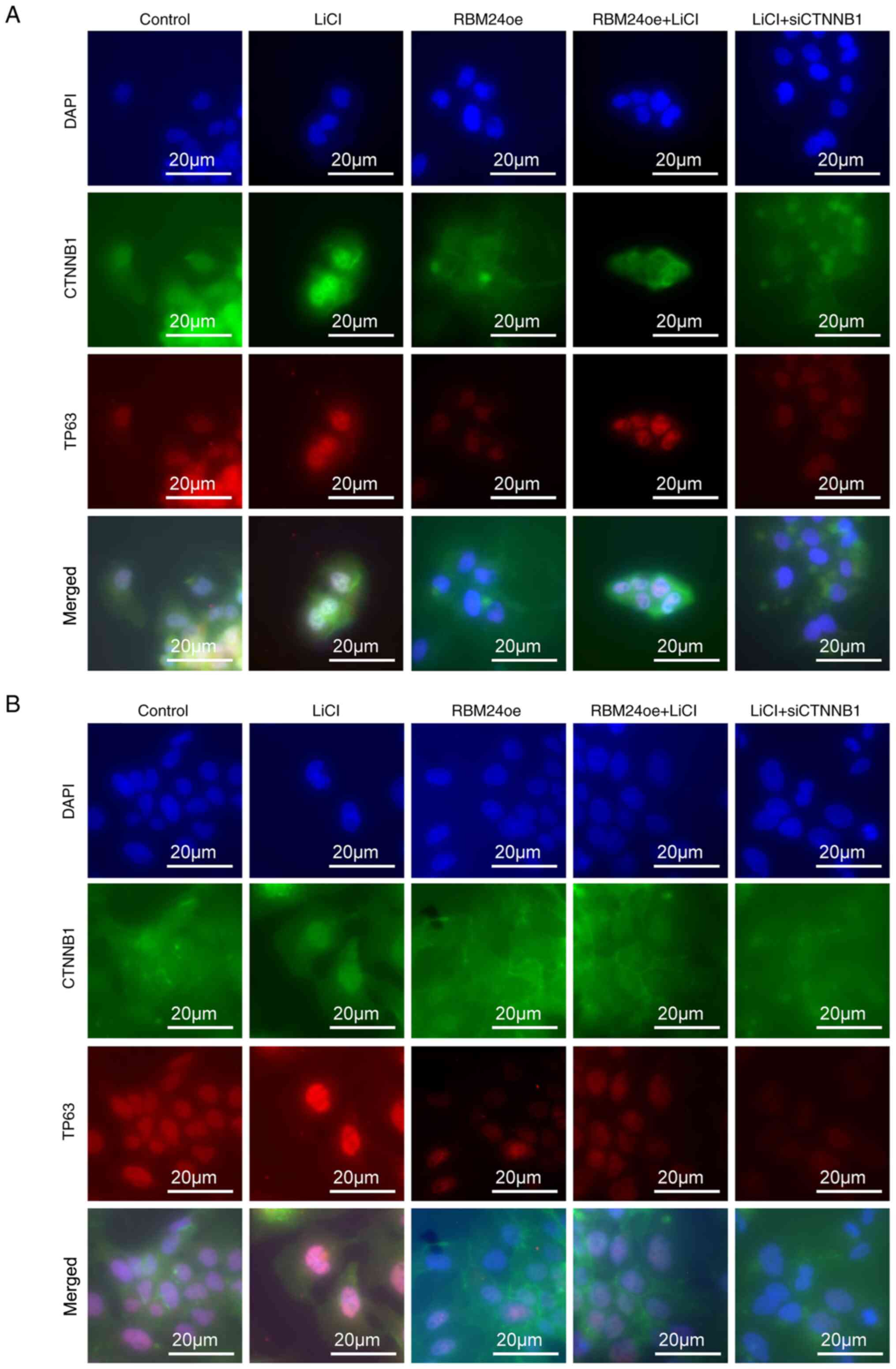
Next, we further validated the effect of RBM24 on
the nuclear localization of CTNNB1 using immunocytochemistry. As
previously described previously (20), we could demonstrate that the
LiCl-treated liver cancer cells strongly induce the CTNNB1
translocation and TP63 expression, However, the RBM24
overexpressing cells showed suppressed nuclear translocation of
CTNNB1 and expression of TP63 (Fig.
5). In addition, we demonstrated that knockdown of CTNNB1
significantly suppressed TP63 expression in the LiCl-treated cells.
Taken together, we suggest that RBM24 interacts with CTNNB1 and
inhibits its nuclear translocation and suppress TP63
expression.
Discussion
Several recent studies have conducted in
vitro and in vivo assays and have demonstrated that RBPs
exhibit strong tumor suppression potential in various cancer
patients (4,5,21–23).
Additionally, RBM24 overexpression decreased prostate cancer cell
growth, indicating a tumor-suppressive role in prostate cancer in
association with the long non-coding RNA HAND2-AS1 (4). Mechanistically, RBM24 binds to
AAU/U-rich elements in target mRNAs and regulates oncogene TP63
gene expression via regulating mRNA stability (11,12,24). In
the present study, we first validated that RBM24 exhibited strong
tumor-suppressive potential in liver cancer. These results
consistent with the tumor-suppressive role of RBM24 in other cancer
types, including lung, prostate, and nasopharyngeal carcinomas
(4,5,10). In
fact, we observed that HepG2 cells did not show the suppression of
sphere formation by RBM24 expression, which may indicate the cell
type-specific regulation of RBM24.
TP63 expression has been shown to be regulated by
TP53 and CTNNB1 (13). In
particular, TP63 expression may be upregulated in the tumor cells
containing a non-functional TP53 and an activated β-catenin
pathway, thereby favoring tumor progression (25). Moreover, p53 regulates RBM24,
facilitating cell cycle arrest, and CTNNB1 and TP63 have been
implicated in the maintenance of stemness of cancer stem cells in
tumor cells (11,26–28). We
observed that RBM24 decreased TP63 expression but increased
caspase3 expression in tumor cells, as described previously
(12,29); however, we failed to detect any
significant change in CTNNB1 expression related to that of TP63.
This suggests a novel mechanism that, in addition to controlling
CTNNB1 expression, RBM24 interacts with CTNNB1 affecting its
binding to TP63 promoter (13).
Accordingly, in our experiments, we detected a decrease in CTNNB1
nuclear translocation after RBM24 overexpression; this could affect
the interaction between CTNNB1 and the TP63 promoter, leading to a
change in TP63 expression through interaction with RBM24.
Collectively, we suggest that RBM24 functions as a
tumor suppressor in liver cancer cells, which is related to the
interaction between TP63 and CTNNB1. RBM24 could be a potential
therapeutic target for treating the patients with liver cancer.
Acknowledgements
The authors acknowledge Dr Young Nyun Park (Yonsei
University, Seoul, Korea) for providing the LX-2 cells. The authors
would also like to acknowledge Dr Gu-Choul Shin (Sung-Kyun-Kwan
University, Gyeonggi, Korea) for providing the L-02 cells.
Funding
The present study was supported by The Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Science, ICT & Future Planning (grant
nos. NRF-2019R1I1A1A01061167, 2019R1A5A2026045,
NRF-2017R1E1A1A01074733, NRF-2017M3A9B6061509 and
NRF-2017M3C9A6047620).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SUM and HGW conceived, designed and performed all
experiments and wrote the manuscript. SUM and JHK analyzed the
data. All authors have read and approved the manuscript. SUM and
HGW confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Finn RS: Emerging targeted strategies in
advanced hepatocellular carcinoma. Semin Liver Dis. 33 (Suppl
1):S11–S19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Daher S, Massarwa M, Benson AA and Khoury
T: Current and future treatment of hepatocellular carcinoma: An
updated comprehensive review. J Clin Transl Hepatol. 6:69–78. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei P, Yang J, Zhang D, Cui M and Li L:
lncRNA HAND2-AS1 regulates prostate cancer cell growth through
targeting the miR-106a-5p/RBM24 axis. OncoTargets Ther.
13:4523–4531. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hua WF, Zhong Q, Xia TL, Chen Q, Zhang MY,
Zhou AJ, Tu ZW, Qu C, Li MZ, Xia YF, et al: RBM24 suppresses cancer
progression by upregulating miR-25 to target MALAT1 in
nasopharyngeal carcinoma. Cell Death Dis. 7:e23522016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kiledjian M, Wang X and Liebhaber SA:
Identification of two KH domain proteins in the alpha-globin mRNP
stability complex. EMBO J. 14:4357–4364. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Collier B, Goobar-Larsson L, Sokolowski M
and Schwartz S: Translational inhibition in vitro of human
papillomavirus type 16 L2 mRNA mediated through interaction with
heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1
and 2. J Biol Chem. 273:22648–22656. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Krecic AM and Swanson MS: hnRNP complexes:
Composition, structure, and function. Curr Opin Cell Biol.
11:363–371. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wurth L: Versatility of RNA-binding
proteins in cancer. Comp Funct Genomics. 2012:1785252012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang D, Ma Y, Ma Z, Liu S, Sun L, Li J,
Zhao F, Li Y, Zhang J, Li S, et al: Circular RNA SMARCA5 suppressed
non small cell lung cancer progression by regulating miR 670
5p/RBM24 axis. Acta Biochim Biophys Sin (Shanghai). 52:1071–1080.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang Y, Zhang M, Qian Y, Xu E, Zhang J
and Chen X: Rbm24, an RNA-binding protein and a target of p53,
regulates p21 expression via mRNA stability. J Biol Chem.
289:3164–3175. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu E, Zhang J, Zhang M, Jiang Y, Cho SJ
and Chen X: RNA-binding protein RBM24 regulates p63 expression via
mRNA stability. Mol Cancer Res. 12:359–369. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ruptier C, De Gaspéris A, Ansieau S,
Granjon A, Tanière P, Lafosse I, Shi H, Petitjean A,
Taranchon-Clermont E, Tribollet V, et al: TP63 P2 promoter
functional analysis identifies β-catenin as a key regulator of
ΔNp63 expression. Oncogene. 30:4656–4665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Massion PP, Taflan PM, Jamshedur Rahman
SM, Yildiz P, Shyr Y, Edgerton ME, Westfall MD, Roberts JR,
Pietenpol JA, Carbone DP, et al: Significance of p63 amplification
and overexpression in lung cancer development and prognosis. Cancer
Res. 63:7113–7121. 2003.PubMed/NCBI
|
|
15
|
Nekulova M, Holcakova J, Coates P and
Vojtesek B: The role of p63 in cancer, stem cells and cancer stem
cells. Cell Mol Biol Lett. 16:296–327. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Y, Chen J, Yu Q, Ji T, Zhang B, Xu
J, Dai Y, Xie Y, Lin H, Liang X, et al: Phosphorylated ERK is a
potential prognostic biomarker for Sorafenib response in
hepatocellular carcinoma. Cancer Med. 6:2787–2795. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chaleshi V, Asadzadeh Aghdaei H, Nourian
M, Iravani S, Jalaeikhoo H, Rajaeinejad M, Khoshdel AR and Naghoosi
H: Association of MALAT1 expression in gastric carcinoma and the
significance of its clinicopathologic features in an Iranian
patient. Gastroenterol Hepatol Bed Bench. 14:108–114.
2021.PubMed/NCBI
|
|
18
|
Seo HR: Roles of tumor microenvironment in
hepatocellular carcinoma. Curr Med Chem. 11:82–93. 2015.
|
|
19
|
Song Y, Kim SH, Kim KM, Choi EK, Kim J and
Seo HR: Activated hepatic stellate cells play pivotal roles in
hepatocellular carcinoma cell chemoresistance and migration in
multicellular tumor spheroids. Sci Rep. 6:367502016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Carotenuto P, Fassan M, Pandolfo R, Lampis
A, Vicentini C, Cascione L, Paulus-Hock V, Boulter L, Guest R,
Quagliata L, et al: Wnt signalling modulates
transcribed-ultraconserved regions in hepatobiliary cancers. Gut.
66:1268–1277. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oh JJ, Taschereau EO, Koegel AK, Ginther
CL, Rotow JK, Isfahani KZ and Slamon DJ: RBM5/H37 tumor suppressor,
located at the lung cancer hot spot 3p21.3, alters expression of
genes involved in metastasis. Lung Cancer. 70:253–262. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sutherland LC, Wang K and Robinson AG:
RBM5 as a putative tumor suppressor gene for lung cancer. J Thorac
Oncol. 5:294–298. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mourtada-Maarabouni M, Keen J, Clark J,
Cooper CS and Williams GT: Candidate tumor suppressor
LUCA-15/RBM5/H37 modulates expression of apoptosis and cell cycle
genes. Exp Cell Res. 312:1745–1752. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shu L, Yan W and Chen X: RNPC1, an
RNA-binding protein and a target of the p53 family, is required for
maintaining the stability of the basal and stress-induced p21
transcript. Genes Dev. 20:2961–2972. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Drewelus I, Göpfert C, Hippel C, Dickmanns
A, Damianitsch K, Pieler T and Dobbelstein M: p63 antagonizes
Wnt-induced transcription. Cell Cycle. 9:580–587. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Katoh M and Katoh M: WNT signaling pathway
and stem cell signaling network. Clin Cancer Res. 13:4042–4045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zucchi I, Astigiano S, Bertalot G, Sanzone
S, Cocola C, Pelucchi P, Bertoli G, Stehling M, Barbieri O,
Albertini A, et al: Distinct populations of tumor-initiating cells
derived from a tumor generated by rat mammary cancer stem cells.
Proc Natl Acad Sci USA. 105:16940–16945. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cléry A, Blatter M and Allain FH: RNA
recognition motifs: Boring? Not quite. Curr Opin Struct Biol.
18:290–298. 2008. View Article : Google Scholar
|
|
29
|
Sayan BS, Sayan AE, Yang AL, Aqeilan RI,
Candi E, Cohen GM, Knight RA, Croce CM and Melino G: Cleavage of
the transactivation-inhibitory domain of p63 by caspases enhances
apoptosis. Proc Natl Acad Sci USA. 104:10871–10876. 2007.
View Article : Google Scholar : PubMed/NCBI
|