Introduction
Prostate cancer is the most common non-cutaneous
cancer in men and is a major health problem in developed countries.
It is expected to emerge as a major health concern worldwide as the
average human lifespan increases in developing countries (1). Prostate cancer is often characterized
by asymptomatic slow growth and men with localized prostate cancer
have a high (10-year) survival rate (2). Clinically localized prostate cancer is
a potentially curative stage and is managed by radiation therapy or
radical prostatectomy. However, disseminated tumor cells
(cytokeratin-positive cells) were detected in bone marrow aspirates
in 13% of patients who underwent radical prostatectomy, suggesting
that potential bone metastasis develops during the early stages of
cancer (3). In contrast to localized
prostate cancer, men with metastatic prostate cancer have a poor
survival rate, with a 5-year survival rate of ~30% and a median
survival rate of ~3 years (4,5). In the
locally advanced or metastatic stage, androgen deprivation therapy
(ADT) is the mainstay of treatment and acts by decreasing the
circulating testosterone levels. However, most patients eventually
develop resistance to ADT and progress towards castration-resistant
prostate cancer (CRPC) after 18–36 months (6,7). Among
metastatic organs, bone is the most frequently metastatic site, and
more than 90% of patients with advanced stage harbor bone
metastases prior to 24 months of death (8). Bone metastasis deteriorates patients'
quality of life by causing pain and making bones prone to fracture.
Bone metastasis management based on understanding the developmental
mechanisms is critical in prostate cancer patients, and a few in
vitro models mimicking the bone microenvironment have been
reported (9,10).
Recently, novel androgen receptor axis-targeted
agents (ARATs) have been approved for metastatic castration-naïve
prostate cancer (mCNPC) or non-metastatic castration-resistant
prostate cancer (nmCRPC). The AR axis remains an essential player
in CRPC and maximizes androgen deprivation by blocking AR function
directly with competing antagonists of the cognate ligand DHT or by
reducing intratumoral androgen synthesis with CYP17A1
lyase/hydrolase inhibitors. Abiraterone, a CYP17A1 lyase/hydrolase
inhibitor, blocks androgen synthesis and prolongs survival in
patients with CRPC (11).
Abiraterone is metabolized by 3-beta-hydroxysteroid dehydrogenase 1
(3βHSD1) to delta-4-abiraterone (D4A), which exerts the greatest
antitumor activity among abiraterone and its metabolites. However,
D4A is metabolized by 5α-reductase to 3-keto-5α abiraterone, which
possesses androgenic activity and stimulates prostate cancer
progression. Recently, combination therapy based on abiraterone and
dutasteride (a 5α-reductase inhibitor) has garnered considerable
attention as a CRPC treatment (12).
Although dutasteride is approved for the treatment of BPH and
regresses prostate volume by inhibiting dual 5α-reductase (type 1
and 2), the risk reduction effect on the development of prostate
cancer has been reported in the REDUCE trial (13). Combination therapy of abiraterone
with dutasteride increases serum levels of D4A and reduces the
levels of 3-keto-5α-abiraterone (12). However, the effects of these
investigational agents, including D4A, on the bone microenvironment
remain unclear, and elucidating them would be pivotal in
investigating the association between the bone microenvironment and
tumor. These observations led us to establish a new in vitro
drug sensitivity testing model that accurately reflects the bone
microenvironment.
In this study, we established a novel in
vitro 3D microenvironment model that mimicked the bone
microenvironment of CRPCs and examined whether it recapitulates the
factors reported previously, including TGF-β. We used this model to
evaluate the drug sensitivity of ARATs and the efficacy of the
combination of abiraterone and dutasteride in the bone
microenvironment.
Materials and methods
Cell culture
The human prostate cancer cell line LNCaP C4-2 (cat.
no. CRL-3314) was purchased from the American Type Culture
Collection. Human mesenchymal stem cells (hMSCs; cat. no. C-12974)
from the bone marrow were obtained from PromoCell. The C4-2 cells
were routinely grown in RPMI-1640 medium (Life Technologies)
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc.), hMSCs were cultured in Mesenchymal Stem
Cell Growth Medium 2 (cat. no. C-28009; PromoCell) and maintained
in humidified incubators (5% CO2). Penicillin G (100
U•ml−1) and streptomycin sulfate (0.1
mg•ml−1) (cat. no. A5955; Sigma-Aldrich; Merck KGaA)
were added to all conditioned media.
Transfection of fluorophores
To quantify cell dynamics non-invasively,
fluorescent proteins (GFP and RFP) were introduced into the cells.
The GFP gene was introduced into C4-2 cells and the RFP gene was
introduced into hMSCs using lentivirus. C4-2 cells were incubated
in RPMI-1640 medium supplemented with 10% FBS in a 10-cm dish and
hMSCs were incubated in Mesenchymal Stem Cell Growth Medium 2 in a
10-cm dish. C4-2 cells were transfected with 6 µg of BLIV 2.0
Reporter: MSCV-Luciferase-EF1α-copGFP-T2A-Puro Lentivector Plasmid
(cat. no. BLIV713PA-1; System Biosciences, LLC) and hMSCs were
transfected with 6 µg of BLIV 2.0 Reporter:
pCDH-CMV-MCS-EF1-RFP-T2A-Puro (cat. no. CD516B-2, System
Biosciences, LLC) and 4 µg of pPACKH1 HIV Lentivector Packaging Kit
(cat. no. LV500A-1; System Biosciences, LLC) using X-tremeGENE HP
DNA Transfection Reagent (cat. no. 6366244001; Roche). After
overnight incubation, the medium was replaced with fresh medium and
the cells were incubated for 3 days. The medium was transferred to
a 15 ml tube, followed by centrifugation at 3,000 rpm for 5 min,
and the supernatant containing lentiviral particles was passed
through a syringe filter (cat. no. SLPES2545S; Hawach Scientific).
C4-2 cells were incubated for 3 days in RPMI-1640 medium
supplemented with 10% FBS containing lentiviral particles and
polybrene (8 µg/ml; cat. no. H9268; Sigma-Aldrich; Merck KGaA).
hMSCs were incubated for 3 days in Mesenchymal Stem Cell Growth
Medium 2 containing lentiviral particles and polybrene. Since the
downstream region of the plasmid contains a puromycin resistance
gene, GFP-expressing or RFP-expressing cells were selected using
puromycin (1 mg/ml; cat. no. A1113802; Thermo Fisher Scientific,
Inc.).
Osteogenesis
To promote osteogenic differentiation, hMSCs were
cultured in Mesenchymal Stem Cell Osteogenic Differentiation medium
(cat. no. C-28013; PromoCell). The medium was changed every 3–4
days for 2 weeks. After differentiation, the cells were fixed with
10% formalin solution, stained with 1% Alizarin Red S (cat. no.
5533-25G, Sigma-Aldrich) to confirm extracellular calcium deposits,
and stained with BCIP/NBT (cat. no. B5655; Sigma-Aldrich; Merck
KGaA) to confirm alkaline phosphatase activity.
Bone microenvironment model of
prostate cancer in 3D culture
RFP-transfected hMSCs were plated (5×104
cells/cm2) onto a chitosan nanofiber-coated culture
plate (Hokkaido Soda Co., Ltd.) and incubated with Mesenchymal Stem
Cell Growth Medium 2. When cells reached 100% confluency, the
medium was changed to Mesenchymal Stem Cell Osteogenic
Differentiation medium (Day 0), followed by 14 days of incubation
to induce human osteoblasts. On day 14, GFP-transfected C4-2 cells
(1.5×103 cells/cm2) were added and
co-cultured with human osteoblasts. On day 15, the medium was
changed to androgen-free, phenol red-free RPMI-1640 medium (Life
Technologies) supplemented with 5% charcoal/dextran-treated fetal
bovine serum (HyClone). Penicillin G (100 U•ml−1) and
streptomycin sulfate (0.1 mg•ml−1) were added to all
conditioned media. The medium was changed every 3–4 days.
Quantification of C4-2 and osteoblast
cells
The growth of C4-2 and osteoblasts in the bone
microenvironment model was quantified using an imaging system,
Cell3 iMager duos (SCREEN Holdings Co., Ltd.) every
3-days. The sum of the green or red intensity area of each well was
determined using the Cell3 iMager duos software version
1.4 rev 2.1, (SCREEN Holdings Co., Ltd.).
Drug sensitivity test using bone
microenvironment model
A total of four ARATs (enzalutamide, apalutamide,
darolutamide, and abiraterone), D4A (abiraterone metabolite with AR
antagonist) and dutasteride (5α-reductase inhibitor) were selected
for drug sensitivity testing using a microenvironment model. All
drugs were purchased from Selleckchem. In the drug sensitivity test
over time, each drug was added to the culture medium at a
concentration of 5 µM dissolved in ethanol from day 15. The final
concentration of ethanol in all the drugs and controls was 0.1%.
Investigational agents were added every time the medium was
changed. In the dose-response curve, each drug was added to the
culture medium at each concentration dissolved in ethanol on day
15. The drug exposure time was 48 h, and the antiproliferative
effect of the drug on C4-2 cells was determined by comparison with
the concentration at 0.01 µM of each drug. CompuSyn software was
used to calculate the combination index (CI) at several effective
doses (CI=1; additive effect, CI<1; synergy effect, CI>1;
antagonistic effect) (14).
Total RNA extraction and quantitative
mRNA expression analysis
Total RNA was extracted using the RNeasy Mini Kit
(Qiagen), according to the manufacturer's protocol. RNA quality and
quantity were determined using a NanoDrop Lite spectrophotometer
(Thermo Fisher Scientific, Inc.). First-strand cDNA was synthesized
from 1 µg of total RNA using the RT2 First Strand Kit
(Qiagen). RNA expression of EMT-related genes was analyzed using
the RT2 Profiler PCR Array Human Epithelial to
Mesenchymal Transition (EMT) (cat. no. PAHS-090ZA; Qiagen), and RNA
expression of osteogenesis-related genes was analyzed using the
RT2 Profiler PCR Array Human Osteogenesis (cat. no.
PAHS-026Z, Qiagen). The expression of AR and PSA was analyzed using
TaqMan Gene Expression Assays (Applied Biosystems Inc.). The TaqMan
MGB probes used in this study were as follows: AR (Hs00171172_m1),
PSA (Hs02576345_m1, GAPDH (Hs02758991_g1). Quantitative real-time
PCR was performed in triplicate using Applied Biosystems
StepOnePlus (Applied Biosystems) according to the manufacturer's
protocol. RNA expression levels were determined using StepOnePlus
software (version 2.2.2; Applied Biosystems) and normalized to
GAPDH expression levels. The following thermocycling conditions
were used: initial denaturation at 95°C for 20 sec, followed by 40
cycles at 95°C for 1 sec and 60°C for 20 sec. RNA expression levels
were determined using the 2−ΔΔCT method (15).
Western blot analysis
Cell samples were collected in RIPA lysis buffer
(Santa Cruz Biotechnology Inc.), and protein concentrations were
determined using a BCA Protein Assay Kit (Takara Bio Inc.). Each
lysate sample (20 µg) was separated by SDS-PAGE and
electro-transferred to a polyvinylidene fluoride (PVDF) membrane.
Following blocking with 5% non-fat milk in TBS with 0.05% Tween-20
(TBST), the membranes were incubated with each primary antibody
overnight at 4°C. The primary antibodies used were as follows:
N-cadherin (1/500 dilution, cat. no. 13116), E-cadherin (1/1,000
dilution, cat. no. 3195), Snail (1/1,000 dilution, cat. no. 3879),
TGF-β (1/1,000 dilution, cat. no. 3709), and GAPDH (1/2,000
dilution, cat. no. 5174), from Cell Signaling Technology. AR
(1/1,000 dilution; cat. no. ab133273) from Abcam. After washing
with TBST, the membranes were incubated with HRP-conjugated
anti-rabbit IgG secondary antibody (1/5,000 dilution, cat. no.
ab6721; Abcam) for 1 h at room temperature. After washing with
TBST, membrane signals were detected using an ECL detection system
(Amersham Imager 600; GE Healthcare Life Sciences). GAPDH was used
for normalization of the protein bands.
Magnetic-activated cell sorting
(MACS)
Highly purified C4-2 cells were isolated from
co-cultured cells using MACS® (Miltenyi Biotec), using
positive selection. Co-cultured cells (1×107) were
centrifuged (200 × g for 4 min) and then resuspended in 100 µl of
MACS buffer (PBS containing 2 mM EDTA and 0.5% FBS). Biotin
anti-human PSMA (FOLH1) antibody (10 µl, cat. no. 342510;
BioLegend) was added and incubated for 5 min at 4°C. Cells were
washed in 2 ml of MACS buffer and centrifuged again at 200 × g for
4 min. The cellular pellet was resuspended in 200 µl of MACS buffer
and 20 µl of anti-biotin microbeads (Miltenyi Biotec). After
incubation for 15 min at 4°C, the cells were washed in 2 ml of MACS
buffer and centrifuged at 200 × g for 4 min. The cellular pellet
was resuspended in 500 µl of MACS buffer loaded onto an LS MACS
column (Miltenyi Biotec) in a magnetic field. Pass-through
(unlabeled) cells were collected, and the column was washed three
times with 500 µl of MACS buffer. The MACS column was removed from
the magnetic stand, and PSMA-positive (labeled) cells were eluted
with 5 ml of MACS buffer. The PSMA-positive cells and pass-through
cells were analyzed by flow cytometry.
Flow cytometry analysis
An allophycocyanin (APC)-conjugated anti-human PSMA
(FOLH1) antibody (cat. no. 342508, BioLegend) was used in this
study. Flow cytometric data were acquired using a CytoFLEX S System
cell (Beckman Coulter) and analyzed using FlowJo software (FlowJo,
LLC).
Liquid chromatography-electrospray
ionization-time-of-flight/mass spectrometry analysis
C4-2 cells and osteoblasts were co-cultured for 24
h, and abiraterone alone or a combination of abiraterone and
dutasteride were added to the culture medium at a concentration of
5 µM dissolved in ethanol on day 15. Cell suspensions were
collected on days 17 and 28 (2 and 13 days after drug addition),
and C4-2 cells were isolated by MACS® using an anti-PSMA
antibody. C4-2 cells (1×105 cells) were washed with PBS
by centrifugation at 200 × g for 5 min. They were then washed twice
under the same conditions. The cell pellets were lysed with 50 µl
of 70% acetonitrile and then centrifuged at 17,4000 × g for 5 min
at 4°C. The resulting supernatants were collected, diluted to 1/20
with 80% methanol in water, and analyzed by liquid chromatography
(LC)-electrospray ionization (ESI)-time-of-flight/mass
spectrometery (TOF-MS). The LC system used was a Nexera
SFC/SFE-HPLC system (Shimadzu). The ESI-TOF/MS system used was
Impact II (Bruker Daltonics). MS was operated in the positive ion
mode. The concentrations of abiraterone and abiraterone
metabolites, including D4A, 3-keto-5α-abiraterone,
3α-OH-5α-abiraterone, and 3β-OH-5α-abiraterone in the supernatant
were measured using the exact mass value of the protonated
molecular ion of each compound.
Pharmacokinetic in vivo validation of
abiraterone and dutasteride combination
Details of this method have been reported elsewhere
(16). Briefly, abiraterone and its
metabolite (D4A, 3-keto-5α-abiraterone) were measured before and
after combination treatment with abiraterone and dutasteride in a
phase II clinical trial in patients with CRPC (UMIN Clinical Trial
Registry: UMIN000027795). Pharmacokinetic data in patients who were
judged as effective in combination therapy were compared with the
data from our in vitro model.
Statistical analysis
All numerical data are presented as the mean ±
standard deviation. Unpaired numerical data were compared using an
unpaired Student's t-test (two groups) or analysis of variance
(ANOVA; more than two groups). We used Tukey test as the post-hoc
test following ANOVA. The synergistic effect of the combination of
drugs was calculated using the CompuSyn software (Combosyn Inc.).
Statistical analysis was performed using JMP software (Pro.13; SAS
Institute, Inc.). P-values were two-sided, and statistical
significance was defined as P<0.05, in all tests.
Results
Bone microenvironment model of
prostate cancer with chitosan fiber matrix 3D culture
To demonstrate the bone microenvironment model of
prostate cancer, prostate cancer cells and osteoblasts were
co-cultured on a microfiber scaffold to examine the characteristics
of the co-culture system. We used a microfiber scaffold composed of
a three-dimensional chitosan fiber matrix. The scaffold was
composed of random fibers with an average diameter of ~200 nm.
Using a three-dimensional microfiber scaffold, cells may
proliferate in a three-dimensional morphology. To recapitulate the
bone metastasis microenvironment of castration-resistant prostate
cancer, we used C4-2, a cell line of castration-resistant prostate
cancer. C4-2 maintains AR activation and signaling through de novo
intratumoral steroidogenesis (17,18).
Osteoblasts were differentiated from human mesenchymal stem cells
(hMSCs), which were collected from the bone marrow using a special
medium. Differentiation of hMSCs into osteoblasts was confirmed by
Alizarin red S staining and alkaline phosphatase staining (Fig. S1). To facilitate separate
quantification of C4-2 and osteoblast cells after co-culture, we
stably transfected C4-2 cells with GFP and hMSCs with RFP using
lentiviral vectors. We used flow cytometry to determine whether GFP
was successfully introduced into C4-2 cells or RFP was introduced
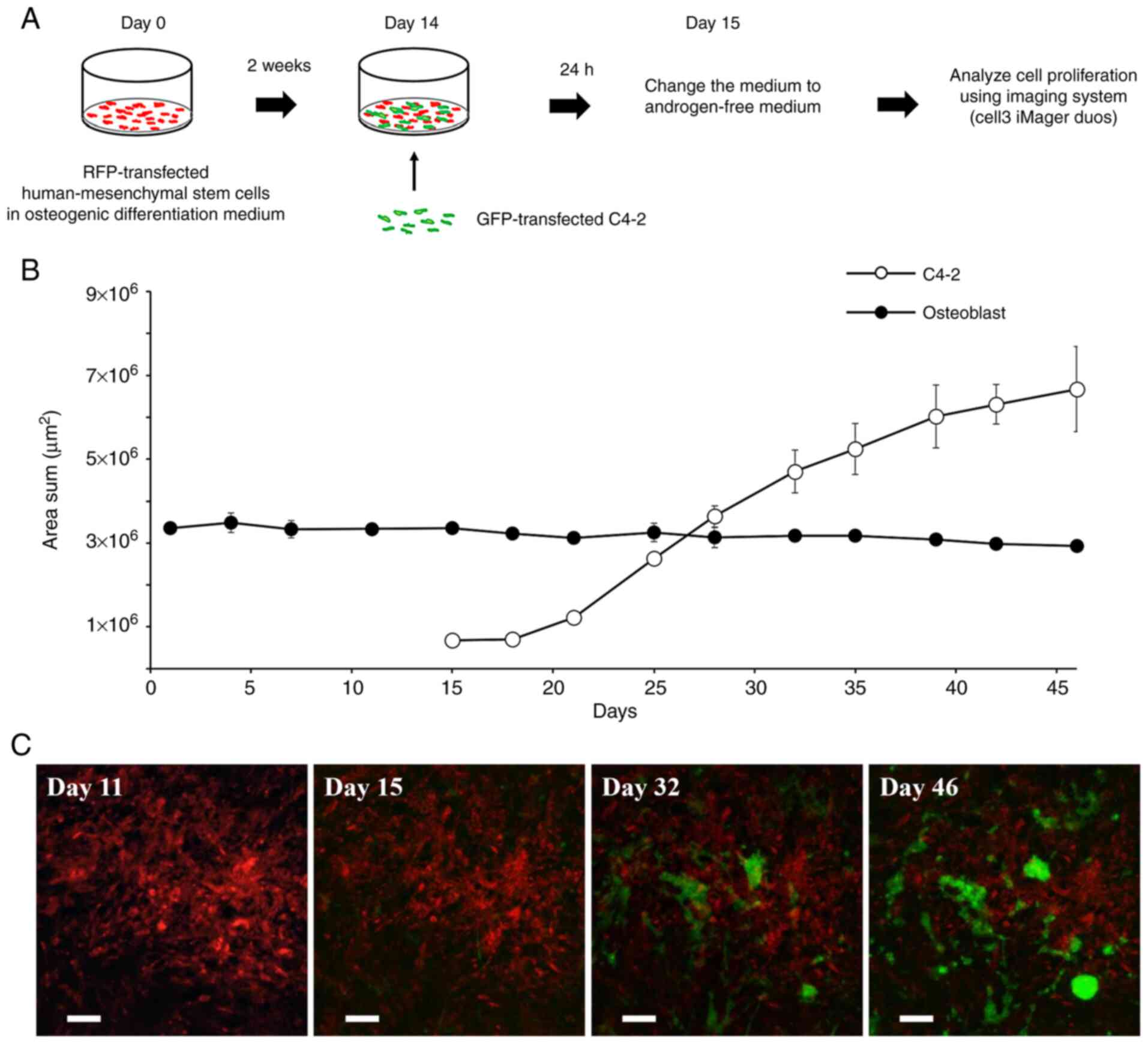
into hMSCs (Fig. S2). Fig. 1-A shows the schema of the bone
microenvironment model. GFP-transferred C4-2 cells and
RFP-transferred osteoblasts differentiated from hMSCs were
co-cultured with chitosan fiber matrix 3D culture. The medium was
changed to androgen-free medium 24 h after starting the co-culture
to reflect the bone metastasis environment of castration-resistant
prostate cancer. The growth of C4-2 cells and osteoblasts was
quantified using a live-cell imaging and analysis system, Cell3
iMager duos (SCREEN). We could non-invasively quantify the static
survival of osteoblasts and could maintain a continuous count of
C4-2 cells for a maximum of 30 days (Fig. 1B). Fluorescence images at days 15,
32, and 46 showed that C4-2 grew to form colonies (Fig. 1C). The C4-2 colonies were physically
in contact with the osteoblasts. We also examined the effects of
co-culturing with osteoblasts. Compared with monoculture,
co-culturing with human osteoblasts demonstrated a significant
growth enhancement on C4-2 cells (t-test, P<0.01, day 52), but
there was no obvious difference in morphology of the C4-2 cells
(Fig. S3).
Drug sensitivity test using bone
microenvironment model
We used this model to evaluate the drug sensitivity
of androgen receptor-axis-targeted agents (ARATs) and D4A
(abiraterone metabolite with AR antagonist). First, we compared the
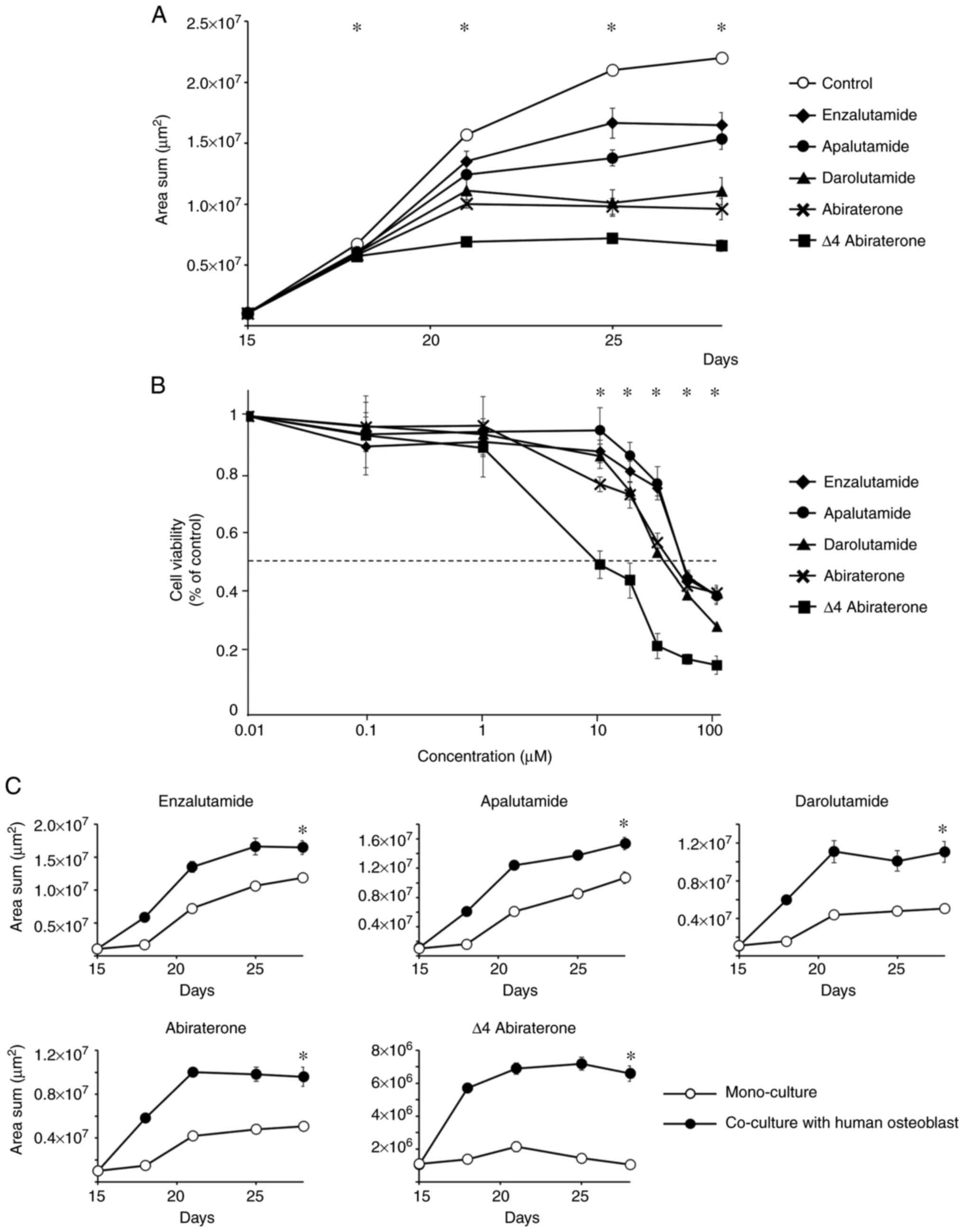
growth curves of the GFP-transfected C4-2 cells (Fig. 2A). Significant difference in growth
inhibition was observed from day 18 to day 25 (ANOVA, P<0.01),
and significant growth inhibition was observed with the addition of
enzalutamide (t-test, P<0.01), apalutamide (P<0.01),
darolutamide (P<0.01), abiraterone (P<0.01), or D4A
(P<0.01) compared to the control on day 28. Significant
differences in growth inhibition were observed among all the
investigational agents except between enzalutamide and apalutamide
(P=0.195) and between darolutamide and abiraterone (P=0.118).
Second, we compared the dose-response curves of each
investigational agent against C4-2 cells (Fig. 2B). Significant difference in growth
inhibition between ARATs was observed at concentrations of 10–100
µM (ANOVA, P<0.01). The 50% inhibition concentration
(IC50) of each drug calculated from the dose-response
curve is shown in Table I. The
IC50 values of each investigational agent were compared,
and significant differences were found among the investigational
agents (t-test, P<0.05), except between enzalutamide and
apalutamide (P=0.982) and between darolutamide and abiraterone
(P=0.106). The results were similar to the drug effects of the
growth curves (Fig. 2A). Next, we
compared the drug sensitivity of C4-2 cells with and without
co-culture with human osteoblasts in chitosan nanofiber-coated 3D
culture plates. Co-culture with human osteoblasts had a inhibitory
effect on the growth of all investigational agents on day 28
(Fig. 2C).
 | Table I.IC50-values of androgen
receptor-axis-targeted agents in the bone microenvironment
model. |
Table I.
IC50-values of androgen
receptor-axis-targeted agents in the bone microenvironment
model.
| Drugs | IC50,
µM |
|---|
| Enzalutamide | 50.85±1.90 |
| Apalutamide | 51.02±1.98 |
| Darolutamide | 35.14±1.26 |
| Abiraterone | 38.85±5.38 |
| Δ4 Abiraterone | 10.47±4.87 |
Analysis of mRNA and protein
expression in C4-2 cells co-cultured with and without
osteoblasts
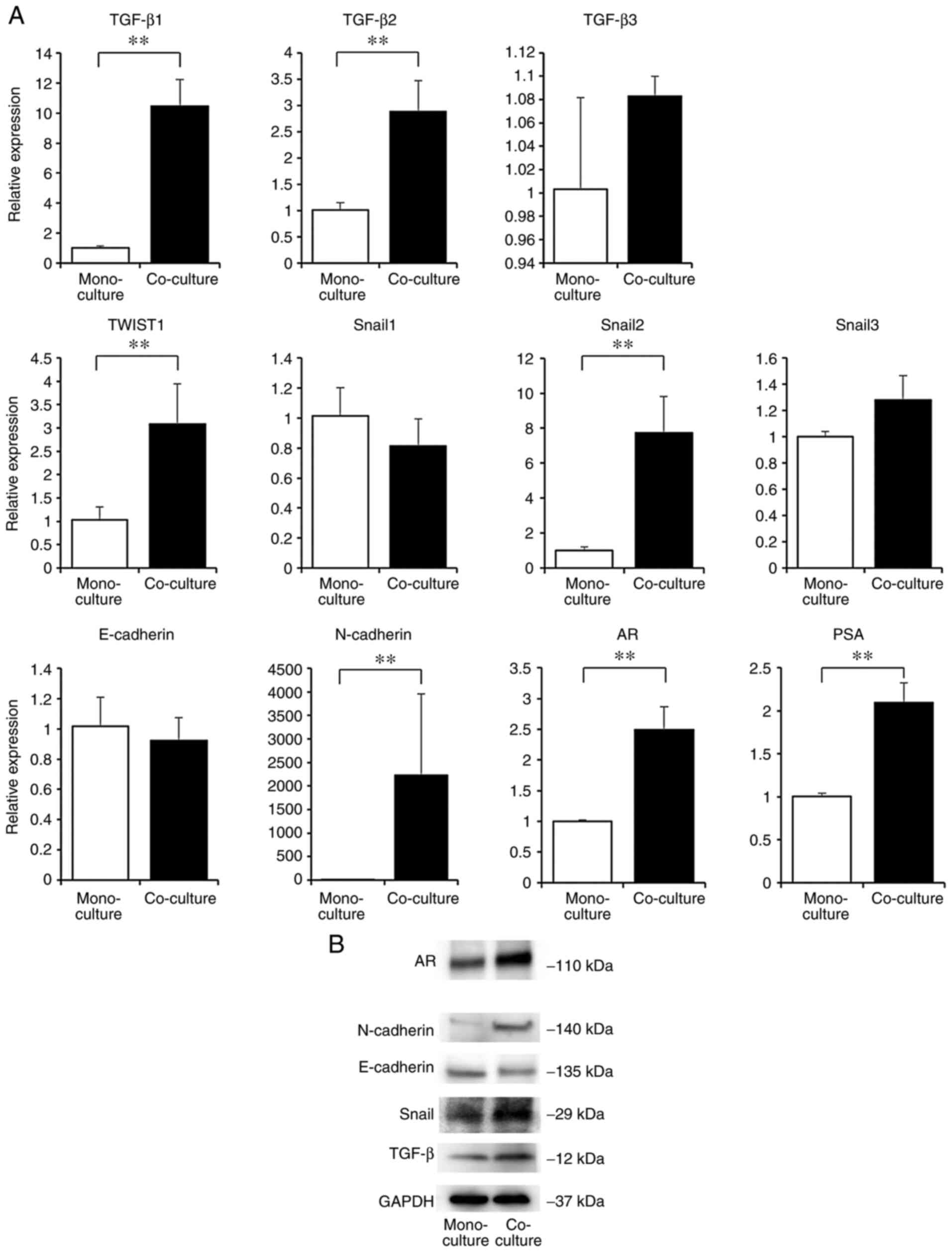
We examined the changes in mRNA and protein
expression in C4-2 cells when co-cultured with osteoblasts in this
model. Highly purified C4-2 cells were isolated from co-cultured
cell suspensions by magnetic-activated cell sorting
(MACS®; Miltenyi Biotec). This was performed using
positive selection using specific binding of anti-PSMA antibodies
to C4-2 cells. The highly specific separation of C4-2 cells was
confirmed by flow cytometry (Fig.
S4). We compared the mRNA expression of C4-2 cells isolated
from co-culture cell suspensions and monocultured C4-2 cells using
RT-PCR (Fig. 3A). We focused on
TGF-β and EMT-related genes, because accumulating evidence suggests
that these molecules play an important role as promoters of tumor
cell survival and development in the bone microenvironment. The
mRNA expression of TGF-β (TGF-β1, TGF-β2) was significantly higher
in the co-culture than in the monoculture. Similarly, the
expression of EMT-related genes (TWIST1, Snail2, and N-cadherin)
was also significantly higher in the co-culture than in the
monoculture. The expression of AR and PSA (downstream gene of AR)
was significantly higher when co-cultured with osteoblasts. The
protein expression of C4-2 cells isolated from co-culture cell
suspensions and monoculture was compared by western blotting
(Fig. 3B). An identical expression
pattern was observed between the mRNA and protein expression of
each gene. We also examined the mRNA expression of osteoblast
stimulatory factors in the co-culture. mRNA expression of bone
morphogenetic protein 2 (BMP2) and vascular endothelial growth
factor (VEGF-A, VEGF-B) were significantly higher in the co-culture
than in the monoculture (Fig.
S5).
Inhibition of tumor growth by
combination therapy with abiraterone and dutasteride
We used this model to evaluate the therapeutic
effects of the combination of abiraterone and dutasteride in the
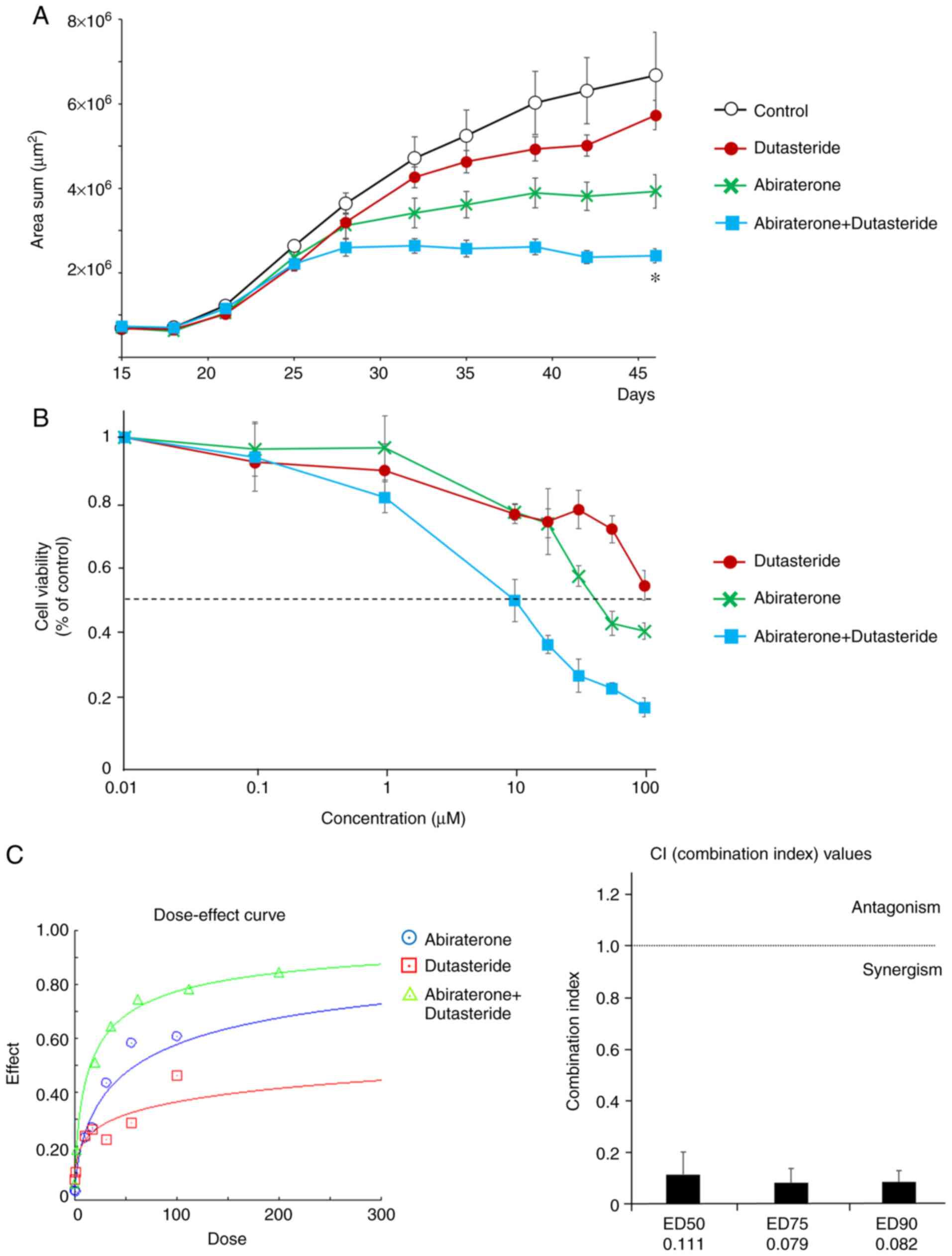
bone microenvironment. First, we compared the growth curves of
GFP-transferred C4-2 cells among abiraterone, dutasteride, and both
combinations (Fig. 4A). The
combination of abiraterone and dutasteride had a greater colony
inhibitory effect on tumor growth (ANOVA, P<0.01, day 46).
Second, we compared the dose-response curves of each
investigational agent against C4-2 cells (Fig. 4B). To examine whether this effect was
additive or synergistic, dose-dependent effects with constant ratio
design and combination index (CI) values were calculated according
to the Chou and Talalay median effect principal (14). Dutasteride synergistically enhanced
the inhibitory effect of abiraterone on the colony growth of C4-2
cells (Fig. 4C). We then examined
the concentrations of abiraterone metabolites in C4-2 cells treated
with abiraterone alone or abiraterone and dutasteride. C4-2 cells
were isolated from cell suspensions on days 17 and 28 (2 and 13
days after drug addition). C4-2 cells were lysed with acetonitrile,
and the supernatants were analyzed using LC-ESI-TOF/MS. The
concentrations of each investigational agent used are listed in
Table II. On day 17, the
combination of dutasteride significantly decreased the
concentration of 3-keto-5α-abiraterone (t-test, P<0.05) and
3β-OH-5α-abiraterone (P<0.01). On day 28, the combination of
dutasteride tended to decrease the concentration of
3-keto-5α-abiraterone (P=0.06) and a significant decrease in
3β-OH-5α-abiraterone (P<0.01). These results suggest that the
combination of abiraterone and dutasteride had a more potent
inhibitory effect on tumor growth by reducing the androgen receptor
agonist activity of 5α-abiraterone. In a phase II study of
abiraterone and dutasteride combination therapy in CRPC patients
(16), effective cases where PSA was
decreased by the combined use of abiraterone and dutasteride showed
a decrease in serum 3-keto-5α-abiraterone concentration (Fig. S6), similar to the results of the
microenvironmental model using the chitosan culture substrate.
 | Table II.Intracellular concentrations of
abiraterone and each metabolite. |
Table II.
Intracellular concentrations of
abiraterone and each metabolite.
| A, Day 17 |
|---|
|
|---|
| Metabolite | Abiraterone | Abiraterone +
Dutasteride | P-value |
|---|
| Abiraterone,
ng/ml | 346.60±68.99 | 351.73±48.41 | NS |
| Δ4 Abiraterone,
ng/ml | 4.54±1.14 | 3.08±0.38 | NS |
|
3-keto-5α-Abiraterone, ng/ml | 3.24±0.91 | 0.91±0.73 |
0.01 |
|
3β-OH-5α-Abiraterone, ng/ml | 8.63±0.43 | 0.99±0.73 | <0.01 |
|
| B, Day
28 |
|
|
Metabolite |
Abiraterone | Abiraterone +
Dutasteride | P-value |
|
| Abiraterone,
ng/ml | 1,228.76±81.91 |
2,125.07±145.86 | <0.01 |
| Δ4 Abiraterone,
ng/ml | 8.04±1.71 | 7.20±0.15 | NS |
|
3-keto-5α-Abiraterone, ng/ml | 2.97±0.81 | 1.73±0.77 |
0.06 |
|
3β-OH-5α-Abiraterone, ng/ml | 24.90±1.86 | 2.20±0.63 | <0.01 |
Discussion
Recently, in vitro culture models using
biomimetic nanofiber scaffolds have been reported (19,20).
Compared with cells cultured on polystyrene culture dishes, cells
cultured on three-dimensional culture scaffolds formed colonies and
maintained their morphology and functions similar to living
organisms for a long time. In terms of the bone environment,
chitosan nanofibers have attracted attention as a novel scaffold
for the repair and regeneration of bone tissue. Long-term culture
of osteoblasts using chitosan nanofiber scaffolds has been reported
to promote osteoblast propagation and maturation by regulating
osteoblast-related osteopontin, osteocalcin, and alkaline
phosphatase expression via runt-related transcription factor 2
(21). We hypothesized that
co-cultured osteoblasts and prostate cancer cells on chitosan
nanofiber scaffolds would enable long-term culture under conditions
similar to those of living organisms.
The bone microenvironment is important for research,
given that bone metastases reflect the clinical picture of advanced
prostate cancer and form a major cause of disease morbidity.
Metastasis of prostate cancer cells to bone is a multi-step process
that involves detachment of cancer cells from the primary site,
migration of cells in the blood or lymph, attachment to bone
tissue, and development of tumors at the site of bone metastasis.
The interaction between prostate cancer cells and osteoblasts is
essential for bone metastasis (22,23).
Prostate cancer cells preferentially migrate to osteoblast-rich
areas of the bone (24,25). The physical contact between prostate
cancer cells and osteoblasts in bone destroys bone structure in the
presence of osteoclasts and develops a mutually enhanced growth
cycle of prostate cancer cells and osteoblasts (26). Excretion of numerous molecular
factors from bone-residing cells further promotes cancer cell
survival and metastatic progress. This phenomenon is known as the
‘feed forward cycle’ or ‘vicious cycle’ where transforming growth
factor-β (TGF-β) plays an essential role as promoter of tumor cell
survival and development in the bone microenvironment (27). TGF-β signaling is a double-edged
sword in cancer. In the early stages of tumorigenesis, TGF-β
signaling acts as a tumor suppressor, while in advanced stages, it
promotes epithelial-to-mesenchymal transition (EMT), invasion, and
metastatic potential (25). TGF-β
expressed in the tumor microenvironment also affects other cell
types, including immune cells and endothelial cells, causing immune
suppression and angiogenesis and promoting metastatic dissemination
of tumor cells (28).
In the present study, we established a novel
microenvironment model that mimics the bone microenvironment of
CRPC, which reflects the bone microenvironment of mCRPC. The
expression of TGF-β was enhanced and promoted EMT in C4-2 cells
co-cultured with osteoblasts. This is supported by previous reports
(29,30) and demonstrates the validity of our
model. Increased expression of TGF-β increases the viability of
C4-2 cells, which is related to drug resistance (31,32).
Co-culture with osteoblasts demonstrated increased expression of AR
and its downstream gene, PSA. TGF-β is involved in the upregulation
of AR and the acquisition of castration resistance (33), which may be responsible for the
difference in resistance to ARATs by co-culture and sensitivity in
drug sensitivity testing. In addition, co-culture with osteoblasts
showed increased expression of osteoblast stimulatory factors, bone
morphogenetic protein 2 (BMP2), and vascular endothelial growth
factor (VEGF-A, VEGF-B) (34). These
results suggest that our model may be useful for understanding the
molecular mechanisms of interaction between prostate cancer cells
and osteoblasts and for the detection of new molecular targets for
the treatment of bone metastasis. We aimed to stablish in
vitro drug sensitivity testing for new ARATs which were
testified to be effective in non-metastatic CRPC patients. These
patients are likely to harbor bone micrometastasis. The gene
expression profile in our model suggest early phase may be
representative of bone micrometastasis, and late phase may
represent clinical bone metastases. Therefore, our model may
include both non-metastatic and metastatic CRPC. In addition, the
combination of abiraterone and dutasteride had a synergistic effect
on the colony growth of C4-2 cells in the microenvironment model.
This could be attributed to the reduction of 3-keto-5α-abiraterone
which acts as an AR agonist. These results provide evidence that
the combination of abiraterone and dutasteride may be a clinically
more potent treatment for prostate cancer than the other ARATs. The
consistent phenomenon that the concentration of
3-keto-5α-abiraterone decreased with the combination of abiraterone
and dutasteride both from effective cases in phase II trials and
from our model using C4-2, means that this model may be valuable
for the assessment of the bone microenvironment in CRPC.
Regarding the previous in vitro model using
chitosan as a 3D substrate, a few publications have reported the
benefit of chitosan in combination with chondroitin acid or
alginate (35–37). Neither chondroitin acid nor alginate
was used in this model. However, these reports indicate that cells
form colonies and grow using scaffolds. We also observed the
induction of EMT by co-culturing osteoblasts and tumor cells in the
absence of chondroitin acid. The advantage of our model is the
co-culture of osteoblasts.
Recently, patient-specific models of solid tumors
using 3D cultures of spheroids and organoids from tumor cells or
biopsies have received much attention (38,39).
Organoids derived from patient tissue have been used for in
vitro screening of drug responsiveness prior to treatment to
determine treatment strategies and predict efficacy. In future
studies, patient-specific 3D models co-cultured with organoids from
patient prostate cancer tissue with osteoblasts using a chitosan
fiber matrix may contribute to tailored medicine.
A limitation of this model is that it does not
include osteoclasts, which are important factors in the bone
microenvironment. Osteoclasts in bone destroy bone structure and
develop a mutually enhanced growth cycle of prostate cancer cells
and osteoblasts. Since bone metastases of prostate cancer are
characterized as osteogenic rather than osteolytic, the lack of
osteoclasts may not be so important for this model. Establishing
mineralization by cancer cells should be the next challenge in
future studies. The other limitation of this in vitro study
is the lack of in vivo study. The mouse model of bone
metastasis using prostate cancer cells has been reported (40), and we would like to eventually
investigate the results of this study using a mouse model. The
other limitation of this model is that it only uses C4-2 cells as
the prostate cancer cell line. This is because it was necessary to
use a cell line to maintain AR activation and signaling through de
novo intratumoral steroidogenesis to mimic the bone
microenvironment of CRPC. Validation using several other CRPC cell
lines, or patient-derived cells (organoids) is needed in future
studies.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms Shizuka Ishii and
Ms Kiyomi Fujita (Department of Urology, Graduate School of
Medicine, Yamaguchi University, Ube, Yamaguchi, Japan) for their
technical support.
Funding
The present study was supported by a Grant-in-Aid
for Scientific Research Foundation B (KAKENHI) from the Japan
Society for the Promotion of Science (grant nos. 17H04330 and
B20H03806) and the Jansen Pharma non-clinical
investigator-initiated study (grant no. ARN-I-17-JPN-001-V01).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MS, MidM, MirM, HH, KU, SO, JM, RI, SY, YY and JH
contributed to the conception and design of this study. HirM, HH,
KU, JM and KT performed the experiments. HidM supervised the study
and contributed to manuscript writing. All authors have read and
approved the final manuscript. HidM and HH confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
Koji Tamada holds stocks in and receives
remuneration from Noile-Immune Biotech Inc., and received lecture
fees from Ono Pharmaceutical, MSD, AstraZeneca, Eli Lilly Japan and
Chugai Pharmaceutical. Hideyasu Matsuyama received funding from
Janssen Pharmaceutical. The other authors declare that they have no
competing interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hamdy FC, Donovan JL, Lane JA, Mason M,
Metcalfe C, Holding P, Davis M, Peters TJ, Turner EL, Martin RM, et
al: 10-Year outcomes after monitoring, surgery, or radiotherapy for
localized prostate cancer. N Engl J Med. 375:1415–1424. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Weckermann D, Polzer B, Ragg T, Blana A,
Schlimok G, Arnholdt H, Bertz S, Harzmann R and Klein CA:
Perioperative activation of disseminated tumor cells in bone marrow
of patients with prostate cancer. J Clin Oncol. 27:1549–1556. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
National Cancer Institute, . SEER Cancer
Stat Facts: Prostate Cancer. 2020.http://seer.cancer.gov/statfacts/html/prost.htmlNovember
14–2020
|
|
5
|
Sweeney CJ, Chen YH, Carducci M, Liu G,
Jarrard DF, Eisenberger M, Wong YN, Hahn N, Kohli M, Cooney MM, et
al: Chemohormonal therapy in metastatic hormone-sensitive prostate
cancer. N Engl J Med. 373:737–746. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wadosky KM and Koochekpour S: Molecular
mechanisms underlying resistance to androgen deprivation therapy in
prostate cancer. Oncotarget. 7:64447–64470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sumanasuriya S and Bono JD: Treatment of
advanced prostate cancer-A review of current therapies and future
promise. Cold Spring Harb Perspect Med. 8:a0306352018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pezaro C, Omlin A, Lorente D, Rodrigues
DN, Ferraldeschi R, Bianchini D, Mukherji D, Riisnaes R, Altavilla
A, Crespo M, et al: Visceral disease in castration-resistant
prostate cancer. Eur Urol. 65:270–273. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shiirevnyamba A, Takahashi T, Shan H,
Ogawa H, Yano S, Kanayama H, Izumi K and Uehara H: Enhancement of
osteoclastogenic activity in osteolytic prostate cancer cells by
physical contact with osteoblasts. Br J Cancer. 104:505–513. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Antunes J, Gaspar VM, Ferreira L, Monteiro
M, Henrique R, Jerónimo C and Mano JF: In-air production of 3D
co-culture tumor spheroid hydrogels for expedited drug screening.
Acta Biomater. 94:392–409. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ryan CJ, Smith MR, Fizazi K, Saad F,
Mulders PF, Sternberg CN, Miller K, Logothetis CJ, Shore ND, Small
EJ, et al: Abiraterone acetate plus prednisone versus placebo plus
prednisone in chemotherapy-naive men with metastatic
castration-resistant prostate cancer (COU-AA-302): Final overall
survival analysis of a randomised, double-blind, placebo-controlled
phase 3 study. Lancet Oncol. 16:152–160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Z, Alyamani M, Li J, Rogacki K, Abazeed
M, Upadhyay SK, Balk SP, Taplin ME, Auchus RJ and Sharifi N:
Redirecting abiraterone metabolism to fine-tune prostate cancer
anti-androgen therapy. Nature. 533:547–551. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Andriole GL, Bostwick DG, Brawley OW,
Gomella LG, Marberger M, Montorsi F, Pettaway CA, Tammela TL,
Teloken C, Tindall DJ, et al: Effect of dutasteride on the risk of
prostate cancer. N Engl J Med. 362:1192–1202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chou TC and Talalay P: Quantitative
analysis of dose-effect relationships: The combined effects of
multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 22:27–55.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsuyama H, Shiota M, Tashiro K, Kanji H,
Horiyama S, Eto M, Egawa S, Haginaka J and Inoue R: Phase II study
of the efficacy of abiraterone acetate with dutasteride for
castration resistant prostate cancer. J Clin Oncol. 39:112. 2021.
View Article : Google Scholar
|
|
17
|
Horoszewicz JS, Leong SS, Kawinski E, Karr
JP, Rosenthal H, Chu TM, Mirand EA and Murphy GP: LNCaP model of
human prostatic carcinoma. Cancer Res. 43:1809–1818.
1983.PubMed/NCBI
|
|
18
|
Cai C, Chen S, Ng P, Bubley GJ, Nelson PS,
Mostaghel EA, Marck B, Matsumoto AM, Simon NI, Wang H, et al:
Intratumoral de novo steroid synthesis activates androgen receptor
in castration-resistant prostate cancer and is upregulated by
treatment with CYP17A1 inhibitors. Cancer Res. 71:6503–6513. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rajendran D, Hussain A, Yip D, Parekh A,
Shrirao A and Cho CH: Long-term liver-specific functions of
hepatocytes in electrospun chitosan nanofiber scaffolds coated with
fibronectin. J Biomed Mater Res A. 105:2119–2128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hussain A, Collins G, Yip D and Cho CH:
Functional 3-D cardiac co-culture model using bioactive chitosan
nanofiber scaffolds. Biotechnol Bioeng. 110:637–647. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ho MH, Liao MH, Lin YL, Lai CH, Lin PI and
Chen RM: Improving effects of chitosan nanofiber scaffolds on
osteoblast proliferation and maturation. Int J Nanomedicine.
9:4293–4304. 2014.PubMed/NCBI
|
|
22
|
Kimura Y, Matsugaki A, Sekita A and Nakano
T: Alteration of osteoblast arrangement via direct attack by cancer
cells: New insights into bone metastasis. Sci Rep. 7:448242017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kingsley LA, Fournier PG, Chirgwin JM and
Guise TA: Molecular biology of bone metastasis. Mol Cancer Ther.
6:2609–2617. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang N, Docherty FE, Brown HK, Reeves KJ,
Fowles AC, Ottewell PD, Dear TN, Holen I, Croucher PI and Eaton CL:
Prostate cancer cells preferentially home to osteoblast-rich areas
in the early stages of bone metastasis: Evidence from in vivo
models. J Bone Miner Res. 29:2688–2696. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Klein CA: Selection and adaptation during
metastatic cancer progression. Nature. 501:365–372. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guise TA, Mohammad KS, Clines G, Stebbins
EG, Wong DH, Higgins LS, Vessella R, Corey E, Padalecki S, Suva L
and Chirgwin JM: Basic mechanisms responsible for osteolytic and
osteoblastic bone metastases. Clin Cancer Res. 12 (Suppl
1):6213S–6216S. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weilbaecher KN, Guise TA and McCauley LK:
Cancer to bone: A fatal attraction. Nat Rev Cancer. 11:411–425.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiechi A, Waning DL, Stayrook KR, Buijs
JT, Guise TA and Mohammad KS: Role of TGF-β in breast cancer bone
metastases. Adv Biosci Biotechnol. 4:15–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pickup M, Novitskiy S and Moses HL: The
roles of TGFβ in the tumour microenvironment. Nat Rev Cancer.
13:788–799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Juárez P and Guise TA: TGF-β in cancer and
bone: Implications for treatment of bone metastases. Bone.
48:23–29. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Montanari M, Rossetti S, Cavaliere C,
D'Aniello C, Malzone MG, Vanacore D, Di Franco R, La Mantia E,
Iovane G, Piscitelli R, et al: Epithelial-mesenchymal transition in
prostate cancer: An overview. Oncotarget. 8:35376–35389. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakazawa M and Kyprianou N:
Epithelial-mesenchymal-transition regulators in prostate cancer:
Androgens and beyond. J Steroid Biochem Mol Biol. 166:84–90. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shiota M, Itsumi M, Takeuchi A, Imada K,
Yokomizo A, Kuruma H, Inokuchi J, Tatsugami K, Uchiumi T, Oda Y and
Naito S: Crosstalk between epithelial-mesenchymal transition and
castration resistance mediated by Twist1/AR signaling in prostate
cancer. Endocr Relat Cancer. 22:889–900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fizazi K, Yang J, Peleg S, Sikes CR,
Kreimann EL, Daliani D, Olive M, Raymond KA, Janus TJ, Logothetis
CJ, et al: Prostate cancer cells-osteoblast interaction shifts
expression of growth/survival-related genes in prostate cancer and
reduces expression of osteoprotegerin in osteoblasts. Clin Cancer
Res. 9:2587–2597. 2003.PubMed/NCBI
|
|
35
|
Xu K, Wang Z, Copland JA, Chakrabarti R
and Florczyk SJ: 3D porous chitosan-chondroitin sulfate scaffolds
promote epithelial to mesenchymal transition in prostate cancer
cells. Biomaterials. 254:1201262020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu K, Ganapathy K, Andl T, Wang Z, Copland
JA, Chakrabarti R and Florczyk SJ: 3D porous chitosan-alginate
scaffold stiffness promotes differential responses in prostate
cancer cell lines. Biomaterials. 217:1193112019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang K, Kievit FM, Florczyk SJ, Stephen ZR
and Zhang M: 3D porous chitosan-alginate scaffolds as an in vitro
model for evaluating nanoparticle-mediated tumor targeting and gene
delivery to prostate cancer. Biomacromolecules. 16:3362–3372. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Elbadawy M, Abugomaa A, Yamawaki H, Usui T
and Sasaki K: Development of prostate cancer organoid culture
models in basic medicine and translational research. Cancers
(Basel). 12:7772020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bartucci M, Ferrari AC, Kim IY, Ploss A,
Yarmush M and Sabaawy HE: Personalized medicine approaches in
prostate cancer employing patient derived 3D organoids and
humanized mice. Front Cell Dev Biol. 4:642016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kuchimaru T, Kataoka N, Nakagawa K,
Isozaki T, Miyabara H, Minegishi M, Kadonosono T and Kizaka-Kondoh
S: A reliable murine model of bone metastasis by injecting cancer
cells through caudal arteries. Nat Commun. 30:29812018. View Article : Google Scholar : PubMed/NCBI
|