Introduction
Acute myeloid leukemia (AML) results from
accumulation of abnormal myeloblasts, most commonly in the bone
marrow, leading to bone marrow failure and death (1). Cellular hypoxia in solid tumors, such
as kidney cancer and colon cancer (2) is also present in hematologic
malignancies, such as AML and lymphoma (3). Although, normal mammalian bone marrow
is relatively hypoxic compared with other tissues, such as liver
and kidney, hematopoietic stem cells can be found in the
lowest-oxygen microenvironment of the bone marrow (4). Leukemic stem cells reside in the most
hypoxic areas within the hematopoietic stem/progenitor cells niche
(5,6). Under hypoxic conditions,
hypoxia-inducible factor 1α (HIF-1α) is transferred into the
nucleus, via the hypoxia response element (HRE)-specific binding
regulating gene expression (7).
HIF-1α regulates target genes, such as vascular
endothelial growth factor, E-cadherin and Bcl-2 involved in several
physiological processes, such as erythropoiesis and angiogenesis,
and in certain pathological processes, including cancer cell
metastasis and cancer stem cell self-renewal (8). It was found that HIF-1α was upregulated
in childhood acute lymphoblastic leukemia bone marrow blasts
(9). HIF-1α expression has been
found to be associated with poor survival prognosis in adult acute
myeloid leukemia (AML) with normal karyotype (10). However, it has also been demonstrated
that HIF-1α can induce AML cell differentiation and inhibit the
progression of AML (11). The role
of hypoxia and HIF remains controversial depending on the study and
model, and hence requires further research.
Abnormal regulation of epigenetic modification in
hematological malignancy can lead to transcriptional silencing of
tumor suppressor genes, such as cyclin-dependent kinase inhibitor
2B [p15(INK4B)] and hypermethylated in cancer 1 (HIC-1) and
increase in abnormal clones (12).
Previous studies have found that epigenetic modification serves an
important role in hypoxia response (13,14). It
has been reported that hypoxia causes demethylation of the CpG
island and expression of Wilms' tumor gene (WT1) mRNA in myeloid
leukemia cells (15). Whether
hypoxia and HIF-1 are involved in other epigenetic modifications
requires further research.
As an α-ketoglutarate and Fe (II)-dependent
oxygenase, Tet methylcytosine dioxygenase 2 (TET2) catalyzes the
conversion of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine
(5-hmC), and 5-hmC leads to the demethylation of cytosine (16). The TET2 gene is expressed in various
human tissues, such as heart and placenta (17), but the highest expression has been
identified in hematopoietic cells (18). Since Delhommeau et al
(19) reported the expression of
TET2 in myeloid malignancies in 2009, extensive attention has been
paid to the significance of this gene in hematologic malignancies.
Recently, it was reported that TET2 also participated in the
regulation of the immune system, TET activity is a biomarker for
predicting the efficiency of anti-programmed cell death protein
1/programmed cell death ligand 1 therapy in solid tumors (20).
The effect of hypoxia on the methylation of AML
cells was observed on 2 levels: Methylation at the genome level and
methylation of specific genes (21).
The p15 (INK4B) gene was chosen as the specific gene for
investigation of methylation in the present study. As a tumor
suppressor gene (22), p15(INK4B)
can affect the cell proliferation cycle by inhibiting the
expression of cyclin-dependent kinase (CDK) 4 and CDK6 (23). In the occurrence and development of
various hematologic malignancies, such as myelodysplastic syndrome
and AML, p15(INK4B) gene inactivation has been demonstrated to be
caused by the abnormal methylation of CpG islands (24). The mechanism of p15(INK4B) gene
methylation remains unclear (25)
and p15(INK4B) gene methylation under hypoxia has not been
reported.
As an important DNA hydroxymethylase and a tumor
suppressor gene, the mechanism of TET2 gene regulation remains
unclear (26). Bioinformatics
analysis revealed two HREs (5′-caCGTG-3′) at −1078-1075 and −12-9
bp of the TET2 gene promoter. Therefore, we hypothesized that
HIF-1α may regulate TET2 gene expression by binding to its promoter
region, hence serving an important role in the epigenetic
regulation.
Based on the above hypothesis, the aim of the
present study is to observe the changes in genome methylation
status under hypoxia in an AML cell line KG-1 and study the effect
of hypoxia and HIF-1α on TET2 gene expression and its function. In
addition, the relevant regulatory mechanism will be explored. The
association between hypoxic metabolism and epigenetic modification
in leukemic cells was revealed by the present study.
Materials and methods
Cell culture and hypoxia
KG-1, a human AML cell line was provided by Procell
Life Science & Technology Co., Ltd. The cells were cultured in
Iscove's modified Dulbecco's medium (Thermo Fisher Scientific,
Inc.) supplemented with 20% FBS (Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin at 37°C. Normoxic cells were
incubated at 21% O2 and 5% CO2. Physical
hypoxic cells were incubated in the hypoxic incubator (Thermo
Fisher Scientific, Inc.), and in an atmosphere of 1/3%
O2, 92/94% N2 and 5% CO2 for 24,
48 and 72 h.
Cell Counting Kit (CCK)-8 assay
Proliferation of KG-1 cells was detected using the
CCK-8 assay (Dojindo Molecular Technologies, Inc.) according to the
manufacturer's instructions. Briefly, the cells were seeded
(5×104 cells/ml) and incubated at 37°C in 96-well plates
for 24, 48 and 72 h. A total of 10 µl CCK-8 solution was added to
the cells and incubated for 2 h in the dark. Absorbance [optical
density (OD) values] was measured at 450 nm using a microplate
reader (Bio-Rad Laboratories, Inc.).
Lactate dehydrogenase (LDH) assay
Cell cultures were collected (5×104
cells/ml) and added to a 96-well plate using an LDH kit (Nanjing
Jiancheng Bioengineering Institute), according to the
manufacturer's instructions. After the sample was added and
incubated, The absorbance was read at 450 nm on a microplate reader
(Bio-Rad Laboratories Inc.).
Annexin V/propidium iodide (PI)
apoptosis detection assay
Both early and late stages of apoptosis were
detected using a FITC Annexin V Apoptosis Detection kit (cat. no.
556547; BD Biosciences). Cells were harvested, resuspended in 100
µl binding buffer, labeled with 5 µl Annexin V-FITC, followed by 5
µl PI and incubated for 15 min at room temperature (25°C) in the
dark. Next, 400 µl binding buffer was added and cells were then
analyzed by flow cytometry within 1 h. Data were analyzed using
FlowJo v.7.6.1; FlowJo, LLC following flow cytometry on a
FACSCalibur flow cytometer (BD Biosciences).
Global DNA methylation assessment
Genomic DNA was extracted from KG-1 cells using a
Genomic DNA kit (cat. no. DP304; Tiangen Biotech Co., Ltd.). The
global DNA methylation level was quantified in 100 ng genomic DNA
using the MethylFlashTM Methylated DNA Quantification kit (cat. no.
P-1034; EpiGentek Group, Inc.), following manufacturer's
instructions. Briefly, the methylated DNA was detected using
capture and detection antibodies according to the manufacturer's
instructions for 5-mC and then quantified calorimetrically by
measuring absorbance at 450 nm using the PowerWave HT Microplate
Spectrophotometer (BioTek Instruments, Inc.). The amount of
methylated DNA was proportional to the OD intensity measured.
Relative quantification was used to calculate the percentage of
5-mC in total DNA as described by the manufacturer's instructions.
Each sample was run in duplicate.
5-hmC level assay
5-hmC levels of genomic DNA were quantified using
the MethylFlash™ Hydroxymethylated DNA Quantification kit (cat. no.
P-1036; EpiGentek Group, Inc.). According to the manufacturer's
instructions, the hydroxymethylated fraction of DNA was detected
through the capture and detection of antibodies, followed by
colorimetric quantification. To quantify the absolute amount of
hydroxymethylated DNA, first, a standard curve was generated
between OD values and the amount of positive control at each
concentration point, the slope of the standard curve was determined
using linear regression, and then the amount of 5-hmC in the total
DNA was calculated using the following formula: 5-hmC%=(Sample
OD-Negative control OD)/(SlopexInput DNA amount) ×100%. Each sample
was run in duplicate.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted using TRIzol®
reagent (Thermo Fisher Scientific, Inc.). An equal amount (3 µg) of
total RNA was used for cDNA synthesis with the MMLV Reverse
Transcription kit (Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. The cDNA was amplified and
quantified using a SYBR Green qPCR kit (cat. no. 330521; Thermo
Fisher Scientific, Inc.). The thermocycling conditions used for
RT-qPCR were as follows: Thermal denaturation for 5 min at 95°C;
amplification for 30 sec at 95°C, 60°C for 30 sec for 40 cycles;
and final extension at 72°C for 10 min. After all PCR reactions are
completed, the temperature was maintained at 4°C. The following
primer sequences were used for RT-qPCR: HIF-1α forward,
5′-TCTCAGAATGAAGTGTACCCTAA-3′ and reverse,
5′-TCACAAATCAGCACCAAGC-3′; TET2 forward,
5′-CCCACAGAGACTTGCACAACAT-3′ and reverse,
5′-CTGGCTCTGCTAACATCCTGAC-3′; and β-actin forward,
5′-TTCCAGCCTTCCTTCCTGGG-3′ and reverse, 5′-TTGCGCTCAGGAGGAGCAAT-3′.
To test the specificity of the PCR reaction, products were
subjected to melting curve analysis and conventional 2% agarose gel
electrophoresis to rule out the synthesis of unspecific products.
All quantitative assays were performed in duplicate. Values
obtained for the target gene expression were normalized to β-actin
and calculated using the 2−ΔΔCq method (27).
Western blotting
KG-1 cells were collected and lysed using RIPA lysis
buffer (Thermo Fisher Scientific, Inc.) containing protease and
phosphatase inhibitors (Roche Applied Science). The supernatants
were collected via centrifugation at 12,000 × g for 15 min at 4°C.
Total protein was quantified using the bicinchoninic acid (BCA)
protein assay kit (Beyotime Institute of Biotechnology). Proteins
were resolved using 10% SDS PAGE, transferred onto PVDF membranes
(mass of protein loaded was 20 µg/lane) and blocked with 5% skimmed
milk for 1.5 h at room temperature. The membranes were incubated
with primary antibodies (all 1:1,000), mouse-anti-HIF-1α monoclonal
antibody (cat. no. H1alpha67; Abcam), rabbit-anti-TET2 polyclonal
antibody (cat. no. ab94580; Abcam) or rabbit-anti-β-actin
monoclonal antibody (cat. no. 4967; Cell Signaling Technology,
Inc.) respectively, overnight at 4°C. Following the primary
incubation, membranes were incubated with secondary antibodies
(1:20,000; cat. no. 926-32211; LI-COR Biosciences) for 1 h at room
temperature. The membranes were visualized using an enhanced
chemiluminescence kit (EMD Millipore). Semi-quantitative analysis
was performed using ImageJv.1.8.0 software (National Institutes of
Health).
Transfection with
lentivirus-HIF-1α-overexpression
Lentiviral vectors encoding HIF-1α overexpression
were generated using the GV358 vector (Shanghai GeneChem Co., Ltd.)
and designated as GV358-HIF-1α. The empty vector (GV358) was used
as a negative control. Lentiviral infection was conducted following
the manufacturer's instructions (2nd generation system). Briefly,
Lentiviral vectors (GV358) encoding containing HIF-1α
overexpression (Shanghai GeneChem Co., Ltd.) and empty control were
transduced into KG-1 cells (Procell Life Science & Technology
Co., Ltd.). A total of 5×104 cells/ml cell suspension
with complete medium (80% IMDM media +20% FBS) was prepared and 2
ml cell suspension/well was inoculated into 6 well plates. Cells
were incubated at 37°C for 24 h until they reached 20% confluence.
During transduction, 1 ml of medium was discarded, and 40 µl of
HiTransG infection solution (Shanghai GeneChem Co., Ltd.) and 40 µl
of lentivirus solution (MOI was 20 according to the pre infection
experiment and the virus titer was 1×108 TU/ml) were
added and cells were cultured at 37°C. After 16 h, the medium was
replaced with fresh medium. Transduction efficiency was observed
via light and fluorescence microscopy (magnification, ×100) after
48 h. The successfully transduced cells had green fluorescence as
there were GFP labelled in the lentivirus particles. Subsequent
experiments were performed 48 h after lentivirus infection. There
were 4 groups in the study: GV358 + normoxia, GV358-HIF-1α +
normoxia, GV358 + hypoxia and GV358-HIF-1α + hypoxia.
Suppression of HIF-1α expression by
3-(5′-hydroxymethyl-2′-furyl)-1-benzylindazole (YC-1)
YC-1 is a potential anticancer agent that suppresses
HIF-1α expression in cancer cells (28). YC-1 (Merck KGaA) was dissolved in
dimethyl sulfoxide (DMSO; Merck KGaA). Cells were incubated with
YC-1 at a final concentration of 10 µmol/l for 24 h before the
induction of both normoxia and hypoxia. There were 4 groups in the
study: DMSO + normoxia, YC-1 + normoxia, DMSO + hypoxia and YC-1 +
hypoxia.
Methylation-specific PCR (MSP)
Genomic DNA was extracted from cells using a Genomic
DNA kit (Tiangen Biotech Co., Ltd.). MSP was performed to evaluate
the methylation status of gene promoters as previously described
(29). Bisulfite conversion of DNA
was performed using the EZ DNA Methylation-Gold kit (cat. no.
D5005; Zymo Research Corp.) according to the manufacturer's
instructions. The primers used for the p15(INK4B) gene methylation
assay were previously described (29). Methylated-specific primers forward,
5′-GCGTTCGTATTTTGCGGTT-3′ and reverse,
5′-CGTACAATAACCGAACGACCGA-3′; unmethylated-specific primers
forward, 5′-TGTGATGTGTTTGTATTTTGTGGTT-3′ and reverse,
5′-CCATACAATAACCAAACAACCAA-3′. Reactions were carried out in a
total volume of 20 µl, containing 1 µl bisulfite-modified DNA, 2 µl
10X PCR buffer, 1.6 µl dNTP mixture (2.5 mM each), 0.5 µl forward
primer (20 µM), 0.5 µl reverse primer (20 µM), 0.1 µl Taq Hot Start
Polymerase (5 U/µl; Tiangen Biotech Co., Ltd.) and 14.3 µl
distilled (d)H2O. Reaction mixtures were denatured at
95°C for 2 min. Amplification was then performed for 35 cycles
(95°C for 30 sec, 60°C for 30 sec and 72°C for 40 sec), followed by
a final extension at 72°C for 5 min. DNA from control subjects was
used as a negative control. Results from duplicate experiments were
used to determine methylation status.
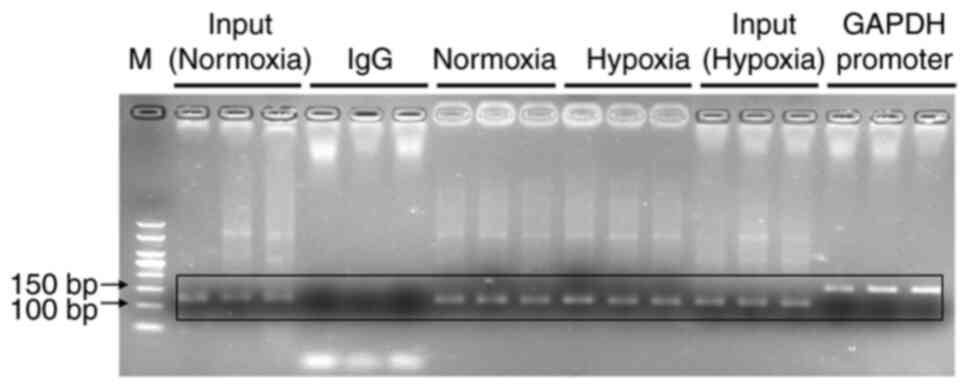
Chromatin immunoprecipitation
(ChIP)-qPCR assays
ChIP-qPCR assays were performed as previously
described (30) and using an
EZ-Magna ChIP A/G kit (EMD Millipore) according to the
manufacturer's instructions. Briefly, KG-1 cells were cross-linked
with 1% paraformaldehyde at 37°C for 10 min and sonicated at 0°C
for 60 sec. The genomic DNA was digested to an average size of
100–1,000 bp. Solubilized chromatin was immunoprecipitated with
antibodies against HIF-1α (5 µg; cat. no. H1alpha67; Abcam) or
negative control IgG antibody (2 µl; cat. no. 2729S; Cell Signaling
Technology, Inc.) at 4°C overnight. Following proteinase K
treatment and crosslink reversal (62°C for 2 h followed by 95°C for
10 min), immunoprecipitated DNA underwent phenol-chloroform
extraction with ethanol precipitation. Immunoprecipitated DNA was
analyzed using RT-qPCR. PCR amplification was conducted using the
precipitated DNA fragment as a template and the relevant primers.
The primers used were as follows: Normoxia and hypoxia groups were
analyzed by PCR using a HRE primer (forward,
5′-GCCTGGCCAATATGGTGAAACC-3′ and reverse,
5′-CGATTCTTCTGCCTCAGCCTC-3′; 111 bp). The positive and negative
control groups were analyzed by PCR using the GAPDH promoter primer
[forward, 5′-TACTAGCGGTTTTACGGGCG-3′ and reverse,
5′-TCGAACAGGAGGAGCAGAGAGCGA-3′ (166 bp)]. The experiments were
performed in triplicate.
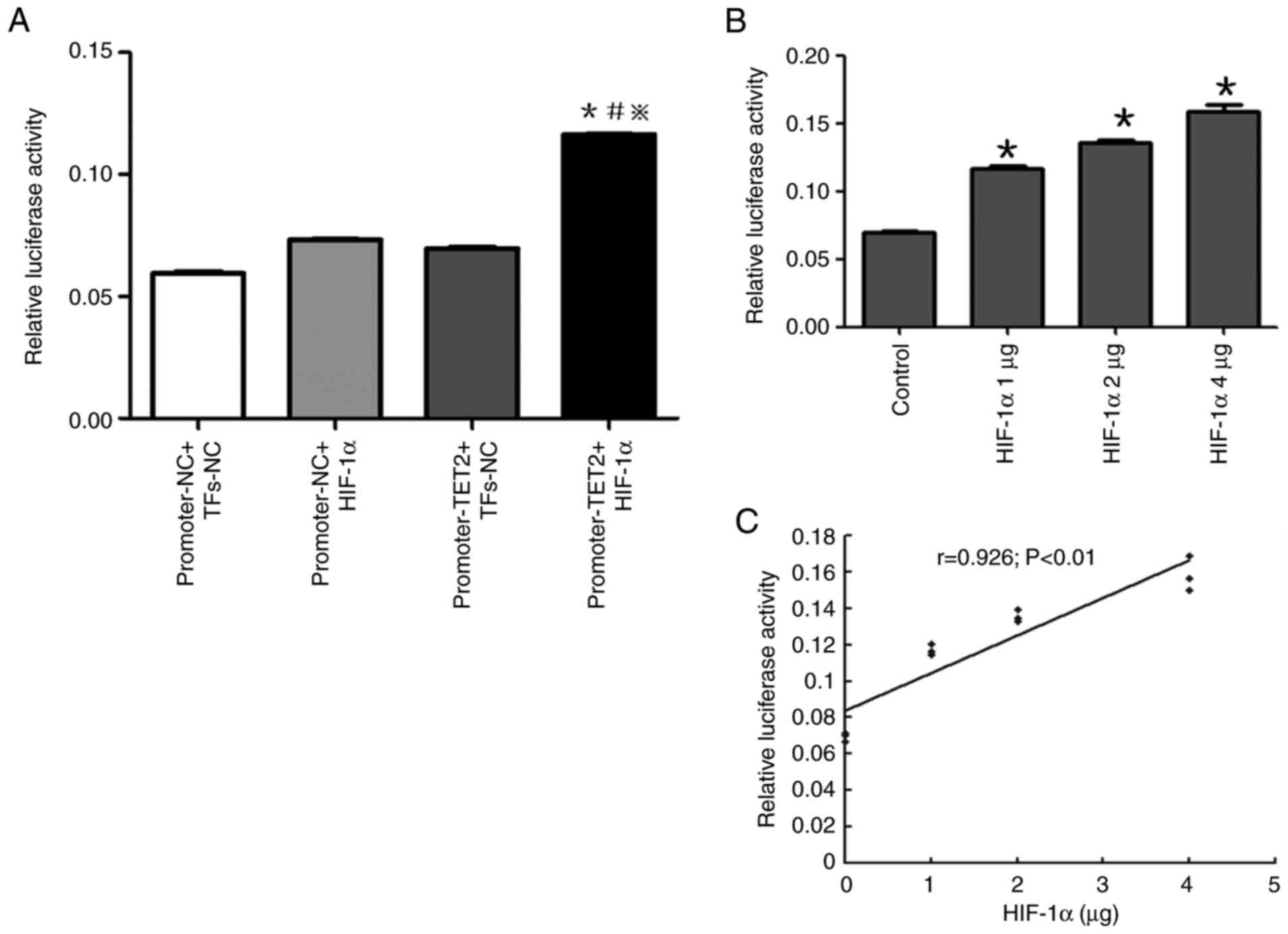
Luciferase reporter assays
GV354-TET2 promoter luciferase reporter and
GV219-HIF-1α overexpression vectors were constructed by GeneChem,
Inc. The GV354-TET2 promoter luciferase reporter vector contained 2
kb 5′-flanking region and 550 bp 5′-untranslated region of the TET2
gene and authenticity was verified by sequencing (data not shown).
For the luciferase reporter assay, KG-1 cells were seeded
(5×104 cells/ml) in a 24-well plate. Transfections were
performed using X-tremeGENE HP DNA Transfection Reagent (Roche
Diagnostics). Each transfection experiment contained 500 ng
GV354-TET2 promoter luciferase reporter vector/empty vector, a
different dose of GV219-HIF-1α overexpression vector (0, 1, 2 and 4
µg)/empty vector and 20 ng Renilla expression vector (Thermo
Fisher Scientific, Inc.). After 48 h of transfection, cells were
harvested and lysed. Then the firefly and Renilla luciferase
activities were measured using the Dual-Glo® Luciferase
Assay System Protocol (Promega Corporation). The Renilla
luciferase activity was set as the internal control.
Statistical analysis
All experiments were repeated 3 times and the data
were presented as the mean ± standard deviation. Statistical
analyses were performed using GraphPad Prism 8 (GraphPad Software,
Inc.) and SPSS 20.0 statistical software package (IBM Corp.).
Comparisons between 2 groups were conducted using the unpaired
Student's t-test and comparisons among multiple groups were
performed using two-way ANOVA followed by the post hoc Tukey's
test. The correlation analysis was performed using Pearson
correlation. P<0.05 was considered to indicate a statistically
significant difference.
Results
Hypoxia increases proliferation,
enhances metabolism and inhibits apoptosis in KG-1 cells
The acute myeloid cell line KG-1 cells were treated
with different oxygen concentrations (21, 3 and 1%) for 24, 48 and
72 h at 37°C. The proliferation, destruction and apoptosis of KG-1
cells were detected by CCK-8 assay, lactate dehydrogenase assay and
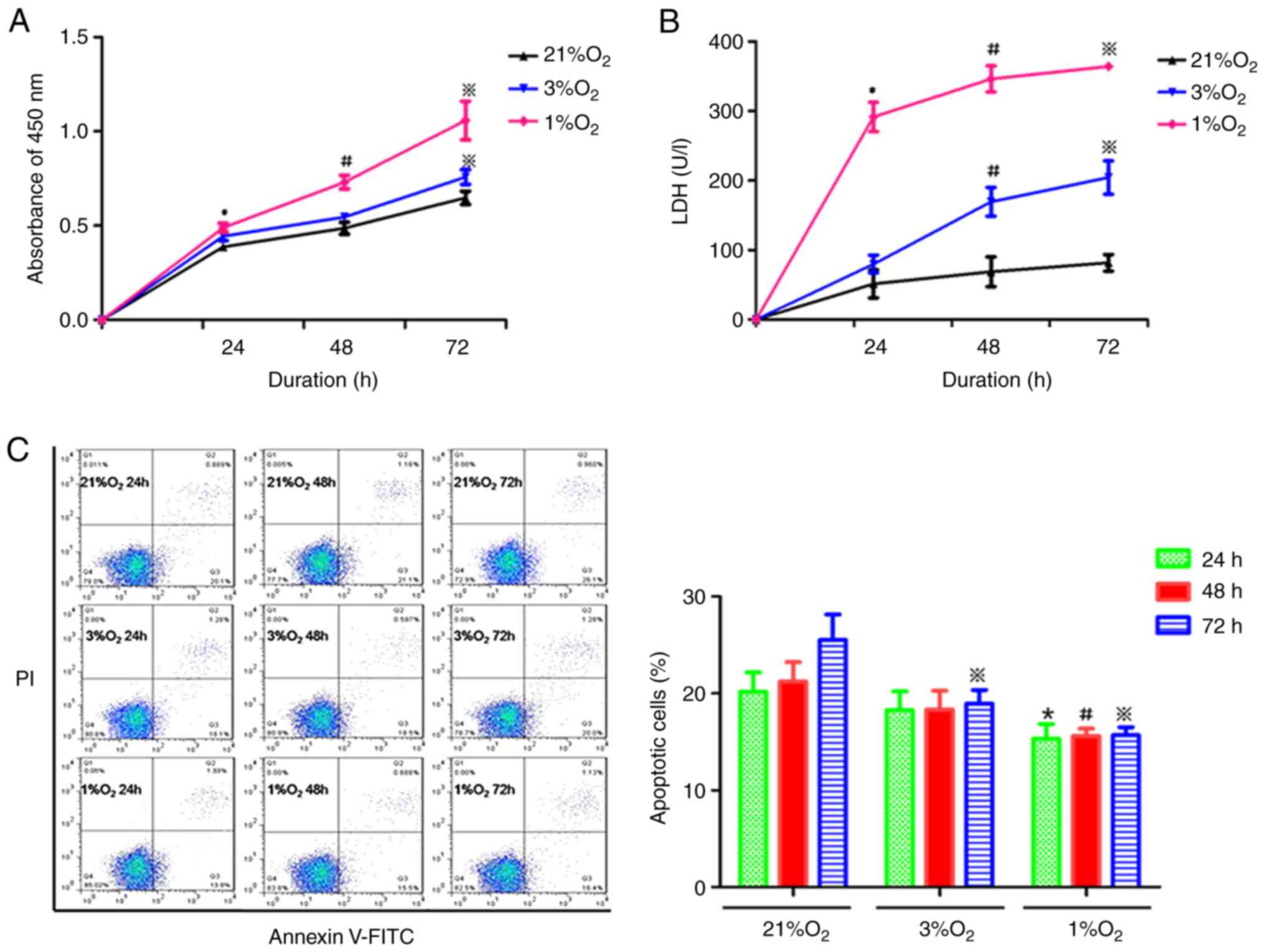
Annexin V-FITC/PI staining, respectively. As shown in Fig. 1A, proliferation was significantly
increased in KG-1 cells under hypoxic compared with normoxia. With
the decrease of oxygen concentration and prolongation of hypoxia,
the proliferation activity of KG-1 cells was gradually enhanced
(Fig. 1A). The LDH content in the
hypoxia cell culture medium was increased compared with the
normoxia medium (Fig. 1B). LDH
content was increased with the prolonged hypoxia time and the
oxygen concentration decreased, suggesting that hypoxia increased
the destruction and metabolism of KG-1 cells. The apoptotic rate of
KG-1 cells was decreased with the decrease in oxygen concentration,
while the duration of hypoxia had little effect on the rate of cell
apoptosis (Fig. 1C). In combination,
these data suggested that hypoxia could increase the proliferation,
enhance the metabolism and inhibit the apoptosis of KG-1 cells.
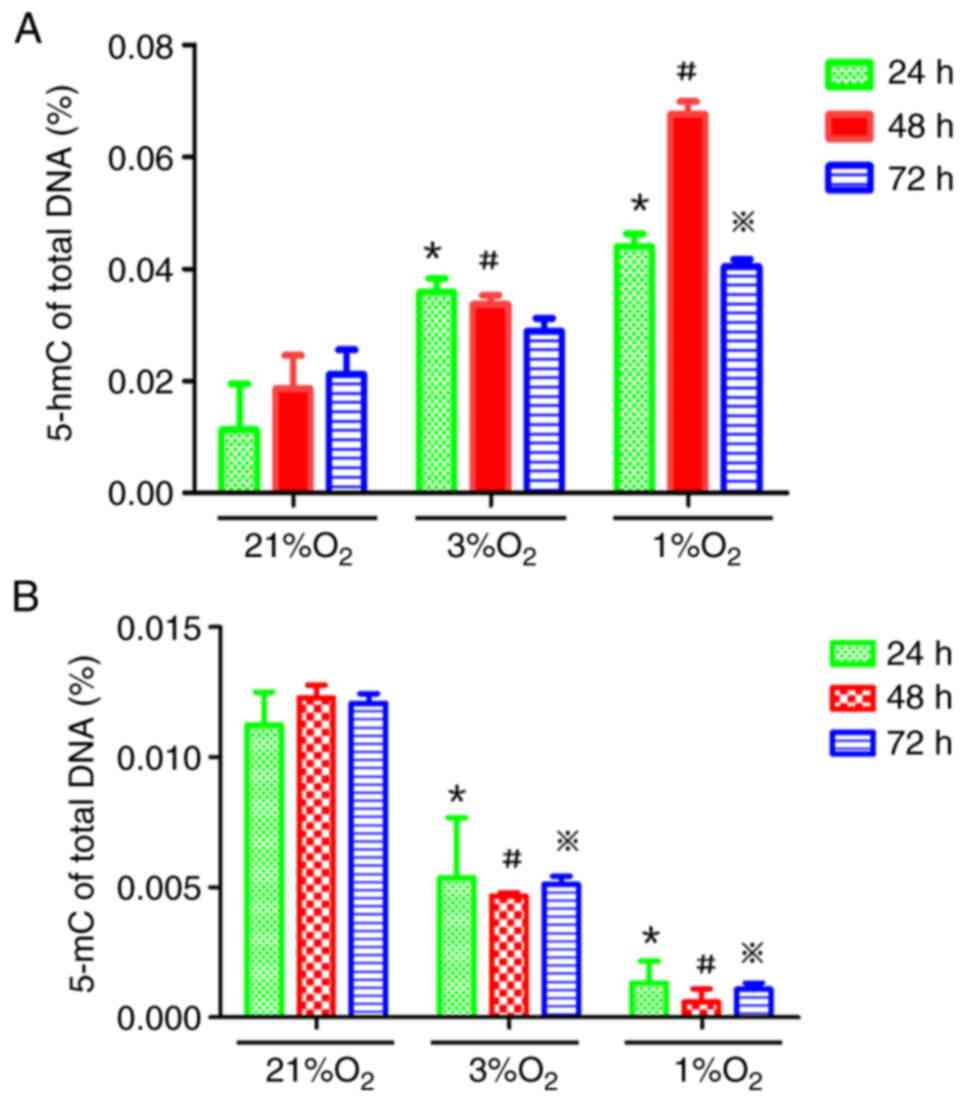
Hypoxia increases global 5-hmC levels
in KG-1 cells
KG-1 cells were treated with different oxygen
concentrations (21, 3 and 1%) for 24, 48 and 72 h and then the
genomic demethylation and methylation status were detected using
5-hmC and 5-mC detection kits, respectively. The 5-hmC content in
the hypoxia group was higher than that in the normoxia group and
the 5-mC content in the hypoxia group was lower than that in the
normoxia group, suggesting that hypoxia could reduce the methylated
status of the genome in KG-1 cells, while hypoxia time under the
same oxygen concentration had little effect on the methylated
status of the genome (Fig. 2A and
B).
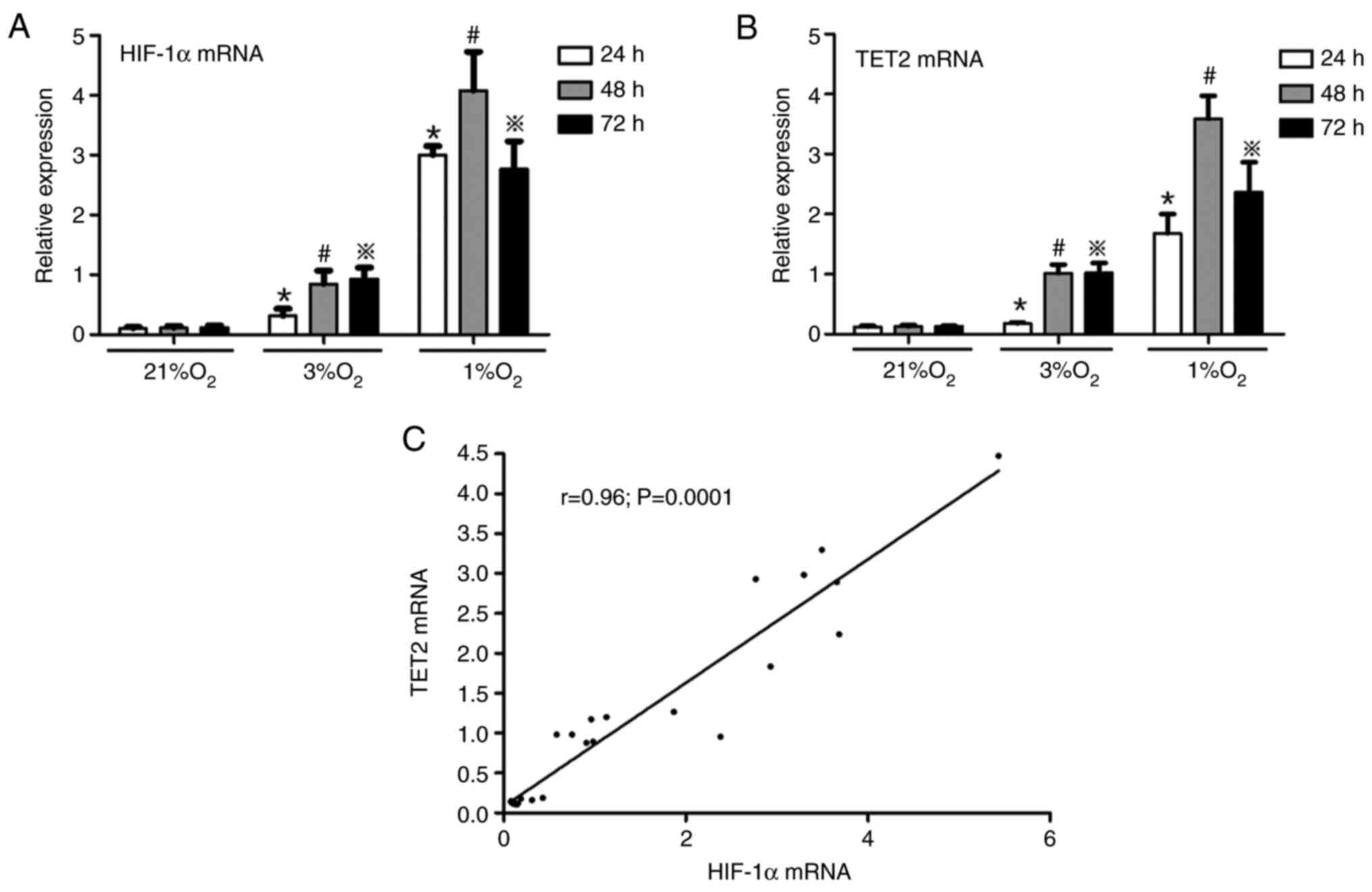
Hypoxia increases HIF-1α and TET2
expression in KG-1 cells
KG-1 cells were treated with different oxygen
concentrations (21, 3 and 1%) for 24, 48 and 72 h, and then the
mRNA and protein levels of HIF-1α and TET2 were investigated by
RT-qPCR and western blotting, respectively. It was demonstrated
that hypoxia promoted the expression of HIF-1α and TET2 mRNA in
KG-1 cells and the mRNA levels of HIF-1α and TET2 were the highest
in the 1% O2 48 h group (Fig.
3A and B). Simultaneously, under the same hypoxia
concentration, with the extension of hypoxia time, the mRNA
expression of HIF-1α and TET2 were increased (Fig. 3A and B). A positive correlation was
identified between the HIF-1α and TET2 mRNA expression levels
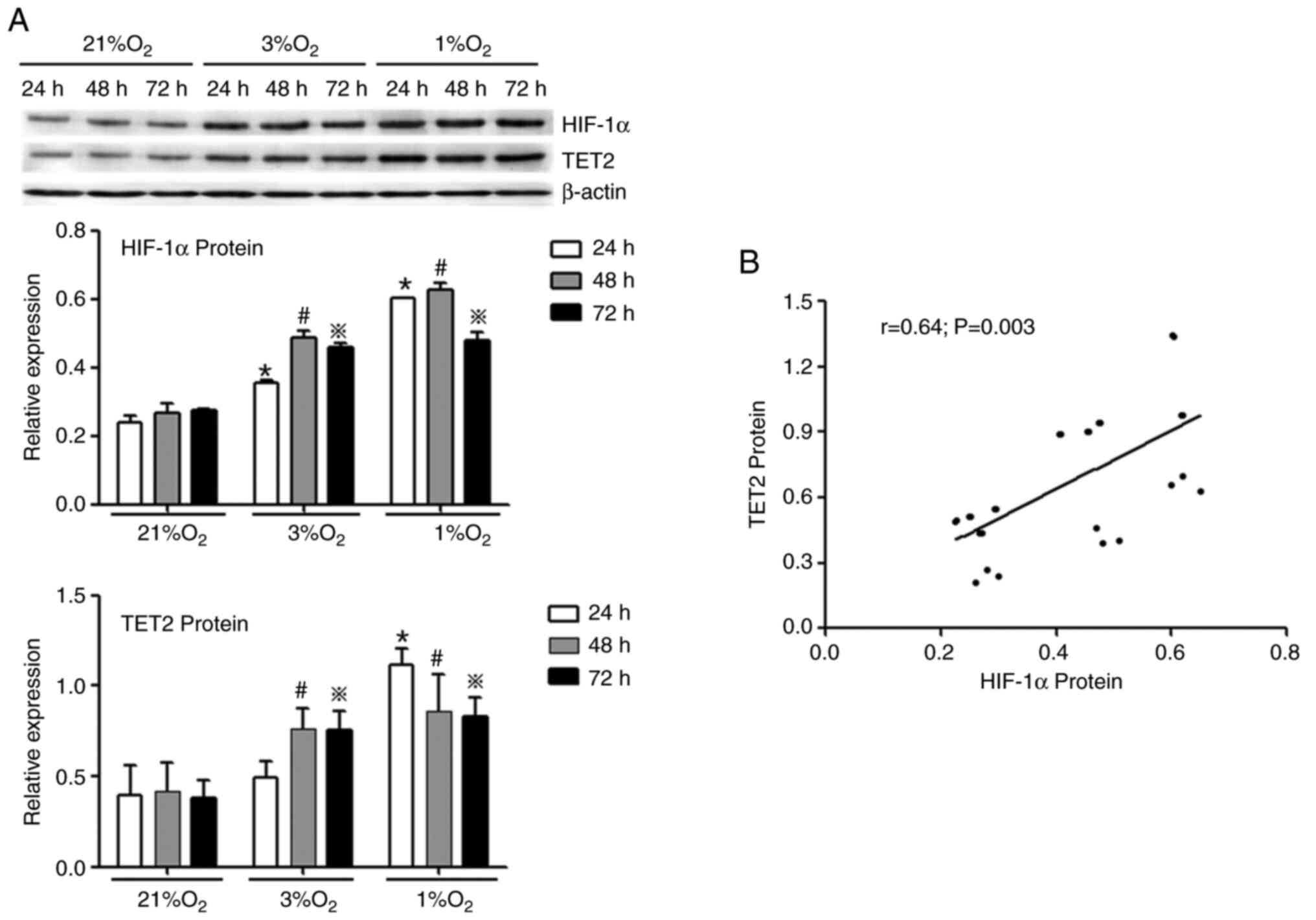
(Fig. 3C). In concert, hypoxia
promoted the protein expression of HIF-1α and TET2 in KG-1 cells
compared with normoxia (Fig. 4A) and
the HIF-1α protein level was positively correlated with the TET2
protein level (Fig. 4B). Overall,
the 1% O2 for 48 h was selected as the hypoxic condition
to be used for subsequent experiments.
HIF-1α overexpression increases TET2
expression, 5-hmC level and p15(INK4B) gene demethylation in KG-1
cells
Lentiviral transfection of HIF-1α eukaryotic
expression plasmid was used to induce HIF-1α overexpression in
cells. The 1% O2 for 48 h was selected for the hypoxic
conditions (1% O2, 94% N2 and 5%
CO2). There were 4 groups: GV358 + normoxia,
GV358-HIF-1α + normoxia, GV358 + hypoxia and GV358-HIF-1α +
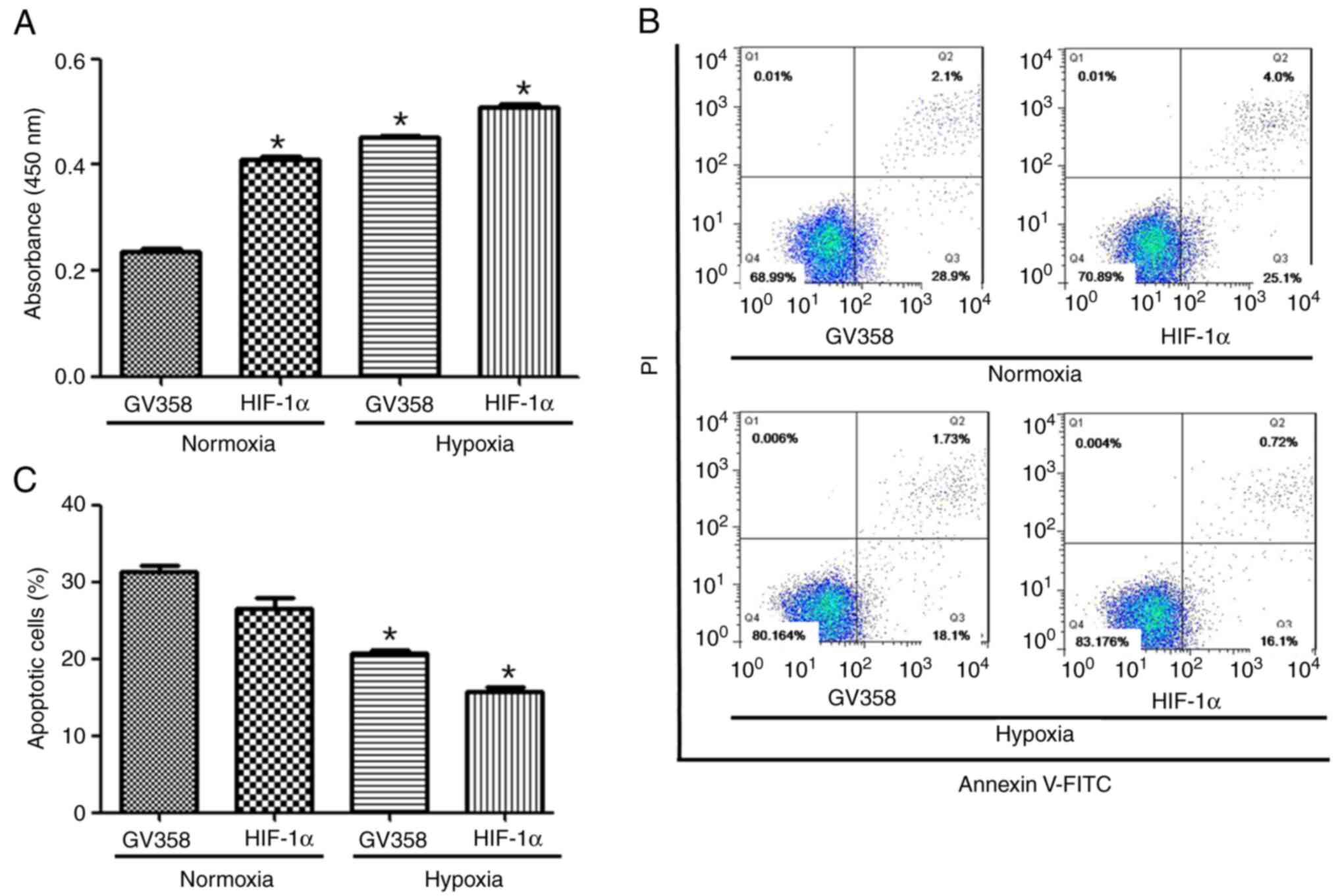
hypoxia. Cell proliferation were increased in the GV358-HIF-1α +
normoxia group compared with the GV358 + normoxia group (Fig. 5A). The proliferation were increased
in HIF-1α-overexpressing cells under both normoxia and hypoxic
conditions compared with non-HIF-1α-overexpressing cells (Fig. 5A). The apoptotic rate of KG-1 cells
was decreased in HIF-1α-overexpressing cells and/or hypoxic
conditions compared with non-HIF-1α-overexpressing cells (Fig. 5B and C).
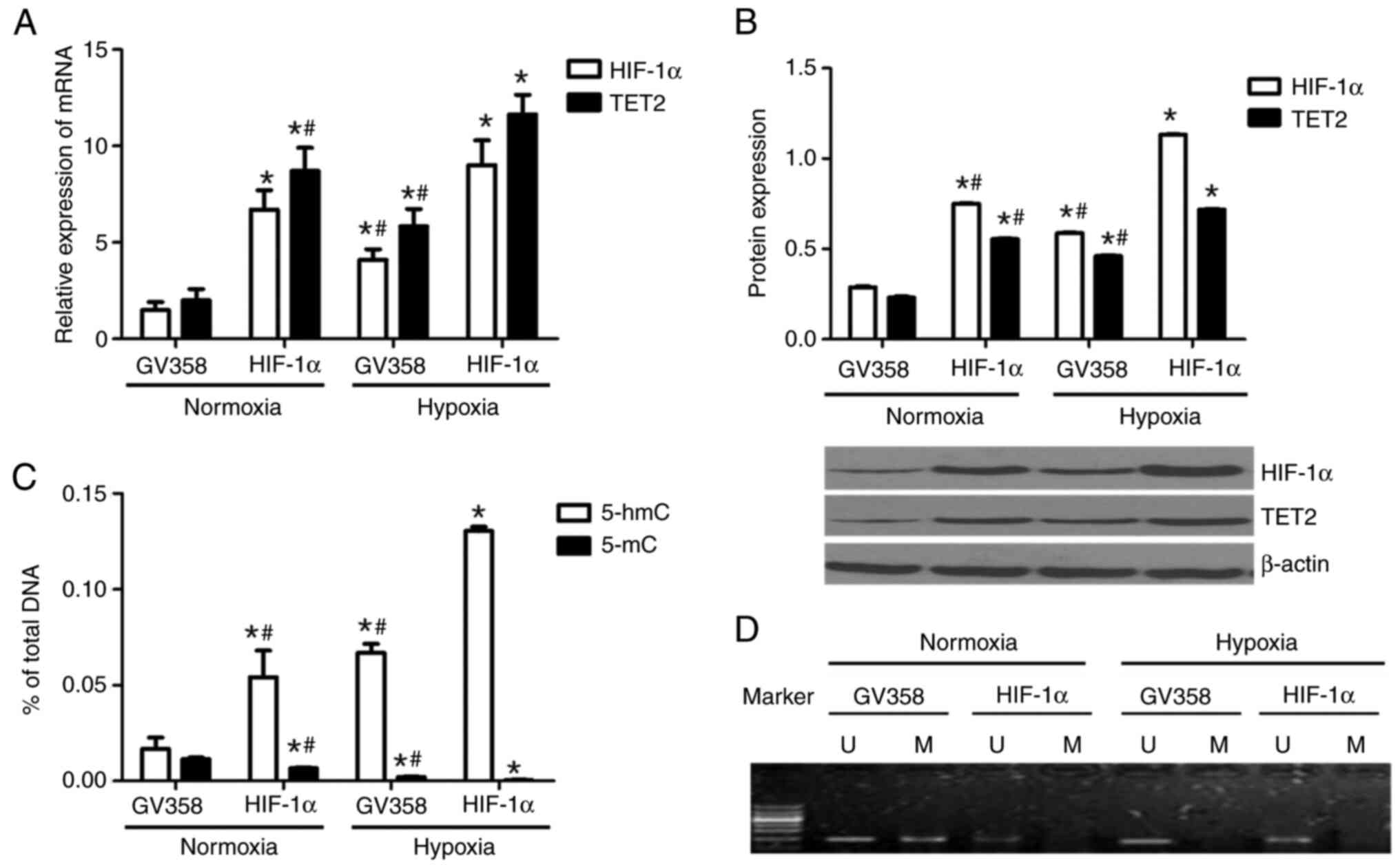
TET2 mRNA and protein levels were increased in the
GV358-HIF-1α + normoxia group compared with the GV358 + normoxia
group (Fig. 6A and B). TET2
expression in the GV358-HIF-1α + hypoxia group was significantly
higher compared with in the GV358-HIF-1α + normoxia group (Fig. 6A and B). Compared with the GV358 +
hypoxia group, TET2 expression in KG-1 cells was also increased in
the GV358-HIF-1α + hypoxia group (Fig.
6A and B). Meanwhile, the HIF-1α mRNA and protein levels were
upregulated in HIF-1α-overexpressing cells under both normoxia and
hypoxic conditions (Fig. 6A and B).
This indicated that the TET2 mRNA and protein levels were
upregulated in HIF-1α-overexpressing cells under both normoxia and
hypoxic conditions (Fig. 6A and
B).
Compared with the GV358 + normoxia group, the
content of 5-hmC in the genome of KG-1 cells was increased in the
GV358-HIF-1α + normoxia group (Fig.
6C). The content of 5-hmC in the GV358-HIF-1α + hypoxia group
was significantly higher compared with the GV358-HIF-1α + normoxia
group (Fig. 6C). Compared with the
GV358 + hypoxia group, the 5-hmC content in the GV358-HIF-1α +
hypoxia group was also increased (Fig.
6C). This indicated that the overexpression of HIF-1α promoted
the demethylation of KG-1 cells under both normoxic and hypoxic
conditions (Fig. 6C).
Compared with the GV358 + normoxia group, the 5-mC
content was decreased in the GV358-HIF-1α + normoxia group
(Fig. 6C). The 5-mC content in the
GV358-HIF-1α + hypoxia group was lower compared with that of the
GV358-HIF-1α+ normoxia group (Fig.
6C). Compared with the GV358 + hypoxia group, 5-mC content was
also decreased in the GV358-HIF-1α + hypoxia group (Fig. 6C).
The p15(INK4B) promoter region of KG-1 cells
demonstrated partial methylation under normoxia and it was
demethylated after hypoxia or/and HIF-1α overexpression (Fig. 6D). In summary, the overexpression of
HIF-1α promoted the demethylation of KG-1 cells by increasing TET2
expression.
HIF-1α suppression decreases TET2
expression, 5-hmC level and p15(INK4B) gene demethylation in KG-1
cells
HIF-1α inhibitor YC-1 (final concentration, 10
µmol/l; 24 h) was added to the cells to inhibit the expression of
HIF-1α. The cells were divided into 4 groups: DMSO + normoxia, YC-1
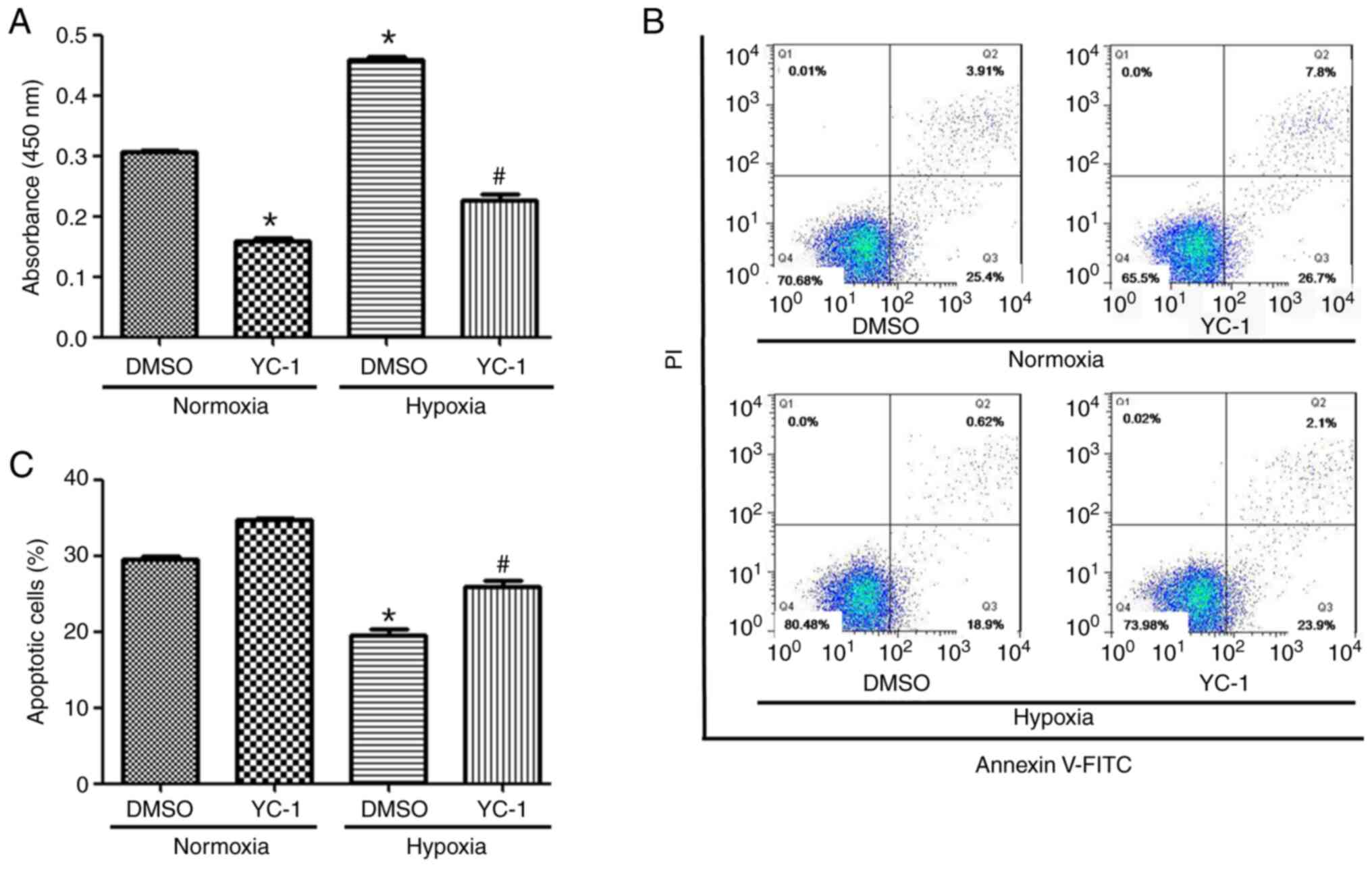
+ normoxia, DMSO + hypoxia and YC-1 + hypoxia. Compared with the
DMSO+ normoxia group, the proliferation of YC-1 + normoxia group
cells was decreased (Fig. 7A).
Compared with the DMSO + hypoxia group, the cell proliferation in
the YC-1+ hypoxia group was also decreased (Fig. 7A). The apoptotic rate of KG-1 cells
was increased in HIF-1α inhibited expressing cells and/or normoxic
conditions compared with non-HIF-1α-inhibited expressing cells
(Fig. 7B and C).
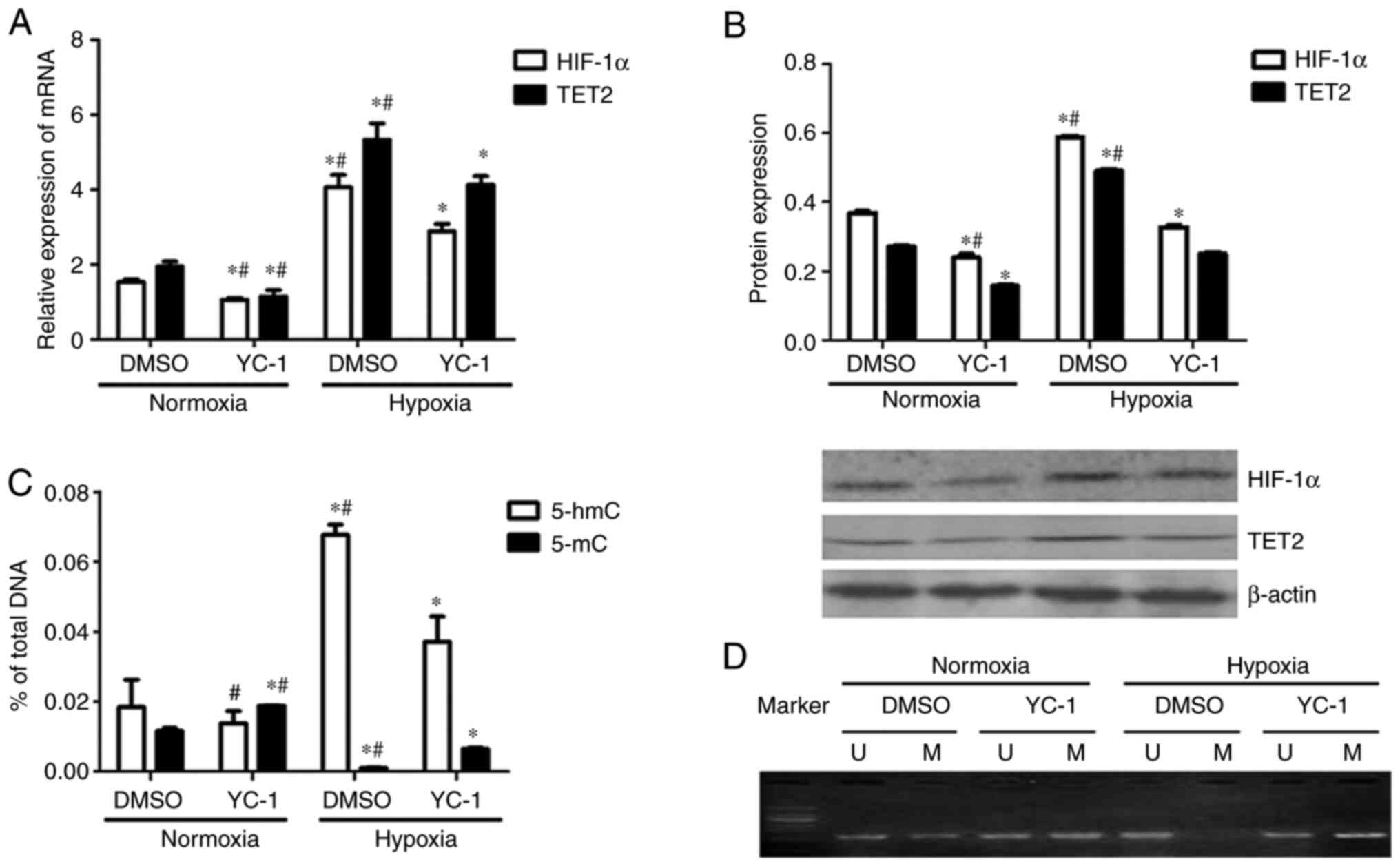
Compared with the DMSO + normoxia group, TET2
expression was decreased in KG-1 cells from the YC-1 + normoxia
group (Fig. 8A and B). Compared with
the DMSO + hypoxia group, TET2 expression in the YC-1 + hypoxia
group was decreased (Fig. 8A and B).
Meanwhile, the HIF-1α mRNA and protein levels were downregulated in
HIF-1α expression inhibited cells under both normoxia and hypoxic
conditions (Fig. 8A and B). This
indicated that the inhibition of HIF-1α expression inhibited the
expression of the TET2 mRNA and protein in KG-1 cells under both
normoxic and hypoxic conditions (Fig. 8A
and B).
Compared with the DMSO + hypoxia group, the 5-hmC
content of the KG-1 cell genome in the YC-1+ hypoxia group was
decreased (Fig. 8C). This indicated
that the inhibition of HIF-1α expression could reduce the
demethylation of KG-1 cells under hypoxic conditions. Compared with
the DMSO+ normoxia group, the 5-mC content of the genome of YC-1 +
normoxia group cells was increased (Fig.
8C). Compared with the DMSO + hypoxia group, the 5-mC content
of the genome was increased in YC-1 + hypoxia group KG-1 cells
(Fig. 8C).
KG-1 cells showed partial methylation of the
promoter region of the p15(INK4B) gene under normoxic conditions,
which was demethylated under hypoxic conditions (Fig. 8D). p15(INK4B) gene promoter
methylation was enhanced when HIF-1α was inhibited by YC-1
(Fig. 8D). Overall, the inhibition
of HIF-1α reduced the demethylation of KG-1 cells by decreasing
TET2 expression.
HIF-1α promotes the transcriptional
activity of the TET2 gene by binding to its promoter region
It was shown by ChIP that HIF-1α bound to the
−1,133-1,022 bp HRE site of the TET2 gene promoter in KG-1 cells
under normoxic or hypoxic conditions (Fig. 9). The transcriptional activity of the
promoter region of the TET2 gene in HIF-1α overexpressing cells was
significantly higher compared with that in all other groups
(Fig. 10A), suggesting that the
overexpression of HIF-1α significantly enhanced the transcriptional
activity of the TET2 promoter region in KG-1 cells.
In addition, different doses of GV219-HIF-1α
eukaryotic expression (0, 1, 2 and 4 µg) and TET2 promoter
luciferase reporter vectors were co-transfected into KG-1 cells and
the relative luciferase activity was detected. It was shown that
the higher the amount of the co-transfected GV219-HIF-1α eukaryotic
expression vector, the higher the transcriptional activity of the
TET2 gene promoter (Fig. 10B), with
a positive correlation identified (Fig.
10C). These results indicated that the overexpression of HIF-1α
promoted the transcriptional activity of the TET2 gene in a
dose-dependent manner.
Discussion
A hypoxic environment has become an essential
condition for maintaining hematopoietic stem cell homeostasis
(31). However, the effects and
mechanism of hypoxia on hematologic malignancy cells are not fully
understood (32). The hypoxic model
can be induced by physical and chemical hypoxia (33). Chemical hypoxia agents, such as
cobalt chloride and deferoxamine, can inhibit the prolyl
hydroxylase domain(PHD) activity by PHD ligand Fe2+
substitution or chelation and then stablize the HIF-1α protein
(33,34). However, due to the fact that the TET2
protease activity is also Fe2+-dependent, physical
hypoxia was selected for the hypoxia model in the present study.
The present study demonstrated that hypoxia promoted the
proliferation and destruction, and inhibited the apoptosis of the
KG-1 AML cell line. The increase of LDH activity in the cell
culture medium in the present study suggested that cells underwent
increased destruction under hypoxic conditions, which was not
inconsistent with the increased proliferation activity of KG-1
cells in a hypoxic environment. Fu et al (35) found that because of the improvement
of the high lactic acid environment at a later stage of cell
culture, CHO cells with human LDH-C gene overexpression exhibited
faster cell proliferation and stronger antiapoptotic ability
compared with LDH-C gene non-overexpressed cells. In the present
study, the effects of different levels and duration of hypoxia on
cell proliferation were firstly observed. According to the
proliferation and apoptosis results, the optimal hypoxic condition
was selected, as ‘1% O2 for 48 h’. This hypoxic
condition was chosen in the present study for subsequent
experimentation of effects of HIF-1α overexpression and expression
inhibition on TET2 expression and function. The findings of the
present study suggested that cell metabolism was accelerated under
hypoxic conditions and that hypoxia may serve certain roles, such
as promoting proliferation and inducing apoptosis in the
pathogenesis of AML.
As an important tumor suppressor gene related to
epigenetic modification, the mechanism of TET2 gene regulation
remains unclear (36). At the
pre-mRNA level, multiple studies have confirmed that microRNAs
[miR-22, miR-29, miR-101 and miR-125] negatively regulate TET2
expression (37,38). At the transcriptional level, the
transcription factor Oct4 positively regulates the expression of
TET2 in mouse embryonic stem cells by binding to the conserved
domains of TET2 (39). At the
posttranslational level, the inhibition of the dvl and axin complex
(IDAX) protein binds directly to the unmethylated CpG dinucleotide
of the promoter region/CpG island of TET2, but then IDAX activates
the caspase leading to the TET2 protein degradation (40). Other studies have found that vitamin
C can restore and enhance TET2 enzymatic activity to suppress
leukemia (41,42). The present study found that hypoxia
promoted the transcriptional expression of the TET2 gene through
the transcription factor HIF-1α, which binds to the HRE of the
promoter region of the TET2 gene, thus affecting the methylation
status and expression of downstream target genes.
In recent years, the link between cellular hypoxia
metabolism and epigenetic regulation has become a new signaling
pathway (43). Certain products in
the process of cellular hypoxia metabolism can affect the opening
or closing of specific cell-cycle arrest, DNA repair and apoptosis
genes, such as p15(INK4B) and Bcl-2 through epigenetic effects
(44,45). Normal and cancer cells have different
metabolites which are used for epigenetic modification (46). Therefore, it is important to
understand which metabolites can regulate gene function and whether
they regulate gene function through epigenetic means or a
combination with transcription factors or other processes, such as
autophagy (47). Methionine
metabolism is essential for Sirtuin 1 (SIRT1)-regulated mouse
embryonic stem cell maintenance and embryonic development, when
SIRT1 is knocked out in mouse embryonic stem cells, the methionine
content is increased and numerous histone methylation markers, such
as (MAT2a) are lost (48). By
constructing a T cell line deficient in LDHA (a key enzyme for
aerobic glycolysis) (49), Brand
et al (49) found that the
histone acetylation and gene expression patterns in wild-type and
LDHA-knockout cells are different. Galligan et al (50) described the existence of Lys and Arg
modifications on histones derived from a glycolytic by-product,
methylglyoxal. These histone posttranslational modifications were
shown to be present during the modulation of chromatin function and
lead to altered gene transcription, which provided a link between
cellular metabolism and the histone code (50).
The abnormal methylation of the p15(INK4B) gene is
more common in the occurrence and development of hematologic
malignancies (51). Hence, the tumor
suppressor gene p15(INK4B) was selected in the present study to
analyze its methylation status. In the present study, methylation
of the promoter region of the p15(INK4B) gene in KG-1 cells was
detected by the MSP method. Among numerous methylation detection
methods, Methylation-Specific PCR (MSP) is likely the most widely
used technique to study DNA methylation of a locus of interest
(52). However, as the degree of
methylation cannot be quantified, its application is limited
(53). In the present study it was
found that under normoxia, the promoter region of the p15(INK4B)
gene was partially methylated and following hypoxia or HIF-1α
overexpression, the promoter region of the p15(INK4B) gene was
demethylated, indicating that the demethylation of this gene was
regulated by HIF-1α. In the present study, the increased
methylation of the p15(INK4B) gene following HIF-1α inhibition
further confirmed this. p15(INK4B) is considered a target gene of
HIF-1α (54), but the specific
mechanism through which HIF-1α regulates p15(INK4B) gene
methylation is not clear. Kroeze et al (55) found that most of the 206 patients
with AML with low 5-hmC exhibited an increased methylation status
of the p15(INK4B) promoter, which was consistent with the results
of the present study. Shimamoto et al (56) found that the methylation rate of the
p15(INK4B) gene in AML reached 51%, which was contrary to the
results of the present study. This difference may be associated
with AML subtypes, detection methods and examined sites.
Methylation is a dynamic and reversible process (57). In the present study, HIF-1α
overexpression caused genome and p15(INK4B) gene demethylation in
KG-1 cells, resulting in the loss of p15(INK4B) gene function and
the malignant proliferation of leukemic cells. However, the
association between the abnormal methylation mechanism of the
p15(INK4B) gene and leukemia requires further study.
The present study investigated the genomic
methylation and change of methylation status in the representative
gene p15(INK4B) during AML cell hypoxia metabolism, as well as the
effect of hypoxia metabolism on the expression of demethylase TET2.
The results of the present study, revealed that hypoxia regulated
the transcriptional expression of TET2 through HIF-1α which, in
turn, affected the methylation status and expression of genes, such
as p15(INK4B) in leukemic cells.
The human acute myeloid leukemia KG-1 cell line was
chosen as the main research object representatively in this study
and other cell lines such as K562, HL-60 and U937 will been chosen
in the follow-up experiments. Different types of leukemia cells
have different tolerance to hypoxia and the mechanism needs to be
further studied. Although much work is still required to fully
understand the role of hypoxia metabolism in leukemia, the present
study shed light on a link between hypoxia metabolism and DNA
demethylation. The development of HIF-1α-specific targeted drugs
should be the focus of further studies.
In conclusion, molecular targeted therapy is
currently used for the treatment of acute leukemia, with an
increasing number of drugs, such as hypomethylating agents,
isocitrate dehydrogenase and Bcl-2 inhibitors being used in the
clinic. The results of the present study indicated that TET2 is an
attractive target gene of HIF-1α transcriptional regulation in AML
cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81500141).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
PH, JL and XLL conceived and designed the present
study. PH, JL, LXZ, GZZ, LP, and QD performed the experiments and
analyzed the data. PH drafted the initial manuscript. JL and LXZ
revised the manuscript. PH and XLL confirmed the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Estey EH: Acute myeloid leukemia: 2019
update on risk-stratification and management. Am J Hematol.
93:1267–1291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brahimi-Horn MC, Chiche J and Pouysségur
J: Hypoxia and cancer. J Mol Med (Berl). 85:1301–1307. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parmar K, Mauch P, Vergilio JA, Sackstein
R and Down JD: Distribution of hematopoietic stem cells in the bone
marrow according to regional hypoxia. Proc Natl Acad Sci USA.
104:5431–5436. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kubota Y, Takubo K and Suda T: Bone marrow
long label-retaining cells reside in the sinusoidal hypoxic niche.
Biochem Biophys Res Commun. 366:335–339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schepers K, Campbell TB and Passegué E:
Normal and leukemic stem cell niches: Insights and therapeutic
opportunities. Cell Stem Cell. 16:254–267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tabe Y and Konopleva M: Advances in
understanding the leukaemia microenvironment. Br J Haematol.
164:767–778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Webb JD, Coleman ML and Pugh CW: Hypoxia,
hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen
sensing. Cell Mol Life Sci. 66:3539–3554. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wellmann S, Guschmann M, Griethe W, Eckert
C, von Stackelberg A, Lottaz C, Moderegger E, Einsiedel HG, Eckardt
KU, Henze G and Seeger K: Activation of HIF pathway in childhood
ALL, prognostic implications of VEGF. Leukemia. 18:926–933. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deeb G, Vaughan MM, McInnis I, Ford LA,
Sait SN, Starostik P, Wetzler M, Mashtare T and Wang ES:
Hypoxia-inducible factor-1α protein expression is associated with
poor survival in normal karyotype adult acute myeloid leukemia.
Leuk Res. 35:579–584. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song LP, Zhang J, Wu SF, Huang Y, Zhao Q,
Cao JP, Wu YL, Wang LS and Chen GQ: Hypoxia-inducible
factor-1alpha-induced differentiation of myeloid leukemic cells is
its transcriptional activity independent. Oncogene. 27:519–527.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Melki JR, Vincent PC and Clark SJ:
Concurrent DNA hypermethylation of multiple genes in acute myeloid
leukemia. Cancer Res. 59:3730–3740. 1999.PubMed/NCBI
|
|
13
|
Liu Q, Liu L, Zhao Y, Zhang J, Wang D,
Chen J, He Y, Wu J, Zhang Z and Liu Z: Hypoxia induces genomic DNA
demethylation through the activation of HIF-1α and transcriptional
upregulation of MAT2A in hepatoma cells. Mol Cancer Ther.
10:1113–1123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bristow RG and Hill RP: Hypoxia and
metabolism: Hypoxia, DNA repair and genetic instability. Nat Rev
Cancer. 8:180–192. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
McCarty G and Loeb DM: Hypoxia-sensitive
epigenetic regulation of an antisense-oriented lncRNA controls WT1
expression in myeloid leukemia cells. PLoS One. 10:e01198372015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tahiliani M, Koh KP, Shen Y, Pastor WA,
Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L and
Rao A: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in
mammalian DNA by MLL partner TET1. Science. 324:930–935. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li W and Liu M: Distribution of
5-hydroxymethylcytosine in different human tissues. J Nucleic
Acids. 2011:8707262011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nikoloski G, Langemeijer SM, Kuiper RP,
Knops R, Massop M, Tönnissen ER, van der Heijden A, Scheele TN,
Vandenberghe P, de Witte T, et al: Somatic mutations of the histone
methyltransferase gene EZH2 in myelodysplastic syndromes. Nat
Genet. 42:665–667. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Delhommeau F, Dupont S, Della Valle V,
James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F,
Alberdi A, et al: Mutation in TET2 in myeloid cancers. N Engl J
Med. 360:2289–2301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu YP, Lv L, Liu Y, Smith MD, Li WC, Tan
XM, Cheng M, Li Z, Bovino M, Aubé J and Xiong Y: Tumor suppressor
TET2 promotes cancer immunity and immunotherapy efficacy. J Clin
Invest. 129:4316–4331. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schumann U, Lee J, Kazan K, Ayliffe M and
Wang MB: DNA-Demethylase regulated genes show
methylation-independent spatiotemporal expression patterns. Front
Plant Sci. 8:14492017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nobori T, Miura K, Wu DJ, Lois A,
Takabayashi K and Carson DA: Deletions of the cyclin-dependent
kinase-4 inhibitor gene in multiple human cancers. Nature.
368:753–756. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hannon GJ and Beach D: p15INK4B is a
potential effector of TGF-beta-induced cell cycle arrest. Nature.
371:257–261. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Herman JG, Jen J, Merlo A and Baylin SB:
Hypermethylation-associated inactivation indicates a tumor
suppressor role for p15INK4B. Cancer Res. 56:722–727.
1996.PubMed/NCBI
|
|
25
|
Tien HF, Tang JH, Tsay W, Liu MC, Lee FY,
Wang CH, Chen YC and Shen MC: Methylation of the p15(INK4B) gene in
myelodysplastic syndrome: It can be detected early at diagnosis or
during disease progression and is highly associated with leukaemic
transformation. Br J Haematol. 112:148–154. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, He N, Wang R, Tian T, Han F, Zhong
C, Zhang C, Hua M, Ji C and Ma D: Analysis of TET2 and EZH2 gene
functions in chromosome instability in acute myeloid leukemia. Sci
Rep. 10:27062020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yeo EJ, Chun YS and Park JW: New
anticancer strategies targeting HIF-1. Biochem Pharmacol.
68:1061–1069. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Herman JG, Graff JR, Myohanen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lan F, Bayliss PE, Rinn JL, Whetstine JR,
Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al: A
histone H3 lysine 27 demethylase regulates animal posterior
development. Nature. 449:689–694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Deynoux M, Sunter N, Hérault O and
Mazurier F: Hypoxia and hypoxia-inducible factors in leukemias.
Front Oncol. 6:412016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cui XY, Skretting G, Jing Y, Sun H,
Sandset PM and Sun L: Hypoxia influences stem cell-like properties
in multidrug resistant K562 leukemic cells. Blood Cells Mol Dis.
51:177–184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Piret JP, Mottet D, Raes M and Michiels C:
CoCl2, a chemical inducer of hypoxia-inducible factor-1,
and hypoxia reduce apoptotic cell death in hepatoma cell line
HepG2. Ann NY Acad Sci. 973:443–447. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee JW, Bae SH, Jeong JW, Kim SH and Kim
KW: Hypoxia-inducible factor (HIF-1) alpha: Its protein stability
and biological functions. Exp Mol Med. 36:1–12. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fu T, Zhang C, Jing Y, Jiang C, Li Z, Wang
S, Ma K, Zhang D, Hou S, Dai J, et al: Regulation of cell growth
and apoptosis through lactate dehydrogenase C over-expression in
Chinese hamster ovary cells. Appl Microbiol Biotechnol.
100:5007–5016. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pasca S, Jurj A, Zdrenghea M and Tomuleasa
C: The potential equivalents of TET2 mutations. Cancers (Basel).
13:14992021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song SJ, Ito K, Ala U, Kats L, Webster K,
Sun SM, Jongen-Lavrencic M, Manova-Todorova K, Teruya-Feldstein J,
Avigan DE, et al: The oncogenic microRNA miR-22 targets the TET2
tumor suppressor to promote hematopoietic stem cell self-renewal
and transformation. Cell Stem Cell. 13:87–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheng J, Guo S, Chen S, Mastriano SJ, Liu
C, D'Alessio AC, Hysolli E, Guo Y, Yao H, Megyola CM, et al: An
extensive network of TET2-targeting MicroRNAs regulates malignant
hematopoiesis. Cell Rep. 5:471–481. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koh KP, Yabuuchi A, Rao S, Huang Y,
Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky
G, et al: Tet1 and Tet2 regulate 5-hydroxymethylcytosine production
and cell lineage specification in mouse embryonic stem cells. Cell
Stem Cell. 8:200–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ko M, An J, Bandukwala HS, Chavez L, Aijö
T, Pastor WA, Segal MF, Li H, Koh KP, Lähdesmäki H, et al:
Modulation of TET2 expression and 5-methylcytosine oxidation by the
CXXC domain protein IDAX. Nature. 497:122–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cimmino L, Dolgalev I, Wang Y, Yoshimi A,
Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, et
al: Restoration of TET2 function blocks aberrant self-renewal and
leukemia progression. Cell. 170:1079–1095.e20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Harjes U: Leukaemia: Beyond the C. Nat Rev
Cancer. 17:5732017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Thienpont B, Steinbacher J, Zhao H, D'Anna
F, Kuchnio A, Ploumakis A, Ghesquière B, Van Dyck L, Boeckx B,
Schoonjans L, et al: Tumor hypoxia causes DNA hypermethylation by
reducing TET activity. Nature. 537:63–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Choudhry H and Harris AL: Advances in
hypoxia-inducible factor biology. Cell Metab. 27:281–298. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lin YT and Wu KJ: Epigenetic regulation of
epithelial-mesenchymal transition: Focusing on hypoxia and TGF-β
signaling. J Biomed Sci. 27:392020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mikirova NA: Bioenergetics of human cancer
cells and normalcells during proliferation and differentiation. Br
J Med Med Res. 7:971–982. 2015. View Article : Google Scholar
|
|
47
|
Iaccarino I and Martins LM: Therapeutic
targets in cancer cell metabolism and death. Cell Death Differ.
18:565–570. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang S, Fang Y, Huang G, Xu X,
Padilla-Banks E, Fan W, Xu Q, Sanderson SM, Foley JF, Dowdy S, et
al: Methionine metabolism is essential for SIRT1-regulated mouse
embryonic stem cell maintenance and embryonic development. EMBO J.
36:3175–3193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Brand A, Singer K, Koehl GE, Kolitzus M,
Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et
al: LDHA-associated lactic acid production blunts tumor
immunosurveillance by T and NK cells. Cell Metab. 24:657–671. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Galligan JJ, Wepy JA, Streeter MD,
Kingsley PJ, Mitchener MM, Wauchope OR, Beavers WN, Rose KL, Wang
T, Spiegel DA and Marnett LJ: Methylglyoxal-derived
posttranslational arginine modifications are abundant histone
marks. Proc Natl Acad Sci USA. 115:9228–9233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Christiansen DH, Andersen MK and
Pedersen-Bjergaard J: Methylation of p15INK4B is common, is
associated with deletion of genes on chromosome arm 7q and predicts
a poor prognosis in therapy-related myelodysplasia and acute
myeloid leukemia. Leukemia. 17:1813–1819. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yokoi K, Yamashita K and Watanabe M:
Analysis of DNA methylation status in bodily fluids for early
detection of cancer. Int J Mol Sci. 18:7352017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ramalho-Carvalho J, Henrique R and
Jerónimo C: Methylation-Specific PCR. Methods Mol Biol.
1708:447–472. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fujii T, Otsuki T, Moriya T, Sakaguchi H,
Kurebayashi J, Yata K, Uno M, Kobayashi T, Kimura T, Jo Y, et al:
Effect of hypoxia on human seminoma cells. Int J Oncol. 20:955–962.
2002.PubMed/NCBI
|
|
55
|
Kroeze LI, Aslanyan MG, van Rooij A,
Koorenhof-Scheele TN, Massop M, Carell T, Boezeman JB, Marie JP,
Halkes CJ, de Witte T, et al: Characterization of acute
myeloidleukemia based on levels of global hydroxymethylation.
Blood. 124:1110–1118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shimamoto T, Ohyashiki JH and Ohyashiki K:
Methylation of p15(INK4b) and E-cadherin genes is independently
correlated with poor prognosis in acute myeloid leukemia. Leuk Res.
29:653–659. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhao L, Duan YT, Lu P, Zhang ZJ, Zheng XK,
Wang JL and Feng WS: Epigenetic targets and their inhibitors in
cancer therapy. Curr Top Med Chem. 18:2395–2419. 2018. View Article : Google Scholar : PubMed/NCBI
|