Introduction
Non-small cell lung cancer (NSCLC) accounts for
>80% of lung cancer cases worldwide according to the statistics
of global cancer in 2018 (1).
Patients with NSCLC are usually treated with surgical resection,
chemotherapy and radiotherapy, and surgical resection is only
appropriate for patients diagnosed at early stages, and resistance
usually develops during the long-term application of chemotherapy
and radiotherapy (2,3). With the emergence of targeted
therapies, survival of certain NSCLC cases, especially patients
diagnosed at advanced stages, has been markedly improved (4,5).
However, as the molecular mechanisms of NSCLC are unclear, there
remains a lack of effective targets for targeted therapy (6,7).
Therefore, an improved understanding of the pathogenesis of NSCLC
is required.
Most cases of NSCLC are associated with tobacco
smoking (8), while ~10% of NSCLC
occurs in never-smokers (9),
suggesting the involvement of other factors, such as molecular
factors, in the pathogenesis of NSCLC. In effect, certain molecular
signaling pathways, such as the TGF-β and KRAS signaling pathways,
have been demonstrated to be potential targets for targeted
therapies of NSCLC, which can be performed to treat NSCLC by
regulating the expression levels of NSCLC-related genes (10,11).
Circular RNAs (circRNAs/circs) are single strand RNA transcripts
closed by covalent bonds (12).
CircRNAs have limited protein-coding capacity; however, they are
involved in cancer biology by regulating gene expression,
suggesting that circRNAs are potential therapeutic targets for
NSCLC (13). In a recent study,
circRNA solute carrier family 26 member 4 (circSLC26A4) has been
reported to be an oncogene in cervical cancer (14); however, to the best of our knowledge,
its role in NSCLC is unknown. Our preliminary microarray analysis
revealed altered expression levels of circSLC26A4 in NSCLC and its
inverse correlation with mature microRNA (miRNA/miR)-15a, which is
a cancer-related miRNA (15). The
present study aimed to explore the potential interaction between
circSLC26A4 and miR-15a in NSCLC.
Materials and methods
Tissue collection
NSCLC tissues and adjacent non-tumor tissues (within
5 cm around tumors) were collected from 64 patients with NSCLC who
underwent surgical resection [40 male and 24 female patients; 28
cases of lung adenocarcinoma (LUAD) and 36 cases of lung squamous
cell carcinoma (LUSC)] at Shanghai Pulmonary Hospital (Shanghai,
China) between June 2016 and May 2019. The patients had an age
range of 45–69 years, with a median age of 56 years. The present
study was approved by the Ethics Committee of Shanghai Pulmonary
Hospital, Tongji University (Shanghai, China). Patients who had
severe clinical disorders or initiated therapies were excluded from
the present study. All patients signed the written informed consent
form. Detailed information of the patients is presented in Table I.
 | Table I.Clinicopathological characteristics of
patients with non-small cell lung cancer (n=64). |
Table I.
Clinicopathological characteristics of
patients with non-small cell lung cancer (n=64).
| Characteristics | No. |
|---|
| Age, years |
|
| ≤55 | 34 |
|
>55 | 30 |
| Sex |
|
| Male | 40 |
|
Female | 24 |
| Smoking history |
|
|
Smoker | 39 |
|
Nonsmoker | 25 |
| Pathological
pattern |
|
| LUAD | 28 |
| LUSC | 36 |
| Lymphatic
metastasis |
|
| No | 24 |
| Yes | 40 |
| Stage |
|
| I+II | 26 |
|
III+IV | 38 |
Therapy and follow-up
The 64 patients with NSCLC were classified into
American Joint Committee on Cancer stage I or II (n=26) and III or
IV (n=38) (16). The 64 patients
were treated with surgical resection, radiotherapy, chemotherapy or
immunotherapy according to their cancer stage and health
conditions. The patients were followed up in a monthly manner until
July 2020. Patient survival conditions were recorded and survival
analysis was performed. All patients completed the follow-up or
died of NSCLC during the follow-up.
NSCLC cells and transfection
The two human NSCLC cell lines, H1793 (LUAD) and DMS
79 (LUSC), obtained from American Type Culture Collection were used
in the present study. Cells were cultured in a mixture composed of
90% RPMI-1640 medium (HyClone; Cytiva) and 10% FBS (Gibco; Thermo
Fisher Scientific, Inc,) with 100 U/ml penicillin and 100 µg/ml
streptomycin. The cell culture conditions were: 37°C, 5%
CO2 and 95% humidity. Cells were collected at a
confluence rate of ~85% for the subsequent experiments. Vector
expressing circSLC26A4 was constructed with pcDNA3.1(+) CircRNA
Mini Vector (Addgene, Inc.) as the backbone. To overexpress
miR-15a, miR-15a mimic (5′-UAGCAGCACAUAAUGGUUUGUG-3′) and
non-specific miRNA [5′-UUCUCCGAACGUGUCACGUTT-3′; used as the
negative control (NC)] were purchased from Sigma-Aldrich (Merck
KGaA). H1793 and DMS 79 cells were counted and 5×107
cells were transfected with 45 nM miRNA or 1 µg expression vector
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 24 h. Empty vector- or miRNA
NC-transfections were performed to serve as NC experiments.
Untransfected cells were used as the control cells. Prior to the
subsequent assays, cells were cultured in fresh medium for another
48 h.
RNA preparation
Ribozol (Invitrogen; Thermo Fisher Scientific, Inc.)
was used to extract total RNA from tissue samples and H1793 and DMS
79 cells. RNA samples were digested with DNase I at 37°C for 2 h to
remove genomic DNAs. RNA integrity was checked using 6% urea-PAGE
gel electrophoresis. The optical density (OD)260/280 ratio of RNA
sample was measured to reflect RNA integrity using NanoDrop 1000
instrument spectrophotometer (Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA (500 ng) was reverse transcribed into cDNA
using PrimeScript RT Master Mix (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. SYBR Green Master Mix
(Bio-Rad Laboratories, Inc.) was used to perform qPCR. The
expression levels of circSLC26A4 were determined using GAPDH as the
internal control. Expression levels of mature miR-15a and miR-15a
precursor were analyzed using the All-in-One™ miRNA qRT-PCR Reagent
kit (GeneCopoeia, Inc.) according to the manufacturer's protocol.
Sequence-specific forward and reverse primers were used to perform
RT and qPCR to determine the expression levels of miR-15a
precursor. To measure the expression levels of mature miR-15a, poly
(A) addition was first performed, followed by using poly (T) as the
reverse primer to perform both RT and qPCR. The conditions of PCR
reaction were initial denaturation at 95°C for 10 min, followed by
40 cycles at 95°C for 15 sec and 60°C for 30 sec. The expression
levels of miRNAs were normalized to the internal reference U6. All
experiments were performed in three technical replicates. The
2−ΔΔCq method was used to normalize Ct values of target
genes to the corresponding internal control (17). The primers used in the present study
were: CircSLC26A4 forward. 5′-TCCAAGTGCTGGTCTCACAG-3′ and reverse,
5′-CCATATCCGACAGGAACTGC-3′; miR-15a precursor forward,
5′-GCCGAGTAGCAGCACACATAA-3′ and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′;
mature miR-15a forward, 5′-TAGCAGCACATAATGG-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; and GAPDH forward,
5′-ATCACTGCCACCCAGAAGAC-3′ and reverse,
5′-TTTCTAGACGGCAGGTCAGG-3′.
Cell Counting Kit-8 (CCK-8) assay
Transfected H1793 and DMS 79 cells were subjected to
cell proliferation analysis using a CCK-8 assay (Sigma-Aldrich;
Merck KGaA). Briefly, 0.1 ml medium containing 4,500 cells was
added into each well of a 96-well plate. H1793 and DMS 79 cells
were cultured at 37°C for 24, 48, 72 and 96 h, and then 10 µl CCK-8
regent was added for 2 h. The measurement of OD values was
performed, and the absorbance was detected at 450 nm.
Statistical analysis
SPSS 22.0 (IBM Corp.) was used to conduct the
statistical analysis of data. The expression levels of circSLC26A4,
mature miR-15a and miR-15a precursor in NSCLC and non-tumor tissues
were presented as average values of three technical replicates.
Data comparisons were performed using a paired t-test. Other
data on cell transfection, RT-qPCR and CCK-8 assay were presented
as the mean ± SD of three biological replicates. The comparisons of
the study groups was conducted using one-way ANOVA followed by
Tukey's post hoc test. Survival analysis was performed by dividing
the 64 patients with NSCLC into high and low circSLC26A4 groups
(n=32; cutoff value, median expression level of circSLC26A4 in
NSCLC tissues), followed by the plotting of Kaplan-Meier survival
curves and the use of a log-rank test to compare survival curves.
Pearson's correlation coefficient was used for correlation
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Altered expression levels of
circSLC26A4, mature miR-15a and miR-15a precursor in NSCLC
tissues
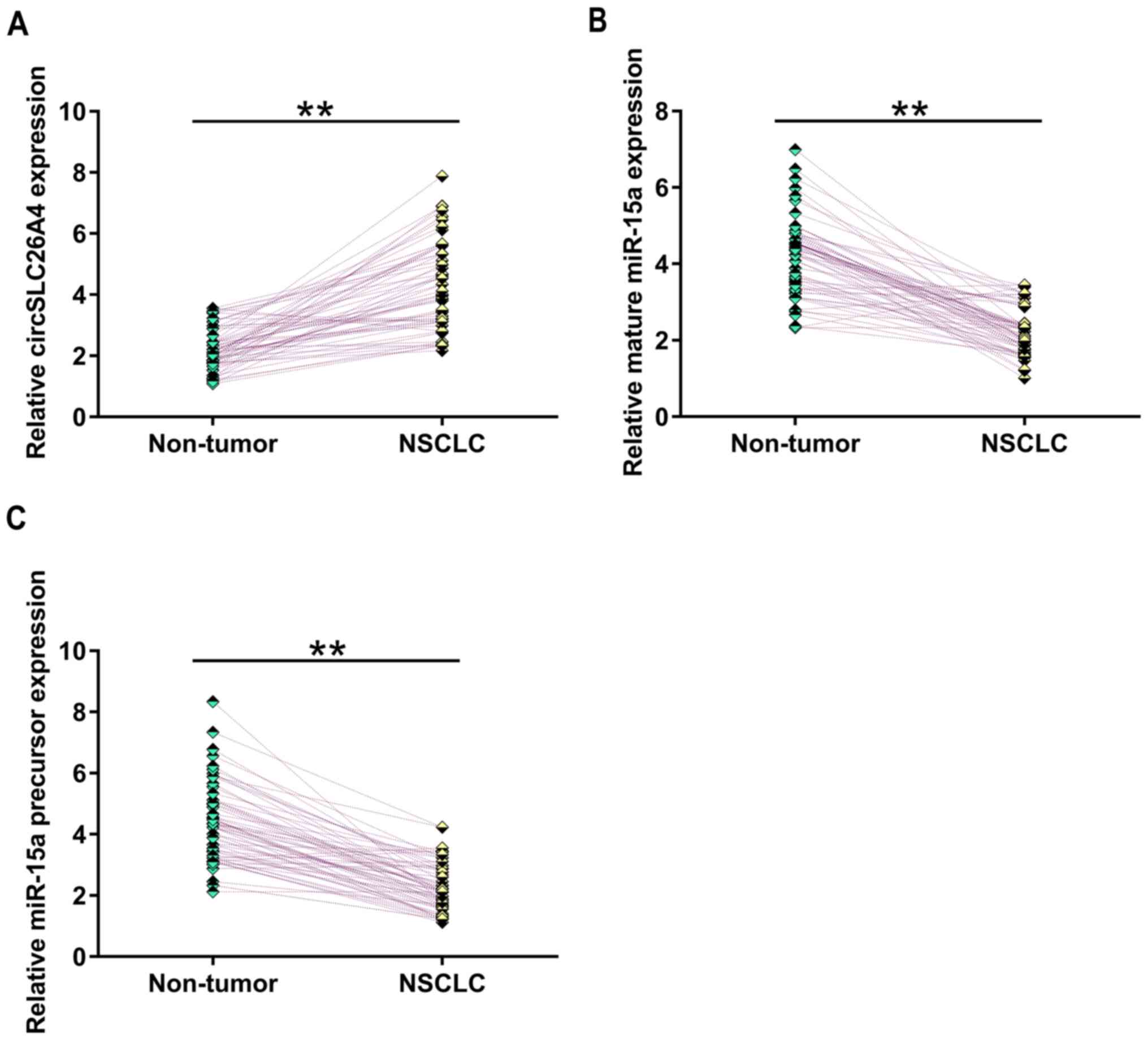
The expression levels of circSLC26A4, mature miR-15a
and miR-15a precursor in NSCLC tissues and adjacent non-tumor
tissues were measured by RT-qPCR. The results revealed that,
compared with that in non-tumor tissues, circSLC26A4 expression was
significantly upregulated in NSCLC tissues (Fig. 1A; P<0.01). Furthermore, the
expression levels of mature miR-15a (Fig. 1B) and miR-15a precursor (Fig. 1B) were significantly downregulated in
NSCLC tissues compared with in non-tumor tissues (P<0.01).
Therefore, circSLC26A4 and miR-15a may be involved in NSCLC.
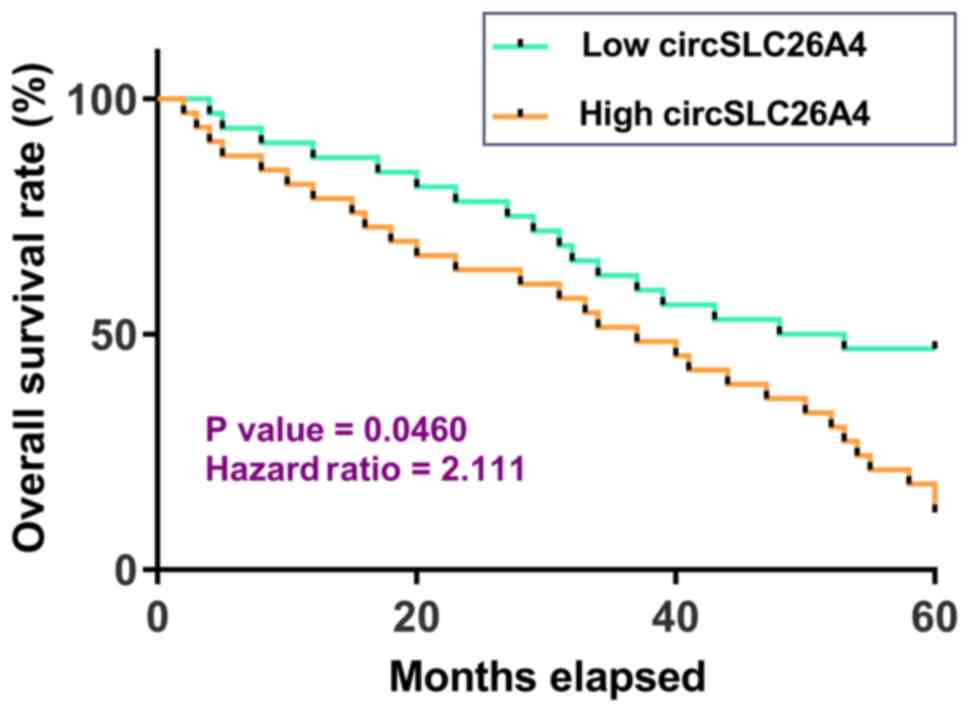
High expression levels of circSLC26A4
in NSCLC tissues are associated with poor survival of patients with
NSCLC
Survival curve analysis revealed that, compared with
patients in the low circSLC26A4 expression group, patients in the
high circSLC26A4 expression group had a significantly lower overall
survival rate. Therefore, high expression levels of circSLC26A4 may
predict poor survival of patients with NSCLC (Fig. 2).
circSLC26A4 suppresses the maturation
of miR-15a in NSCLC
Pearson's correlation coefficient analysis was used
to determine the correlation between circSLC26A4 expression and
mature miR-15a or miR-15a precursor expression in NSCLC tissues. It
was observed that circSLC26A4 expression was inversely and
significantly correlated with the expression levels of mature
miR-15a in NSCLC tissues (Fig. 3A).
However, there was no significant correlation between the
expression levels of circSLC26A4 and miR-15a precursor (Fig. 3B). To further explore the interaction
between them, circSLC26A4 and miR-15a were overexpressed in both
H1793 and DMS 79 cells (P<0.05; Fig.
3C). Overexpression of circSLC26A4 resulted in downregulation
of mature miR-15a expression (P<0.05; Fig. 3D), but not in downregulation of
miR-15a precursor expression (Fig.
3E). These findings suggested that the maturation of miR-15a in
NSCLC cells could be suppressed by circSLC26A4. By contrast,
overexpression of miR-15a exhibited no significant effect on the
expression levels of circSLC26A4 (Fig.
3F).
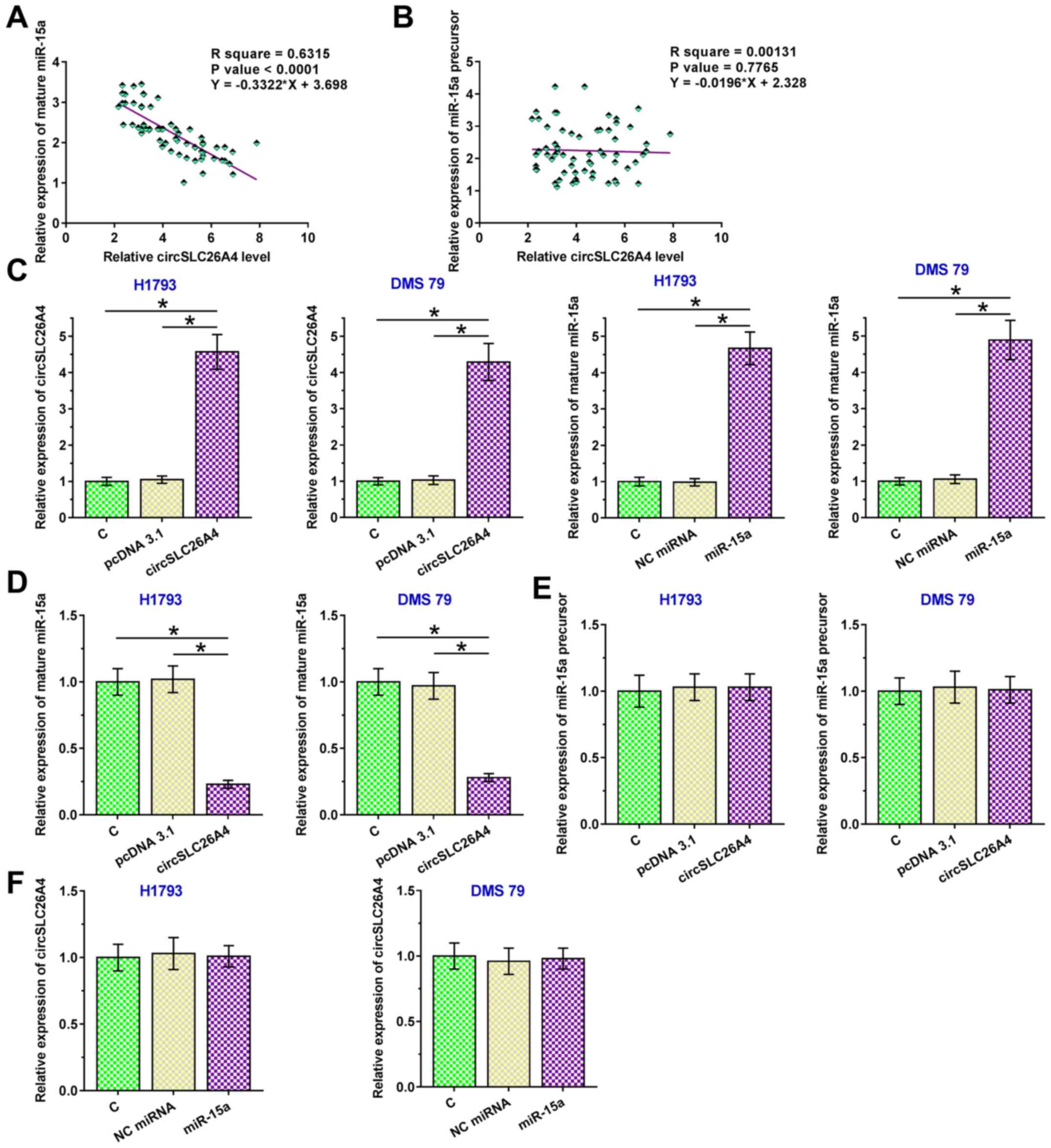 | Figure 3.circSLC26A4 suppresses the maturation
of miR-15a in NSCLC. Pearson's correlation coefficient was used to
analyze the correlations between the expression levels of
circSLC26A4 and (A) mature miR-15a or (B) miR-15a precursor in
NSCLC tissues. (C) To further explore the interaction between
circSLC26A4 and miR-15a, circSLC26A4 expression vector or miR-15a
mimic was transfected into H1793 and DMS 79 cells, and transfection
efficiency was confirmed at 48 h after transfection. The effects of
overexpression of circSLC26A4 on (D) mature miR-15a and (E) miR-15a
precursor expression, and (F) the effects of overexpression of
miR-15a on the expression levels of circSLC26A4 were also analyzed
by reverse transcription-quantitative PCR. Data of three biological
replicates of cell transfection experiments are presented as the
mean ± SD, and one-way ANOVA followed by Tukey's post-hoc test was
used for data comparisons of study groups. *P<0.05. C, control
(untransfected cells); circSLC26A4, circular RNA solute carrier
family 26 member 4; miR-15a, microRNA-15a; miRNA, microRNA; NC,
negative control; NSCLC, non-small cell lung cancer. |
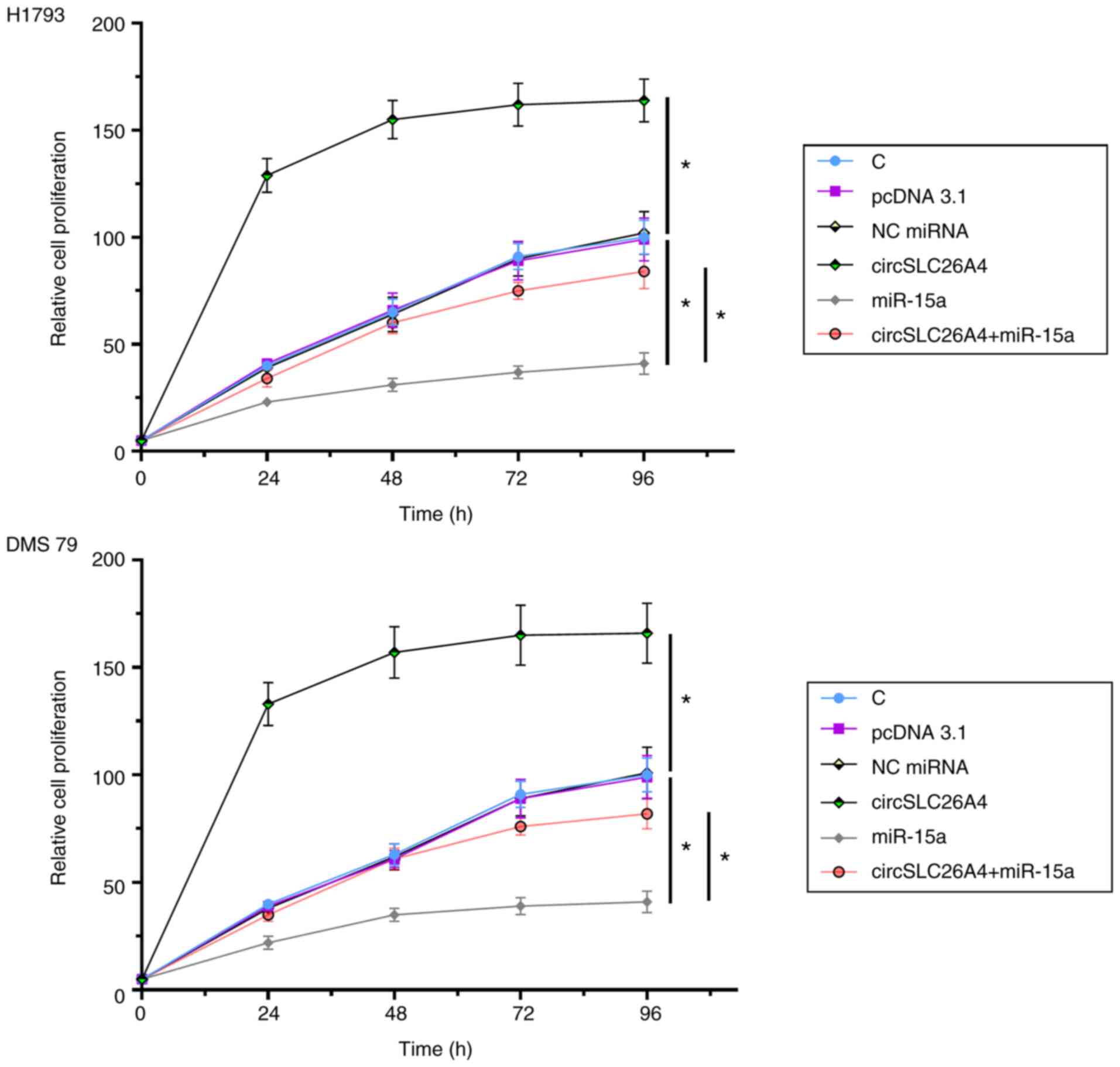
Overexpression of circSLC26A4 promotes
NSCLC cell proliferation via miR-15a
A CCK-8 assay was performed to explore the role of
circSLC26A4 and miR-15a in regulating the proliferation of H1793
and DMS 79 cells. Overexpression of circSLC26A4 significantly
increased NSCLC cell proliferation, while overexpression of miR-15a
decreased cell proliferation compared with their respective NCs. In
addition, overexpression of circSLC26A4 reduced the inhibitory
effects of overexpression of miR-15a on cell proliferation
(P<0.05; Fig. 4).
Discussion
The differential expression of circSLC26A4 in NSCLC
and its interaction with miR-15a were investigated in the present
study. The present data demonstrated that circSLC26A4 expression
was significantly upregulated in NSCLC tissues and it could promote
the proliferation of NSCLC cells by suppressing the maturation of
miR-15a.
In a recent study, circSLC26A4 expression was
reported to be upregulated in cervical cancer, and its high
expression was significantly associated with poor survival of
patients with cervical cancer (14).
In addition, circSLC26A4 promotes the migration and proliferation
of cervical cancer cells by sponging miR-1287-5p to upregulate
HOXA7 (14). However, the regulatory
effects of circSLC26A4 in NSCLC is still unclear. In the present
study, to investigate the role of circSLC26A4 in the development of
NSCLC, the expression levels of circSLC26A4 in NSCLC and adjacent
non-tumor tissues were measured. circSLC26A4 expression was
upregulated in NSCLC tissues. In addition, overexpression of
circSLC26A4 increased the proliferation of both LUAD and LUSC
cells, which are the two major subgroups of NSCLC. Therefore,
circSLC26A4 may promote the proliferation of cancer cells,
resulting in the development and progression of NSCLC.
Even with advances in the diagnosis and treatment of
NSCLC, the prognosis of NSCLC is still poor (18,19).
Goyal et al (19) conducted a
retrospective study of patients with NSCLC diagnosed between 2004
and 2014 using the National Cancer Database, revealing that the
overall 5-year survival rate of patients with stage IV NSCLC is
only ~24%. This is mainly due to the low early diagnostic rate
(19). Furthermore, due to the lack
of sensitive markers, the early diagnosis of NSCLC is unlikely to
be markedly improved in the near future (20). The present study demonstrated that
high expression levels of circSLC26A4 were closely associated with
poor survival of patients, suggesting a potential role of
circSLC26A4 as a prognostic biomarker for NSCLC. Therefore,
evaluation of the expression levels of circSLC26A4 prior to therapy
may assist the determination of treatment approaches, which would
in turn improve patient survival. miR-15a has been reported to be a
tumor suppressor in different types of cancer, including NSCLC
(21,22). One study reported that miR-15a
expression was downregulated in NSCLC tissues and cells, and
overexpression of miR-15a inhibited NSCLC cell proliferation,
migration and invasion (23).
Another study revealed that miR-15a expression is markedly
downregulated in NSCLC tissues, and overexpression of miR-15a
markedly suppresses cell viability, invasion and migration, and
accelerates the apoptosis of NSCLC cells (24). miR-15a not only suppresses tumor
metastasis, but also increases the sensitivity of cancer cells to
chemotherapy (19,20). Consistently, the present study
revealed the downregulation of miR-15a expression in NSCLC and its
inhibitory effects on cell proliferation. Notably, the present
study revealed that overexpression of circSLC26A4 suppressed the
maturation of miR-15a in NSCLC cells. However, to the best of our
knowledge, the underlying mechanism is unknown. To be cleaved into
mature miR-15a, miR-15a precursor should be transported out of
nucleus to enter the cytoplasm (25). The involvement of circSLC26A4 in the
transportation of miR-15a will be explored in our future
studies.
In conclusion, circSLC26A4 expression is upregulated
in NSCLC and may promote cancer cell proliferation by suppressing
the maturation of miR-15a. The current findings may help to
increase the understanding of NSCLC pathogenesis and to identify
potential targets for the treatment of patients with NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Shanghai Science
and Technology Commission Animal Experiment Model Project (grant
no. 19140904102).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QC conducted the experiments and contributed to data
analysis and manuscript writing. JL provided critical guidance for
this manuscript, designed this study, interpreted data, revised
this manuscript, and gave final approval of the version to be
published. HL contributed to data analysis and interpretation. All
authors confirmed the authenticity of the data and read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Shanghai Pulmonary Hospital, Tongji University (Shanghai, China),
and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Adjei AA: Lung cancer worldwide. J Thorac
Oncol. 14:9562019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lance. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jamal-Hanjani M, Wilson GA, McGranahan N,
Brikbak N, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R,
Rosenthal R, et al: Tracking the evolution of non-small-cell lung
cancer. N Engl J Med. 376:2109–2121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chan BA and Hughes BG: Targeted therapy
for non-small cell lung cancer: Current standards and the promise
of the future. Transl Lung Cancer Res. 4:36–54. 2015.PubMed/NCBI
|
|
5
|
Janku F, Stewart DJ and Kurzrock R:
Targeted therapy in non-small-cell lung cancer-is it becoming a
reality? Nat Rev Clin Oncol. 7:401–414. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hirsch FR, Suda K, Wiens J and Bunn PA Jr:
New and emerging targeted treatments in advanced non-small-cell
lung cancer. Lancet. 388:1012–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Joseph AM, Rothman AJ, Almirall D, Begnaud
A, Chiles C, Cinciripini PM, Fu SS, Graham AL, Lindgren BR, Melzer
AC, et al: Lung cancer screening and smoking cessation clinical
trials. SCALE (smoking cessation within the context of lung cancer
screening) collaboration. Am J Respir Crit Care Med. 197:172–182.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saito S, Espinoza-Mercado F, Liu H, Sata
N, Cui X and Soukiasian HJ: Current status of research and
treatment for non-small cell lung cancer in never-smoking females.
Cancer Biol Ther. 18:359–368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eser PÖ and Jänne PA: TGFβ pathway
inhibition in the treatment of non-small cell lung cancer.
Pharmacol Ther. 184:112–130. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tomasini P, Walia P, Labbe C, Jao K and
Leighl NB: Targeting the KRAS pathway in non-small cell lung
cancer. Oncologist. 21:1450–1460. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y,
Fan C, Li X, Li G, Li Y, et al: Circular RNAs (circRNAs) in cancer.
Cancer Lett. 425:134–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: CircRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji F, Du R, Chen T, Zhang M, Zhu Y, Luo X
and Ding Y: Circular RNA circSLC26A4 accelerates cervical cancer
progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids.
19:413–420. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He J: Knocking down miR-15a expression
promotes the occurrence and development and induces the EMT of
NSCLC cells in vitro. Saudi J Biol Sci. 24:1859–1865. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Detterbeck FC, Boffa DJ, Kim AW and Tanoue
LT: The eighth edition lung cancer stage classification. Chest.
151:193–203. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-timequantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stokes WA, Bronsert MR, Meguid RA, Blum
MG, Jones BL, Koshy M, Sher DJ, Louie AV, Palma DA, Senan S, et al:
Post-treatment mortality after surgery and stereotactic body
radiotherapy for early-stage non-small-cell lung cancer. J Clin
Oncol. 36:642–651. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goyal G, Kommalapati A, Bartley AC,
Gunderson TM, Adjei AA and Go RS: Association between hospital
volume and mortality of patients with metastatic non-small cell
lung cancer. Lung Cancer. 122:214–219. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Postmus PE, Kerr KM, Oudkerk M, Senan S,
Waller DA, Vansteenkiste J, Escriu C and Peters S; ESMO Guidelines
Committee, : Early and locally advanced non-small-cell lung cancer
(NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 28 (Suppl-4):iv1–iv21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lan F, Yue X, Ren G, Li H, Ping L, Wang Y
and Xia T: miR-15a/16 enhances radiation sensitivity of non-small
cell lung cancer cells by targeting the TLR1/NF-κB signaling
pathway. Int J Radiat Oncol Biol Phys. 91:73–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bozok Çetintaş V, Tetik Vardarlı A, Düzgün
Z, Tezcanlı Kaymaz B, Açıkgöz E, Aktuğ H, Kosova Can B, Gündüz C
and Eroğlu Z: miR-15a enhances the anticancer effects of cisplatin
in the resistant non-small cell lung cancer cells. Tumour Biol.
37:1739–1751. 2016. View Article : Google Scholar
|
|
23
|
Guo S, Li M, Li J and Lv Y: Inhibition
mechanism of lung cancer cell metastasis through targeted
regulation of Smad3 by miR-15a. Oncol Lett. 19:1516–1522.
2020.PubMed/NCBI
|
|
24
|
Yang T, Thakur A, Chen T, Chen T, Yang L,
Lei G, Liang Y, Zhang S, Ren H and Chen M: MicroRNA-15a induces
cell apoptosis and inhibits metastasis by targeting BCL2L2 in
non-small cell lung cancer. Tumour Biol. 36:4357–4365. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pickering BF, Yu D and Van Dyke MW:
Nucleolin protein interacts with microprocessor complex to affect
biogenesis of microRNAs 15a and 16. J Biol Chem. 286:44095–44103.
2011. View Article : Google Scholar : PubMed/NCBI
|