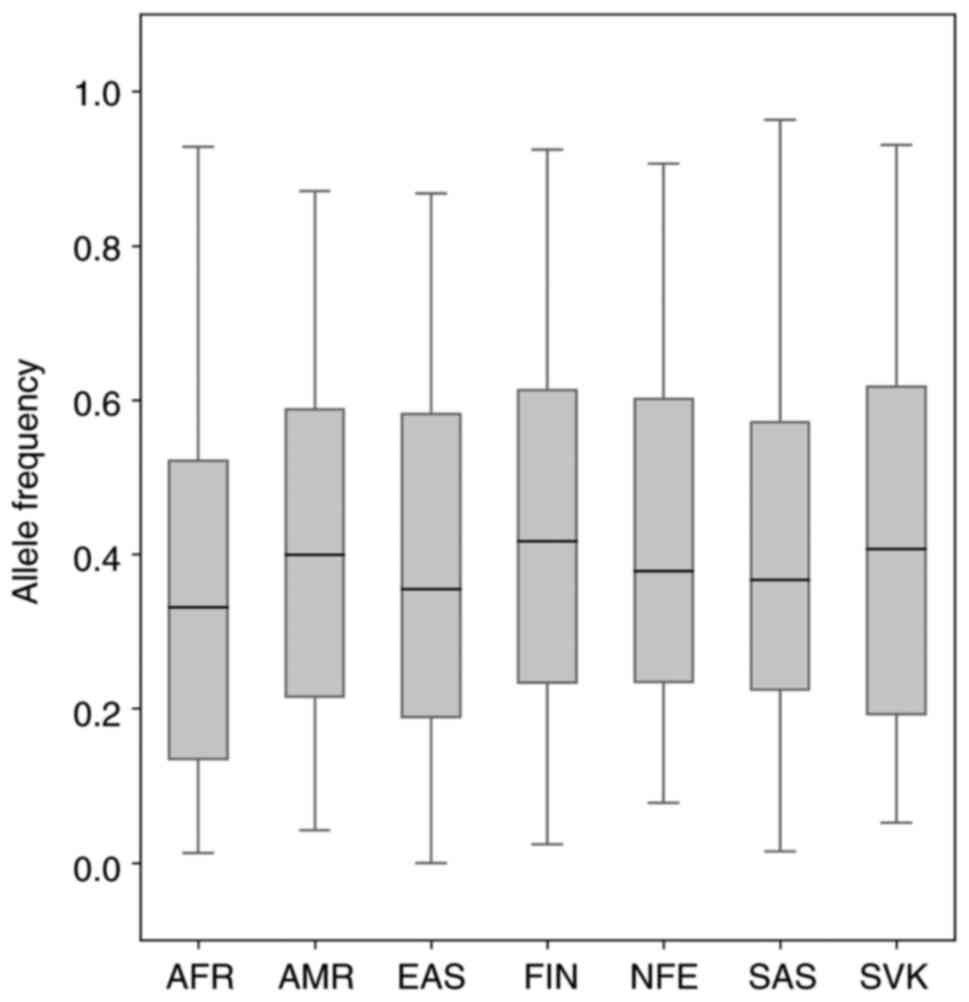
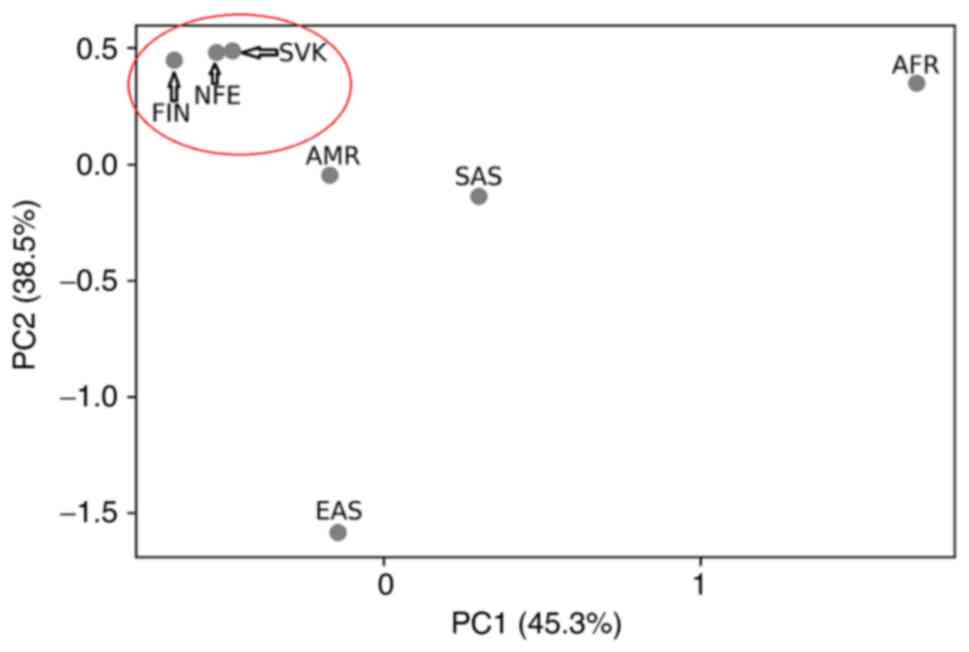
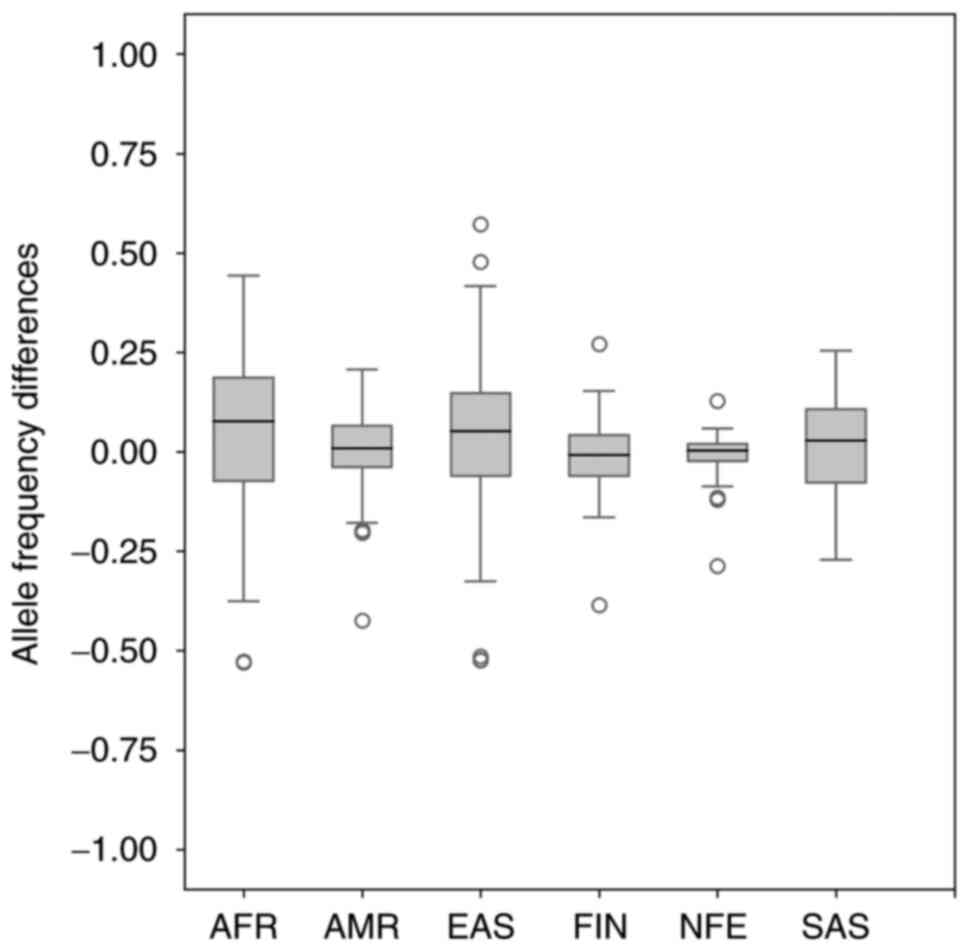
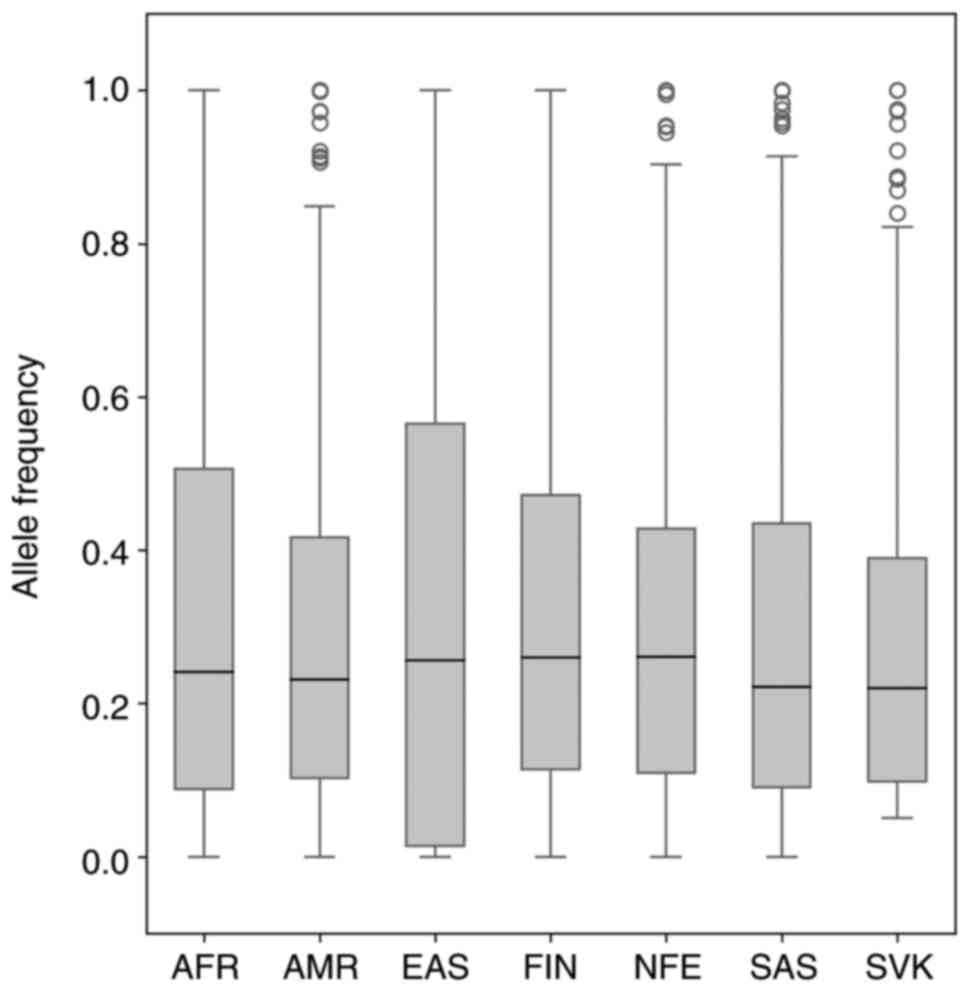
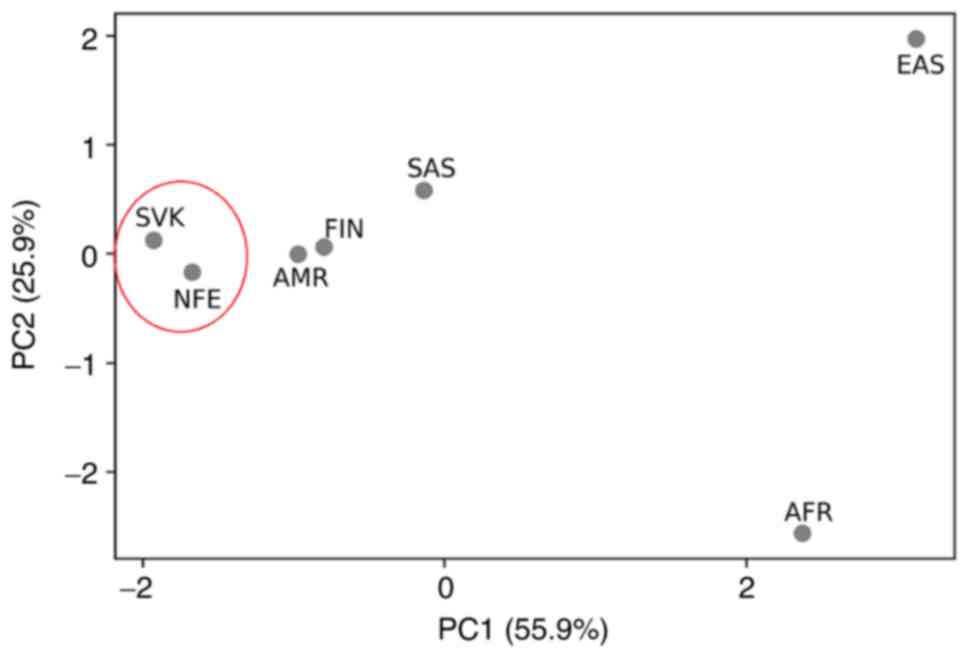
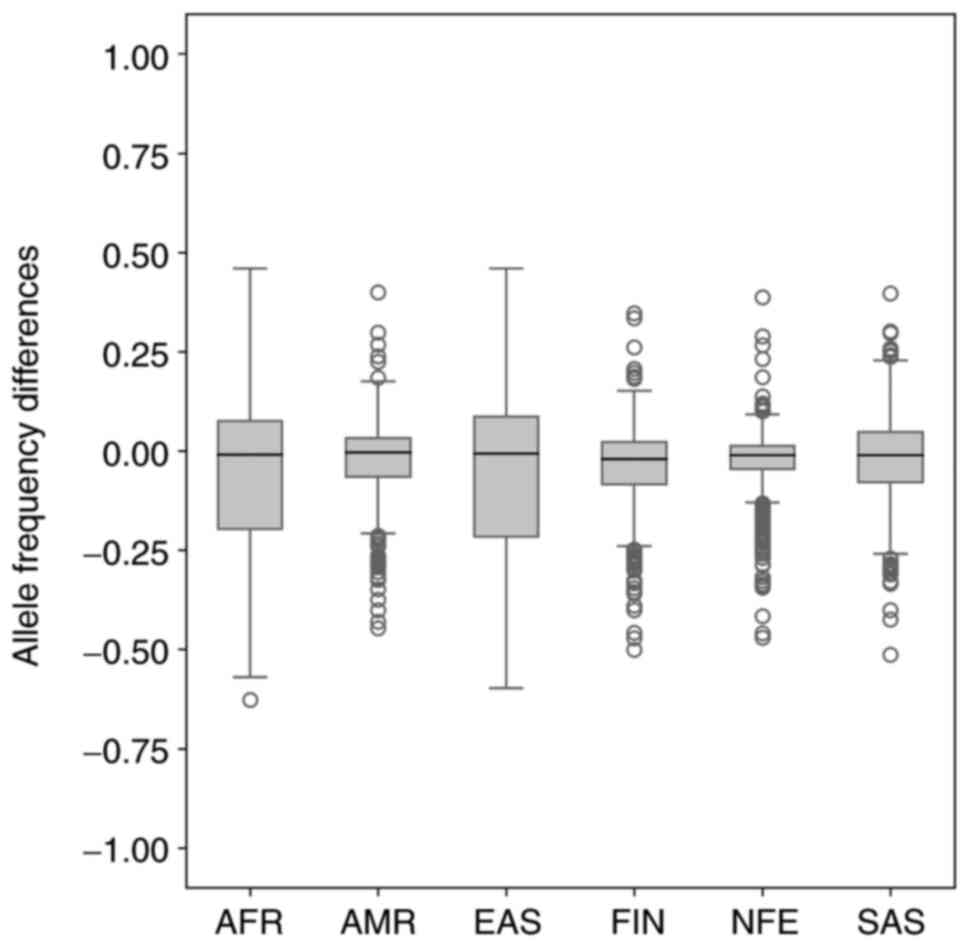
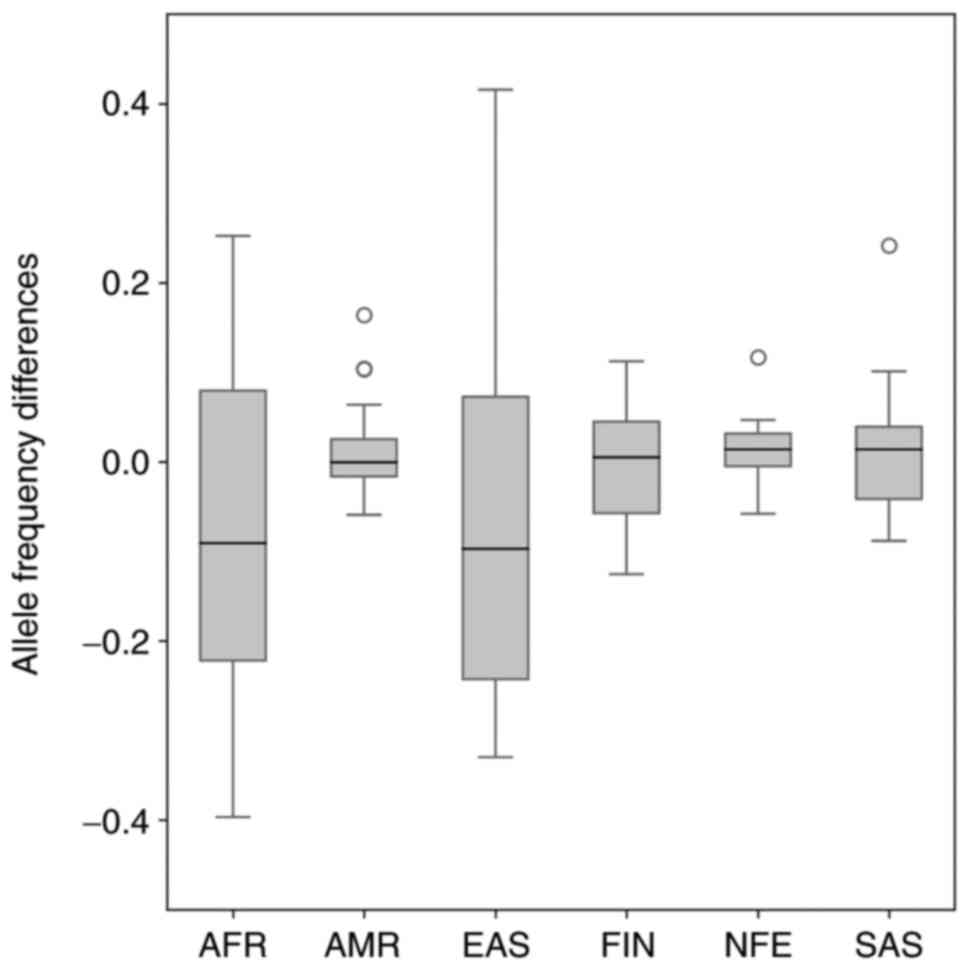
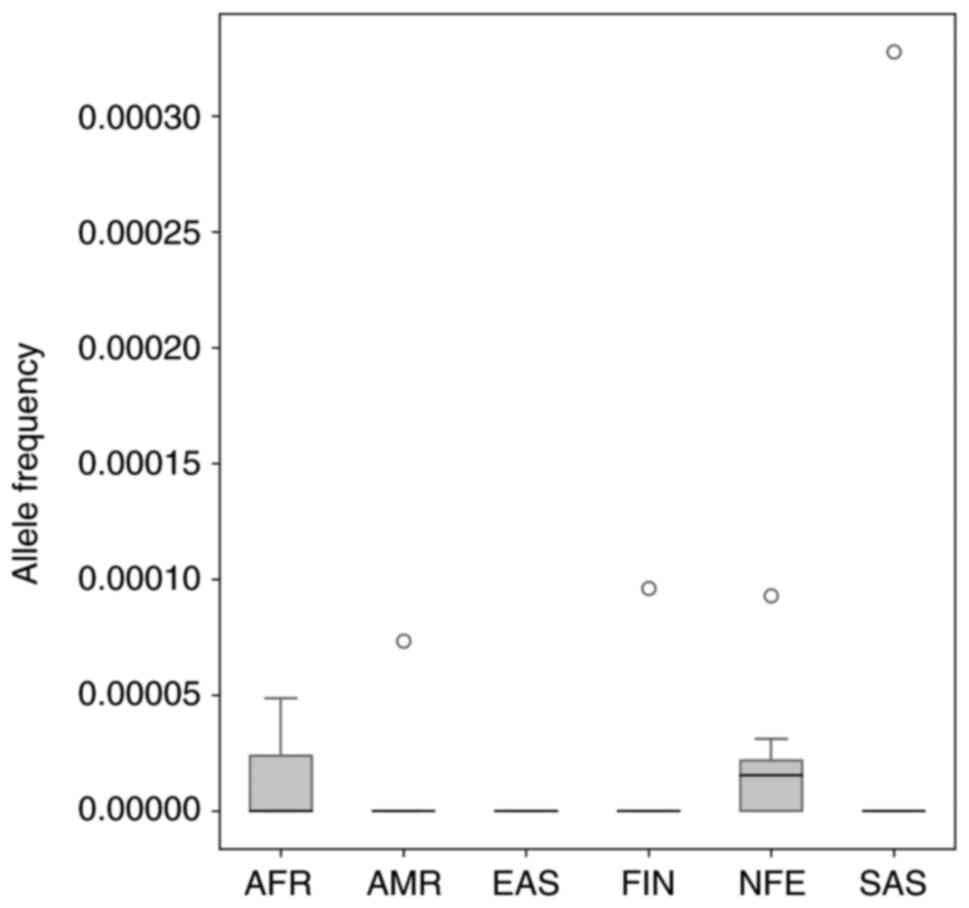
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global Cancer Observatory, . International
Agency for Research on Cancer. (Lyon, France). 2020.https://gco.iarc.fr/today/data/factsheets/populations/703-slovakia-fact-sheets.pdfNovember
9–2020
|
|
3
|
Thanikachalam K and Khan G: Colorectal
cancer and nutrition. Nutrients. 11:1642019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rawla P, Sunkara T and Barsouk A:
Epidemiology of colorectal cancer: Incidence, mortality, survival,
and risk factors. Prz Gastroenterol. 14:89–103. 2019.PubMed/NCBI
|
|
5
|
Cai S, Li Y, Ding Y, Chen K and Jin M:
Alcohol drinking and the risk of colorectal cancer death: A
meta-analysis. Eur J Cancer Prev. 23:532–539. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dashti SG, Buchanan DD, Jayasekara H,
Ouakrim DA, Clendenning M, Rosty C, Winship IM, Macrae FA, Giles
GG, Parry S, et al: Alcohol consumption and the risk of colorectal
cancer for mismatch repair gene mutation carriers. Cancer Epidemiol
Biomarkers Prev. 26:366–375. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Botteri E, Iodice S, Bagnardi V, Raimondi
S, Lowenfels AB and Maisonneuve P: Smoking and colorectal cancer: A
meta-analysis. JAMA. 300:2765–2778. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Limsui D, Vierkant RA, Tillmans LS, Wang
AH, Weisenberger DJ, Laird PW, Lynch CF, Anderson KE, French AJ,
Haile RW, et al: Cigarette smoking and colorectal cancer risk by
molecularly defined subtypes. J Natl Cancer Inst. 102:1012–1022.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ordóñez-Mena JM, Walter V, Schöttker B,
Jenab M, O'Doherty MG, Kee F, Bueno-de-Mesquita B, Peeters PH,
Stricker BH, Ruiter R, et al: Impact of prediagnostic smoking and
smoking cessation on colorectal cancer prognosis: A meta-analysis
of individual patient data from cohorts within the CHANCES
consortium. Ann Oncol. 29:472–483. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thrift AP, Gong J, Peters U, Chang-Claude
J, Rudolph A, Slattery ML, Chan AT, Locke AE, Kahali B, Justice AE,
et al: Mendelian randomization study of body mass index and
colorectal cancer risk. Cancer Epidemiol Biomarkers Prev.
24:1024–1031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gharahkhani P, Ong JS, An J, Law MH,
Whiteman DC, Neale RE and MacGregor S: Effect of increased body
mass index on risk of diagnosis or death from cancer. Br J Cancer.
120:565–570. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu Y, Kweon SS, Tanikawa C, Jia WH, Xiang
YB, Cai Q, Zeng C, Schmit SL, Shin A, Matsuo K, et al: Large-scale
genome-wide association study of east asians identifies loci
associated with risk for colorectal cancer. Gastroenterology.
156:1455–1466. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng C, Matsuda K, Jia WH, Chang J, Kweon
SS, Xiang YB, Shin A, Jee SH, Kim DH, Zhang B, et al:
Identification of susceptibility loci and genes for colorectal
cancer risk. Gastroenterology. 150:1633–1645. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Al-Tassan NA, Whiffin N, Hosking FJ,
Palles C, Farrington SM, Dobbins SE, Harris R, Gorman M, Tenesa A,
Meyer BF, et al: A new GWAS and meta-analysis with 1000Genomes
imputation identifies novel risk variants for colorectal cancer.
Sci Rep. 5:104422015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang B, Jia WH, Matsuda K, Kweon SS,
Matsuo K, Xiang YB, Shin A, Jee SH, Kim DH, Cai Q, et al:
Large-scale genetic study in East Asians identifies six new loci
associated with colorectal cancer risk. Nat Genet. 46:533–542.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takahashi Y, Sugimachi K, Yamamoto K,
Niida A, Shimamura T, Sato T, Watanabe M, Tanaka J, Kudo S,
Sugihara K, et al: Japanese genome-wide association study
identifies a significant colorectal cancer susceptibility locus at
chromosome 10p14. Cancer Sci. 108:2239–2247. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schmit SL, Edlund CK, Schumacher FR, Gong
J, Harrison TA, Huyghe JR, Qu C, Melas M, Van Den Berg DJ, Wang H,
et al: Novel common genetic susceptibility loci for colorectal
cancer. J Natl Cancer Inst. 111:146–157. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Biller LH, Syngal S and Yurgelun MB:
Recent advances in lynch syndrome. Fam Cancer. 18:211–219. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yurgelun MB and Hampel H: Recent advances
in lynch syndrome: Diagnosis, treatment, and cancer prevention. Am
Soc Clin Oncol Educ Book. 38:101–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Møller P, Seppälä T, Bernstein I,
Holinski-Feder E, Sala P, Evans DG, Lindblom A, Macrae F, Blanco I,
Sijmons R, et al: Incidence of and survival after subsequent
cancers in carriers of pathogenic MMR variants with previous
cancer: A report from the prospective lynch syndrome database. Gut.
66:1657–1664. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Møller P, Seppälä TT, Bernstein I,
Holinski-Feder E, Sala P, Evans DG, Lindblom A, Macrae F, Blanco I,
Sijmons RH, et al: Cancer risk and survival in carriers by gene and
gender up to 75 years of age: A report from the prospective lynch
syndrome database. Gut. 67:1306–1316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Soares BL, Brant AC, Gomes R, Pastor T,
Schneider NB, Ribeiro-Dos-Santos Â, de Assumpção PP, Achatz MI,
Ashton-Prolla P and Moreira MA: Screening for germline mutations in
mismatch repair genes in patients with lynch syndrome by next
generation sequencing. Fam Cancer. 17:387–394. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cox VL, Bamashmos AA, Foo WC, Gupta S,
Yedururi S, Garg N and Kang HC: Lynch syndrome: Genomics update and
imaging review. Radiographics. 38:483–499. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Le S, Ansari U, Mumtaz A, Malik K, Patel
P, Doyle A and Khachemoune A: Lynch syndrome and muir-torre
syndrome: An update and review on the genetics, epidemiology, and
management of two related disorders. Dermatol Online J.
23:130302017.
|
|
26
|
Peltomäki P: Update on lynch syndrome
genomics. Fam Cancer. 15:385–393. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duraturo F, Liccardo R, Cavallo A, De Rosa
M, Grosso M and Izzo P: Association of low-risk MSH3 and MSH2
variant alleles with Lynch syndrome: Probability of synergistic
effects. Int J Cancer. 129:1643–1650. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuiper RP, Vissers LELM, Venkatachalam R,
Bodmer D, Hoenselaar E, Goossens M, Haufe A, Kamping E, Niessen RC,
Hogervorst FB, et al: Recurrence and variability of germline EPCAM
deletions in Lynch syndrome. Hum Mutat. 32:407–414. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shah SN, Hile SE and Eckert KA: Defective
mismatch repair, microsatellite mutation bias, and variability in
clinical cancer phenotypes. Cancer Res. 70:431–435. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Martin-Morales L, Rofes P, Diaz-Rubio E,
Llovet P, Lorca V, Bando I, Perez-Segura P, de la Hoya M, Garre P,
Garcia-Barberan V and Caldes T: Novel genetic mutations detected by
multigene panel are associated with hereditary colorectal cancer
predisposition. PLoS One. 13:e02038852018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Budis J, Gazdarica J, Radvanszky J,
Harsanyova M, Gazdaricova I, Strieskova L, Frno R, Duris F, Minarik
G, Sekelska M, et al: Non-invasive prenatal testing as a valuable
source of population specific allelic frequencies. J Biotechnol.
299:72–78. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pös O, Budis J, Kubiritova Z, Kucharik M,
Duris F, Radvanszky J and Szemes T: Identification of structural
variation from NGS-Based non-invasive prenatal testing. Int J Mol
Sci. 20:44032019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu S, Huang S, Chen F, Zhao L, Yuan Y,
Francis SS, Fang L, Li Z, Lin L, Liu R, et al: Genomic analyses
from non-invasive prenatal testing reveal genetic associations,
patterns of viral infections, and Chinese population history. Cell.
175:347–359. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tran NH, Vo TB, Nguyen VT, Tran NT, Trinh
THN, Pham HAT, Dao THT, Nguyen NM, Van YLT, Tran VU, et al: Genetic
profiling of Vietnamese population from large-scale genomic
analysis of non-invasive prenatal testing data. Sci Rep.
10:191422020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pös O, Budiš J and Szemes T: Recent trends
in prenatal genetic screening and testing. F1000Res. 8:F10002019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Minarik G, Repiska G, Hyblova M, Nagyova
E, Soltys K, Budis J, Duris F, Sysak R, Bujalkova MG, Vlkova-Izrael
B, et al: Utilization of benchtop next generation sequencing
platforms ion torrent PGM and MiSeq in noninvasive prenatal testing
for chromosome 21 trisomy and testing of impact of in silico and
physical size selection on its analytical performance. PLoS One.
10:e01448112015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Beyene J and Pare G: Statistical genetics
with application to population-based study design: A primer for
clinicians. Eur Heart J. 35:495–500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu L, Huang Y, Fang X, Liu C, Deng W,
Zhong C, Xu J, Xu D and Yuan Y: A novel and reliable method to
detect microsatellite instability in colorectal cancer by
next-generation sequencing. J Mol Diagn. 20:225–231. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yurgelun MB, Allen B, Kaldate RR, Bowles
KR, Judkins T, Kaushik P, Roa BB, Wenstrup RJ, Hartman AR and
Syngal S: Identification of a variety of mutations in cancer
predisposition genes in patients with suspected lynch syndrome.
Gastroenterology. 149:604–613. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Valle L, de Voer RM, Goldberg Y, Sjursen
W, Försti A, Ruiz-Ponte C, Caldés T, Garré P, Olsen MF, Nordling M,
et al: Update on genetic predisposition to colorectal cancer and
polyposis. Mol Aspects Med. 69:10–26. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Budiš J, Kucharík M, Ďuriš F, Gazdarica J,
Zrubcová M, Ficek A, Szemes T, Brejová B and Radvanszky J: Dante:
Genotyping of known complex and expanded short tandem repeats.
Bioinformatics. 35:1310–1317. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiao S, Peters U, Berndt S, Brenner H,
Butterbach K, Caan BJ, Carlson CS, Chan AT, Chang-Claude J, Chanock
S, et al: Estimating the heritability of colorectal cancer. Hum Mol
Genet. 23:3898–3905. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Law PJ, Timofeeva M, Fernandez-Rozadilla
C, Broderick P, Studd J, Fernandez-Tajes J, Farrington S, Svinti V,
Palles C, Orlando G, et al: Association analyses identify 31 new
risk loci for colorectal cancer susceptibility. Nat Commun.
10:21542019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang K, Civan J, Mukherjee S, Patel F and
Yang H: Genetic variations in colorectal cancer risk and clinical
outcome. World J Gastroenterol. 20:4167–4177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hofer P, Hagmann M, Brezina S, Dolejsi E,
Mach K, Leeb G, Baierl A, Buch S, Sutterlüty-Fall H, Karner-Hanusch
J, et al: Bayesian and frequentist analysis of an Austrian
genome-wide association study of colorectal cancer and advanced
adenomas. Oncotarget. 8:98623–98634. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang H, Schmit SL, Haiman CA, Keku TO,
Kato I, Palmer JR, van den Berg D, Wilkins LR, Burnett T, Conti DV,
et al: Novel colon cancer susceptibility variants identified from a
genome-wide association study in African Americans. Int J Cancer.
140:2728–2733. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Closa A, Cordero D, Sanz-Pamplona R, Solé
X, Crous-Bou M, Paré-Brunet L, Berenguer A, Guino E, Lopez-Doriga
A, Guardiola J, et al: Identification of candidate susceptibility
genes for colorectal cancer through eQTL analysis. Carcinogenesis.
35:2039–2046. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dunlop MG, Dobbins SE, Farrington SM,
Jones AM, Palles C, Whiffin N, Tenesa A, Spain S, Broderick P, Ooi
LY, et al: Common variation near CDKN1A, POLD3 and SHROOM2
influences colorectal cancer risk. Nat Genet. 44:770–776. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang H, Haiman CA, Burnett T, Fortini BK,
Kolonel LN, Henderson BE, Signorello LB, Blot WJ, Keku TO, Berndt
SI, et al: Fine-mapping of genome-wide association study-identified
risk loci for colorectal cancer in African Americans. Hum Mol
Genet. 22:5048–5055. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hong SN, Park C, Kim JI, Kim DH, Kim HC,
Chang DK, Rhee PL, Kim JJ, Rhee JC, Son HJ and Kim YH: Colorectal
cancer-susceptibility single-nucleotide polymorphisms in Korean
population. J Gastroenterol Hepatol. 30:849–857. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Moazeni-Roodi A, Ghavami S, Ansari H and
Hashemi M: Association between the flap endonuclease 1 gene
polymorphisms and cancer susceptibility: An updated meta-analysis.
J Cell Biochem. 120:13583–13597. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chou AK, Shen MY, Chen FY, Hsiao CL, Shih
LC, Chang WS, Tsai CW, Ying TH, Wu MH, Huang CY and Bau DT: The
association of flap endonuclease 1 genotypes with the
susceptibility of endometriosis. Cancer Genomics Proteomics.
14:455–460. 2017.PubMed/NCBI
|
|
53
|
Kubiritova Z, Gyuraszova M, Nagyova E,
Hyblova M, Harsanyova M, Budis J, Hekel R, Gazdarica J, Duris F,
Kadasi L, et al: On the critical evaluation and confirmation of
germline sequence variants identified using massively parallel
sequencing. J Biotechnol. 298:64–75. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Trost B, Engchuan W, Nguyen CM,
Thiruvahindrapuram B, Dolzhenko E, Backstrom I, Mirceta M, Mojarad
BA, Yin Y, Dov A, et al: Genome-wide detection of tandem DNA
repeats that are expanded in autism. Nature. 586:80–86. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mahmood S, Sivoňová M, Matáková T, Dobrota
D, Wsólová L, Dzian A, et al: Association of EGF and p53 gene
polymorphisms and colorectal cancer risk in the Slovak population.
Cent Eur J Med. 9:405–416. 2014.
|
|
56
|
Škereňová M, Halašová E, Matáková T,
Jesenská L, Jurečeková J, Šarlinová M, Čierny D and Dobrota D: Low
variability and stable frequency of common haplotypes of the tp53
gene region in colorectal cancer patients in a Slovak population.
Anticancer Res. 37:1901–1907. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kašubová I, Kalman M, Jašek K, Burjanivová
T, Malicherová B, Vaňochová A, Meršaková S, Lasabová Z and Plank L:
Stratification of patients with colorectal cancer without the
recorded family history. Oncol Lett. 17:3649–3656. 2019.PubMed/NCBI
|
|
58
|
Jia WH, Zhang B, Matsuo K, Shin A, Xiang
YB, Jee SH, Kim DH, Ren Z, Cai Q, Long J, et al: Genome-wide
association analyses in East Asians identify new susceptibility
loci for colorectal cancer. Nat Genet. 45:191–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Peters U, Jiao S, Schumacher FR, Hutter
CM, Aragaki AK, Baron JA, Berndt SI, Bézieau S, Brenner H,
Butterbach K, et al: Identification of genetic susceptibility loci
for colorectal tumors in a genome-wide meta-analysis.
Gastroenterology. 144:799–807. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Whiffin N, Hosking FJ, Farrington SM,
Palles C, Dobbins SE, Zgaga L, Lloyd A, Kinnersley B, Gorman M,
Tenes A, et al: Identification of susceptibility loci for
colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet.
23:4729–4737. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Houlston RS, Cheadle J, Dobbins SE, Tenesa
A, Jones AM, Howarth K, Spain SL, Broderick P, Domingo E,
Farrington S, et al: Meta-analysis of three genome-wide association
studies identifies susceptibility loci for colorectal cancer at
1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 42:973–977. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Orlando G, Law PJ, Palin K, Tuupanen S,
Gylfe A, Hänninen UA, Cajuso T, Tanskanen T, Kondelin J, Kaasinen
E, et al: Variation at 2q35 (PNKD and TMBIM1) influences colorectal
cancer risk and identifies a pleiotropic effect with inflammatory
bowel disease. Hum Mol Genet. 25:2349–2359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Schmit SL, Schumacher FR, Edlund CK, Conti
DV, Raskin L, Lejbkowicz F, Pinchev M, Rennert HS, Jenkins MA,
Hopper JL, et al: A novel colorectal cancer risk locus at 4q32.2
identified from an international genome-wide association study.
Carcinogenesis. 35:2512–2519. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Peters U, Hutter CM, Hsu L, Schumacher FR,
Conti DV, Carlson CS, Edlund CK, Haile RW, Gallinger S, Zanke BW,
et al: Meta-analysis of new genome-wide association studies of
colorectal cancer risk. Hum Genet. 131:217–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tomlinson IPM, Webb E, Carvajal-Carmona L,
Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A,
Sullivan K, et al: A genome-wide association study identifies
colorectal cancer susceptibility loci on chromosomes 10p14 and
8q23.3. Nat Genet. 40:623–630. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
66
|
Tenesa A, Farrington SM, Prendergast JGD,
Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E,
Cetnarskyj R, Cartwright N, et al: Genome-wide association scan
identifies a colorectal cancer susceptibility locus on 11q23 and
replicates risk loci at 8q24 and 18q21. Nat Genet. 40:631–637.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tomlinson IPM, Carvajal-Carmona LG,
Dobbins SE, Tenesa A, Jones AM, Howarth K, Palles C, Broderick P,
Jaeger EEM, Farrington S, et al: Multiple common susceptibility
variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part
of the missing heritability of colorectal cancer. PLoS Genet.
7:e10021052011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
COGENT Study, ; Houlston RS, Webb E,
Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I,
Vijayakrishnan J, Sullivan K, et al: Meta-analysis of genome-wide
association data identifies four new susceptibility loci for
colorectal cancer. Nat Genet. 40:1426–1435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Broderick P, Carvajal-Carmona L, Pittman
AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K,
Fielding S, et al: A genome-wide association study shows that
common alleles of SMAD7 influence colorectal cancer risk. Nat
Genet. 39:1315–1317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Schumacher FR, Schmit SL, Jiao S, Edlund
CK, Wang H, Zhang B, Hsu L, Huang SC, Fischer CP, Harju JF, et al:
Genome-wide association study of colorectal cancer identifies six
new susceptibility loci. Nat Commun. 6:71382015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Haiman CA, Le Marchand L, Yamamato J,
Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D and Henderson BE: A
common genetic risk factor for colorectal and prostate cancer. Nat
Genet. 39:954–956. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tomlinson I, Webb E, Carvajal-Carmona L,
Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood
W, et al: A genome-wide association scan of tag SNPs identifies a
susceptibility variant for colorectal cancer at 8q24.21. Nat Genet.
39:984–988. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hutter CM, Slattery ML, Duggan DJ,
Muehling J, Curtin K, Hsu L, Beresford SA, Rajkovic A, Sarto GE,
Marshall JR, et al: Characterization of the association between
8q24 and colon cancer: Gene-environment exploration and
meta-analysis. BMC Cancer. 10:6702010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Cui R, Okada Y, Jang SG, Ku JL, Park JG,
Kamatani Y, Hosono N, Tsunoda T, Kumar V, Tanikawa C, et al: Common
variant in 6q26-q27 is associated with distal colon cancer in an
Asian population. Gut. 60:799–805. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang H, Burnett T, Kono S, Haiman CA,
Iwasaki M, Wilkens LR, Loo LW, Van Den Berg D, Kolonel LN,
Henderson BE, et al: Trans-ethnic genome-wide association study of
colorectal cancer identifies a new susceptibility locus in VTI1A.
Nat Commun. 5:46132014. View Article : Google Scholar : PubMed/NCBI
|